Abstract
Ribosomopathies compose a collection of disorders in which genetic abnormalities cause impaired ribosome biogenesis and function, resulting in specific clinical phenotypes. Congenital mutations in RPS19 and other genes encoding ribosomal proteins cause Diamond-Blackfan anemia, a disorder characterized by hypoplastic, macrocytic anemia. Mutations in other genes required for normal ribosome biogenesis have been implicated in other rare congenital syndromes, Schwachman-Diamond syndrome, dyskeratosis congenita, cartilage hair hypoplasia, and Treacher Collins syndrome. In addition, the 5q− syndrome, a subtype of myelodysplastic syndrome, is caused by a somatically acquired deletion of chromosome 5q, which leads to haploinsufficiency of the ribosomal protein RPS14 and an erythroid phenotype highly similar to Diamond-Blackfan anemia. Acquired abnormalities in ribosome function have been implicated more broadly in human malignancies. The p53 pathway provides a surveillance mechanism for protein translation as well as genome integrity and is activated by defects in ribosome biogenesis; this pathway appears to be a critical mediator of many of the clinical features of ribosomopathies. Elucidation of the mechanisms whereby selective abnormalities in ribosome biogenesis cause specific clinical syndromes will hopefully lead to novel therapeutic strategies for these diseases.
Identification of ribosomal abnormalities in human disease
In 1999, Draptchinskaia et al reported recurrent mutations in a ribosomal protein gene, RPS19, in patients with Diamond-Blackfan anemia (DBA), a rare congenital bone marrow failure syndrome with a striking erythroid defect.1 Since that initial discovery, mutations in a number of ribosomal proteins have been identified in up to 50% of patients with DBA. Moreover, other congenital syndromes have been linked to defective ribosome biogenesis, including Schwachman-Diamond syndrome (SDS), X-linked dyskeratosis congenita (DKC), cartilage hair hypoplasia (CHH), and Treacher Collins syndrome (TCS).2 In the 5q− syndrome, a subtype of adult myelodysplastic syndrome, acquired haploinsufficiency for RPS14 resulting from an interstitial chromosomal deletion causes a severe refractory anemia.3 Each of these disorders is associated with specific defects in ribosome biogenesis, which cause distinct clinical phenotypes, most often involving bone marrow failure and/or craniofacial or other skeletal defects. The disorders of ribosome dysfunction have become collectively known as ribosomopathies.
Overview of ribosome biogenesis
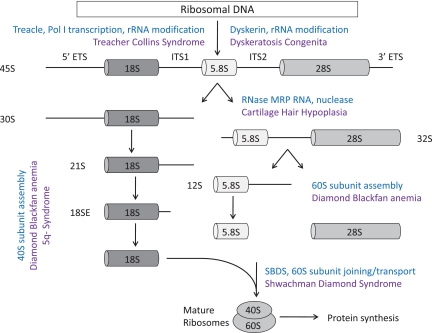
The eukaryotic ribosome is composed of 40S and 60S subunits, which associate to form the translationally active 80S ribosome. This process requires the coordinated synthesis of 4 ribosomal RNAs (rRNAs), approximately 80 core ribosomal proteins, more than 150 associated proteins, and approximately 70 small nucleolar RNAs (snoRNAs).4,5 The 40S subunit contains 18S rRNA and the 60S subunit contains 28S, 5.8S, and 5S rRNAs. In eukaryotes, assembly of rRNA and ribosomal proteins, along with associated proteins and snoRNAs, occurs in the nucleolus, leading to the production of pre-60S and pre-40S preribosomal particles. These particles are exported to the cytoplasm where the final steps in assembly and maturation of ribosomes occur.6 A simplified overview of this process is outlined in Figure 1.
Figure 1.
Simplified schematic of eukaryotic ribosome biogenesis. Adapted from Liu and Ellis2 with permission.
Mature rRNAs are produced from precursor rRNAs after a series of chemical modifications and nucleolytic cleavages. Whereas RNA polymerase III transcribes the 5S rRNA, RNA polymerase I transcribes a 45S precursor transcript that is processed into the 5.8S, 18S, and 28S rRNAs. Ribosomal proteins assemble onto precursor rRNA transcripts and facilitate a series of endonucleolytic and exonucleolytic cleavages required for generation of the mature rRNAs. Haploinsufficiencies for distinct ribosomal proteins have been linked to defects at distinct steps in pre-rRNA processing, which are outlined in Figure 1.2 Several ribosomal proteins have extraribosomal functions, including replication and DNA repair, so mutations in ribosomal proteins may have effects that are independent of the protein translation machinery.7,8
Clinical syndromes
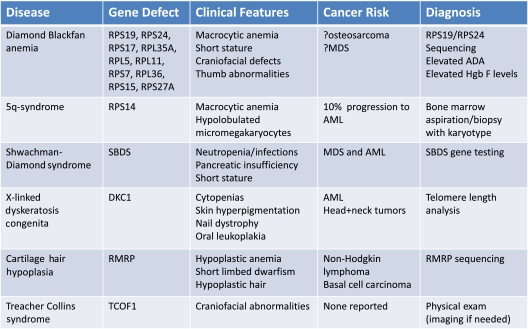
Although there is an abundance of genetic and experimental evidence that mutations in ribosomal genes cause impaired erythropoiesis in DBA and the 5q− syndrome, ribosome dysfunction may also play a role in other congenital syndromes, which are summarized in Table 1. Clinical images of selected physical abnormalities seen in these syndromes are shown in Figure 2.
Table 1.
Summary of known and suspected ribosomopathies with details about clinical characteristics, cancer risk, and diagnostic work-up
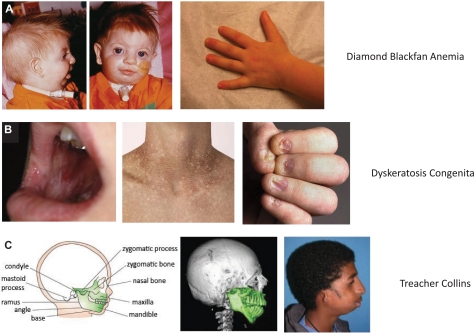
Figure 2.
Selected physical abnormalities seen in ribosomopathies. (A) Left panel: Patient with DBA, illustrating the characteristic craniofacial abnormalities (courtesy of J. Lipton). Note the absent lower eyelashes, deformed external ears, and micrognathia. Second panel: An example of the triphalangeal thumb seen in DBA patients (courtesy of E. Atsidaftos and E. Muir). First panel: Reprinted from Lipton and Ellis9 with permission. (B) These panels illustrate the diagnostic triad of DKC, including dystrophic fingernails, lacy/reticular pigmentation on neck and trunk, and oral leukoplakia (courtesy of B. Alter). Reprinted from Savage and Alter10 with permission. (C) The craniofacial abnormalities of a patient with TCS are illustrated (courtesy of M. R. Passos Bueno). Note the down slanting palpebral fissures, malar and maxillary hypoplasia, and malformation of the ears. Reprinted from Passos-Bueno et al11 with permission.
Diamond-Blackfan anemia
DBA was originally described by Josephs in a review of anemia in infancy and childhood in 193612 and later categorized by Diamond and Blackfan as a congenital hypoplastic anemia.12,13 The disorder is characterized by anemia, macrocytosis, reticulocytopenia, and a selective decrease or absence of erythroid precursors in an otherwise normocellular bone marrow.9 The incidence is estimated to be 4 to 5 cases per million live births, and there are 595 patients currently registered in the Diamond-Blackfan Anemia Registry of North America (A. Vlachos, Schneider Children's Hospital, Division of Hematology/Oncology, personal written communication, January 2010). The majority of patients are diagnosed in the first year of life, with pallor and lethargy being the most common presenting symptoms. There is often a family history of the disease, and approximately 45% of cases are thought to be autosomal dominant.14 Other notable characteristics of the disease include an elevated red blood cell adenosine deaminase level, the presence of fetal membrane antigen “i,” and a range of physical abnormalities seen in 40% to 62% of patients and ranging from short stature to thumb abnormalities to cardiac defects.9 Current therapies include steroids and chronic transfusions, with the only definitive treatment being bone marrow transplantation.15
Since the initial description in 1999 by Draptchinskaia, mutations in RPS19, RPS24, RPS17, and RPL35A have been identified in approximately one-third of patients with DBA.16–19 More recently, Gazda et al identified mutations in RPL5 and RPL11 in an additional 11.4% of patients and noted that mutations in RPL5 were associated with a higher frequency of physical abnormalities, including cleft lip and/or palate, whereas mutations in RPL11 had more isolated thumb abnormalities compared with patients with mutations in RPS19.20 Subsequent work by the Czech DBA registry revealed that, in the 10 patients with either a RPL5 or RPL11 mutation, there was a thumb abnormality, whereas none of the 7 patients with RPS19 mutations in the registry exhibited such an anomaly.21 This may indicate that mutations in different ribosomal genes lead to distinct clinical phenotypes. A large-scale screen of RP genes in the DBA population has also revealed candidate mutations in RPS7, RPL36, RPS15, and RPS27A.20 Despite extensive investigation, mutations have not been discovered in any genes other than those encoding ribosomal proteins for patients with DBA.
