Abstract
Multiple endocrine neoplasia type 1 (MEN1) is a familial tumor syndrome linked to mutation of the MEN1 gene, which encodes a tumor suppressor, menin. We previously reported that menin upregulates the caspase 8 expression and promotes TNF-α-induced apoptosis. However, it remains unclear how menin upregulates caspase 8 expression and whether menin-mediated caspase 8 expression plays a role in repressing MEN1 development. Here we show that menin binds the 5’-untranslated region (5’-UTR) of the Caspase 8 locus in vivo and activates transcription of a reporter gene through the 5’-UTR. Menin directly binds the 5’-UTR in a sequence-independent manner in vitro. Moreover, Men1 ablation in cells reduces acetylation of histones H3 and H4 at the 5’-UTR of the caspase 8 locus bound by menin in vivo. Notably, the MEN1-derived menin point mutants lose their ability to bind the caspase 8 locus and fail to induce caspase 8 expression and TNF-α-mediated apoptosis. Consistent with these observations, the expression level of caspase 8 is markedly reduced in insulinomas from Men1+/− mice. Together, our results indicate that menin enhances the caspase 8 expression by binding the caspase 8 locus, and these results also suggest that menin suppresses MEN1 tumorigenesis, at least in part, by upregulating caspase 8 expression.
Multiple endocrine neoplasia type 1 (MEN1), an inherited tumor syndrome, is caused by mutation of the tumor suppressor gene, Men 1 (1,2), which encodes a protein of 610 amino acid residues, menin (3,4). Due to lack of conserved structural domains, the molecular basis for menin to act as a tumor suppressor is largely unknown. Mice with heterozygous Men1 inactivation develop a spectrum of endocrine tumors similar to those observed in patients with MEN1 syndrome (5–7). Homozygous Men1 disruption in mice leads to embryonic lethality on embryonic days 11.5–13.5 with a variety of developmental defects including aberrant organogenesis of the multiple organs such as the neural tube, the heart and the liver (8). Men1+/− mice start to develop pancreatic insulinoma at approximately the sixth month after birth, and during the process the remaining normal allele of Men1 is lost in the tumor, resulting in loss of heterozygosity (LOH) of Men1 (5,7).
Menin contains several nuclear localization signals in its C-terminal part (9,10) and regulates expression of multiple genes including Hoxc8, p27Kip1, p18Ink4c, telomerase and IGFBP-2 (11–16). Menin interacts in vitro with multiple transcription factors, such as NFκB, Smad3 and JunD (17–19). It has been reported that menin physically binds to the loci of Hoxc8, p27Kip1, p18Ink4c, and telomerase (11,12,15,20). Menin interacts with histone methyltransferase (HMT) complex containing mixed lineage leukemia (MLL) protein(11,21) and promotes histone H3 lysine 4 (H3K4) trimethylation at the loci of genes such as Hoxa9, p18Ink4c, and p27Kip1 (12,15,16,22,23). It has also been reported that menin, by interacting with histone deacetylases (HDACs), suppresses the JunD-mediated transcription of a reporter gene, whereas Tricostatin A (TSA), an HDAC inhibitor, abrogates menin-mediated repression on gene transcription (24,25). Thus, menin may regulate the gene expression by influencing the chromatin structure, including modifications of histones.
We previously demonstrated that caspase 8 expression was downregulated in menin-null MEFs, whereas complementation of menin restored the high level of caspase 8 expression (26). Caspase 8 is a crucial component in the apoptosis pathway induced by death-related receptors (27,28). Targeted caspase 8 disruption in mice leads to defects in apoptosis of lymphocytes as well as embryonic lethality (28). Because the caspase 8 is reduced in several types of tumors (29–31), caspase 8 may play a role in suppressing tumorigenesis by potentiating death ligand-induced apoptosis. In agreement with this, caspase 8 expression is silenced due to DNA hypermethylation at the locus in neuroblastomas, (29–31) leading to resistance of the tumor cells to death ligand-induced apoptosis. (32–34).
However, it remains unclear how menin regulates caspase 8 expression and whether the menin-dependent caspase 8 expression is relevant to tumorigenesis in MEN1 syndrome. To address these questions, we have shown that menin specifically binds the 5’-UTR of the caspase 8 locus in vivo, and this Menin-5’-UTR binding is correlated with an enhanced histone acetylation at the caspase 8 locus. The DNA fragment bound by menin in vivo also mediates menin-dependent transcriptional activation in vitro. Importantly, we have also shown that MEN1-derived menin point mutants not only lose their ability to bind the caspase 8 locus and induce caspase 8 expression, but also fail to potentiate TNF-α-induced apoptosis. Moreover, caspase 8 expression is markedly decreased in islets or insulinomas in Men1+/− mice. These results suggest that menin suppresses MEN1 tumorigenesis, at least in part, through upregulating caspase 8 expression.
EXPERIMENTAL PROCEDURES
Plasmid Construction
pMX-menin and pcDNA-menin were constructed by inserting PCR amplified human menin cDNA (U93236) into the BamH I/NotI site of pMX-puro and pcDNA3 vectors respectively as previously described (13,35). To generate L22R and A242V point mutations of menin, pMX-menin and pcDNA-menin were used as a template, and mutations were introduced to the template by the Quick Change Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). To express the GST-menin, the PCR-amplified cDNA was cloned into the BamH I/Not I site of pGEX-6P (GE Healthcare, Piscataway, NJ), and the protein was expressed in E. coli strain BL21 (DE3) as a GST-tagged protein. pGEX-F1, F2 and F3 expressing the N-terminal part, the middle and the C-terminal part of menin, respectively, were generated using pGEX-menin as a template, and expressed and purified as previously described (35). To construct pcas-Luc, genomic DNA covering −2935 to +623 of the caspase 8 locus was amplified from mouse genomic DNA, and cloned into the KpnI and SmaI site of the pGL3-basic vector (Promega, Madison, WI). Additionally, the mutant pcasM-Luc was constructed by cloning the PCR-amplified genomic DNA from −2935 to +40 of the caspase 8 locus, which lacked most of the 5’-UTR sequence, to the KpnI and XhoI site of the pGL3-basic vector.
Cell Culture, Generation of Recombinant Retroviruses, and Complementation of Menin-null MEFs
Human embryonic kidney (HEK) 293 cells and E-NX cells, which were derived from 293 cells by transfecting the cells with individual cDNAs expressing retrovirus-packaging proteins (36) were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% (v/v) fetal calf serum, penicillin (100 units/ml), and streptomycin (100 µg/ml) and used for packaging recombinant retroviruses as previously described (37). Mouse embryonic fibroblast (MEF) cell lines were generated from Men1ΔN3-8/+ mice heterozygous for the Men1 locus (7,37), and cultured in DMEM with 10%(v/v) fetal calf serum, penicillin (100 units/ml), streptomycin (100units/ml), 1% MEM nonessential amino-acid and 1% L-Glutamine. Menin-null MEFs complemented with menin or its mutants were established as previously described (26). Briefly, control retroviruses, menin or its mutants-expressing retroviruses were used to infect Men1−/− MEFs. The resulting cells were selected with 2µg/ml puromycin for 2 days.
Transfection and Luciferase Assays
E-NX cells were transfected using calcium phosphate precipitation method. For luciferase assay, 4 ×105 cells were seeded per well in 6-well plates on day 0. On day1, cells were transfected with reporter constructs, control pcDNA, pcDNA-menin, L22R or A242V as indicated. pCMV-β-Gal expressing the LacZ gene (Clontech, Mountain View, CA) was co-transfected as an internal control to normalize the luciferase activity. After overnight incubation, cells were switched to a normal medium. On day 3, cells were harvested for luciferase (Promega, Madison, WI, USA) and β-galactosidase (β-Gal) assays (Clontech, Mountain View, CA), following manufacturer’s instructions. Both Luciferase and β-galactosidase activities were measured by TR717 Microplate Luminometer (EG&G Berthold, Midway Lane Oak Ridge, TN).
