Abstract
Purpose
The role of the microenvironment during the initiation and progression of carcinogenesis is now realized to be of critical importance, both for enhanced understanding of fundamental cancer biology, as well as exploiting this source of relatively new knowledge for improved molecular diagnostics and therapeutics.
Methods
This review focuses on: (1) the approaches of preparing and analyzing secreted proteins, (2) the contribution of tumor microenvironment elements in cancer, and (3) the potential molecular targets for cancer therapy.
Results
The microenvironment of a tumor is an integral part of its physiology, structure, and function. It is an essential aspect of the tumor proper, since it supplies a nurturing environment for the malignant process. A fundamental deranged relationship between tumor and stromal cells is essential for tumor cell growth, progression, and development of life threatening metastasis. Improved understanding of this interaction may provide new and valuable clinical targets for cancer management, as well as risk assessment and prevention. Non-malignant cells and secreted proteins from tumor and stromal cells are active participants in cancer progression.
Conclusions
Monitoring the change in the tumor micro-environment via molecular and cellular profiles as tumor progresses would be vital for identifying cell or protein targets for cancer prevention and therapy.
Keywords: Tumor, Microenvironment, Secreted proteins, Extracellular matrix, Molecular targets, Therapy
Introduction
Cancer is the number one cause of death in the United States for people less than 75 years old. Every year, more than 11 million people are diagnosed with cancer throughout the world and it may likely increase to 16 million by 2020. In 2005, cancer accounted for 7.6 million deaths from a total of 58 million deaths worldwide [1]. As a result, a re-evaluation of our basic assumptions concerning the nature of cancer and how to better assess risk, prevent, and medically manage is a high priority.
Cancer development has been defined as a multistep process in which somatic cells first undergo an initiating event (i.e., environmental insult) and then a second or promoting event. Both events accumulate genetic modifications. The fact that cancer cells have mutated genomes is well established [2]. A carcinogen or mutagen, for instance from tobacco smoke, when inhaled in sufficient quantity and duration may act as an adduct forming an unwanted bond on DNA and potentially mutating a gene. However, many cancers will develop as a result of a chronic inflammatory state due to infections.
For instance, this is commonly seen with Hepatitis B and C, which can be a harbinger for Hepatocellular Carcinoma. Gastric infection from Helicobacter pylori, will increase gastric cancer risk by 75%, and is the second most common type of cancer globally [3]. A few other clinical examples illustrating the association of chronic inflammation and increased cancer risk include: inflammatory bowel disease (ulcerative colitis, Crohn's disease) and colon cancer; cervical infection (human papillomavirus) and cervical cancer, and chronic reflux esophagitis resulting in Barrett's esophagus that is high risk for esophageal carcinoma. In all cases, these chronic inflammatory conditions help to establish a tumor microenvironment full of deranged proliferative signaling networks, which is largely orchestrated by inflammatory cells and is an indispensable participant in the neoplastic process [4].
The tumor microenvironment was lately recognized as the product of a developing crosstalk between different cells types. For instance, in epithelial tumors these cells include the invasive carcinoma and its stromal elements. Critical stromal elements include cancer-associated fibroblasts, which provide an essential communication network via secretion of growth factors and chemokines, inducing an altered ECM thus providing additional oncogenic signals enhancing cancer-cell proliferation and invasion [5]. Active contribution of tumor-associated stromal cells to cancer progression has been recognized [2, 6]. Stromal elements consists of the extracellular matrix (ECM) as well as fibroblasts of various phenotypes, and a scaffold composed of immune and inflammatory cells, blood and lymph vessels, and nerves.
For tumors to progress and develop into life threatening entities, they must develop four critical abilities. First, the ability to move, second the capacity to degrade tissue matrix (ECM), third the aptitude to survive in blood and finally the physical quality of being able to establish itself in a new tissue environment. But how exactly do cancer cells acquire these traits? Recent scientific evidence points towards cancer cells using activated transcription factors from development/embryology programs, thus gaining pleiotrophic abilities. The microenvironment is of critical importance for success in this process [7].
The microenvironment of cancer cells provide the necessary signals that turn on the transcription factors. Thus, it is the stromal or non-malignant cells that induce the requisite transcription programs allowing the necessary mesenchymal phenotypes to invade distant tissues and establish a new environment. The cancer cells must then shut down the transcription factor programs and reconvert from mesenchymal to epithelial cells, thus recreating themselves from the core of primary tumor cells. It is felt that the role of the microenvironment is essential in all of these steps [7].
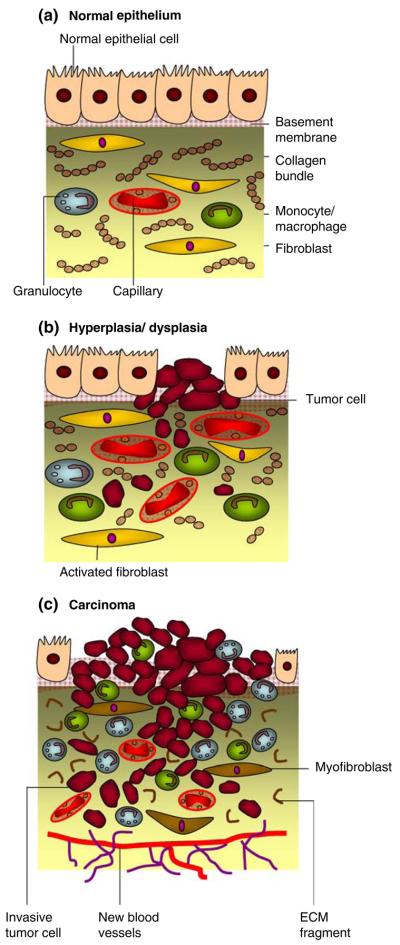
Tumor cells directly secrete a variety of proteins that include growth factors and ECM-degrading proteinases or induce the host to elaborate biomolecules that are able to degrade the matrix and its component adhesion molecules (Fig. 1). The matrix degradation takes place in a region close to the tumor cell surface, where the amount of the active degradative enzymes outbalances the natural proteinase inhibitors present in the matrix or that secreted by normal cells [8]. Proteins secreted by tumor cell into the ECM microenvironment are therefore involved in cell adhesion, motility, intercellular communication and invasion. The knowledge and control of the direct microenvironment within a growing tumor is as important as the corresponding knowledge and control of the abnormal behavior of epithelial cells within that tumor.
Fig. 1. Carcinogenesis.
a A well-differentiated stratified epithelium is separated from the stromal compartment by a well delineated basement membrane. The stromal compartment essentially consists of fibroblasts, leukocytes (monocytes and macrophages), collagen bundles and mature blood vessels. b During carcinogenesis, both epithelial and stromal components initially undergo modifications that promote epithelial cell propagation and mutation. Fibroblast becomes activated while the number of macrophages increases. Transient angiogenesis occurs with new vasculature similar to the one with normal epithelia. c Proliferation of epithelial cells with the development of an activated stroma is associated with the progression to carcinoma. Hence, extracellular-matrix components such as collagen are degraded, the number of inflammatory cells increases and fibroblasts differentiate to myofibroblasts resulting in the expression of growth factors, matrix components and degrading proteases. Angiogenesis is maintained resulting in the development of new blood vessels. Cells and molecular components of the microenvironment are targets for chemopreventive or therapeutic agents at all steps shown
Secreted proteins are of particular importance because they play a key role in a disease state or the biological pathway leading to disease, and their identification and characterization may lead to the understanding of disease patterns. In cancer, the understanding of the interactions between tumor and stroma is needed for the development of more effective therapies. The analysis of secreted proteins for proteomics studies is usually a challenge because proteins are generally secreted at low concentration in the culture media, which makes their recovery difficult due to dynamic range (concentration) constraints. Accordingly, very few studies have been published to date analyzing this important class of proteins that may help in the better understanding of tumor progression.