Mutations in RPS19 and in RPS24, the first 2 mutations identified in DBA, impair pre-rRNA processing of the 18S rRNA, which leads to decreased production of the 40S ribosomal subunit.22–25 Moreover, there is evidence that depletion of any of the RPS proteins causes a reduction in the amount of free 40S subunits and a significant reduction in the amount of mature 80S ribosomes (with the exception of RPS25, which has not been shown to be mutated in DBA).4 In contrast, when RPL proteins, including RPL35A, are depleted, the amount of the 60S subunit is reduced, as is the level of mature 80S ribosomes.4 Overall, it is clear that mutations in a range of ribosomal proteins ultimately lead to a decrease in the number of mature ribosomes, which would be expected to have a number of consequences for a cell.
5q− syndrome
The 5q− syndrome was first described in 1974 by van Den Berghe in 3 patients with refractory anemia and an interstitial deletion of the long arm of chromosome 5.26 Patients with the 5q− syndrome, an independent subtype of myelodysplastic syndrome (MDS) in the World Health Organization classification system in which del(5q) is the sole cytogenetic abnormality, characteristically have a severe macrocytic anemia, normal/elevated platelets with hypolobulated micromegakaryocytes, and a relatively low rate of progression to acute myeloid leukemia (AML) compared with other types of MDS.27 Lenalidomide, a derivative of the immunomodulatory agent thalidomide, is highly effective for patients with the 5q− syndrome. In a phase 2 trial in low-risk MDS patients with 5q deletions, lenalidomide treatment decreased transfusion requirement in 76% of patients, and 61% of patients had a complete cytogenetic response.28 Lenalidomide has many biologic activities, including the promotion of erythropoiesis, modulation of cytokine production, inhibition of 2 phosphatases on 5q (CDC25C, and PP2A) and the activation of Rac1 and RhoA.28–32
RPS14 was identified as a 5q− syndrome gene in an RNA interference screen of each gene within the 5q− syndrome common deleted region.3,33 In patients with the 5q− syndrome, 1 allele of RPS14 is deleted,3 and haploinsufficient expression of RPS14 has been confirmed in patient samples.3,34 Decreased expression of RPS14 causes impaired erythropoiesis, with relative preservation of the other lineages. Conversely, overexpression of RPS14 in samples from patients with the 5q− syndrome rescues erythropoiesis. It has also been shown that RPS14 deficiency causes a block in the processing of the 18S rRNA and the formation of the 40S ribosomal subunit.3 A murine model has now been developed in which coordinate deletion of loci syntenic with the common deleted region of the 5q− syndrome, including haploinsufficiency for Rps14, recapitulates the macrocytic anemia characteristic of the disease.35 Acquired haploinsufficiency for RPS14 in the 5q− syndrome is therefore analogous to the inactivating mutations in RPS19 and other ribosomal genes in DBA.
Heterozygous deletions of chromosome 5q in MDS are large, and haploinsufficiency for multiple genes probably contributes to the phenotype of the 5q− syndrome.33 For example, haploinsufficiency for miR-145 and miR-146a may contribute to thrombocytosis, and haploinsufficiency for EGR1 may contribute to the clonal advantage of the del(5q) clone.36–39 Nevertheless, in vitro and in vivo studies indicate that the erythroid defect, the aspect of the 5q− syndrome phenotype most analogous to DBA, is caused by RPS14 haploinsufficiency.3,35
Schwachman-Diamond syndrome
SDS was first reported in 1964 by Schwachman et al in a group of 5 children being followed in a cystic fibrosis clinic at Harvard University.40 SDS is an autosomal recessive disease, with an incidence estimated at 1 in 50 000 births, characterized by exocrine pancreatic insufficiency, ineffective hematopoiesis, and an increased risk of leukemia. Patients often present with steatorrhea and failure to thrive from the pancreatic insufficiency. Patients may also present with serious infections as a result of neutropenia, the most common hematologic problem in SDS. Other signs may include anemia, thrombocytopenia, short stature, and skeletal abnormalities.41 Patients with SDS are also at an increased risk for myelodysplasia and malignant transformation. Supportive measures for patients with SDS include pancreatic enzymes, antibiotics, transfusions, and granulocyte colony-stimulating factor. The only definitive therapy is bone marrow transplantation.42
In 2003, Boocock et al reported causal mutations in the SBDS gene, named after Schwachman-Bodian-Diamond.43 Approximately 90% of SDS patients have been found to have biallelic mutations in SBDS.43 It was also shown that 75% of these mutations are the result of a gene conversion with an adjacent pseudogene, SBDSP, which shares 97% homology with SBDS.43 The 250-amino acid SBDS protein is highly conserved through evolution, and the SBDS mRNA is ubiquitously expressed. The structure and function of SBDS are not yet known, but mounting evidence suggests that the gene plays a role in ribosome biogenesis and RNA processing.44 In patient bone marrow samples, translation-related gene expression is the most prominently regulated gene cluster compared with a number of other pathways known to be defective in various marrow failure conditions, including hematopoietic transcription factors and telomere maintenance.45 SDS cells have also been noted to have abnormal expression of multiple genes involved in ribosome biogenesis and rRNA and mRNA processing and to have decreased expression of several ribosomal protein genes involved in cell growth and survival, including RPS9, RPS20, RPL6, RPL15, RPL22, RPL23, and RPL29.45 Furthermore, SBDS has been shown to cosediment with the 60S ribosomal precursor subunit in sucrose gradients and to associate with the 28S rRNA, which is a component of the 60S subunit.44
Although these findings provide evidence for a role of SBDS in ribosome biogenesis, SBDS is a multifunctional protein, and nonribosomal activities may play a dominant role in the clinical phenotype. In particular, SBDS has been shown to have a role in stabilizing the mitotic spindle,46,47 which might indicate a role for SBDS in proliferation and/or chromosome segregation, thereby contributing to chromosomal instability and some component of bone marrow failure.
Dyskeratosis congenita
The first case report that linked the classic triad of abnormal skin pigmentation, oral leukoplakia, and nail dystrophy was in 1910.48 Subsequently, bone marrow failure and a range of other abnormalities were identified in similar patients, and the disorder became known as DKC.49 Patients usually present during the first decade of life with skin hyperpigmentation and nail changes. Almost 90% of patients will eventually develop a peripheral cytopenia of 1 or more lineages, and bone marrow failure is the major cause of death. The estimated prevalence of DKC is 1 case in 1 million people, and the male-to-female ratio is approximately 3:1.50,51 Patients with DKC have a high risk of developing leukemia, solid tumors, and pulmonary fibrosis. Management includes steroids and supportive measures; the only curative option is stem cell transplantation, but the success rate of stem cell transplantation is limited because of a high prevalence of fatal pulmonary complications.10
DKC is a heterogeneous disorder, but in all characterized cases, the causative mutations are in components of the telomerase complex. Telomerase is an enzyme complex composed of telomerase reverse transcriptase (TERT), telomerase RNA (TERC), and dyskerin, which adds specific DNA sequence repeats to the ends of chromosomes and counters some of the normal shortening that occurs during DNA replication.52 X-linked DKC, which has a more severe phenotype compared with the autosomal dominant form of DKC, is caused by a mutation in DKC1, which encodes dyskerin.53 One of the functions of dyskerin is to act as a nucleolar protein associated with the snoRNPs involved in rRNA modification. Dyskerin associates with a specific group of snoRNPs known as H/ACA, which function in the pseudo-uridylation of rRNAs.2 However, the functional consequences of this defect in pseudo-uridylation remain unclear.