Real-Time RT-PCR Quantification, Northern Blotting and Gelshift Assay
Exponentially growing MEF cells were seeded at 2×105 cells /100mm dish. After two-day culture, total RNA was isolated with a QIAGEN RNeasy® Mini Kit. Real-Time RT-PCR reaction was performed using the ABI Prism 7900HT Real-Time PCR system, with 5µg of total RNA as a template. Taqman Probes for caspase 8, Hoxc8 and GAPDH were purchased from Applied Biosystems (Mm00802247_ml, Mm00439369_m1 and Mm99999915_gl, Foster City, CA). Northern Blotting for identifying menin-induced genes was performed as previously described (35,37). Briefly, total RNA was isolated from exponentially growing MEFs using cesium chloride ultracentrifuge method. RNA (20µg) was separated on agarose gel and transferred to Hybon N+ membrane. The membranes were further incubated with the labeled caspase 8 probe or the GAPDH probe, followed by autoradiography. Gelshift assays were carried out as previously described with a double stranded 5’-UTR of the caspase 8 gene (+208 to +623) (35,38). The probe was radio-labeled with γ-32P-ATP and T4 polynucleotide kinase. (38)
Cyclic amplification and selection target (CASTing)
To amplify the potential menin-binding DNA sequence using CASTing (39), a pool of 60-mer nucleotides with 20-nucleotide random sequences in the middle part were synthesized, converted to double stranded DNA, and labeled with γ-32P-ATP. The labeled probe was incubated with each of the following proteins: GST-menin fusion protein, GST-F1 (amino acid 1–218), F2 (amino acid 219–395), F3 (amino acid 396–610), and Smad3, as described for gelshift assay. The reactions were separated on 4% native polyacrylamide gel (35). The gel, after completion of electrophoresis, was stained with colloid Coomassie Blue, followed by autoradiography. The subregion of the gel with the dsDNA probe bound by menin protein or Smad3, as assessed by comparing the Coomassie-stained protein bands on gel and the corresponding autoradiography, was excised and used to extract DNA. Using primers corresponding to the 5’-end and 3’-end sequences of the dsDNA, the purified DNA was further PCR amplified and utilized as the DNA probe for the next round of gelshift assays with either menin or Smad3, which specifically binds to DNA (40). After four rounds of amplification, PCR-amplified dsDNA was subjected to incubation with 500ng GST-menin, GST-F1, F2, or F3 or Smad3 in the presence of 1 µg poly (dI-dC). The reaction was further separated on 4% native polyacrylamide gel and followed by autoradiograph.
Western Blotting and Detection of Apoptotic Cells
MEFs were seeded at 2 × 105 cells /100mm dish on day 0. On day 2, cells were harvested and processed for Western blotting as previously described (35). Briefly, on day 0, MEFs (2 ×105 cells) were seeded in a 100mm dish. After two days of culture, cells were harvested. The whole cell lysate (50µg protein) was separated on SDS-PAGE gel and transferred to Hybond-C+ membranes. Antibodies used for Western blotting were anti-actin (sc-1615, Santa Cruz Biotech, Santa Cruz, CA), anti-menin (BL342, Bethyl Laboratory, Montgomery, TX), anti-caspase 8 (ALX-804-447-C100, Alexis Biochemicals, Lausen, Switzerland), horse radish peroxidase conjugated goat anti-rabbit IgG and sheep anti-mouse IgG (A-6154 and A-5906, Sigma, St. Louis MO).
For analyzing TNF-α induced apoptosis, MEFs were seeded at a density of 5 ×104 cells per well in 6-well dishes. On day 2, cells were treated with or without TNF-α (30ng/ml)/actinomycin D (20ng/ml). After 24 hr treatment, cells were collected for propidium iodide (PI) staining. The stained cells were subjected to Fluorescence Activating Cell Scanning analysis (FACS) (BD Biosciences LSR cytometer, San Jose, CA).
Chromatin Immunoprecipitation (ChIP) Assay
MEFs were seeded at 106 cells/100 mm dish on day 0, harvested and analyzed on day 1 with a ChIP assay kit (Upstate Biotechnology, Lake Placid, NY) according to manufacturer’s instructions. Briefly, chromatin DNA was cross-linked to proteins by formaldehyde and sheared by pulsed ultrasonication. Sheared DNA/protein complex was incubated with the antibody or control IgG overnight. Antibodies used for ChIP were anti-menin (BL342, Bethyl Laboratory, Montgomery, TX), anti-acetytelated histone H3 antibody and anti-acetylated histone H4 (Upstate, Lake Placid, NY). Antibody-precipitated DNA/protein complex was reverse-cross-linked, followed by phenol/chloroform extraction, and the precipitated DNA was used as template for conventional PCR or Taqman PCR. Primers for the 5’-UTR Taqman PCR were 5’-CCCGAGCTGGAGTTGTGA-3 ’, 5 ’-CCGCTGGTGGGAAAGGAA-3’, and the Taqman probe was 5 ’-TCACCTGTGGCCGAGTAC-3; Primers for the island were 5’-TGAGCAGTTGGCTTCTAGTCCA-3’, 5’-GCAGACCAGGAAGCTTGGTTT-3’ and the Taqman probe was 5 ’-CCAGGCTTCTGACCAATCCAGAC ACAATTA-3’.
Detection of mRNA and Protein Levels of Caspase 8 in islets/insulinomas in Men1+/− and Men1+/+ Mice
Islets were isolated using Ficoll gradient centrifugation as described previously (12). RNA was extracted from Men1+/+ or Men1+/− islets using Trizol (Invitrogen, Carlsbad, CA) and was reverse-transcribed using Retroscript Kit (Ambion Inc, Austin, TX, USA). Primers for RT-PCR analysis of caspase 8 mRNA were 5 ’-ACAATGCCCAGATTTCTCCCTAC-3’ and 5 ’-CAGACAGTATCCCCGAGGTTTG-3’. Each RT-PCR reaction was repeated at least 3 times. Islet protein were extracted using triple detergent lysis buffer (GE Healthcare, Piscataway, NJ) and processed for Western blotting using an anti-caspase 8 antibody (Alexis Biochemical, 3B10).
RESULTS
Menin Binds to the 5’-UTR of Caspase 8 Locus in vivo
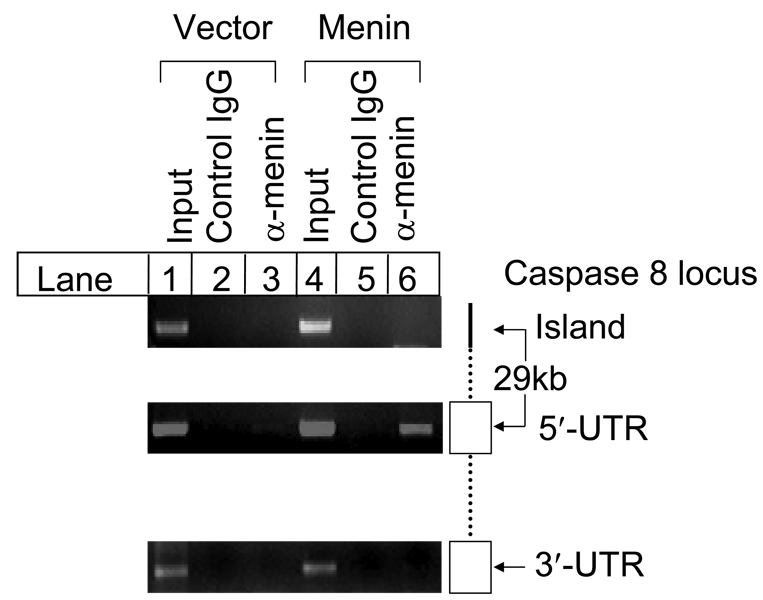
We have previously shown that menin increases caspase 8 expression specifically (26). We sought to determine whether menin binds caspase 8 locus in vivo, and if it does, whether the binding is DNA-sequence specific or not. Thus, we performed chromatin immunoprecipitation (ChIP) assay in Menin-null MEFs complemented with vector or menin to address these questions. Three pairs of primers used for ChIP target three distinct regions of the caspase 8 locus, including 5’-UTR, 3’-UTR, and a region containing a ~80 bp fragment that was highly conserved between human and murine genomic DNA with 78% identity at the 5’ end of the caspase 8 locus, which was named as “island”. ChIP assay indicates that anti-menin antibody, but not the control IgG, pulled down the 5’-UTR fragment in menin-expressing cells (Fig.1, lanes 5 and 6, middle panel), while anti-menin antibody failed to pull down the DNA fragment in vector cells (Fig. 1, lane 3). In contrast, menin failed to pull down the island and 3’-UTR regions in either menin-expressing or menin-null cells (Fig. 1, lanes 3 and 6, top and bottom panels). These results demonstrate that menin specifically associated with the 5’-UTR region of the caspase 8 locus in vivo.
Fig. 1.