Although some recent reviews have focused on the contribution of cancer development for different cell types present in the tumor microenvironment [9-11], the identification and targeting of proteins secreted by cancer cells into the tumor microenvironment remains relatively unexplored. Secreted protein signatures in cancer provide important information that may be an aid to earlier diagnosis, improved disease monitoring as well as enhanced assessment of the efficacy of therapy along with rationale for required revisions of therapy. This review focuses on: (1) the approaches of preparing and analyzing secreted proteins, (2) the contribution of secreted proteins in tumor progression, and (3) the identification of secreted biomarkers for targets in tumor therapy.
Methods for preparing secreted proteins
The subset of proteins occurring in the conditioned media from cultured cells was previously called “secretome” [12]. A concrete examination of the composition of this subproteome reveals that the term secretome does not comprise only those proteins secreted through the classical pathways (i.e., Golgi and endoplasmic reticulum). The secretome in an extensive sense consists of proteins released through various mechanisms including classical secretion, secretion through non-classical pathways, and released through secretion of exosomes. Hitherto, proteomic approaches to analyze the secretome detected only a fraction of proteins secreted from the cell because many secreted proteins are expressed only by specialized cell types, have an induced expression during particular cellular response or are expressed during specific stages of development.
Detection and identification of protein secreted by cells into the ECM, has turned out to be a challenge for several reasons. In vitro, cells are grown in media rich in salts and supplemented with serum proteins. Furthermore, a very small amount of proteins are secreted by cells into the extra-cellular microenvironment. To detect this category of proteins, the growth culture media should be free of any contaminant proteins. Consequently, cells are usually grown in serum-free media with the eventual consequence of cell death. Prior to cell incubation in serum-free media, the washing step may be adjusted so that the remaining cytosolic proteins and serum are washed away without cell stressing [13-17]. The confluence of cells grown in flask or on plate are then controlled to keep enough space for cell growth, thus avoiding cell deaths due to saturation (Table 1).
Table 1.
Preparation and analysis of secreted proteins
| Mediuma | Incubation time (hours) |
Conf.b (%) |
Protein Recoveryc |
Protein concentrationd | Analyzing method | Cell type | References |
|---|---|---|---|---|---|---|---|
| Argine and lysine-free | 24 | 80 | Filtration | MWCO filter | SILAC-SDS-PAGE-LC-MS/MS | Pancreatic duct epithelial ceUs (HPDE) | Gronborg et al. [19] |
| Serum-free | Pancreatic ductal adenocarcinoma cells (Panl) | ||||||
| Methionine and cysteine-free | 2 | NA | Filtration | Precipitation (ethanol) | LC-MS/MS | Hepathocarcinoma cells (HepG2) | Zwickl et al. [20] |
| Serum and phenol-free | 24–18 | NA | Centrifugation | Ultrafiltration | 2DE-MALDI/MS | Murine myeloid cells (J774) | Chevallet et al. [13] |
| Adsorption | 2DE-LC-MS/MS | ||||||
| Phase extraction (phenol) | |||||||
| Precipitation (TCA) | |||||||
| Serum and phenol-free | 72 | 75 | Centrifugation | MWCO filter | ICAT-LC-MS/MS | Prostate cancer cells (LNCaP) | Martin et al. [80] |
| Serum-free | 48 | 60–70 | Centrifugation | MWCO filter | 2DE-MALDI/MS | Colon carcinoma cells (SW480) | Volmer et al. [12] |
| Filtration | |||||||
| Serum-free | 48 | NA | Centrifugation | Precipitation (TCA) | 2DE-MALDI/MS | Glioblastoma cells (LN-Z308) | Khwaja et al. [18] |
| ICAT-2DE-LC-MS/MS | |||||||
| Serum-free | 36 | 60-70 | Centrifugation | MWCO filter | SDS-PAGE-LC-MS/MS | Melanoma cells (UM) | Pardo et al. [16] |
| Filtration | Precipitation (TCA/acetone) | ||||||
| Serum-free | 30 | 60-70 | Filtration | SPE (tC2) | LC-MS/MS | Melanoma cells (WM 266-4) | Mbeunkui et al. [14] |
| Speed vacuum | Osteosarcoma cells (OHS) | ||||||
| Breast cancer cells (MA11) | |||||||
| Serum-free | 16 | 60 | Centrifugation | SPE (tC2) | LC-MS/MS | Breast cancer cells (MCF10A, MCF10AT, MCF10 DCIS.com, MCF10CA cl. D) | Mbeunkui et al. [15] |
| Filtration | Speed vacuum | SDS-PAGE-Immunoassay | Human tumor xenografts (MT-1) | ||||
| MWCO filter |
Modification of cell growth medium. Components added or removed from the medium
Cell confluence in the growth medium prior to harvesting
Methods for recovering secreted proteins free of floating or dead cells
Methods for concentrating proteins from the growth medium
Even a very small number of dead cells are able to release an amount of proteins that far exceeds the amount of proteins really secreted. Cell viability in serum-free media is assessed prior to secreted protein preparation either by trypane blue exclusion method [12, 15] or by assessing the presence of the most abundant cytosolic proteins in the culture media [14]. Due to the high concentration of serum protein (5–10%) supplemented in the culture media prior to starvation; serum-free medium is not always completely free of serum [14]. Hence, cells easily tolerate the transition from serum supplemented to serum-free media and the expression of stress proteins is minimized. The incubation time in serum-free media depends on each cell type and growth rate (Table 1).
Once cell growth conditions and incubation times in serum-free media are optimized, the conditioned media are recovered free of any floating cells and debris using various methods. Floating cells are removed from the conditioned media by low speed centrifugation [12, 15, 18], by filtration [14, 19, 20] or by combination of both methods [13, 17]. The concentration of secreted proteins in the conditioned media can go down to the ng/ml range and to concentrate to an amount detectable by proteomic methods is problematic. Methods such as precipitation, ultrafiltration using a molecular weigh cutoff filter, adsorption to a resin, phase extraction and speed vacuum are commonly used.
Precipitation
Precipitation is a method widely used for the recovery of biomolecules like proteins. It is generally induced by adding salts or organic solvents to the sample, or by altering its pH leading to the change of protein conformation. Ammonium sulfate is a salt commonly used for precipitation because of its high solubility and is rather inexpensive. It is an efficient method for quantitative protein recovery but it is accompanied by loss of many proteins [21]. This method is recommended for enzyme purification because the extraction media can be buffered or stabilizing agents can be added to maintain the maximum enzyme activity. The organic solvent, acetone is also frequently used for protein precipitation. It is an easy to perform concentration and desalting method that results in a good recovery. However, not all proteins precipitate and non-proteins like DNA/ RNA and glycans also precipitate, therefore the pellet is sometimes hard to resolubilize. This shortcoming can be overcome by combining trichloroacetic acid (TCA) and acetone precipitation in which proteins are first precipitated with TCA and traces of TCA removed with acetone [21].