Patients with the autosomal dominant form of DKC have been found to have mutations in TERC, and 2 families with the autosomal recessive form of DKC have been found to have mutation in TERT, suggesting that disruption of the telomerase complex alone could result in defective hematopoiesis.54 Also of note, mutations in the TERT gene have been identified in some patients with aplastic anemia and short telomeres.55 Further confusing the picture is a hypomorphic Dkc1 mutant mouse, which demonstrated impaired ribosomal RNA pseudo-uridylation before the onset of clinical features of DKC, whereas reductions in telomere length became evident only in later generations.56 Although it is clear that telomeres play a significant role in the pathogenesis of DKC, there may be a distinct contribution from ribosomal defects. Ongoing work examining genotype/phenotype correlations and basic mechanisms will hopefully elucidate the true contributions of telomerase activity and impaired ribosomal biogenesis to the pathophysiology of DKC.
Cartilage hair hypoplasia
CHH was first described in several Amish families by McKusick et al in 1965 as a form of short-limbed dwarfism resulting from skeletal dysplasia.57 The incidence in Old Order Amish population is 1 in 1300 newborns57 and in people of Finnish descent, 1 in 20 000.58 Outside of these populations, the condition is rare and the specific incidence is not known.59 The autosomal recessive syndrome is also characterized by hypoplastic hair, immune dysfunction, and an increased predisposition to malignancies, especially non-Hodgkin lymphoma and basal cell carcinoma.41 Hematologic abnormalities can include a macrocytic anemia and lymphopenia. Management is again supportive, with stem cell transplantation being the only curative option. Transplantation is particularly indicated in patients with severe T-cell immunodeficiency with or without concomitant B-cell disease, although transplantation does not correct the skeletal abnormalities.60
In 2001, Ridanpää et al identified mutations in the untranslated RMRP gene as causative for this pleiotropic disease, with an ancient founder mutation in Finland.61 RMRP encodes the RNA component of the mitochondrial RNA processing complex (RNase MRP), which is primarily located in the nucleolus and is classified as a snoRNA. snoRNAs form small nucleolar ribonucleoprotein complexes (snoRNPs) within the nucleolus and are involved in various steps in the synthesis of ribosomal RNA.62 One of the important functions of the RNase MRP complex is to cleave the precursor rRNA, which contributes to the maturation of the 5′ end of the 5.8S rRNA. RNase MRP also shares protein subunits and contains a structurally related RNA subunit with another snoRNP, which is involved in tRNA precursor maturation.
Mutations in RMRP have also been associated with anauxetic dysplasia, an autosomal recessive growth disorder without marrow involvement.63 Thiel et al showed that, in cells overexpressing the RMRP mutants seen in anauxetic dysplasia, there were decreased cyclin A2 levels.63 In contrast, cells with the CHH founder RMRP mutation had significantly increased levels of cyclin B2 mRNA.63 Cyclin B2 is known to contribute to chromosomal instability through alterations of the mitotic spindle checkpoint,64 which suggests another possible explanation for the bone marrow dysfunction seen in CHH. Recent work by Maida et al has revealed a connection between RMRP and TERT.65 The clinical heterogeneity seen in CHH and in anauxetic dysplasia might be attributable to differences in the magnitude and type of alteration in ribosomal assembly, telomere dysfunction, and cyclin B2 levels caused by various RMRP mutations.63
Treacher Collins syndrome
In 1900, the British ophthalmologist Edward Treacher Collins described the essential features of a syndrome, which included characteristic craniofacial changes66 that arise from symmetrically and bilaterally diminished growth of the structures derived from the first and second pharyngeal arch, groove, and pouch.67 TCS is now known to be an autosomal dominant condition with an estimated incidence of 1 in 50 000 live births.68 Patients often have complications from the craniofacial dystosis, which can include issues with airway, swallowing, brain development, and hearing.67 Management of these patients requires a multidisciplinary approach involving craniofacial surgeons, orthodontists, ophthalmologists, otolaryngologists, and speech pathologists.67
In 1996, TCOF1 was identified as the gene responsible for TCS; TCOF1 encodes a protein known as treacle.69 Immunofluorescence studies demonstrated that treacle colocalizes with upstream binding factor and RNA polymerase I in the nucleolus and that treacle is a constituent of one of the preribosomal ribonucleoprotein (pre-RNP) complexes. Further data have shown that treacle is essential for the transcription of ribosomal DNA and that it may also play a role in the methylation of rRNA.70 Mice haploinsufficient for Tcof1 exhibit diminished production of ribosomes, and this deficiency correlates with decreased proliferation of both neural ectoderm and neural crest cells.71 Interestingly, a study by Jones et al showed that chemical and genetic inhibition of p53 activity in these mice can prevent the craniofacial abnormalities.72 This is of special note, and the link between p53 and ribosomopathies is discussed in greater detail later in this review.
Although the craniofacial defects in TCS are highly similar to those seen in some cases of DBA and there is a suggestion of a ribosomal basis for the pathophysiology, there are no hematologic abnormalities associated with TCS. This adds yet another fascinating wrinkle to our understanding of the ribosomopathies.
Diagnosis of ribosomopathies: current and future strategies
When a child is first suspected of having a bone marrow failure syndrome, he or she undergoes a personal and family history, physical examination, and a series of radiographic and laboratory evaluations. The laboratory data includes a CBC with differential, smear review, reticulocyte count, erythropoietin levels, and hemoglobin electrophoresis to assess the severity of anemia and the level of ineffective erythropoiesis. Ultimately, a bone marrow aspiration and biopsy is required. In addition, there are now specific diagnostic tests for the ribosomopathies ranging from SBDS gene testing for SDS and RMRP sequencing in CHH. These tests are summarized in Table 1. In the future, given the shared ribosomal pathogenesis of these disorders, other diagnostic tests might be developed. These may include sequencing of the relevant ribosomal genes or analysis of aberrant preribosomal RNA precursor accumulation by quantitative RT-PCR. After the diagnosis of an inherited bone marrow failure syndrome, patients and families can be referred to a number of resources. Useful links can be found at www.marrowfailure.cancer.gov.
Cellular consequences of ribosomal haploinsufficiency
It is clear that mutations in genes encoding ribosomal proteins or other proteins involved in ribosome biogenesis cause specific phenotypes, restricted to particular cell types, despite the universal expression of ribosomal proteins and the ubiquitous requirement for protein synthesis. Specifically, defects in ribosome biogenesis or function appear to be capable of causing anemia and other hematologic phenotypes, defects in growth and development, and congenital anomalies, such as craniofacial defects and thumb. The fundamental question of how a mutation in a ribosomal protein, which would be expected to have widespread and diverse effects throughout an organism, can lead to such selective defects is not fully answered, but possible mechanisms are outlined in this section.
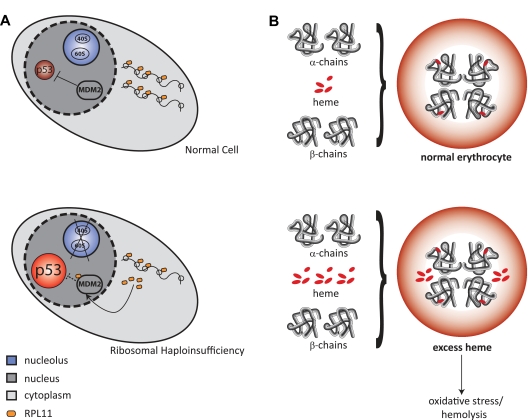
A leading hypothesis, depicted in Figure 3A, posits that ribosomal haploinsufficiency leads to disrupted ribosome biogenesis and an accumulation of free ribosomal proteins that bind MDM2, a repressor of p53. The consequent activation of p53 leads to apoptosis and cell-cycle arrest, which ultimately leads to anemia.74,75 Another potential mechanism, depicted in Figure 3B, is that defective maturation of ribosomal subunits could delay translation of globin genes, resulting in a relative excess of free heme, which would also lead to erythroid-specific apoptosis and anemia.76 Both of these potential mechanisms are discussed in more detail in “Ribosomal haploinsufficiency in human disease.” Alternate mechanisms include pathogenic functions for the aberrantly accumulated ribosomal precursors and aberrant translation by defective ribosomes.77 Further understanding of the molecular basis of ribosomopathies should lead to more insights into the clinical phenotypes of these diseases and to the development of novel therapeutic strategies.
Figure 3.