Menin specifically binds to the caspase 8 locus in vivo. Menin-null or menin-expressing MEFs were seeded at 1×106 cells per 100mm dish on day 0. On day 1, cells were processed to cross-link the DNA/protein complexes, which were incubated with anti-menin antibody or rabbit IgG control. On day 2, DNA/complexes were precipitated with protein A agarose beads, followed by reverse-crosslinking. Purified DNA was used for PCR templates with 0.1% input as positive controls. A pair of primers covering 5’-UTR, +208 to +623 from transcription start site (TSS), were used for PCR amplification
We have previously shown that menin binds dsDNA in vitro (35). However, it remains unknown whether menin binds the 5’-UTR of caspase 8 in vitro, and if it does, whether menin binds it in a DNA sequence-dependent manner. To address this question, we amplified the 5’-UTR and end-labeled it with 32Phosphate (32P). The labeled probe was used to perform gelshift assay to determine whether the probe binds the C-terminus of menin (GST-menin fusion protein, GST-F3, amino acid 396–610, Fig. 2A), which was previously shown to bind dsDNA (35). Our results clearly show that GST-F3 binds the 5’-UTR DNA fragment (Fig. 2A, lane 3 and 4). We further tested whether the menin-DNA binding is affected by non-specific dsDNA. As shown in Fig. 2A, even the lowest amount of poly (dI-dC) (0.125µg), a homopolymer of non-specific dsDNA, markedly blocked GST-F3 binding to the 5’-UTR DNA fragment (Fig. 2A, lanes 5–18), suggesting that C-terminal menin (residues 396–610) binds the 5 ’-UTR in a sequence-independent manner.
Fig. 2.
Menin binds the 5’-UTR of caspase 8 gene in a DNA sequence-independent manner. (A) Menin binding to the 5’-UTR was competed away by poly (dI-dC). 5’-UTR fragment, +208 to +623, was end-labeled by [γ-32P] ATP, and incubated with or without 500ng GST-F3 for gelshift assay. GST-F3 and DNA interaction was also competed by 0, 0.125, 0.25, 0.5 or 1.0 µg poly dI-dC. After 10 min incubation, DNA-protein complexes were separated and detected, as described in EXPERIMENTAL PROCEDURES. (B) Selection of potential menin or Smad3 binding DNA sequences from a pool of double stranded oligonucleotides. Menin or its subfragmentsand, including GST-F1 (amino acid 1–218), GST-F2 (amino acid 219–395) and GST-F3 (amino acid 396–610), Smad3 were incubated with a pool of oligonucleotides in the presence of ploy (dI-dC) (1µg). The DNA-protein complexes were separated on 4% native polyacrylamide gel for 4 rounds of amplifications, as described in EXPERIMENTAL PROCEDURES.
However, we could not rule out the possibility that menin might prefer to bind some DNA sequences with a relatively high affinity. To address this question, we applied cyclic amplification and selection of targets (CASTing) (39), a robust method, to screen for any potential menin binding sequences. To this end, a pool of double stranded oligonucleotides containing random sequence in the middle part were end-labeled with 32P. To be comprehensive, we determined whether full-length GST-menin fusion protein as well as the GST fusion proteins containing three fragments of menin, F1 (amino acid 1–218), F2 (amino acid 219–395) or F3 (amino acid 396–610) bind to the pool of the dsDNAs. GST-Smad3, known to bind DNA sequence specifically (40), was used as a positive control. The four GST-menin fusion proteins and Smad3 were incubated with the radio-labeled nucleotides, and subjected to gelshift assay. The gel was stained with sensitive colloid Coomassie blue dye, and the band corresponding to either menin or Smad3 proteins was excised and DNA was extracted.
The recovered DNA was amplified by PCR and used for the next round of gelshift assay using menin and Smad3 fusion proteins. Our results showed that after the fourth round of amplification, the amplified oligonucleotides (dsDNA) selected by Smad3 were specifically bound by Smad3 (Fig. 2B, lane 6), consistent with the previous report (40). In contrast, all of the various menin fusion proteins, including F3, failed to select any specific sequence (Fig. 2B, lanes 2–5). Taken together, these results strongly suggest that menin cannot bind DNA sequence specifically even though it has been shown to bind caspase 8 locus specifically in vivo (Fig. 1). Thus, it is very likely that menin is recruited to the caspase 8 locus through interacting with other DNA sequence-specific transcription factors and/or co-regulators.
Menin Stimulates Caspase 8 Transcription Activity Through its Binding Sites
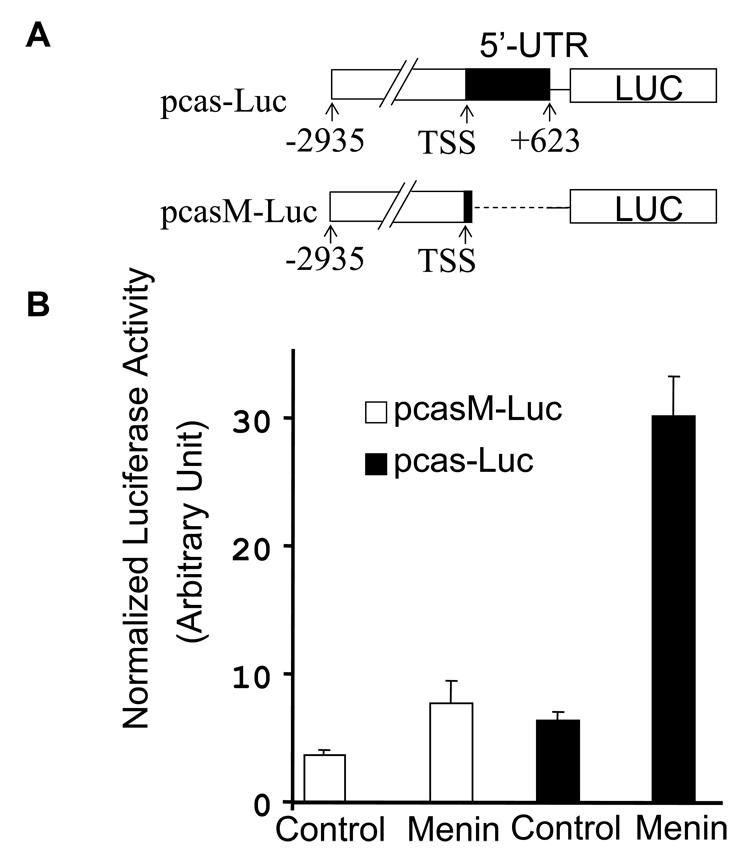
Menin binds the 5’-UTR region specifically in vivo, however, it is unclear if menin activates caspase 8 transcription through the menin-bound fragment. To address this question, we cloned the 5’ part of the caspase 8 gene containing the 5’-UTR and to the upstream of a luciferase reporter gene (Fig. 3A, pcas-luc). A luciferase reporter without menin binding site was used as a control (Fig. 3A, pcasM-luc). The resulting constructs were cotransfected into cells with or without menin cDNA, followed by luciferase and β-galactosidase assays. The results indicate that menin increased expression of the luciferase reporter driven by the menin-binding sequence by five folds, whereas the absence of the menin-binding sequence abolished menin-mediated induction of reporter expression (pcasM-Luc) (Fig. 3B). These results suggest that menin can activate reporter expression through its binding sequence, 5’-UTR, further indicating that menin regulates caspase 8 transcription by binding caspase 8 locus. It is conceivable that additional cellular factors must be required for menin-dependent expression of caspase 8 since menin does not bind DNA specifically by itself.
Fig. 3.
The 5’-UTR of caspase 8 locus, which is bound by menin in vivo, mediates menin-dependent transcription activity. (A) A diagram of constructs used for the luciferase reporter gene. TSS, transcriptional starting site. (B) Menin induces the activity of the reporter gene driven by the promoter including the menin binding fragment, the 5’-UTR (+208 to +623) of the caspase 8 gene. E-NX cells (4 ×105 cells per well) were seeded in 6-well plates on day 0. On day 1, cells in each well were cotransfected with 1µg pcas-Luc or pcasM-Luc, or 62.5ng pCMV-β-Gal and 1 µg vector control or menin expressing construct (pcDNA-menin), using calcium phosphate precipitation method. On day 3, cells were harvested for luciferase and β-galactosidase assays. All the luciferase activity was normalized to the β- galactosidase activity and presented as an average of duplicate samples.
Menin Enhances Acetylation of Histones in the Caspase 8 Locus
It has previously been reported that menin interacts with MLL complex, and menin –MLL complex methylates histone H3 lysine 4 (H3K4) (11,21). Thus, we attempted to test if MLL associates with caspase 8 locus, especially in the region bound by menin. The ChIP assay failed to detect any significant binding of MLL to the 5’-UTR and other regions of the caspase 8 locus (data not shown). Using antibodies against di- or tri-methylated lysine 4 of histone 3, we did not observe any enrichment of the modified histones around the caspase 8 locus in the presence of menin. These data suggest that menin may regulate expression of caspase 8 in a H3K4 methylation-independent manner.