Ultrafiltration and adsorption
Ultrafiltration is a membrane filtration process that separates particles based on their molecular weight, while adsorption is a more convenient method for salt-free concentration of small quantities of proteins for mass spectrometry and 2-DE analyses. Ultrafiltration is extensively used for secreted proteins concentration [12, 19, 22] because salts and low molecular weigh particles have been removed. It performs well in terms of purity and protein quantity but it is labor demanding to get the concentrated volume. The filters are always blocked and low molecular weigh proteins and polypeptides are lost. Adsorption is valuable for the concentration of large volume of conditioned media from cell grown in vitro. Solid-phase extraction (SPE) is the most cost-effective and versatile method for protein concentration from plasma, urine or conditioned media. The range of chemistries for SPE includes reversed phase, normal phase, ion exchange and other sorbents for special applications.
Following the methods described above for secreted protein preparation, low molecular weigh peptides are removed. Even if they are still present in the preparation, they are dismissed as noise, biological trash or too small and unstable to be biologically relevant [23]. The analysis of this class of peptides called peptidome is a rich source of cancer-specific diagnostic information because it is a recording of the extracellular enzymatic events taking place in the tumor microenvironment. Peptidomics analysis may lead to the identification of the parent protein, the determination of the fragment sizes and cleavage ends, posttranslational modifications, the quantification of the peptide and the nature of the carrier protein in which it is bound. It is a relatively new field in proteomics analysis and secreted biomarkers discovery; and hopefully it should be more sensitive and specific than conventional approaches [24].
Heavy isotope labeling
An alternative method for secreted protein preparation without worry about contamination from dead cells is to metabolically label proteins synthesized during a limited period of incubation [20]. In fact, cells are incubated in serum-free media supplemented with isotope labeled amino acids for a short time, and then washed and re-incubated in serum-free media. The conditioned media is then collected and secreted protein concentrated as described above. Proteins are run on 2D gel and those detected by autoradiography are synthesized by living cells during the period of metabolic labeling [25]. Secreted proteins are then identified by comparison with controls. This method is indeed intelligible but it suffers from its potential applicability in the secretome analysis. Metabolic turnover and the protein expression time depend on each cell type and the physiological status of cells. Moreover protein released into the medium via classical or non-classical (not signal peptide triggered protein secretion, e.g, estimated from SecretomeP 2.0 Server) pathways need a longer incubation time to be detected incorporating the isotopic labeled amino acids.
Methods for analyzing secreted protein
A series of separation, chromatography, electrophoresis and mass spectrometry-based methods, are available to analyze and elucidate secreted proteins. The 2D gel electrophoresis (2DE) has become a standard method for the analysis of the proteome in which protein mixtures are separated according to their charge, by isoelectric focusing (IEF) in the first dimension, and their molecular weight with SDS-PAGE in the second dimension. This approach coupled to mass spectrometry, is a widely used method for differential protein expression study in cancers [26]. However, performing 2DE involves many steps ranging from cell culture up to the stained gel and identification of the protein spot. Moreover, the main difficulty associated with this approach is protein precipitation at their isoelectric point during IEF, and reproducibility.
Additionally, 2DE was conceived more than 30 years ago. This technology has been useful for low-complexity protein mixtures but never matured into a comprehensive and accurate proteomics technology. The introduction of high-sensitivity protein identification by MS at first seemed to help 2D gel analysis, but subsequently revealed that the thousands of spots seen in the gel maps are actually variants of a few hundred of the most abundant proteins. Recently, it has also become clear that quantitation of even these proteins is far from accurate because of spot overlap. Accordingly, “biomarkers” found by these technologies tend to be the same regardless of the system under investigation [27].
Shotgun methods are thus a powerful alternative to gels. In these methods, proteins are digested with proteases into a more complex peptide mixture and then analyzed by LC–MS/MS. LC–MS/MS couples peptide fractionation with mass spectrometry to generate a complete record of the proteome of fluid samples [28, 29] and secreted proteins [14].
Electrospray ionization (ESI) and matrix-assisted laser desorption/ionization (MALDI), are the common techniques used to volatilize and ionize proteins or peptides for mass spectrometry analysis [30]. The main difference between these two techniques is the nature state of the analytes, which is liquid for the ESI and solid for MALDI. Therefore, MALDI–MS is generally used for the analysis of simple peptide mixtures, while ESI–MS is desired for the analysis of complex samples. Ion trap, time-of-flight (TOF), quadrupole and Fourier transform cyclotron (FT–MS) analyzers are the basic types of mass analyzers commonly used in proteomics analysis.
MS technology with low resolving power, especially in the form of the so-called surface enhanced laser desorption and ionization (SELDI) method, caught the imagination of clinicians a few years ago. This approach involves measuring a MALDI spectrum of proteins from the body fluid of a patient and then employs machine-learning-based computation to differentiate disease and healthy states. However, from a mass-spectrometric point of view, SELDI boils down to simple MALDI spectra of very complex mixtures and would be expected to only yield a subset of the most abundant low-mass peptides and protein fragments. Such species could still have proven sufficient to classify patient samples. However, as the scientific community demanded identification of the peaks comprising the SELDI patterns, these usually turned out to belong to non-specific proteins unlikely to be directly associated with the disease [27].
Even though SELDI and LC–MS/MS are technically competent for secreted proteins analysis, they remain basically qualitative approaches for secreted biomarkers discovery because peptide peaks are identified solely as present or absent in normal and cancerous specimens. In addition, very small quantities of many proteins or polypeptides are probably present in body fluids and cannot routinely be detected by SELDI or LC–MS/MS, which almost all immunoassays can detect easily. Quantitative proteomics approaches for the identification of secreted biomarkers can be coupled to immunoassays such as the traditional ELISA-based and Western blot approaches for validation [15]. Described technologies such as isotope-coded affinity tagging (ICAT) [31, 32] and stable isotope labeling with amino acids in cell culture (SILAC) [33, 34] can be combined with mass spectrometry approaches to produce efficient procedures to quantitatively analyze cancer secretomes.
Role of secreted proteins in cancer progression
Cell–cell interactions and cell–ECM interactions are known to be important during oncogenesis, and the characterization of associated proteins is essential to illuminating the primary molecular mechanism of the disease [35]. Likewise, extracellular proteins related to pre-malignancy transformation offer potential targets for early detection of cancer. The ECM is extensively modified and remodeled by proteases either secreted by non-neoplastic and neoplastic cells or localized at the cell surface. Thus, important changes in cell–cell and cell–ECM interactions arise, and new warning signs are created from the cells surface. These signals have an effect on gene expression and eventually influence critical cell behaviors such as proliferation, survival, differentiation and motility.
Proteases expressed in the extracellular milieu, and particularly matrix metalloproteinases can target ECM proteins and non-ECM proteins such as growth factors, cytokines, cell-associated molecules and growth factor receptors. For that reason, the activity of these proteases in cancer is very complex and includes tumor promoting as well as tumor suppressive effects. The balance between proteases and protease inhibitors is a vital determinant in cancer development because both an absence and an excess of proteolysis could have a negative effect on angiogenesis [36].
At the time of diagnosis, cancer has already metastasized beyond the size of initiation in more than 34% of breast cancer patients. Unfortunately, patient survival rates drop about fivefold for those diagnosed with distal spread of the disease, as compared with localized diseases [37]. The role of secreted proteins and proteases in cancer progression is very complex and diverse. Wnt proteins, for example, are a large family of secreted glycoproteins involved in the genesis of breast cancer [38]. Adipokines are another family of proteins secreted by adipose tissue and their participation in the development of breast cancers, for which obesity has been established as a risk factor, has been investigated [39, 40]. Agents targeting some secreted molecules are currently in clinical trials for cancer or have been approved by the food and drug administration (Table 2).