Potential mechanisms for the cellular consequences of ribosomal haploinsufficiency. (A) Top panel: Normal cell in unstressed conditions, with unperturbed ribosome biogenesis and steady levels of p53. Bottom panel: Ribosomal haploinsufficiency leads to up-regulation of rpL11, which binds to MDM2 causing p53 activation, which results in apoptosis and cell-cycle arrest. (B) Top panel: Normal hemoglobin synthesis, with the coordinated production of heme and globin. Bottom panel: Relative excess of free heme leads to oxidative stress and hemolysis through a variety of mechanisms.73
Ribosomal haploinsufficiency in human disease
RPS19 was the first ribosomal gene implicated in human disease and is the most frequently mutated gene in DBA with a total of 77 mutations having been described.78 The majority are whole gene deletions, translocations, or truncating mutations (nonsense or frameshift) universally present in only a single allele.16,78 One group of missense mutations in RPS19 alters the nucleolar localization signal with a resulting dramatic decrease in protein expression.16 This suggests that haploinsufficiency for a ribosomal gene in the nucleolus is the basis for the pathology.78 Cmejlova et al showed that the level of translation was on average 48% to 73% of controls in both unstimulated and phytohemagglutinin-stimulated DBA lymphocytes irrespective of mutations in RPS19.79 In the DBA patients who have been found to have RPL35A mutations, haploinsufficiency of RPL35A is inferred based on genomic deletions or a nonsense splicing defect in 1 allele.19 In the 5q− syndrome, 1 allele of RPS14 is deleted, without mutation or epigenetic silencing of the intact allele, resulting in haploinsufficient expression of RPS14.3,34 To date, there is no evidence of biallelic inactivation of any ribosomal genes in human disease.
In vitro and in vivo models of ribosomal haploinsufficiency
The central hematopoietic defects in DBA are thought to be the hypoproliferation of erythroid cells and the enhanced sensitivity of hematopoietic progenitors to apoptosis.80 Both in vitro and in vivo models of ribosomal haploinsufficiency recapitulate the hematopoietic phenotype of DBA. These models have informed the biology of ribosomopathies and will be critical resources for the development of and testing of novel therapies. A limitation to note, particularly with the in vitro models, is that the degree of knockdown of ribosomal proteins is often greater than 50%; therefore, these are not perfect models for studying haploinsufficiency. It is very possible that the mechanism and phenotype of these disorders are influenced by the precise level of ribosomal activity, which is why ongoing work to generate optimal models will be essential for further progress in the field.
Decreased expression of RPS19 in vitro is sufficient to phenocopy many aspects of the DBA phenotype. Knockdown of RPS19 using RNA interference in primary human hematopoietic progenitor cells from normal persons causes a severe defect in the differentiation and proliferation of erythroid progenitor cells.81,82 Similarly, primary CD34+ cells from DBA patients have a higher number of apoptotic cells compared with normal CD34+ cells,83 and mononuclear cells derived from patients with DBA fail to proliferate in response to erythropoietin and form smaller erythroid colonies in methylcellulose compared with normal patients.84 The hematopoietic defect caused by RPS19 deficiency can be rescued by forced overexpression of RPS19. Expression of a RPS19 transgene by retroviral vectors in CD34+ bone marrow cells from DBA patients significantly improved erythropoiesis.85 In addition, the erythroid defect can be partially rescued by treatment with corticosteroids, the primary pharmacologic treatment for DBA.82
Both zebrafish and murine models have been generated with mutations in or decreased expression of ribosomal genes. Two groups have knocked down expression of RPS19 in zebrafish using morpholinos with similar findings.86,87 The RPS19 morphants have hematopoietic and developmental abnormalities that resemble DBA, and this phenotype is partially rescued by treatment with corticosteroids.86 As with RNA interference studies, morpholinos may decrease ribosomal gene expression by greater than 50%, potentially resulting in more severe hematopoietic defects.
Genetically engineered mice have the potential to model ribosomal haploinsufficiency more precisely. An initial Rps19 knockout mouse model was lethal in the homozygous state and did not have a DBA phenotype in the heterozygous state, although the heterozygous mice did not have the expected decline in RPS19 levels.88 Murine models now exist with heterozygous missense mutations in Rps19 or Rps20 or conditional deletion of Rps6. The RPS19 and RPS20 mice have only a mild macrocytic anemia, although this relatively benign phenotype may be explained by the fact that the mutations cause hypomorphic alleles rather than true haploinsufficiency.74 Mice with conditional deletion of a set of genes on 5q, including Rps14, develop a severe macrocytic anemia, consistent with the phenotype of the 5q− syndrome and DBA.35
Studies using the RPS6 mice provide further insight into the biology of ribosome dysfunction. Conditional homozygous deletion of RPS6 using CD4-Cre abolished T-cell development. In contrast, haploinsufficiency did not have any effect on T-cell maturation in the thymus. However, once T cells were activated and induced to proliferate, T cells with RPS6 haploinsufficiency were unable to proliferate.89 Thus, cells with ribosomal haploinsufficiency may synthesize intact ribosomes sufficiently when the cells proliferate slowly but have more defective ribosome biogenesis in states of rapid proliferation, including hematopoiesis, which might explain some of the lineage specificity seen in ribosomopathies.
The role of p53
Evidence from a variety of model systems indicates that p53 is activated by ribosome dysfunction. Among its myriad of roles, p53 is known to play a fundamental role in the surveillance of protein translation.90 The murine double minute 2 protein (MDM2, or HDM2 in humans) functions as a link between ribosome biogenesis and the p53 pathway. MDM2 is a central regulator of p53, acting as a ubiquitin ligase that leads to the degradation of p53.91 MDM2 has also been shown to bind specifically to several free ribosomal proteins, including RPL5, RPL23, RPL11, RPS7, and RPL26.92–98 In an elegant series of experiments, nucleolar disruption, experimentally induced by treatment with actinomycin D, was shown to lead to the release of RPL11 and other ribosomal proteins into the nucleoplasm, the binding of RPL11 to MDM2, the inhibition of MDM2 activity, and the consequent accumulation of p53.75,95 A schematic view of this pathway is shown in Figure 3A.
In the Rps19 mutant mouse model described previously, induction of p53 and p53 target genes were identified in the hyperpigmented foot pads of the mice. These mice also had a decreased hematocrit and increased MCV. When the Rps19 mutant mouse line had 1 allele of p53 genetically inactivated, there was an increase in red blood cell count and decrease in MCV. Homozygous inactivation of p53 in Rps19 mutant mice fully corrected the hematologic phenotype.74 Furthermore, when the mice with the conditional deletion of a set of genes found in the common deleted region of the 5q− syndrome were crossed with p53 null mice, there was a complete rescue of the erythroid phenotype.35 These findings indicate that p53 is critical for the macrocytic anemia caused by ribosomal dysfunction.
Studies in mice with conditional inactivation of Rps6 have further elucidated the mechanism of MDM2-mediated induction of p53 by ribosomal haploinsufficiency.75 Conditional deletion of Rps6 in murine liver inhibited 40S (but not 60S) ribosomal biogenesis, and the liver in affected mice did not regenerate normally after partial hepatectomy resulting from cell-cycle arrest.99 The increased free RPL11 that was generated by the mechanism shown in Figure 3A was found to be the result of the increased translation of rpL11 mRNAs through derepression of 5′-TOP mRNA translation.75 If specific agents that target the signaling components involved in rpL11 up-regulation and/or that regulate 5′-TOP mRNA translation could be developed, there is the potential to alleviate the block in ribosomal biogenesis without involving other pathways involved in p53 induction.
Balance of heme and globin
An alternative or potentially complementary hypothesis for the erythroid specificity of DBA involves an imbalance between heme and globin production, as represented in Figure 3B. Erythroid progenitor cells are notable for their incredibly rapid rate of proliferation, doubling every 12 to 24 hours,100 and for their need to synthesize globin at a tremendous pace. This elevated rate of proliferation requires a very high level of ribosome biogenesis and ribosomal activity. Cells with impaired ribosome biogenesis may not be able to maintain the level of globin synthesis required by erythroid progenitor cells, which raises the possibility that the erythroid phenotype seen in many ribosomopathies is the result of a decrease in globin synthesis, which leads to a relative excess of free heme resulting in apoptosis and anemia.
In a murine model with conditional inactivation of FLVCR, a heme export protein, animals develop a severe macrocytic anemia, indicating that erythroid heme toxicity can cause a phenotype similar to ribosomal haploinsufficiency.101 FLVCR is the receptor for the feline leukemia virus, subgroup C (FeLV-C), which causes a red cell aplasia in cats.101 Pro-erythroblasts may need FLVCR both to prevent heme toxicity and to preserve adequate heme supply. Homozygous conditional inactivation of FLVCR in the murine bone marrow causes a macrocytic anemia, reticulocytopenia, and maturation arrest at the pro-erythroblast stage, phenocopying the hematologic phenotype in DBA patients. Of note, the mutant mouse embryos were also smaller than controls and had digit abnormalities, wide-spaced eyes, and flattened facies. The conditionally deleted FLVCR−/− mice were also noted to accumulate iron.76
These experiments indicate that mutations in ribosomal proteins could cause defective ribosomal subunit maturation, leading to delayed globin translation and a relative excess of free heme, with FLVCR acting as a safety valve. This hypothesis implies that the severity of the heme toxicity may modulate the clinical phenotype of patients with ribosomal haploinsufficiency, although accumulation of heme in erythroid progenitor cells from patients with DBA has not been examined. To date, no mutations have been identified in FLVCR in any of the ribosomopathies.