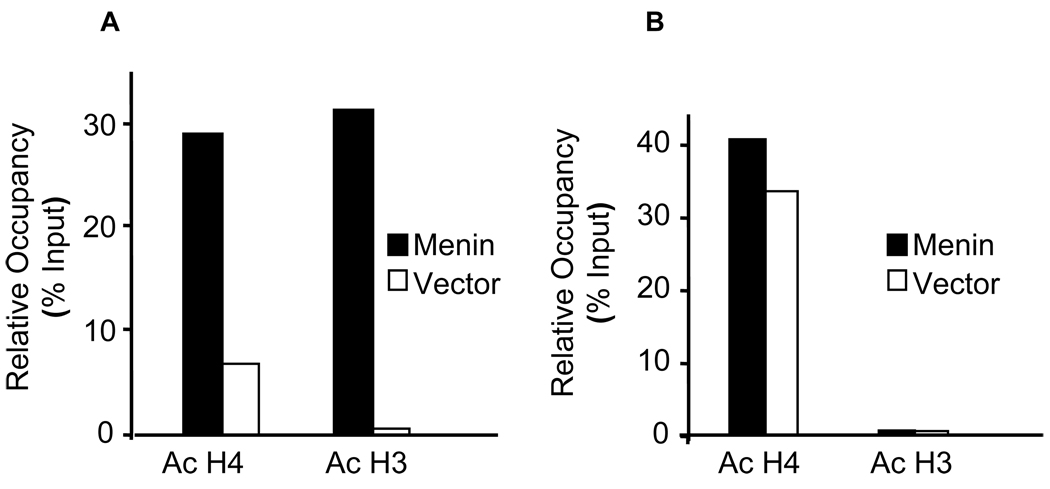
Thus, we turned our attention to the potential impact of menin on histone acetylation, which also might regulate gene expression in the caspase 8 locus. To this end, we performed ChIP assay using anti-acetylated H3 or H4 antibodies to detect the histone acetylation on 5’-UTR and the island in menin-null cells complemented with either menin or vector. The results indicate that the level of acetylated H4 at the 5’-UTR increased about 4.5 folds in menin-expressing cells as compared to that in menin-null cells (Fig. 4A). Notably, the amount of acetylated H3 in the 5’-UTR region was 38 times higher than that of the menin-null cells (Fig. 4A). In contrast, although a high level of acetylated histone H4 was detected in the island region (Fig. 4B, 40% of the input) in menin-expressing cells, loss of menin only slightly reduced the level of acetylated H4 (Fig. 4B, 35% of the input). Furthermore, the amount of acetylated H3 in the island was barely detectable in both menin-expressing and menin-null cells (Fig. 4B). Together, these results demonstrate that menin facilitates acetylation of both histones H3 and H4 at the menin-binding 5’-UTR region, but not in the island without the menin binding site.
Fig. 4.
Menin promotes the histone acetylation at the 5’-UTR of the caspase 8 locus. Menin-null MEFs complemented with vector or menin were used for ChIP assays as described in EXPERIMENTAL PROCEDURES. The cross-linked DNA/protein complexes were extracted, and the purified DNA was used for real-time PCR with Taqman probes detecting the 5’-UTR or the island sequence. Association of acetylated histone H3 (Ac H3) and histone H4 (Ac H4) to the caspase 8 5’-UTR (A) or the island sequence (B) was expressed as the percentage of input (Relative Occupancy). The results were representative of three independent experiments.
Treating menin-null MEFs with vaproic acid, an HDAC inhibitor similar to TSA (41), markedly increased expression of Hoxc8, another known menin-target gene (P. La, data not shown). However, in the present study, increasing concentrations of valproic acid did not increase the expression of casp8 (P. La, data not shown), suggesting that menin-related acetylation of histones H3 and H4 surrounding the 5’-UTR may not be the rate-limiting factor for menin-mediated induction of caspase 8 expression.
MEN1-Derived Point Mutants Lose Their Ability to Induce Caspase 8 Expression and Sensitivity to TNF-α-induced Apoptosis
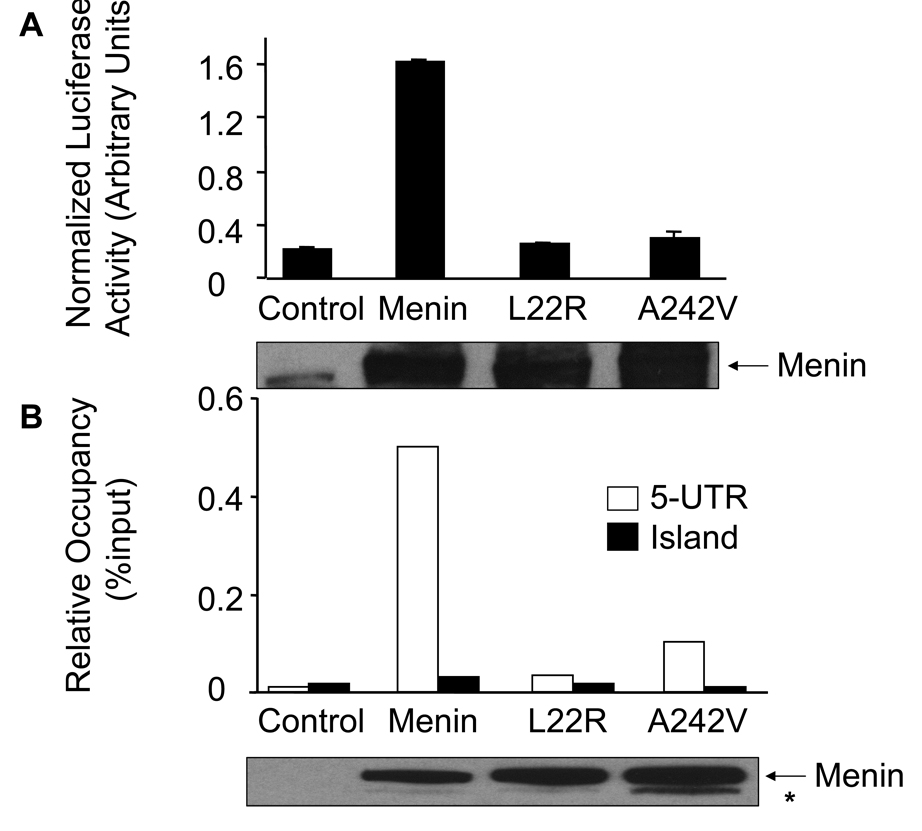
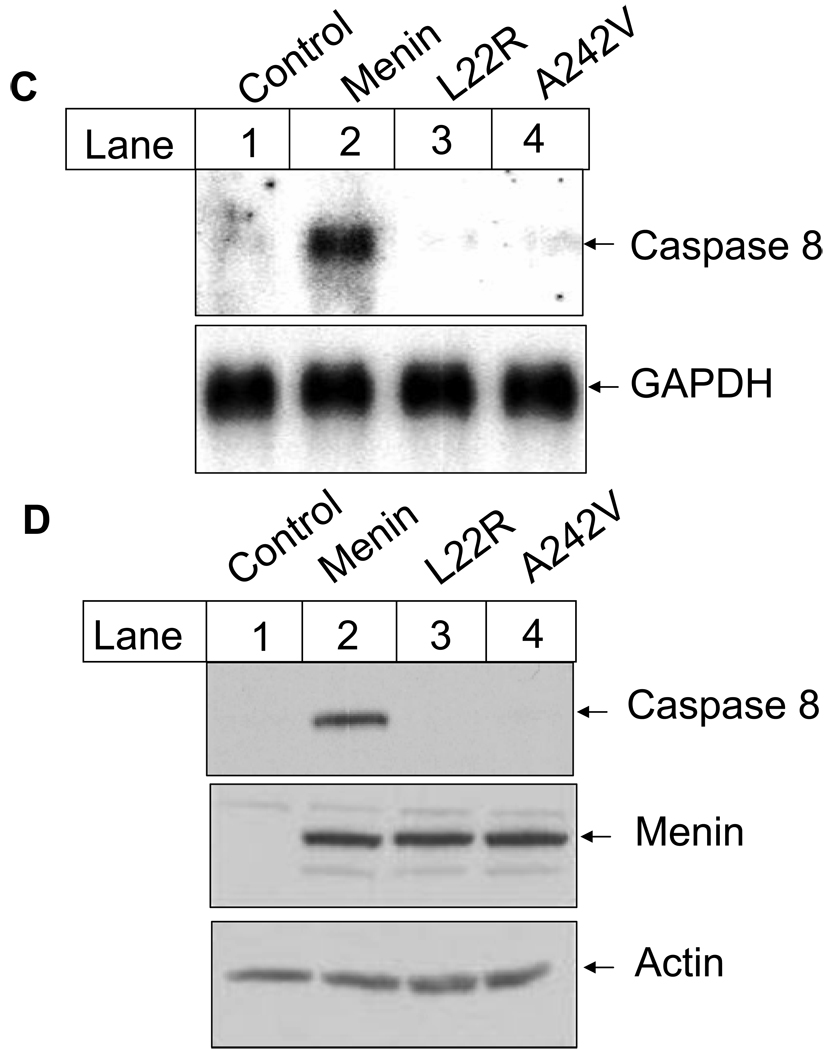
Menin is mutated in MEN1 patients, and Men1 +/− mice have a spectrum of tumors similar to that of MEN1 patients (7). Thus, we wondered if menin-dependent caspase 8 expression contributes to the repression of MEN1 tumorigenesis. We determined whether MEN1-derived menin mutations compromise the ability of menin to induce caspase 8 expression and TNF-α-mediated apoptosis. Two menin point mutants, L22R and A242V, were tested for their ability to promote caspase 8 expression. We performed the luciferase assay using the reporter driven by the caspase 8 5’-UTR, pcas-Luc, in cotransfection with either control, wild type menin or one of the mutants, L22R or A242V. Western blotting confirmed that menin, L22R and A242V were all expressed at a similar level in the transfected cells. Wild type menin induced the luciferase expression by 7.5 folds, while the L22R and A242V mutants failed to stimulate expression of the luciferase reporter gene (Fig. 5A).