Table 2.
Therapeutic targeted molecules in the tumor microenvironment
| Molecular target | Drug name | Clinical triala | References |
|---|---|---|---|
| Basic Wbroblast growth factor | Interferon-alpha | II and III | Kerbel and Folkman [81] |
| Endothelial microtubules | Combretastatin A4 phosphate | II | Young and Chaplin [82] |
| Endothelin A receptor | Atrasentan | II | Jimeno and Carducci [83] |
| Fibroblast activation protein | Sibrotuzumab | I | Scott et al. [75] |
| Hypoxia-inducible factor 1α | 2-Methoxyestradiol (2-ME2) | I | Mooberry [84] |
| Integrin α5β1 | Volociximab (M200) | II | Jin and Varner [85] |
| Integrin ανβ3 | Vitaxin | II | Gutheil et al. [86]; Jin and Varner [85] |
| Integrin ανβ3 and ανβ5 | Cilengitide (EMD121974) | I and II | Burke et al. [87] |
| MMP-2, MMP-9 and MMP-12 | Neovastat (AE-941) | III | Gingras et al. [88] |
| NFκB, TNFα, IL-6 and VEGF | Thalidomid (Thalidomide) | III | Bartlett et al. [89], Sleijfer et al. [90] |
| NFκB, TNFα, IL-6 and VEGF | CC-5013 (Revimid) | II and III | Bartlett et al. [89], Sleijfer et al. [90] |
| PDGFR, c-Kit and Tyrosine kinases | Gleevec (Imatinib Mesylate) | FDA-approved | Druker et al. [91], Pietras et al. [92] |
| Protein kinase cβ | Enzastaurin (LY-317615) | II | GraV et al. [93] |
| Tyrosine kinase, VEGFR, and CSF-1R | SU11248 | II | Mendel et al. [94], Pietras and Hanahan [95] |
| Vascular endothelial growth factor | Avastin (Bevacizumab) | FDA-approved | Ferrara et al. [96], Hurwitz et al. [97] |
| Vascular endothelial growth factor | Neovastat (AE-941) | III | Gingras et al. [88] |
| Vascular endothelial growth factor A | VEGF-Trap | I and II | Holash et al. [98] |
| Vascular endothelial growth factor receptor | Vatalanib (PTK787/ZK 222584) | II and III | Manley et al. [99]; Morgan et al. [100] |
| Vascular endothelial growth factor receptor 2 | Vandetanib (ZD6474) | I and II | Wedge et al. [101] |
MMP matrix metalloproteinase, VEGF vascular endothelial growth factor, VEGFR vascular endothelial growth factor receptor, NFκB nuclear factor of κB, TNF tumor necrosis factor, IL interleukin, PDGFR platelet-derived growth factor receptor, CSF-1R colony-stimulating factor receptor
Phase of therapeutic agents in clinical trial or approved by FDA
Osteopontin
Osteopontin (OPN) is a secreted glycophosphoprotein that is involved in physiologic and pathologic processes. It has been shown to play a role in various developmental processes and tissue differentiation [41, 42] and in wound repair [43]. OPN is over expressed in several human cancers and has been associated with metastasis and poor prognosis in breast cancer patients [44]. The mechanisms by which OPN can affect tumor aggressiveness are not yet completely established, but several evidences show that it is involved in different processes associated with malignancy such as increased cellular migratory and invasion behaviors, increased metastasis, protection from apoptosis, and induction of tumor-associated inflammatory cells [45-47]. Experimental studies have shown that OPN can interact with a diverse range of factors such as cell surface receptors like integrins and CD44 [48], secreted metalloproteinase inducer [49], and growth factor/receptor pathways [50, 51]. The interactions of OPN with these factors are evidence of its role in malignancy.
Galectin-3
The galectins consist of a family of beta-galactoside-binding proteins, characterized by their affinity for beta-galactosides and by a conserved sequence in the carbohydrate recognition domain that bind to the carbohydrate portion of cell surface glycoproteins or glycolipids. Galectin-3 is a multifunctional protein that is localized and functions in the cytoplasm, cell membrane, nucleus, and the extracellular milieu. Galectin-3 expression is related to neoplastic transformation and progression toward metastasis in breast [52], colon, stomach and thyroid [53]. Additionally, down-regulation of galectin-3 expression in human breast and colon carcinoma resulted in a considerable decrease of the meta-static potential [54]. Consistently, galectin-3 binding protein (90 K) was found to be strongly expressed in the aggressive cell lines (MCF10DCIS.com and MCF10CA cl. D) and not or less expressed in the non-tumor cell line (MCF10A) of the breast cancer series MCF10 [15]. Its expression was also significant in the secretome of three unrelated cancer cell lines, MA11 in breast cancer, WM266-4 in melanoma and OHS in osteosarcoma [14].
Transforming growth factor-β (TGF-β)
Transforming growth factor-β is a secreted polypeptide expressed through receptor serine/threonine kinases and the intracellular Smad. TGF-β was discovered as a secreted protein that plays a dual role in regulating cell proliferation and eliciting normal transformation of fibroblasts [55, 56]. It is a pluripotent cytokine that is known to inhibit tumor growth at the early stage but it can also promote advanced tumor cell invasiveness and metastasis at the later stage [56]. TGF-β inhibits proliferation and induces apoptosis in diverse cell types, and the accumulation of loss-of-function mutations in the TGF-β receptor or Smad genes in various human cancers classify its pathway as a tumor suppressor in humans [57]. TGF-β promotes tumor cell survival, invasiveness and metastasis by targeting fibroblasts, myofibroblasts, and immune cells in the tumor microenvironment. The mechanisms by which TGF-β regulates the function of numerous cells that participate in tumor progression have been investigated [58].
Matrix metalloproteinases
The ECM is regarded as a barrier to tumor progression. Cleavage of its components by proteases is assumed to remove the physical obstruction and allow cell migration and invasion. The tumor microenvironment is expansively modified and remodeled by proteases and as a result of this activity, important changes in cell–cell and cell–ECM interactions occur and new signals are generated from the cell surface. A correlation between increased MMP expression and a tumor cell's ability to invade neighboring tissue has been established [59]. MMPs can target many other non-ECM proteins such as growth factors, growth factor receptors, cytokines and cell-associated molecules. The participation of these proteases in tumor progression is far more complex than primarily anticipated.
For instance, MMP-7 expressed by malignant breast epithelial cells not only can cleave matrix components in the tumor microenvironment resulting in basement membrane structures disruption [60], but can also cleave the cell adhesion molecule E-cadherin, resulting in the disruption of breast epithelial cell-cell junctions [61]. In addition, MMPs are expressed in the extracellular milieu as inactive proforms (zymogen forms) that become activated throughout a variety of mechanisms that regularly involve collaboration among a number of MMPs families. Thus, overexpression of these proteases documented in tumors by immunohistochemistry does not essentially mean an increase in proteolitic activity, because most antibodies available to detect these proteases do not distinguish between their active and inactive forms [62].
Tissue inhibitor of metalloproteinases (TIMPs), endogenous inhibitors of MMPs, provides control of MMP activity in vivo under normal physiological circumstances. Therefore, the balance between MMPs and TIMPs is a critical determinant in cancer progression and metastasis. Actually, the concept that proteases are necessary to degrade the ECM and allow endothelial cells to invade neighboring tissues appears to be more complex than initially predicted. In angiogenesis, both an absence and an excess of proteolytic activity could have a negative outcome [36]. Moreover, an association between a poor prognosis in cancer patients and high levels of TIMPs like TIMP-2 and plasminogen activator inhibitor-1 has been reported in many cancer types [63]. Increasing evidence suggests that TIMP-2 might be a multifunctional protein harboring an inhibitory and stimulatory effect on cancer progression.