Ribosomal dysfunction and cancer
All of the pediatric bone marrow failure syndromes linked to ribosomal dysfunction appear to have an increased incidence of cancer, although the type and frequency vary considerably, adding yet another intriguing piece to the story.
For patients with SDS, a 2005 review of the French Severe Chronic Neutropenia Registry estimated the risk of MDS or AML at 19% at 20 years and 36% at 30 years.102 A recent report from the National Cancer Institute dyskeratosis congenita cohort revealed a ratio of observed to expected cancers of 11-fold compared with the general population with the observed/expected ratios being significantly elevated for tongue cancer and AML.103 Of the 123 Finnish patients with CHH who were followed for a mean 19.2 years, 14 cases of cancer were diagnosed. Non-Hodgkin lymphoma was the most frequent tumor, with 9 cases reported, followed by squamous cell carcinoma, leukemia, and Hodgkin lymphoma. Nine of the 14 tumors were in patients younger than 45 years. In addition, 10 patients in the cohort developed basal cell carcinoma of the skin.104 The authors concluded that there was a 7-fold increase overall cancer rate in patients with CHH compared with the normal population, with the risk of non-Hodgkin lymphoma and skin cancer being even higher. A predisposition for cancer has not been reported in TCS, which is not associated with hematologic manifestations.
For DBA, the predisposition to cancer is less clear. In the latest update from the DBA Registry, of the 568 patients registered, there were 15 cancers in 13 patients. These included 4 cases of MDS/AML, 3 cases of osteosarcoma, 2 colon cancers, 1 soft tissue sarcoma, 2 squamous cell carcinomas (1 oral and 1 vaginal), 2 breast cancers, and 1 uterine cancer (A. Vlachos, Schneider Children's Hospital, Division of Hematology/Oncology, personal written communication, January 2010).
An animal model that ties together ribosomopathies and tumorogenesis was developed in 2004 by Amsterdam et al.105 They generated several hundred lines of zebrafish, each of which was heterozygous for an embryonic lethal recessive mutation. Eleven of the 12 lines with elevated cancer incidence had a mutation in different types of ribosomal protein genes.105 Of note, the distribution and penetrance of cancers in these zebrafish lines phenocopy the malignancies in p53 null zebrafish. Further work indicates that the tumors in zebrafish with ribosomal haploinsufficiency lose expression of p53.106
Studies in human cancer specimens have also yielded some tantalizing clues. The gene encoding nucleophosphomin (NPM1 or B23) has been show to bind to p53 and its associated proteins, including MDM2.107 NPM1 has also been shown to be mutated in up to 60% of normal karyotype adult AML107 and to be involved in certain cases of anaplastic large cell lymphoma, MDS, and acute promyelocytic leukemia.108 NPM1 is an abundant protein located in the nucleolus and functions as an RNA-binding nucleolar phosphoprotein involved in preribosomal assembly. It also serves as a shuttle between the nucleoplasm and the cytoplasm for both nucleic acids and proteins.107 NPM1 heterozygous mice develop a hematologic syndrome similar to MDS.109
Although provocative links have been drawn between bone marrow failure syndromes, ribosome dysfunction, p53 activation, and tumor development, key mechanisms remain incompletely understood.
In conclusion, disease-causing mutations in a collection of congenital and acquired syndromes result from defective ribosome biogenesis and function. In at least some of these cases, ribosomal dysfunction appears to be central to the molecular pathology of the disorder. Different defects in ribosome biogenesis have now been associated with distinct clinical phenotypes. These phenotypes, including impaired erythropoiesis and craniofacial abnormalities, have been linked to p53 activation. The identification of additional disease alleles and characterization of their functional effects will further inform the biology of ribosomopathies and will hopefully lead to novel therapeutic strategies for the treatment of ribosomopathies.
Acknowledgments
The authors thank Eva Atsidaftos (Diamond-Blackfan Anemia Registry), Ellen Muir (Diamond-Blackfan Anemia Surveillance and Awareness Program), Blanche Alter, Jeffrey Lipton, Adrianna Vlachos, and Maria Rita Passos Bueno for clinical images and registry data; Gretchen Jones and Rachel Murphy for assistance with figure generation; and Steven Ellis for critical review of this manuscript.
This work was supported by the National Institutes of Health (R01 HL82945).
The authors regret the omission of many important references because of length constraints.
Authorship
Contribution: A.N. and B.L.E. wrote the manuscript.
Conflict-of-interest disclosure: B.L.E. received research funding from GlaxoSmithKline. A.N. declares no competing financial interests.
Correspondence: Benjamin L. Ebert, Brigham and Women's Hospital, Karp Research Bldg, 1 Blackfan Cir, 5th Fl, CHRB 05.211, Boston, MA 02115; e-mail: bebert@partners.org.
References
- 1.Draptchinskaia N, Gustavsson P, Andersson B, et al. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat Genet. 1999;21(2):169–175. doi: 10.1038/5951. [DOI] [PubMed] [Google Scholar]
- 2.Liu JM, Ellis SR. Ribosomes and marrow failure: coincidental association or molecular paradigm? Blood. 2006;107(12):4583–4588. doi: 10.1182/blood-2005-12-4831. [DOI] [PubMed] [Google Scholar]
- 3.Ebert BL, Pretz J, Bosco J, et al. Identification of RPS14 as a 5q− syndrome gene by RNA interference screen. Nature. 2008;451(7176):335–339. doi: 10.1038/nature06494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robledo S, Idol RA, Crimmins DL, Ladenson JH, Mason PJ, Bessler M. The role of human ribosomal proteins in the maturation of rRNA and ribosome production. RNA. 2008;14(9):1918–1929. doi: 10.1261/rna.1132008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doudna JA, Rath VL. Structure and function of the eukaryotic ribosome: the next frontier. Cell. 2002;109(2):153–156. doi: 10.1016/s0092-8674(02)00725-0. [DOI] [PubMed] [Google Scholar]
- 6.Henras AK, Soudet J, Gérus M, et al. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci. 2008;65(15):2334–2359. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wool IG. Extraribosomal functions of ribosomal proteins. Trends Biochem Sci. 1996;21(5):164–165. [PubMed] [Google Scholar]
- 8.Warner JR, McIntosh KB. How common are extraribosomal functions of ribosomal proteins? Mol Cell. 2009;34(1):3–11. doi: 10.1016/j.molcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lipton JM, Ellis SR. Diamond-Blackfan anemia: diagnosis, treatment, and molecular pathogenesis. Hematol Oncol Clin North Am. 2009;23(2):261–282. doi: 10.1016/j.hoc.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Savage SA, Alter BP. Dyskeratosis congenita. Hematol Oncol Clin North Am. 2009;23(2):215–231. doi: 10.1016/j.hoc.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Passos-Bueno MR, Ornelas CC, Fanganiello RD. Syndromes of the first and second pharyngeal arches: a review. Am J Med Genet A. 2009;149(8):1853–1859. doi: 10.1002/ajmg.a.32950. [DOI] [PubMed] [Google Scholar]
- 12.Josephs HW. Anaemia of infancy and early childhood. Medicine. 1936;15:307. [Google Scholar]
- 13.Diamond LK, Blackfan KD. Hypoplastic anemia. Am J Dis Child. 1938;56:464. [Google Scholar]
- 14.Orfali KA, Ohene-Abuakwa Y, Ball SE. Diamond-Blackfan anaemia in the UK: clinical and genetic heterogeneity. Br J Haematol. 2004;125(2):243–252. doi: 10.1111/j.1365-2141.2004.04890.x. [DOI] [PubMed] [Google Scholar]