Fig. 5.
MEN1-related menin point mutants reduce their ability to induce expression of caspase 8 in vitro and in vivo. (A) Luciferase assays were performed as described in Fig. 3B and in EXPERIMENTAL PROCEDURES. These data were representative of three independent experiments. Expression of wild type and mutant menin was detected using Western blotting (bottom panel).
(B) MEN1-related point mutants of menin failed to bind the 5’-UTR of the caspase 8 locus. The various cells were used for ChIP assay as indicated in Fig. 4. The experiment was performed independently three times. Expression of wild type and mutant menin was detected using Western blotting (bottom panel). Asterisk (*) represents likely a proteolyzed menin product.
(C) Menin mutants fail to induce caspase 8 expression at the mRNA level. Cells described in Fig. 5B were used for Northern blotting with probes for murine caspase 8 and GAPDH. Total RNA was isolated from exponentially growing MEFs using cesium chloride ultracentrifuge method. RNA (20µg) was separated on agarose gel and transferred to Hybon N+ membrane. The membranes were further incubated with the labeled caspase 8 probe or the GAPDH probe, followed by autoradiography.
(D) Menin point mutants fail to restore caspase 8 expression at the protein level. Menin-null MEFs or MEFs expressing either wild type menin or its mutants were used. Caspase 8 and menin were detected using corresponding primary antibodies and secondary antibodies (top and middle panel), as described in EXPERIMENTAL PROCEDURES. To confirm the equal load of protein for caspase 8 and menin, the same amount of total protein (50µg) was also subjected to the detection with anti-actin antibody (bottom panel). It appears that the affinity of the anti-menin antibody to the wild type menin or the mutants was similar.
We further determined if L22R and A242V also fail to bind the caspase 8 locus in vivo, using quantitative ChIP assay. To this end, menin-null MEFs were transduced with vector retroviruses, or retroviruses expressing either wild type menin, L22R or A242V. The resulting cell lines were used for ChIP assay. The results indicate that wild type menin binds t h e 5 ’-UTR of caspase 8 locus, as expected (Fig. 5B), though L22R and A242R fail to bind the 5’-UTR (p=0.02 and 0.03, respectively, as compared to the wild type menin). It is noteworthy that both wild type and menin mutants fail to bind the island sequence, strongly suggesting the specificity of menin binding to the caspase 8 locus (Fig. 5B). Together, these results demonstrate that menin associates with the 5’-UTR and activates caspase 8 expression, whereas the menin mutants lose the ability to associate with the 5’-UTR, and fail to activate the luciferase reporter expression. It is possible that the mutants failed to be recruited or stabilized to the caspase 8 locus, as the steady level of the mutant proteins is similar to that of the wild type menin (Fig. 5B, bottom panel).
To further test if MEN1-derived menin point mutants, L22R and A242V, lose their ability to induce caspase 8 expression, we examined the effect of the two mutants on the endogenous caspase 8 gene expression. Thus, we analyzed the mRNA and protein levels of caspase 8 in vector cells as well as cells expressing either wild type menin or one of the mutants. Northern blotting shows that complementation of the menin-null cells with wild type menin markedly increased the caspase 8 mRNA level (Fig. 5C, lane 2). In contrast, point mutants, L22R and A242V, completely lost their ability to promote caspase 8 expression at the mRNA level (Fig. 5C, lanes 3–4). Consistent with the Northern blotting results, Western Blotting also shows that only wild type menin promoted expression of caspase 8 at the protein level (Fig. 5D, lane 2), and the two menin point mutants failed to do so. As a control, wild type along with the two menin point mutants were all expressed at a similar protein level (Fig. 5D, lanes 2–4, middle panel). Together, these results demonstrate that menin binds to the 5’-UTR region in vivo and stimulates caspase 8 expression, whereas MEN1-derived point mutants lose the ability to associate with the 5’-UTR and thus fail to induce gene expression.
The response of menin-null MEFs or MEFs expressing wild type menin, mutants L22R or A242V to TNF-α-induced apoptosis was also examined. Cells were treated with TNF-α and followed by staining with propidium iodide and flow cytometry analysis. The results indicate that wild type menin potentiated TNF-α-induced apoptosis by 28% (Fig. 6), as previously reported (26). In contrast, A242V failed to potentiate TNF-α-induced apoptosis (Fig. 6, 12%). Notably, TNF-α-induced apoptosis was even repressed in cells expressing L22R (Fig. 6, 2%), and reason for this is unclear. It is possible that the mutant menin inhibits one of components in the TNF-α pathway including TNF-α receptors. Together, these results indicate menin upregulates caspase 8 expression and potentiates TNF-α-induced apoptosis, and the MEN1-derived menin point mutants failed to promote TNF-α-induced apoptosis. These findings strongly suggest that menin may control the threshold of TNF-α or other death ligand-induced apoptosis through upregulating caspase 8 expression, leading to repressing MEN1 tumorigenesis.
Fig. 6.
Menin potentiates the TNF-α-induced apoptosis. Menin-null MEFs or MEFs expressing wild type menin, mutants L22R or A242V were used for analyzing TNF-α-induced apoptosis. Cells were seeded at a density of 5 × 104 cells per well in 6-well dishes. On day 2, cells were treated with or without TNF-α (30 ng/mL) / actinomycin D (20 ng/mL). After 24 hr treatment, the cells were collected for propidium iodide (PI) staining, followed by FACS analysis.
The Reduction of Caspase 8 Expression in Mouse MEN1 Insulinomas
To further test if menin-mediated caspase 8 expression contributes to menin suppressing MEN1 tumorigenesis, we monitored the caspase 8 expression in Men1 +/− mice that develop a tumor syndrome similar to human MEN1 tumor syndrome. It has been reported that ~62 % Men1+/− mice develop hyperplasia but not adenoma in pancreatic islets at an early age (5,42). At an age of 9.5 months, most of Men1 +/− mice develop insulinomas in which the normal allele of Men1 was lost (loss of heterozygosity) (7).
To determine the caspase 8 expression during MEN1 tumorigenesis, pancreatic islets or insulinomas were isolated from age-matched Men1+/− or Men1+/+ mice. The isolated islets or insulinomas were used to prepare RNA and proteins for evaluating caspase 8 expression using RT-PCR and western blotting. At an age of 4.5 months, islets from both Men1 +/− and Men1 +/+ mice expressed a similar level of caspase 8 (Fig. 7A, lanes 2 and 4, top panel). From the age of 6.5 months to 9.5 months, however, the caspase 8 mRNA level was progressively reduced islets/insulinomas in Men1+/− mice, as compared to those in wild type mice (Fig. 7A, lanes 2 and 4, middle and bottom panel). These results are consistent with the notion that when insulinomas develop after 6 months of age and LOH occurred, caspase 8 expression was markedly reduced due to loss of Men1. In agreement with this explanation, the caspase 8 protein level was also markedly reduced in islets/insulinomas from Men1 +/− mice at age of 9.5 months, as compared with that from Men1 +/+ mice (Fig. 7B). However, due to the technical restrain, the ideal demonstration of menin LOH and reduced caspase 8 expression in the same islet cells from Men1+/− mice remains to be done. Together, these results suggest that caspase 8 may play a crucial role in menin-mediated suppression of MEN1 tumorigenesis since loss of Men1 in aged Men1 +/− mice was precisely correlated with the reduction of caspase 8 expression.
Fig. 7.
Caspase 8 expression is reduced in insulinomas in Men1+/− mice. (A) mRNA level of caspase 8 was decreased in insulinomas in Men1+/− mice. Islets or insulinomas were isolated from Men1+/− and Men1+/+ mice at different ages, and total RNA was extracted from pooled islets and/or insulinomas, and subjected to the semi-quantitative RT-PCR analysis. (B) Protein level of caspase 8 was decreased in insulinomas in Men1+/− mice at the age of 9.5 month. Total proteins were isolated from islets and/or insulinomas using the triple detergent lysis buffer, separated on SDS-PAGE gel and detected with either anti-caspase 8 or anti-actin antibodies, using an ECL kit.