Targeting the tumor microenvironment for cancer therapy
Recent data indicate that primary dysfunction in the tumor microenvironment, in addition to epithelial malfunction, can be critical for carcinogenesis and formation of metastasis. As discussed earlier, a chronic inflammatory state can serve to promote cancer initiation, in a wide variety of organs. Additionally, anti-inflammatory medications in the form of non-steroidal anti-inflammatory (NSAIDs) or aspirin have been shown to dramatically lower the risk of cancer for patients with certain chronic inflammatory conditions [64, 65]. These findings imply a convincing case for targeting the tumor microenvironment for cancer therapy and possible preventive strategies.
Tumor cells are surrounded by non-tumor/stromal cells that can contribute both positive and negative signals to the tumor. Non-tumor cells are more likely to be genetically stable compared to tumor cells, which are known to be genetically unstable. Thus, given their inherent genetic instability, tumor cells are more likely to mutate and acquire or develop resistance to medication. Accordingly, non-tumor cells may serve as therapy targets in the tumor microenvironment but, a drawback is the delicate balance between their tumor-inhibitory and tumor-promotion activities, as well as the normal function of the non-cancer related stromal cells. Ideally, therapeutic approaches should remove the cancer promoting properties while retaining the normal physiologic role of these specialized stromal cells.
To date, there is a wealth of information about specific targets for cancer therapy in the tumor microenvironment [10, 11]. The remodeling of the ECM is arbitrated in an orchestrated way by several families of matrix-degrading enzymes such as MMPs, proteases of cysteine and serine, as well as endoglycosidases [66]. The inhibition of enzymes such as cysteine proteases, cathepsins and heparanase, offers the possibility to block multiple processes in the tumor microenvironment [67, 68]. However, some MMP enzymes can also release proteins such as tumastin, endostatin, and angiostatin which inhibit tumorigenesis [69]. Certain criteria for instance, cytostatic and cytotoxic effects of drugs, have to be considered when targeting the tumor microenvironment for cancer therapy. Integrins, a family of heterodimeric receptors are also therapeutic targets in the extracellular milieu. Several of them are upregulated in cancer, and are expressed on endothelial and tumor cells [70]. β1-integrin appears to be one of the promising therapeutic targets because β1-integrin blocking antibodies have been shown to annul cell adhesion-mediated drug resistance (CAM-DR) [71], and during tumorigenesis ECM remodeling could contribute to CAM-DR by changing the integrin repertoire and affecting the local ECM composition [72].
Fibroblasts are another key component of the tissue stroma and play a primary role in cancer development and progression at all stages. Carcinoma-associated fibroblasts (CAFs) are activated fibroblasts that are allied with malignant tumors and often express α-smooth muscle actin as a maker. Critical functions of CAFs include: deposition of ECM, regulation of epithelial differentiation, regulation of inflammation, and wound healing. Fibroblasts are also an important source of ECM-degrading proteases (MMPs), therefore highlighting their critical role in the regulation of ECM turnover. Through the secretion of growth factors (i.e., hepatocyte growth factor (HGF), insulin-like GF (IGF), nerve GF, EGF, FEF2, WNT1) proliferative signals are induced within adjacent epithelial cells. Thus, fibroblasts are involved with direct mesenchymal-epithelial cell interactions. Fibroblasts also serve roles in immune system modulation, following tissue injury, through secretion of cytokines (IL-1) and chemokines (monocyte chemostatic protein 1 (MCP1)) [5]. Transforming growth factor β is one of the fibroblast-supplied factors involved in the suppression of epithelial cell transformation [58]. Molecules enriched in the process of fibroblast activation such as the fibroblast activation protein (FAP) [73], the hepatocyte growth factor [58] and cathepsin K [74], can be good selective targets in the tumor microenvironment with some recent evidences in preclinical models and a phase 1 clinical trial for an anti-FAP antibody [75].
Signal transducers and activators of transcription (STATs), nuclear transcription factor (NFκB) and hypoxiainducible factor 1α (HIFIα) are attractive therapeutic targets in the microenvironment, as they are known to be closely involved in regulating inflammation, wound healing and angiogenesis. STATs phosphorylation, which is required for transcriptional activity, is regulated by kinases, phosphatases and binding proteins, which are good targets for cancer therapy [76]. STAT3 is able to increase the expression of fibrinogen genes in lung cancer cells and fibrinogen overexpression can enhance tumor cell growth, metastasis and invasion [77]. Moreover, the transcriptional activity of STAT3 is upregulated during chronic inflammation in mouse lung in vivo and the STAT3 activation is found in many human lung cancers [77].
NFκB is also an important target for chemoprevention directed at the tumor microenvironment for its critical role and associated proteins role in many aspects of inflammation [78]. New cancer drug development directed at the regulation of NFκB function is a promising field of investigation and will certainly generate newer agents for chemoprevention. HIF1 is a key regulator in the control of tissue homeostasis and is a key target for cancer prevention and treatment. HIF1α subunit is actually the target for drug development as the chemopreventive agent sulphoraphane for instance controls the HIF1 role in endothelial cells by inhibiting the expression of HIF1α and HIF1-regulated genes [9, 79].
Challenges and perspectives
The microenvironment is essential for the ultimate goal of this predator we call cancer, which is the invasion metastasis cascade. If cancer did not spread, successful medical management would have been achieved many years ago. The invasion metastasis cascade consists of the following critical steps: (1) local invasion, (2) intravasation or the invasion of blood vessels, (3) transport of the cancer cell through the blood to a distant site, (4) extravasation or escape of the cancer cell from the blood to tissue, and (5) adaptation of the relocated cancer cell to the new local environment, continued growth, and thus becoming life threatening. The tumor microenvironment is critical in achieving these cascading steps [7].
Thus, the microenvironment is an essential intrinsic part of the tumor itself. These findings raise the hope that therapeutic targeting of cellular and molecular components involved in events occurring in the tumor microenvironment might be relevant. The first step toward this aim is the identification of molecular elements and signaling pathways in the tumor microenvironment for each cancer type. In vitro identification of molecular components in the tumor microenvironment without contamination from the cell cytosol has been a challenge and techniques for enrichment and analysis have continuously been improved. Monitoring changes of the microenvironment molecular profile through tumor progression maybe helpful for identifying cell or protein targets that have undergone modification. Additional questions relate to how the tumor microenvironment varies across cancer types. Scientific studies summarized in this review illustrate how the tumor microenvironment contributes to cancer development and progression, as well as the role of secreted proteins and their unique technical preparations, to enhance possible biomarker discovery or drug target elucidation.