- 15.Vlachos A, Federman N, Reyes-Haley C, Abramson J, Lipton JM. Hematopoietic stem cell transplantation for Diamond-Blackfan anemia: a report from the Diamond-Blackfan Anemia Registry. Bone Marrow Transplant. 2001;27(4):381–386. doi: 10.1038/sj.bmt.1702784. [DOI] [PubMed] [Google Scholar]
- 16.Da Costa L, Tchernia G, Gascard P, et al. Nucleolar localization of RPS19 protein in normal cells and mislocalization due to mutations in the nucleolar localization signals in 2 Diamond-Blackfan anemia patients: potential insights into pathophysiology. Blood. 2003;101(12):5039–5045. doi: 10.1182/blood-2002-12-3878. [DOI] [PubMed] [Google Scholar]
- 17.Gazda HT, Grabowska A, Merida-Long LB, et al. Ribosomal protein S24 gene is mutated in Diamond-Blackfan anemia. Am J Hum Genet. 2006;79(6):1110–1118. doi: 10.1086/510020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cmejla R, Cmejlova J, Handrkova H, Petrak J, Pospisilova D. Ribosomal protein S17 gene (RPS17) is mutated in Diamond-Blackfan anemia. Hum Mutat. 2007;28(12):1178–1182. doi: 10.1002/humu.20608. [DOI] [PubMed] [Google Scholar]
- 19.Farrar JE, Nater M, Caywood E, et al. Abnormalities of the large ribosomal subunit protein, Rpl35a, in Diamond-Blackfan anemia. Blood. 2008;112(5):1582–1592. doi: 10.1182/blood-2008-02-140012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gazda HT, Sheen MR, Vlachos A, et al. Ribosomal protein L5 and L11 mutations are associated with cleft palate and abnormal thumbs in Diamond-Blackfan anemia patients. Am J Hum Genet. 2008;83(6):769–780. doi: 10.1016/j.ajhg.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cmejla R, Cmejlova J, Handrkova H, et al. Identification of mutations in the ribosomal protein L5 (RPL5) and ribosomal protein L11 (RPL11) genes in Czech patients with Diamond-Blackfan anemia. Hum Mutat. 2009;30(3):321–327. doi: 10.1002/humu.20874. [DOI] [PubMed] [Google Scholar]
- 22.Choesmel V, Bacqueville D, Rouquette J, et al. Impaired ribosome biogenesis in Diamond-Blackfan anemia. Blood. 2007;109(3):1275–1283. doi: 10.1182/blood-2006-07-038372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flygare J, Aspesi A, Bailey JC, et al. Human RPS19, the gene mutated in Diamond-Blackfan anemia, encodes a ribosomal protein required for the maturation of 40S ribosomal subunits. Blood. 2007;109(3):980–986. doi: 10.1182/blood-2006-07-038232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Idol RA, Robiedo S, Du H, et al. Cells depleted for RPS19, a protein associated with Diamond-Blackfan anemia, show defects in 18S ribosomal RNA synthesis and small ribosomal subunit production. Blood Cells Mol Dis. 2007;39(1):35–43. doi: 10.1016/j.bcmd.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 25.Choesmel V, Fribourg S, Aguissa-Touré AH, et al. Mutation of ribosomal protein RPS24 in Diamond-Blackfan anemia results in a ribosome biogenesis disorder. Hum Mol Genet. 2008;17(9):1253–1263. doi: 10.1093/hmg/ddn015. [DOI] [PubMed] [Google Scholar]
- 26.Van den Berghe H, Cassiman JJ, David G, Fryns JP, Michaux JL, Sokal G. Distinct haematological disorder with deletion of long arm of no. 5 chromosome. Nature. 1974;251(5474):437–438. doi: 10.1038/251437a0. [DOI] [PubMed] [Google Scholar]
- 27.Vardiman JW, Harris NL, Brunning RD. The World Health Organization (WHO) classification of the myeloid neoplasms. Blood. 2002;100(7):2292–2302. doi: 10.1182/blood-2002-04-1199. [DOI] [PubMed] [Google Scholar]
- 28.List A, Dewald G, Bennett J, et al. Lenalidomide in the myelodysplastic syndrome with chromosome 5q deletion. N Engl J Med. 2006;355(14):1456–1465. doi: 10.1056/NEJMoa061292. [DOI] [PubMed] [Google Scholar]
- 29.Ebert BL, Galili N, Tanayo P, et al. An erythroid differentiation signature predicts response to lenalidomide in myelodysplastic syndrome. PLoS Med. 2008;5(2):e35. doi: 10.1371/journal.pmed.0050035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bartlett JB, Dredge K, Dalgleish AG. The evolution of thalidomide and its IMiD derivatives as anticancer agents. Nat Rev Cancer. 2004;4(4):314–322. doi: 10.1038/nrc1323. [DOI] [PubMed] [Google Scholar]
- 31.Wei S, Chen X, Rocha K, et al. A critical role for phosphatase haplodeficiency in the selective suppression of deletion 5q MDS by lenalidomide. Proc Natl Acad Sci U S A. 2009;106(31):12974–12979. doi: 10.1073/pnas.0811267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu Y, Ferguson GD, Mercurio F, et al. Immunomodulatory drugs reorganize cytoskeleton by modulating Rho GTPases. Blood. 2009;114(2):338–345. doi: 10.1182/blood-2009-02-200543. [DOI] [PubMed] [Google Scholar]
- 33.Ebert BL. Deletion 5q in myelodysplastic syndrome: a paradigm for the study of hemizygous deletions in cancer. Leukemia. 2009;23(7):1252–1256. doi: 10.1038/leu.2009.53. [DOI] [PubMed] [Google Scholar]
- 34.Pellagatti A, Hellström-Lindberg E, Giagounidis A, et al. Haploinsufficiency of RPS14 in 5q− syndrome is associated with deregulation of ribosomal- and translation-related genes. Br J Haematol. 2008;142(1):57–64. doi: 10.1111/j.1365-2141.2008.07178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barlow JL, Drynan LF, Hewett DR, et al. A p53-dependent mechanism underlies macrocytic anemia in a mouse model of human 5q− syndrome. Nat Med. 2010;16(1):59–66. doi: 10.1038/nm.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Min IM, Pietramaggiori G, Kim FS, Passegué E, Stevenson KE, Wagers AJ. The transcription factor EGR1 controls both the proliferation and localization of hematopoietic stem cells. Cell Stem Cell. 2008;2(4):380–391. doi: 10.1016/j.stem.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 37.Joslin JM, Fernald AA, Tennant TR, et al. Haplo-insufficiency of EGR1, a candidate gene in the del(5q), leads to the development of myeloid disorders. Blood. 2007;110(2):719–726. doi: 10.1182/blood-2007-01-068809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Starczynowski DT, Kuchenbauer F, Argiropoulos B, et al. Identification of miR-145 and miR-146a as mediators of the 5q− syndrome phenotype. Nat Med. 16(1):49–58. doi: 10.1038/nm.2054. [DOI] [PubMed] [Google Scholar]
- 39.Kumar M, Narla A, Nonami A, et al. Coordinate loss of a microRNA Mir 145 and a protein-coding gene RPS14 cooperate in the pathogenesis of 5q− syndrome [abstract]. Blood (ASH Annual Meeting Abstracts) 2009;114(22) Abstract 947. [Google Scholar]
- 40.Schwachman H, Diamond LK, Oski FA, Khaw KT. The syndrome of pancreatic insufficiency and bone marrow dysfunction. J Pediatr. 1964;65:645–663. doi: 10.1016/s0022-3476(64)80150-5. [DOI] [PubMed] [Google Scholar]
- 41.Ganapathi KA, Shimamura A. Ribosomal dysfunction and inherited marrow failure. Br J Haematol. 2008;141(3):376–387. doi: 10.1111/j.1365-2141.2008.07095.x. [DOI] [PubMed] [Google Scholar]
- 42.Burroughs L, Woolfrey A, Shimamura A. Schwachman-Diamond syndrome: a review of the clinical presentation, molecular pathogenesis, diagnosis, and treatment. Hematol Oncol Clin North Am. 2009;23(2):233–248. doi: 10.1016/j.hoc.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boocock GR, Morrioson JA, Popovic M, et al. Mutations in SBDS are associated with Schwachman-Diamond syndrome. Nat Genet. 2003;33(1):97–101. doi: 10.1038/ng1062. [DOI] [PubMed] [Google Scholar]
- 44.Ganapathi KA, Austin KM, Lee CS, et al. The human Schwachman-Diamond syndrome protein, SBDS, associates with ribosomal RNA. Blood. 2007;110(5):1458–1465. doi: 10.1182/blood-2007-02-075184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rujkijyanont P, Adams SL, Beyene J, Dror Y. Bone marrow cells from patients with Schwachman-Diamond syndrome abnormally express genes involved in ribosome biogenesis and RNA processing. Br J Haematol. 2009;145(6):806–815. doi: 10.1111/j.1365-2141.2009.07692.x. [DOI] [PubMed] [Google Scholar]