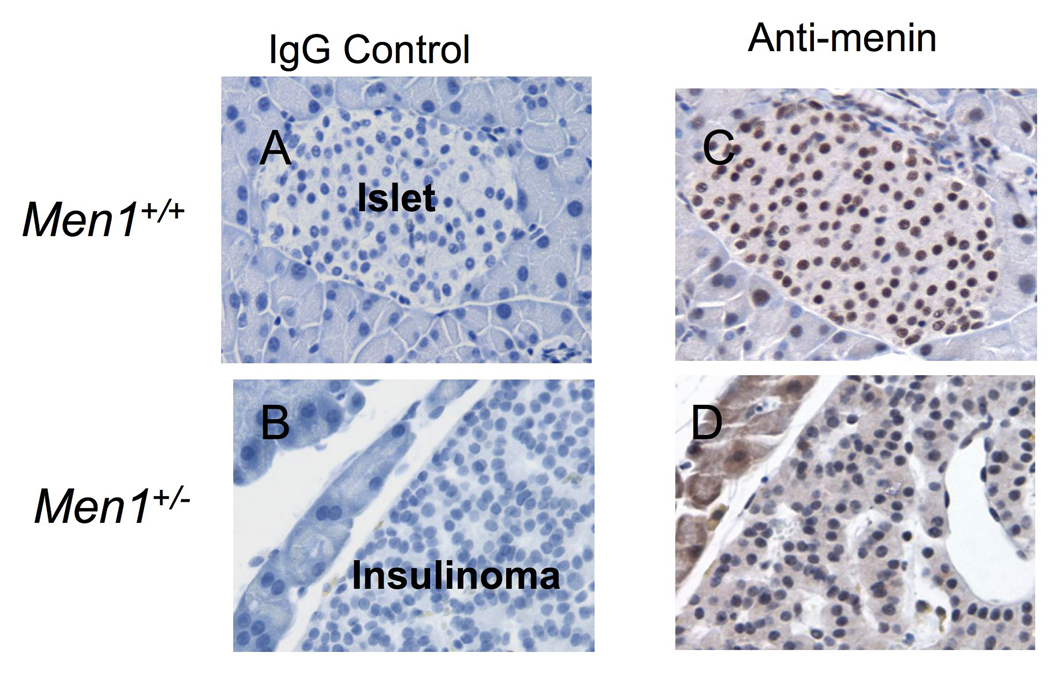
To further evaluate the role of menin as a tumor suppressor in pancreatic islets, we determined menin expression in normal islets from wild type Men1+/+ mice or in insulinoma cells from Men1+/− mutant mice, using immunohistochemistry staining. Menin was readily detectable in the nucleus of normal islet cells (Fig. 8C). In contrast, most of insulinoma cells lost menin expression (Fig. 8D). These results are consistent with other reports that demonstrated loss of heterozygosity at the Men1 locus in insulinomas from Men1+/− mice (5,7), suggesting that menin functions as a tumor suppressor.
Fig. 8.
Insulinoma cells from Men1+/− mice lose menin expression. Men1+/+ or Men1+/− mice (9 months old) were sacrificed and pancreas were collected from the corresponding mice and prepared for immunostaining for menin expression. A & B, Negative control primary antibody (control IgG) was used to stain the islet from Men1+/+ mice (A) or the insulinoma from Men1+/− mice (B). C & D, Islets from Men1+/+ mice (C) or insulinoma from Men1+/− mice (D), respectively, were stained with the anti-menin antibody (Bethyl Lab). Samples were fixed in 10% neutral buffered formalin, embedded in paraffin and sectioned at 4 to 6 µm. Slides were pretreated by incubating in antigen retrieval solution (Vector Laboratories) in a boiling water bath prior to treatment with 30% hydrogen peroxide to quench endogenous peroxidases. Non-specific protein binding was blocked using PBS-0.5%Tween 20 with 5% newborn calf serum and 5% dry milk for 1 hour at room temperature. The slides were stained with either the control IgG (A and B) or with the anti-menin antibody (Bethyl Lab, C and D) in PBS-0.25% Tween 20 with 4% BSA and 5% newborn calf serum. A secondary antibody from the Vecta Elite kit (Vector Laboratories) was used with DAB as the chromogen. Slides were then counterstained with Harris Hematoxylin.
DISCUSSION
We and others have previously shown that ectopic expression of menin causes apoptosis in vitro (26,43,44). Consistent with the role of menin in apoptosis, menin also induces expression of caspase 8, a proapoptotic protease (27). However, it was poorly understood how menin induces caspase 8 expression, and whether menin-mediated caspase 8 expression is crucial for suppressing MEN1 tumorigenesis. Our current findings demonstrate that menin specifically binds the caspase 8 locus in vivo and stimulates transcription of the reporter gene driven by the menin-binding DNA fragment from the caspase 8 locus. Importantly, MEN1-derived menin point mutants lose their ability to induce caspase 8 expression and TNF-α-induced apoptosis. Caspase 8 expression is markedly decreased in islets or insulinomas from Men1+/− mice. Thus, menin-dependent caspase 8 expression may sensitize death ligand-induced apoptosis in normal islets, and the dysregulation of caspase 8 expression caused by MEN1 mutations may in part contribute to the development of MEN 1 syndrome.
Menin Specifically Binds the Caspase 8 Locus in vivo to regulate its expression
Menin regulates transcription of a number of endogenous genes including hTERT, Hoxc8, p27Kip1, p18Ink4c and IGFBP-2(11–15,45). Menin has also been shown to bind the loci of most of these genes in vivo as demonstrated by ChIP assays. We have demonstrated that menin specifically binds to the 5’-UTR of the caspase 8 locus, and the menin-bound sequence also mediates menin-induced transcription of the reporter gene. Our previous studies have shown that menin binds dsDNA (35). We have now further demonstrated that menin binds dsDNA in a sequence-independent manner, as shown by CASTing assay (Fig. 2B). Since menin specifically binds to the 5’-UTR of the caspase 8 gene in vivo (Fig. 1) but binds dsDNA non-specifically in vitro, these results suggest that menin may be recruited directly or indirectly by sequence specific DNA binding transcription factors or coregulators to the caspase 8 locus. In agreement with the reasoning, comparison of the DNA sequences from these menin bound fragments from caspase 8, p18Ink4c, p27Kip1, and Hoxa9 did not show significant homology (data not shown).
Consistent with this notion, a recent report shows that menin interacts with ligand-activated estrogen receptor to stimulate transcription of an endogenous gene (46). Thus, it is likely that menin interacts with a transcription factor or a coregulator, and then targets the caspase 8 locus to modulate caspase 8 transcription. We have previously shown that two patches of positively charged residues in the C-terminus of menin mediate binding to dsDNA, and mutations of these residues reduce caspase 8 expression in cells (10,35) It is likely that menin-DNA interaction, albeit non-specific, is crucial for stabilizing the transcriptional machinery at the caspase 8 locus. Since mutations in mutants L22R and A242V reside in the N-terminal and middle parts of menin, outside of the DNA-binding C-terminus, these mutants may not directly affect the ability of menin to bind DNA . Nevertheless, we cannot rule out that the mutations can indirectly affect the DNA binding.
As menin interacts with H3K4 methyltransferases (11,21) and affects the H3K4 methylation at the Hoxa9 locus (22,23), we analyzed the MLL related H3K4 methylation at the caspase 8 gene. The ChIP assay did not show significant impact of menin on the H3K4 modification. This is consistent with a recent report that a substantial number of menin target genes may not be co-regulated by MLL (47). Instead, we found that menin enhances H3 and H4 acetylation specifically at caspase 8 5’-UTR, which is bound by menin. However, menin does not affect the status of histones H3 and H4 acetylation at the island that is further upstream of the 5’-UTR, and menin does not bind the island sequence in vivo. These results suggest that menin may bind to the 5’-UTR and affect the acetylation of the histones surrounding the menin-binding region, resulting in activation of caspase 8 transcription.
It is unclear how menin regulates acetylation of the histones, which is usually correlated with transcriptional activation (48). It is likely that menin directly or indirectly affects the activity of histone acetyltransferases, but not HDACs, at the caspase 8 locus, since HDAC inhibitor, valproic acid (VPA), did not affect caspase 8 expression (P. L., data not shown). On the other hand, it is also possible that the impact of menin on histone acetylation is not a rate-limiting factor in regulating caspase 8 transcription.