References
- 1.World Health Organization 2006 [Google Scholar]
- 2.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 3.Kuper H, Adami HO, Trichopoulos D. Infections as a major preventable cause of human cancer. J Intern Med. 2000;248(3):171–183. doi: 10.1046/j.1365-2796.2000.00742.x. [DOI] [PubMed] [Google Scholar]
- 4.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420(6917):860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6(5):392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 6.Liotta LA, Kohn EC. The microenvironment of the tumour-host interface. Nature. 2001;411(6835):375–379. doi: 10.1038/35077241. [DOI] [PubMed] [Google Scholar]
- 7.Weinberg RA. Garland Science. Taylor & Francis Group, LLC; 2007. The biology of cancer. [Google Scholar]
- 8.Handsley MM, Edwards DR. Metalloproteinases and their inhibitors in tumor angiogenesis. Int J Cancer. 2005;115(6):849–860. doi: 10.1002/ijc.20945. [DOI] [PubMed] [Google Scholar]
- 9.Albini A, Sporn MB. The tumour microenvironment as a target for chemoprevention. Nat Rev Cancer. 2007;7(2):139–147. doi: 10.1038/nrc2067. [DOI] [PubMed] [Google Scholar]
- 10.Joyce JA. Therapeutic targeting of the tumor microenvironment. Cancer Cell. 2005;7(6):513–520. doi: 10.1016/j.ccr.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 11.Mueller MM, Fusenig NE. Friends or foes—bipolar effects of the tumour stroma in cancer. Nat Rev Cancer. 2004;4(11):839–849. doi: 10.1038/nrc1477. [DOI] [PubMed] [Google Scholar]
- 12.Volmer MW, Radacz Y, Hahn SA, et al. Tumor suppressor Smad4 mediates downregulation of the anti-adhesive invasion-promoting matricellular protein SPARC: landscaping activity of Smad4 as revealed by a “secretome” analysis. Proteomics. 2004;4(5):1324–1334. doi: 10.1002/pmic.200300703. [DOI] [PubMed] [Google Scholar]
- 13.Chevallet M, Diemer H, Van Dorssealer A, Villiers C, Rabilloud T. Toward a better analysis of secreted proteins: the example of the myeloid cells secretome. Proteomics. 2007;7(11):1757–1770. doi: 10.1002/pmic.200601024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mbeunkui F, Fodstad O, Pannell LK. Secretory protein enrichment and analysis: an optimized approach applied on cancer cell lines using 2D LC–MS/MS. J Proteome Res. 2006;5(4):899–906. doi: 10.1021/pr050375p. [DOI] [PubMed] [Google Scholar]
- 15.Mbeunkui F, Metge BJ, Shevde LA, Pannell LK. Identification of differentially secreted biomarkers using LC-MS/MS in isogenic cell lines representing a progression of breast cancer. J Proteome Res. 2007;6(8):2993–3002. doi: 10.1021/pr060629m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pardo M, Garcia A, Antrobus R, Blanco MJ, Dwek RA, Zitzmann N. Biomarker discovery from uveal melanoma secretomes: identification of gp100 and cathepsin D in patient serum. J Proteome Res. 2007;6(7):2802–2811. doi: 10.1021/pr070021t. [DOI] [PubMed] [Google Scholar]
- 17.Volmer MW, Stuhler K, Zapatka M, et al. Differential proteome analysis of conditioned media to detect Smad4 regulated secreted biomarkers in colon cancer. Proteomics. 2005;5(10):2587–2601. doi: 10.1002/pmic.200401188. [DOI] [PubMed] [Google Scholar]
- 18.Khwaja FW, Svoboda P, Reed M, Pohl J, Pyrzynska B, Van Meir EG. Proteomic identification of the wt-p53-regulated tumor cell secretome. Oncogene. 2006;25(58):7650–7661. doi: 10.1038/sj.onc.1209969. [DOI] [PubMed] [Google Scholar]
- 19.Gronborg M, Kristiansen TZ, Iwahori A, et al. Biomarker discovery from pancreatic cancer secretome using a differential proteomic approach. Mol Cell Proteomics. 2006;5(1):157–171. doi: 10.1074/mcp.M500178-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Zwickl H, Traxler E, Staettner S, et al. A novel technique to specifically analyze the secretome of cells and tissues. Electrophoresis. 2005;26(14):2779–2785. doi: 10.1002/elps.200410387. [DOI] [PubMed] [Google Scholar]
- 21.Jiang L, He L, Fountoulakis M. Comparison of protein precipitation methods for sample preparation prior to proteomic analysis. J Chromatogr A. 2004;1023(2):317–320. doi: 10.1016/j.chroma.2003.10.029. [DOI] [PubMed] [Google Scholar]
- 22.Schwarz K, Fiedler T, Fischer RJ, Bahl H. A standard operating procedure (SOP) for the preparation of intra- and extra-cellular proteins of Clostridium acetobutylicum for proteome analysis. J Microbiol Methods. 2007;68(2):396–402. doi: 10.1016/j.mimet.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 23.Diamandis EP. Point: Proteomic patterns in biological fluids: do they represent the future of cancer diagnostics? Clin Chem. 2003;49(8):1272–1275. doi: 10.1373/49.8.1272. [DOI] [PubMed] [Google Scholar]
- 24.Villanueva J, Shaffer DR, Philip J, et al. Differential exoprotease activities confer tumor-specific serum peptidome patterns. J Clin Invest. 2006;116(1):271–284. doi: 10.1172/JCI26022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerner C, Vejda S, Gelbmann D, et al. Concomitant determination of absolute values of cellular protein amounts, synthesis rates, and turnover rates by quantitative proteome profiling. Mol Cell Proteomics. 2002;1(7):528–537. doi: 10.1074/mcp.m200026-mcp200. [DOI] [PubMed] [Google Scholar]
- 26.Hanash S, Brichory F, Beer D. A proteomic approach to the identification of lung cancer markers. Dis Markers. 2001;17(4):295–300. doi: 10.1155/2001/657605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mann M, Kelleher NL. Special feature: precision proteomics: the case for high resolution and high mass accuracy. Proc Natl Acad Sci USA. 2008 doi: 10.1073/pnas.0800788105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gronborg M, Bunkenborg J, Kristiansen TZ, et al. Comprehensive proteomic analysis of human pancreatic juice. J Proteome Res. 2004;3(5):1042–1055. doi: 10.1021/pr0499085. [DOI] [PubMed] [Google Scholar]
- 29.Kristiansen TZ, Bunkenborg J, Gronborg M, et al. A proteomic analysis of human bile. Mol Cell Proteomics. 2004;3(7):715–728. doi: 10.1074/mcp.M400015-MCP200. [DOI] [PubMed] [Google Scholar]
- 30.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989;246(4926):64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 31.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17(10):994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 32.Gygi SP, Rist B, Griffin TJ, Eng J, Aebersold R. Proteome analysis of low-abundance proteins using multidimensional chromatography and isotope-coded affinity tags. J Proteome Res. 2002;1(1):47–54. doi: 10.1021/pr015509n. [DOI] [PubMed] [Google Scholar]