- 46.Austin KM, Gupta ML, Coats SA, et al. Mitotic spindle destabilization and genomic instability in Schwachman-Diamond syndrome. J Clin Invest. 2008;118(4):1511–1518. doi: 10.1172/JCI33764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Orelio C, Verkuijlen P, Geissler J, van den Berg TK, Kuijpers TW. SBDS expression and localization at the mitotic spindle in human myeloid progenitors. PLoS ONE. 2009;4(9):e7084. doi: 10.1371/journal.pone.0007084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zinsser F. Atrophia Cutis Reticularis cum Pigmentations, Dystrophia Unguium et Leukoplakis oris (Poikioodermia atrophicans vascularis Jacobi). Ikonographia Dermatologica. 1910;5:219–223. [Google Scholar]
- 49.Walne AJ, Dokal I. Dyskeratosis congenita: a historical perspective. Mech Ageing Dev. 2008;129(1):48–59. doi: 10.1016/j.mad.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 50.Dokal I, Vulliamy T. Dyskeratosis congenita: its link to telomerase and aplastic anaemia. Blood Rev. 2003;17(4):217–225. doi: 10.1016/s0268-960x(03)00020-1. [DOI] [PubMed] [Google Scholar]
- 51.Handley TP, McCaul JA, Ogden GR. Dyskeratosis congenita. Oral Oncol. 2006;42(4):331–336. doi: 10.1016/j.oraloncology.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 52.Kirwan M, Dokal I. Dyskeratosis congenita: a genetic disorder of many faces. Clin Genet. 2008;73(2):103–112. doi: 10.1111/j.1399-0004.2007.00923.x. [DOI] [PubMed] [Google Scholar]
- 53.Heiss NS, Knight SW, Vulliamy TJ, et al. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat Genet. 1998;19(1):32–38. doi: 10.1038/ng0598-32. [DOI] [PubMed] [Google Scholar]
- 54.Marrone A, Walne A, Tamary H, et al. Telomerase reverse-transcriptase homozygous mutations in autosomal recessive dyskeratosis congenita and Hoyeraal-Hreidarsson syndrome. Blood. 2007;110(13):4198–4205. doi: 10.1182/blood-2006-12-062851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamaguchi H, Calado RT, Ly H, et al. Mutations in TERT, the gene for telomerase reverse transcriptase, in aplastic anemia. N Engl J Med. 2005;352(14):1413–1424. doi: 10.1056/NEJMoa042980. [DOI] [PubMed] [Google Scholar]
- 56.Ruggero D, Grisendi S, Piazza F, et al. Dyskeratosis congenita and cancer in mice deficient in ribosomal RNA modification. Science. 2003;299(5604):259–262. doi: 10.1126/science.1079447. [DOI] [PubMed] [Google Scholar]
- 57.McKusick VA, Eldridge R, Hostetler JA, Ruangwit U, Egeland JA. Dwarfism in the Amish: II. Cartilage-hair hypoplasia. Bull Johns Hopkins Hosp. 1965;116:285–326. [PubMed] [Google Scholar]
- 58.Mäkitie O. Cartilage-hair hypoplasia in Finland: epidemiological and genetic aspects of 107 patients. J Med Genet. 1992;29(9):652–655. doi: 10.1136/jmg.29.9.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ridanpää M, Sistonen P, Rockas S, Rimoin DL, Mäkitie O, Kaitila I. Worldwide mutation spectrum in cartilage-hair hypoplasia: ancient founder origin of the major70A→G mutation of the untranslated RMRP. Eur J Hum Genet. 2002;10(7):439–447. doi: 10.1038/sj.ejhg.5200824. [DOI] [PubMed] [Google Scholar]
- 60.Berthet F, Siegrist CA, Ozsahin H, et al. Bone marrow transplantation in cartilage-hair hypoplasia: correction of the immunodeficiency but not of the chondrodysplasia. Eur J Pediatr. 1996;155(4):286–290. doi: 10.1007/BF02002714. [DOI] [PubMed] [Google Scholar]
- 61.Ridanpää M, van Eenennaam H, Pelin K, et al. Mutations in the RNA component of RNase MRP cause a pleiotropic human disease, cartilage-hair hypoplasia. Cell. 2001;104(2):195–203. doi: 10.1016/s0092-8674(01)00205-7. [DOI] [PubMed] [Google Scholar]
- 62.Welting TJ, van Venrooij WJ, Pruijn GJ. Mutual interactions between subunits of the human RNase MRP ribonucleoprotein complex. Nucleic Acids Res. 2004;32(7):2138–2146. doi: 10.1093/nar/gkh539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thiel CT, Horn D, Zabel B, et al. Severely incapacitating mutations in patients with extreme short stature identify RNA-processing endoribonuclease RMRP as an essential cell growth regulator. Am J Hum Genet. 2005;77(5):795–806. doi: 10.1086/497708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sarafan-Vasseur N, Lamy A, Bourguignon J, et al. Overexpression of B-type cyclins alters chromosomal segregation. Oncogene. 2002;21(13):2051–2057. doi: 10.1038/sj.onc.1205257. [DOI] [PubMed] [Google Scholar]
- 65.Maida Y, Yasukawa M, Furuuchi M, et al. An RNA-dependent RNA polymerase formed by TERT and the RMRP RNA. Nature. 2009;461(7261):230–235. doi: 10.1038/nature08283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Treacher Collins E. Case with symmetrical congenital notches in the outer part of each lower lid and defective development of the malar bones. Trans Ophthalmol Soc UK. 1900;20:90. [Google Scholar]
- 67.Sakai D, Trainor PA. Treacher Collins syndrome: unmasking the role of Tcof1/treacle. Int J Biochem Cell Biol. 2009;41(6):1229–1232. doi: 10.1016/j.biocel.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Posnick JC, Ruiz RL. Treacher Collins syndrome: current evaluation, treatment, and future directions. Cleft Palate Craniofac J. 2000;37(5):434. doi: 10.1597/1545-1569(2000)037<0434:TCSCET>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 69.Treacher Collins Syndrome Collaborative Group. Positional cloning of a gene involved in the pathogenesis of Treacher Collins syndrome. Nat Genet. 1996;12(2):130–136. doi: 10.1038/ng0296-130. [DOI] [PubMed] [Google Scholar]
- 70.Valdez BC, Henning D, So RB, Dixon J, Dixon MJ. The Treacher Collins syndrome (TCOF1) gene product is involved in ribosomal DNA gene transcription by interacting with upstream binding factor. Proc Natl Acad Sci U S A. 2004;101(29):10709–10714. doi: 10.1073/pnas.0402492101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dixon J, Jones NC, Sandell LL, et al. Tcof1/Treacle is required for neural crest cell formation and proliferation deficiencies that cause craniofacial abnormalities. Proc Natl Acad Sci U S A. 2006;103(36):13403–13408. doi: 10.1073/pnas.0603730103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jones NC, Lynn ML, Gaudenz K, et al. Prevention of the neurocristopathy Treacher Collins syndrome through inhibition of p53 function. Nat Med. 2008;14(2):125–133. doi: 10.1038/nm1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kumar S, Bandyopadhyay U. Free heme toxicity and its detoxification systems in human. Toxicol Lett. 2005;157(3):175–188. doi: 10.1016/j.toxlet.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 74.McGowan KA, Li JZ, Park CY, et al. Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nat Genet. 2008;40(8):963–970. doi: 10.1038/ng.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fumagalli S, Di Cara A, Neb-Gulati A, et al. Absence of nucleolar disruption after impairment of 40S ribosome biogenesis reveals an rpL11-translation-dependent mechanism of p53 induction. Nat Cell Biol. 2009;11(4):501–508. doi: 10.1038/ncb1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Keel SB, Doty RT, Yang Z, et al. A heme export protein is required for red blood cell differentiation and iron homeostasis. Science. 2008;319(5864):825–828. doi: 10.1126/science.1151133. [DOI] [PubMed] [Google Scholar]
- 77.Blázquez-Domingo M, Grech G, von Lindern M. Translation initiation factor 4E inhibits differentiation of erythroid progenitors. Mol Cell Biol. 2005;25(19):8496–8506. doi: 10.1128/MCB.25.19.8496-8506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Campagnoli MF, Ramenghi U, Armiraglio M, et al. RPS19 mutations in patients with Diamond-Blackfan anemia. Hum Mutat. 2008;29(7):911–920. doi: 10.1002/humu.20752. [DOI] [PubMed] [Google Scholar]