Menin May Suppress MEN1 Tumorigenesis Through Upregulating Caspase 8 and Potentiating Death Ligand-Induced Apoptosis
Caspase 8 plays a pivotal role in apoptosis induced by death ligands including TNF-α (49). Expression of caspase 8 is silenced by hypermethylation of DNA in the caspase 8 locus in neuroblastomas. Re-expression of caspase 8 sensitizes the tumor cells to death ligand-induced apoptosis (32–34), suggesting a potential role for caspase 8 in suppressing the development of neuroblastomas. We have previously shown that menin enhances caspase 8 expression, caspase 8 enzymatic activity, and apoptosis in MEFs upon treatment with TNF-α (26). However, it remains unclear whether menin-mediated caspase 8 expression contributes to suppressing MEN1 tumorigenesis.
Thus, crucial questions remain as to whether menin mutations derived from MEN1 patients affect caspase 8 expression and sensitivity to TNF-α-induced apoptosis. Our current findings indicate that the tested missense point mutants of menin not only fail to upregulate caspase 8 mRNA and protein but also lose menin’s ability to potentiate TNF-α-induced apoptosis. Consistent with these results, the menin mutants, as compared to the wild type menin, also failed to bind the caspase 8 locus in vivo, and lost their ability to upregulate reporter expression in vitro.
We observed that caspase 8 expression in MEN1 insulinomas was progressively decreased, following the trend of the development of insulinomas in Men1+/− mice. These results indicate that accompanying the LOH of Men1, which takes place in most of the insulinomas (7), caspase 8 expression also decreases. This suggests that loss of menin expression in MEN1 insulinomas leads to a reduction of caspase 8 expression. These observations enforce the notion that menin-dependent caspase 8 expression plays a crucial role in repressing MEN1 tumorigenesis. Supporting this conclusion, caspase 8 is frequently silenced in neuroblastomas and ectopic expression of caspase 8 in the tumor cells render the cells sensitive to death ligand-induced apoptosis (29,32–34).
Thus, our results are consistent with a model in which menin normally binds to the caspase 8 locus, alters chromatin structure and increases caspase 8 expression. The upregulated level of caspase 8 may potentiate death ligand-mediated apoptosis in endocrine cells, resulting in suppression of MEN1 tumorigenesis. Since menin is also actively involved in repressing cell proliferation through upregulating p18Ink4c, p27Kip1 and repressing CDK2 and Cdc7/ASK activity (12,15,16,50) and genome stability (37,51), menin-dependent caspase 8 expression may coordinate with other menin functions in suppressing MEN1 tumorigenesis.
As targeted disruption of the caspase-8 locus in mice resulted in embryonic lethality, cardiac deformations, the neural tube defects and deficiency in proliferation of T lymphocytes (28,52), it remains unclear whether the non-apoptotic function of caspase 8 is also involved in suppressing MEN1-related tumorigenesis. Investigation of the precise role of caspase 8 in MEN1 development using conditional caspase 8 knock out mice is currently underway.
Acknowledgments
This work is in part supported by NIH grants (R01 CA113962 and R01 CA100912 to XH). We appreciate the valuable comments from other members of our laboratories.
The abbreviations used are
- MEN 1
multiple endocrine neoplasia type 1
- 5’-UTR
5’-untranslated region
- 3’-UTR
3’-untranslated region
- TNF-α
tumor necrosis factor α
- MEFs
mouse embryo fibroblasts
- MLL
mixed lineage leukemia protein
- IGFBP-2
insulin-like growth factor binding protein 2
- HDAC
histone deacetylase
- LOH
loss of heterozygosity
- HMT
histone methyltransferase
- TSA
Tricostatin A
- VPA
valproic acid
- β-Gal
β-galactosidase
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- CASTing
cyclic amplification and selection target
- PI
propidium iodide
- FACS
Fluorescence activating cell scanning analysis
- ChIP
chromatin immunoprecipitation
- Ac H3
acetylated histone H3
- Ac H4
acetylated histone H4
REFERENCES
- 1.Marx SJ, Stratakis CA. J Intern Med. 2005;257(1):2–5. doi: 10.1111/j.1365-2796.2004.01419.x. [DOI] [PubMed] [Google Scholar]
- 2.Pannett AA, Thakker RV. Endocr Relat Cancer. 1999;6(4):449–473. doi: 10.1677/erc.0.0060449. [DOI] [PubMed] [Google Scholar]
- 3.Chandrasekharappa SC, Guru SC, Manickam P, Olufemi SE, Collins FS, Emmert-Buck MR, Debelenko LV, Zhuang Z, Lubensky IA, Liotta LA, Crabtree JS, Wang Y, Roe BA, Weisemann J, Boguski MS, Agarwal SK, Kester MB, Kim YS, Heppner C, Dong Q, Spiegel AM, Burns AL, Marx SJ. Science. 1997;276(5311):404–407. doi: 10.1126/science.276.5311.404. [DOI] [PubMed] [Google Scholar]
- 4.Lemmens I, Van de Ven WJ, Kas K, Zhang CX, Giraud S, Wautot V, Buisson N, De Witte K, Salandre J, Lenoir G, Pugeat M, Calender A, Parente F, Quincey D, Gaudray P, De Wit MJ, Lips CJ, Hoppener JW, Khodaei S, Grant AL, Weber G, Kytola S, Teh BT, Farnebo F, Thakker RV, et al. Hum Mol Genet. 1997;6(7):1177–1183. doi: 10.1093/hmg/6.7.1177. [DOI] [PubMed] [Google Scholar]
- 5.Bertolino P, Tong WM, Galendo D, Wang ZQ, Zhang CX. Mol Endocrinol. 2003;17(9):1880–1892. doi: 10.1210/me.2003-0154. [DOI] [PubMed] [Google Scholar]
- 6.Biondi CA, Gartside MG, Waring P, Loffler KA, Stark MS, Magnuson MA, Kay GF, Hayward NK. Mol Cell Biol. 2004;24(8):3125–3131. doi: 10.1128/MCB.24.8.3125-3131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crabtree JS, Scacheri PC, Ward JM, Garrett-Beal L, Emmert-Buck MR, Edgemon KA, Lorang D, Libutti SK, Chandrasekharappa SC, Marx SJ, Spiegel AM, Collins FS. Proc Natl Acad Sci U S A. 2001;98(3):1118–1123. doi: 10.1073/pnas.98.3.1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bertolino P, Radovanovic I, Casse H, Aguzzi A, Wang ZQ, Zhang CX. Mech Dev. 2003;120(5):549–560. doi: 10.1016/s0925-4773(03)00039-x. [DOI] [PubMed] [Google Scholar]
- 9.Guru SC, Goldsmith PK, Burns AL, Marx SJ, Spiegel AM, Collins FS, Chandrasekharappa SC. Proc Natl Acad Sci U S A. 1998;95(4):1630–1634. doi: 10.1073/pnas.95.4.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.La P, Desmond A, Hou Z, Silva AC, Schnepp RW, Hua X. Oncogene. 2006 doi: 10.1038/sj.onc.1209400. [DOI] [PubMed] [Google Scholar]
- 11.Hughes CM, Rozenblatt-Rosen O, Milne TA, Copeland TD, Levine SS, Lee JC, Hayes DN, Shanmugam KS, Bhattacharjee A, Biondi CA, Kay GF, Hayward NK, Hess JL, Meyerson M. Mol Cell. 2004;13(4):587–597. doi: 10.1016/s1097-2765(04)00081-4. [DOI] [PubMed] [Google Scholar]
- 12.Karnik SK, Hughes CM, Gu X, Rozenblatt-Rosen O, McLean GW, Xiong Y, Meyerson M, Kim SK. Proc Natl Acad Sci U S A. 2005;102(41):14659–14664. doi: 10.1073/pnas.0503484102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.La P, Schnepp RW, C DP, A CS, Hua X. Endocrinology. 2004;145(7):3443–3450. doi: 10.1210/en.2004-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin S, Elledge SJ. Cell. 2003 June 27;113:881–889. doi: 10.1016/s0092-8674(03)00430-6. [DOI] [PubMed] [Google Scholar]