- 33.Amanchy R, Kalume DE, Pandey A. Stable isotope labeling with amino acids in cell culture (SILAC) for studying dynamics of protein abundance and posttranslational modifications. Sci STKE. 2005;2005(267):l2. doi: 10.1126/stke.2672005pl2. [DOI] [PubMed] [Google Scholar]
- 34.Ong SE, Blagoev B, Kratchmarova I, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1(5):376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 35.DeClerck YA, Mercurio AM, Stack MS, et al. Proteases, extracellular matrix, and cancer: a workshop of the path B study section. Am J Pathol. 2004;164(4):1131–1139. doi: 10.1016/S0002-9440(10)63200-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Devy L, Blacher S, Grignet-Debrus C, et al. The pro- or antiangiogenic effect of plasminogen activator inhibitor 1 is dose dependent. FASEB J. 2002;16(2):147–154. doi: 10.1096/fj.01-0552com. [DOI] [PubMed] [Google Scholar]
- 37.Etzioni R, Urban N, Ramsey S, et al. The case for early detection. Nat Rev Cancer. 2003;3(4):243–252. doi: 10.1038/nrc1041. [DOI] [PubMed] [Google Scholar]
- 38.Li Y, Hively WP, Varmus HE. Use of MMTV-Wnt-1 transgenic mice for studying the genetic basis of breast cancer. Oncogene. 2000;19(8):1002–1009. doi: 10.1038/sj.onc.1203273. [DOI] [PubMed] [Google Scholar]
- 39.Harvie M, Hooper L, Howell AH. Central obesity and breast cancer risk: a systematic review. Obes Rev. 2003;4(3):157–173. doi: 10.1046/j.1467-789x.2003.00108.x. [DOI] [PubMed] [Google Scholar]
- 40.Rose DP, Komninou D, Stephenson GD. Obesity, adipocytokines, and insulin resistance in breast cancer. Obes Rev. 2004;5(3):153–165. doi: 10.1111/j.1467-789X.2004.00142.x. [DOI] [PubMed] [Google Scholar]
- 41.Nemir M, Bhattacharyya D, Li X, Singh K, Mukherjee AB, Mukherjee BB. Targeted inhibition of osteopontin expression in the mammary gland causes abnormal morphogenesis and lactation deficiency. J Biol Chem. 2000;275(2):969–976. doi: 10.1074/jbc.275.2.969. [DOI] [PubMed] [Google Scholar]
- 42.Rittling SR, Matsumoto HN, McKee MD, et al. Mice lacking osteopontin show normal development and bone structure but display altered osteoclast formation in vitro. J Bone Miner Res. 1998;13(7):1101–1111. doi: 10.1359/jbmr.1998.13.7.1101. [DOI] [PubMed] [Google Scholar]
- 43.Liaw L, Birk DE, Ballas CB, Whitsitt JS, Davidson JM, Hogan BL. Altered wound healing in mice lacking a functional osteopontin gene (spp1) J Clin Invest. 1998;101(7):1468–1478. doi: 10.1172/JCI1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rudland PS, Platt-Higgins A, El Tanani M, et al. Prognostic significance of the metastasis-associated protein osteopontin in human breast cancer. Cancer Res. 2002;62(12):3417–3427. [PubMed] [Google Scholar]
- 45.El Tanani MK, Campbell FC, Kurisetty V, Jin D, McCann M, Rudland PS. The regulation and role of osteopontin in malignant transformation and cancer. Cytokine Growth Factor Rev. 2006;17(6):463–474. doi: 10.1016/j.cytogfr.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 46.Tuck AB, Chambers AF. The role of osteopontin in breast cancer: clinical and experimental studies. J Mammary Gland Biol Neoplasia. 2001;6(4):419–429. doi: 10.1023/a:1014734930781. [DOI] [PubMed] [Google Scholar]
- 47.Weber GF. The metastasis gene osteopontin: a candidate target for cancer therapy. Biochim Biophys Acta. 2001;1552(2):61–85. doi: 10.1016/s0304-419x(01)00037-3. [DOI] [PubMed] [Google Scholar]
- 48.Mangala LS, Fok JY, Zorrilla-Calancha IR, Verma A, Mehta K. Tissue transglutaminase expression promotes cell attachment, invasion and survival in breast cancer cells. Oncogene. 2007;26(17):2459–2470. doi: 10.1038/sj.onc.1210035. [DOI] [PubMed] [Google Scholar]
- 49.Mi Z, Oliver T, Guo H, Gao C, Kuo PC. Thrombin-cleaved COOH(−) terminal osteopontin peptide binds with cyclophilin C to CD147 in murine breast cancer cells. Cancer Res. 2007;67(9):4088–4097. doi: 10.1158/0008-5472.CAN-06-4066. [DOI] [PubMed] [Google Scholar]
- 50.Tuck AB, Elliott BE, Hota C, Tremblay E, Chambers AF. Osteopontin-induced, integrin-dependent migration of human mammary epithelial cells involves activation of the hepatocyte growth factor receptor (Met) J Cell Biochem. 2000;78(3):465–475. doi: 10.1002/1097-4644(20000901)78:3<465::aid-jcb11>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 51.Tuck AB, Hota C, Wilson SM, Chambers AF. Osteopontin-induced migration of human mammary epithelial cells involves activation of EGF receptor and multiple signal transduction pathways. Oncogene. 2003;22(8):1198–1205. doi: 10.1038/sj.onc.1206209. [DOI] [PubMed] [Google Scholar]
- 52.Matarrese P, Fusco O, Tinari N, et al. Galectin-3 overexpression protects from apoptosis by improving cell adhesion properties. Int J Cancer. 2000;85(4):545–554. [PubMed] [Google Scholar]
- 53.Inohara H, Honjo Y, Yoshii T, et al. Expression of galectin-3 in fine-needle aspirates as a diagnostic marker differentiating benign from malignant thyroid neoplasms. Cancer. 1999;85(11):2475–2484. [PubMed] [Google Scholar]
- 54.Honjo Y, Nangia-Makker P, Inohara H, Raz A. Down-regulation of galectin-3 suppresses tumorigenicity of human breast carcinoma cells. Clin Cancer Res. 2001;7(3):661–668. [PubMed] [Google Scholar]
- 55.Moses HL, Branum EL, Proper JA, Robinson RA. Transforming growth factor production by chemically transformed cells. Cancer Res. 1981;41(7):2842–2848. [PubMed] [Google Scholar]
- 56.Roberts AB, Wakefield LM. The two faces of transforming growth factor beta in carcinogenesis. Proc Natl Acad Sci USA. 2003;100(15):8621–8623. doi: 10.1073/pnas.1633291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Levy L, Hill CS. Alterations in components of the TGF-beta superfamily signaling pathways in human cancer. Cytokine Growth Factor Rev. 2006;17(1–2):41–58. doi: 10.1016/j.cytogfr.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 58.Bhowmick NA, Chytil A, Plieth D, et al. TGF-beta signaling in fibroblasts modulates the oncogenic potential of adjacent epithelia. Science. 2004;303(5659):848–851. doi: 10.1126/science.1090922. [DOI] [PubMed] [Google Scholar]
- 59.Nakajima M, Welch DR, Belloni PN, Nicolson GL. Degradation of basement membrane type IV collagen and lung subendothelial matrix by rat mammary adenocarcinoma cell clones of differing metastatic potentials. Cancer Res. 1987;47(18):4869–4876. [PubMed] [Google Scholar]
- 60.Fingleton B, Vargo-Gogola T, Crawford HC, Matrisian LM. Matrilysin [MMP-7] expression selects for cells with reduced sensitivity to apoptosis. Neoplasia. 2001;3(6):459–468. doi: 10.1038/sj.neo.7900190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Noe V, Fingleton B, Jacobs K, et al. Release of an invasion promoter E-cadherin fragment by matrilysin and stromelysin-1. J Cell Sci. 2001;114(Pt 1):111–118. doi: 10.1242/jcs.114.1.111. [DOI] [PubMed] [Google Scholar]