- 79.Cmejlova J, Dolezalova L, Pospilova D, Petrtylova K, Petrak J, Cmejla R. Translational efficiency in patients with Diamond-Blackfan anemia. Haematologica. 2006;91(11):1456–1464. [PubMed] [Google Scholar]
- 80.Perdahl EB, Naprstek BL, Wallace WC, Lipton JM. Erythroid failure in Diamond-Blackfan anemia is characterized by apoptosis. Blood. 1994;83(3):645–650. [PubMed] [Google Scholar]
- 81.Flygare J, Kiefer T, Miyake K, et al. Deficiency of ribosomal protein S19 in CD34+ cells generated by siRNA blocks erythroid development and mimics defects seen in Diamond-Blackfan anemia. Blood. 2005;105(12):4627–4634. doi: 10.1182/blood-2004-08-3115. [DOI] [PubMed] [Google Scholar]
- 82.Ebert BL, Lee MM, Pretz JL, et al. An RNA interference model of RPS19 deficiency in Diamond-Blackfan anemia recapitulates defective hematopoiesis and rescue by dexamethasone: identification of dexamethasone-responsive genes by microarray. Blood. 2005;105(12):4620–4626. doi: 10.1182/blood-2004-08-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Miyake K, Utsugisawa T, Flygare J, et al. Ribosomal protein S19 deficiency leads to reduced proliferation and increased apoptosis but does not affect terminal erythroid differentiation in a cell line model of Diamond-Blackfan anemia. Stem Cells. 2008;26(2):323–329. doi: 10.1634/stemcells.2007-0569. [DOI] [PubMed] [Google Scholar]
- 84.Ohene-Abuakwa Y, Orfali KA, Marius C, Ball SE. Two-phase culture in Diamond-Blackfan anemia: localization of erythroid defect. Blood. 2005;105(2):838–846. doi: 10.1182/blood-2004-03-1016. [DOI] [PubMed] [Google Scholar]
- 85.Hamaguchi I, Ooka A, Brun A, Richter J, Dahl N, Karlsson S. Gene transfer improves erythroid development in ribosomal protein S19-deficient Diamond-Blackfan anemia. Blood. 2002;100(8):2724–2731. doi: 10.1182/blood.V100.8.2724. [DOI] [PubMed] [Google Scholar]
- 86.Danilova N, Sakamoto KM, Lin S. Ribosomal protein S19 deficiency in zebrafish leads to developmental abnormalities and defective erythropoiesis through activation of p53 protein family. Blood. 2008;112(13):5228–5237. doi: 10.1182/blood-2008-01-132290. [DOI] [PubMed] [Google Scholar]
- 87.Uechi T, Nakajima Y, Chakraborty A, Torihara H, Higa S, Kenmochi N. Deficiency of ribosomal protein S19 during early embryogenesis leads to reduction of erythrocytes in a zebrafish model of Diamond-Blackfan anemia. Hum Mol Genet. 2008;17(20):3204–3211. doi: 10.1093/hmg/ddn216. [DOI] [PubMed] [Google Scholar]
- 88.Matsson H, Davey EJ, Fröjmark AS, et al. Erythropoiesis in the Rps19 disrupted mouse: analysis of erythropoietin response and biochemical markers for Diamond-Blackfan anemia. Blood Cells Mol Dis. 2006;36(2):259–264. doi: 10.1016/j.bcmd.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 89.Sulic S, Panic L, Barkic M, Mercep M, Uzelac M, Volarevic S. Inactivation of S6 ribosomal protein gene in T lymphocytes activates a p53-dependent checkpoint response. Genes Dev. 2005;19(24):3070–3082. doi: 10.1101/gad.359305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Constantinou C, Elia A, Clemens MJ. Activation of p53 stimulates proteasome-dependent truncation of eIF4E-binding protein 1 (4E-BP1). Biol Cell. 2008;100(5):279–289. doi: 10.1042/BC20070121. [DOI] [PubMed] [Google Scholar]
- 91.Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J Biol Chem. 2000;275(12):8945–8951. doi: 10.1074/jbc.275.12.8945. [DOI] [PubMed] [Google Scholar]
- 92.Dai MS, Lu H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J Biol Chem. 2004;279(43):44475–44482. doi: 10.1074/jbc.M403722200. [DOI] [PubMed] [Google Scholar]
- 93.Dai MS, Zeng SX, Jin Y, Sun XX, David L, Lu H. Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol Cell Biol. 2004;24(17):7654–7668. doi: 10.1128/MCB.24.17.7654-7668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jin A, Itahana K, O'Keefe K, Zhang Y. Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Mol Cell Biol. 2004;24(17):7669–7680. doi: 10.1128/MCB.24.17.7669-7680.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, Vousden KH. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell. 2003;3(6):577–587. doi: 10.1016/s1535-6108(03)00134-x. [DOI] [PubMed] [Google Scholar]
- 96.Zhang Y, Wolf GW, Bhat K, et al. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol Cell Biol. 2003;23(23):8902–8912. doi: 10.1128/MCB.23.23.8902-8912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen D, Zhang Z, Li M, et al. Ribosomal protein S7 as a novel modulator of p53-MDM2 interaction: binding to MDM2, stabilization of p53 protein, and activation of p53 function. Oncogene. 2007;26(35):5029–5037. doi: 10.1038/sj.onc.1210327. [DOI] [PubMed] [Google Scholar]
- 98.Ofir-Rosenfeld Y, Boggs K, Michael D, Kastan MB, Oren M. Mdm2 regulates p53 mRNA translation through inhibitory interactions with ribosomal protein L26. Mol Cell. 2008;32(2):180–189. doi: 10.1016/j.molcel.2008.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Volarevic S, Stewart MJ, Ledermann B, et al. Proliferation, but not growth, blocked by conditional deletion of 40S ribosomal protein S6. Science. 2000;288(5473):2045–2047. doi: 10.1126/science.288.5473.2045. [DOI] [PubMed] [Google Scholar]
- 100.Lajtha LG, Oliver R. A kinetic model of the erythron. Proc R Soc Med. 1961;54:369–371. [PubMed] [Google Scholar]
- 101.Quigley JG, Yang Z, Worthington MT, et al. Identification of a human heme exporter that is essential for erythropoiesis. Cell. 2004;118(6):757–766. doi: 10.1016/j.cell.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 102.Donadieu J, Leblanc T, Bader Meunier B, et al. Analysis of risk factors for myelodysplasias, leukemias and death from infection among patients with congenital neutropenia: experience of the French Severe Chronic Neutropenia Study Group. Haematologica. 2005;90(1):45–53. [PubMed] [Google Scholar]
- 103.Alter BP, Giri N, Savage SA, Rosenberg PS. Cancer in dyskeratosis congenita. Blood. 2009;113(26):6549–6557. doi: 10.1182/blood-2008-12-192880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Taskinen M, Ranki A, Pukkala E, Jeskanen L, Kaitila I, Mäkitie O. Extended follow-up of the Finnish cartilage-hair hypoplasia cohort confirms high incidence of non-Hodgkin lymphoma and basal cell carcinoma. Am J Med Genet A. 2008;146(18):2370–2375. doi: 10.1002/ajmg.a.32478. [DOI] [PubMed] [Google Scholar]
- 105.Amsterdam A, Sadler KC, Lai K, et al. Many ribosomal protein genes are cancer genes in zebrafish. PLoS Biol. 2004;2(5):E139. doi: 10.1371/journal.pbio.0020139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.MacInnes AW, Amsterdam A, Whittaker CA, Hopkins N, Lees J. Loss of p53 synthesis in zebrafish tumors with ribosomal protein gene mutations. Proc Natl Acad Sci U S A. 2008;105(30):10408–10413. doi: 10.1073/pnas.0805036105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Grummitt CG, Townsley FM, Johnson CM, Warren AJ, Bycroft M. Structural consequences of nucleophosmin mutations in acute myeloid leukemia. J Biol Chem. 2008;283(34):23326–23332. doi: 10.1074/jbc.M801706200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Naoe T, Suzuki T, Kiyoi H, Urano T. Nucleophosmin: a versatile molecule associated with hematological malignancies. Cancer Sci. 2006;97(10):963–969. doi: 10.1111/j.1349-7006.2006.00270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Grisendi S, Bernardi R, Rossi M, et al. Role of nucleophosmin in embryonic development and tumorigenesis. Nature. 2005;437(7055):147–153. doi: 10.1038/nature03915. [DOI] [PubMed] [Google Scholar]