- 15.Milne TA, Hughes CM, Lloyd R, Yang Z, Rozenblatt-Rosen O, Dou Y, Schnepp RW, Krankel C, Livolsi VA, Gibbs D, Hua X, Roeder RG, Meyerson M, Hess JL. Proc Natl Acad Sci U S A. 2005;102(3):749–754. doi: 10.1073/pnas.0408836102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schnepp RW, Chen YX, Wang H, Cash T, Silva A, Diehl JA, Brown E, Hua X. Cancer Res. 2006;66(11):5707–5715. doi: 10.1158/0008-5472.CAN-05-4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agarwal SK, Guru SC, Heppner C, Erdos MR, Collins RM, Park SY, Saggar S, Chandrasekharappa SC, Collins FS, Spiegel AM, Marx SJ, Burns AL. Cell. 1999;96(1):143–152. doi: 10.1016/s0092-8674(00)80967-8. [DOI] [PubMed] [Google Scholar]
- 18.Heppner C, Bilimoria KY, Agarwal SK, Kester M, Whitty LJ, Guru SC, Chandrasekharappa SC, Collins FS, Spiegel AM, Marx SJ, Burns AL. Oncogene. 2001;20(36):4917–4925. doi: 10.1038/sj.onc.1204529. [DOI] [PubMed] [Google Scholar]
- 19.Kaji H, Canaff L, Lebrun JJ, Goltzman D, Hendy GN. Proc Natl Acad Sci U S A. 2001;98(7):3837–3842. doi: 10.1073/pnas.061358098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin SY, Elledge SJ. Cell. 2003;113(7):881–889. doi: 10.1016/s0092-8674(03)00430-6. [DOI] [PubMed] [Google Scholar]
- 21.Yokoyama A, Wang Z, Wysocka J, Sanyal M, Aufiero DJ, Kitabayashi I, Herr W, Cleary ML. Mol Cell Biol. 2004;24(13):5639–5649. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen YX, Yan J, Keeshan K, Tubbs AT, Wang H, Silva A, Brown EJ, Hess JL, Pear WS, Hua X. Proc Natl Acad Sci U S A. 2006;103(4):1018–1023. doi: 10.1073/pnas.0510347103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yokoyama A, Somervaille TC, Smith KS, Rozenblatt-Rosen O, Meyerson M, Cleary ML. Cell. 2005;123(2):207–218. doi: 10.1016/j.cell.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 24.Gobl AE, Berg M, Lopez-Egido JR, Oberg K, Skogseid B, Westin G. Biochim Biophys Acta. 1999;1447(1):51–56. doi: 10.1016/s0167-4781(99)00132-3. [DOI] [PubMed] [Google Scholar]
- 25.Kim H, Lee JE, Cho EJ, Liu JO, Youn HD. Cancer Res. 2003;63(19):6135–6139. [PubMed] [Google Scholar]
- 26.Schnepp RW, Mao H, Sykes SM, Zong WX, Silva A, La P, Hua X. J Biol Chem. 2004;279(11):10685–10691. doi: 10.1074/jbc.M308073200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen G, Goeddel DV. Science. 2002;296(5573):1634–1635. doi: 10.1126/science.1071924. [DOI] [PubMed] [Google Scholar]
- 28.Varfolomeev EE, Schuchmann M, Luria V, Chiannilkulchai N, Beckmann JS, Mett IL, Rebrikov D, Brodianski VM, Kemper OC, Kollet O, Lapidot T, Soffer D, Sobe T, Avraham KB, Goncharov T, Holtmann H, Lonai P, Wallach D. Immunity. 1998;9(2):267–276. doi: 10.1016/s1074-7613(00)80609-3. [DOI] [PubMed] [Google Scholar]
- 29.Takita J, Yang HW, Bessho F, Hanada R, Yamamoto K, Kidd V, Teitz T, Wei T, Hayashi Y. Med Pediatr Oncol. 2000;35(6):541–543. doi: 10.1002/1096-911x(20001201)35:6<541::aid-mpo9>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 30.Teitz T, Lahti JM, Kidd VJ. J Mol Med. 2001;79(8):428–436. doi: 10.1007/s001090100233. [DOI] [PubMed] [Google Scholar]
- 31.Teitz T, Wei T, Valentine MB, Vanin EF, Grenet J, Valentine VA, Behm FG, Look AT, Lahti JM, Kidd VJ. Nat Med. 2000;6(5):529–535. doi: 10.1038/75007. [DOI] [PubMed] [Google Scholar]
- 32.Hopkins-Donaldson S, Bodmer JL, Bourloud KB, Brognara CB, Tschopp J, Gross N. Cancer Res. 2000;60(16):4315–4319. [PubMed] [Google Scholar]
- 33.Hopkins-Donaldson S, Bodmer JL, Bourloud KB, Brognara CB, Tschopp J, Gross N. Med Pediatr Oncol. 2000;35(6):608–611. doi: 10.1002/1096-911x(20001201)35:6<608::aid-mpo25>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 34.Hopkins-Donaldson S, Ziegler A, Kurtz S, Bigosch C, Kandioler D, Ludwig C, Zangemeister-Wittke U, Stahel R. Cell Death Differ. 2003;10(3):356–364. doi: 10.1038/sj.cdd.4401157. [DOI] [PubMed] [Google Scholar]
- 35.La P, Silva AC, Hou Z, Wang H, Schnepp RW, Yan N, Shi Y, Hua X. J Biol Chem. 2004;279(47):49045–49054. doi: 10.1074/jbc.M409358200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grignani F, Kinsella T, Mencarelli A, Valtieri M, Riganelli D, Grignani F, Lanfrancone L, Peschle C, Nolan GP, Pelicci PG. Cancer Res. 1998;58(1):14–19. [PubMed] [Google Scholar]
- 37.Jin S, Mao H, Schnepp RW, Sykes SM, Silva AC, D'Andrea AD, Hua X. Cancer Res. 2003;63(14):4204–4210. [PubMed] [Google Scholar]
- 38.Paull TT, Cortez D, Bowers B, Elledge SJ, Gellert M. Proc Natl Acad Sci U S A. 2001;98(11):6086–6091. doi: 10.1073/pnas.111125998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ouellette MM, Chen J, Wright WE, Shay JW. Oncogene. 1992;7(6):1075–1081. [PubMed] [Google Scholar]
- 40.Zawel L, Dai JL, Buckhaults P, Zhou S, Kinzler KW, Vogelstein B, Kern SE. Mol Cell. 1998;1(4):611–617. doi: 10.1016/s1097-2765(00)80061-1. [DOI] [PubMed] [Google Scholar]
- 41.Gurvich N, Tsygankova OM, Meinkoth JL, Klein PS. Cancer Res. 2004;64(3):1079–1086. doi: 10.1158/0008-5472.can-03-0799. [DOI] [PubMed] [Google Scholar]
- 42.Bertolino P, Tong WM, Herrera PL, Casse H, Zhang CX, Wang ZQ. Cancer Res. 2003;63(16):4836–4841. [PubMed] [Google Scholar]
- 43.Hussein N, Casse H, Fontaniere S, Morera AM, Asensio MJ, Bakeli S, Lu JL, Coste I, Clemente ND, Bertolino P, Zhang CX. Eur J Cancer. 2007;43(2):402–414. doi: 10.1016/j.ejca.2006.08.038. [DOI] [PubMed] [Google Scholar]
- 44.Sayo Y, Murao K, Imachi H, Cao WM, Sato M, Dobashi H, Wong NC, Ishida T. Endocrinology. 2002;143(6):2437–2440. doi: 10.1210/endo.143.6.8950. [DOI] [PubMed] [Google Scholar]
- 45.Hua X, Liu X, Ansari DO, Lodish HF. Genes Dev. 1998;12(19):3084–3095. doi: 10.1101/gad.12.19.3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dreijerink KM, Mulder KW, Winkler GS, Hoppener JW, Lips CJ, Timmers HT. Cancer Res. 2006;66(9):4929–4935. doi: 10.1158/0008-5472.CAN-05-4461. [DOI] [PubMed] [Google Scholar]
- 47.Scacheri PC, Davis S, Odom DT, Crawford GE, Perkins S, Halawi MJ, Agarwal SK, Marx SJ, Spiegel AM, Meltzer PS, Collins FS. PLoS Genet. 2006;2(4):e51. doi: 10.1371/journal.pgen.0020051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carrozza MJ, Utley RT, Workman JL, Cote J. Trends Genet. 2003;19(6):321–329. doi: 10.1016/S0168-9525(03)00115-X. [DOI] [PubMed] [Google Scholar]
- 49.Kruidering M, Evan GI. IUBMB Life. 2000;50(2):85–90. doi: 10.1080/713803693. [DOI] [PubMed] [Google Scholar]
- 50.Schnepp RW, Hou Z, Wang H, Petersen C, Silva A, Masai H, Hua X. Cancer Res. 2004;64(18):6791–6796. doi: 10.1158/0008-5472.CAN-04-0724. [DOI] [PubMed] [Google Scholar]
- 51.Farley SM, Chen G, Guo S, Wang M, A J, Lee F, Lee F, Sawicki M. J Surg Res. 2006;133(1):29–37. doi: 10.1016/j.jss.2006.02.021. [DOI] [PubMed] [Google Scholar]
- 52.Salmena L, Lemmers B, Hakem A, Matysiak-Zablocki E, Murakami K, Au PY, Berry DM, Tamblyn L, Shehabeldin A, Migon E, Wakeham A, Bouchard D, Yeh WC, McGlade JC, Ohashi PS, Hakem R. Genes Dev. 2003;17(7):883–895. doi: 10.1101/gad.1063703. [DOI] [PMC free article] [PubMed] [Google Scholar]