- 62.Coussens LM, Fingleton B, Matrisian LM. Matrix metalloproteinase inhibitors and cancer: trials and tribulations. Science. 2002;295(5564):2387–2392. doi: 10.1126/science.1067100. [DOI] [PubMed] [Google Scholar]
- 63.Remacle A, McCarthy K, Noel A, et al. High levels of TIMP-2 correlate with adverse prognosis in breast cancer. Int J Cancer. 2000;89(2):118–121. doi: 10.1002/(sici)1097-0215(20000320)89:2<118::aid-ijc3>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 64.Garcia-Rodriguez LA, Huerta-Alvarez C. Reduced risk of colorectal cancer among long-term users of aspirin and nonaspirin nonsteroidal antiinflammatory drugs. Epidemiology. 2001;12(1):88–93. doi: 10.1097/00001648-200101000-00015. [DOI] [PubMed] [Google Scholar]
- 65.Koki AT, Masferrer JL. Celecoxib: a specific COX-2 inhibitor with anticancer properties. Cancer Control. 2002;9(2 Suppl):28–35. doi: 10.1177/107327480200902S04. [DOI] [PubMed] [Google Scholar]
- 66.Carmeliet P. Angiogenesis in health and disease. Nat Med. 2003;9(6):653–660. doi: 10.1038/nm0603-653. [DOI] [PubMed] [Google Scholar]
- 67.Joyce JA, Baruch A, Chehade K, et al. Cathepsin cysteine proteases are effectors of invasive growth and angiogenesis during multistage tumorigenesis. Cancer Cell. 2004;5(5):443–453. doi: 10.1016/s1535-6108(04)00111-4. [DOI] [PubMed] [Google Scholar]
- 68.Joyce JA, Freeman C, Meyer-Morse N, Parish CR, Hanahan D. A functional heparan sulfate mimetic implicates both heparanase and heparan sulfate in tumor angiogenesis and invasion in a mouse model of multistage cancer. Oncogene. 2005;24(25):4037–4051. doi: 10.1038/sj.onc.1208602. [DOI] [PubMed] [Google Scholar]
- 69.Hamano Y, Zeisberg M, Sugimoto H, et al. Physiological levels of tumstatin, a fragment of collagen IV alpha3 chain, are generated by MMP-9 proteolysis and suppress angiogenesis via alphaV beta3 integrin. Cancer Cell. 2003;3(6):589–601. doi: 10.1016/s1535-6108(03)00133-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guo W, Giancotti FG. Integrin signalling during tumour progression. Nat Rev Mol Cell Biol. 2004;5(10):816–826. doi: 10.1038/nrm1490. [DOI] [PubMed] [Google Scholar]
- 71.Sethi T, Rintoul RC, Moore SM, et al. Extracellular matrix proteins protect small cell lung cancer cells against apoptosis: a mechanism for small cell lung cancer growth and drug resistance in vivo. Nat Med. 1999;5(6):662–668. doi: 10.1038/9511. [DOI] [PubMed] [Google Scholar]
- 72.Morin PJ. Drug resistance and the microenvironment: nature and nurture. Drug Resist Updat. 2003;6(4):169–172. doi: 10.1016/s1368-7646(03)00059-1. [DOI] [PubMed] [Google Scholar]
- 73.Park JE, Lenter MC, Zimmermann RN, Garin-Chesa P, Old LJ, Rettig WJ. Fibroblast activation protein, a dual specificity serine protease expressed in reactive human tumor stromal fibroblasts. J Biol Chem. 1999;274(51):36505–36512. doi: 10.1074/jbc.274.51.36505. [DOI] [PubMed] [Google Scholar]
- 74.Allinen M, Beroukhim R, Cai L, et al. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell. 2004;6(1):17–32. doi: 10.1016/j.ccr.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 75.Scott AM, Wiseman G, Welt S, et al. A Phase I dose-escalation study of sibrotuzumab in patients with advanced or meta-static fibroblast activation protein-positive cancer. Clin Cancer Res. 2003;9(5):1639–1647. [PubMed] [Google Scholar]
- 76.Yu H, Jove R. The STATs of cancer-new molecular targets come of age. Nat Rev Cancer. 2004;4(2):97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 77.Dauer DJ, Ferraro B, Song L, et al. Stat3 regulates genes common to both wound healing and cancer. Oncogene. 2005;24(21):3397–3408. doi: 10.1038/sj.onc.1208469. [DOI] [PubMed] [Google Scholar]
- 78.Karin M. NF-kappaB and cancer: mechanisms and targets. Mol Carcinog. 2006;45(6):355–361. doi: 10.1002/mc.20217. [DOI] [PubMed] [Google Scholar]
- 79.Bertl E, Bartsch H, Gerhauser C. Inhibition of angiogenesis and endothelial cell functions are novel sulforaphane-mediated mechanisms in chemoprevention. Mol Cancer Ther. 2006;5(3):575–585. doi: 10.1158/1535-7163.MCT-05-0324. [DOI] [PubMed] [Google Scholar]
- 80.Martin DB, Gifford DR, Wright ME, et al. Quantitative proteomic analysis of proteins released by neoplastic prostate epithelium. Cancer Res. 2004;64(1):347–355. doi: 10.1158/0008-5472.can-03-2062. [DOI] [PubMed] [Google Scholar]
- 81.Kerbel R, Folkman J. Clinical translation of angiogenesis inhibitors. Nat Rev Cancer. 2002;2(10):727–739. doi: 10.1038/nrc905. [DOI] [PubMed] [Google Scholar]
- 82.Young SL, Chaplin DJ. Combretastatin A4 phosphate: background and current clinical status. Expert Opin Investig Drugs. 2004;13(9):1171–1182. doi: 10.1517/13543784.13.9.1171. [DOI] [PubMed] [Google Scholar]
- 83.Jimeno A, Carducci M. Atrasentan: targeting the endothelin axis in prostate cancer. Expert Opin Investig Drugs. 2004;13(12):1631–1640. doi: 10.1517/13543784.13.12.1631. [DOI] [PubMed] [Google Scholar]
- 84.Mooberry SL. New insights into 2-methoxyestradiol, a promising antiangiogenic and antitumor agent. Curr Opin Oncol. 2003;15(6):425–430. doi: 10.1097/00001622-200311000-00004. [DOI] [PubMed] [Google Scholar]
- 85.Jin H, Varner J. Integrins: roles in cancer development and as treatment targets. Br J Cancer. 2004;90(3):561–565. doi: 10.1038/sj.bjc.6601576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gutheil JC, Campbell TN, Pierce PR, et al. Targeted anti-angiogenic therapy for cancer using Vitaxin: a humanized monoclonal antibody to the integrin alphavbeta3. Clin Cancer Res. 2000;6(8):3056–3061. [PubMed] [Google Scholar]
- 87.Burke PA, DeNardo SJ, Miers LA, Lamborn KR, Matzku S, DeNardo GL. Cilengitide targeting of alpha(v)beta(3) integrin receptor synergizes with radioimmunotherapy to increase efficacy and apoptosis in breast cancer xenografts. Cancer Res. 2002;62(15):4263–4272. [PubMed] [Google Scholar]
- 88.Gingras D, Boivin D, Deckers C, Gendron S, Barthomeuf C, Beliveau R. Neovastat-a novel antiangiogenic drug for cancer therapy. Anticancer Drugs. 2003;14(2):91–96. doi: 10.1097/00001813-200302000-00001. [DOI] [PubMed] [Google Scholar]
- 89.Bartlett JB, Dredge K, Dalgleish AG. The evolution of thalidomide and its IMiD derivatives as anticancer agents. Nat Rev Cancer. 2004;4(4):314–322. doi: 10.1038/nrc1323. [DOI] [PubMed] [Google Scholar]
- 90.Sleijfer S, Kruit WH, Stoter G. Thalidomide in solid tumours: the resurrection of an old drug. Eur J Cancer. 2004;40(16):2377–2382. doi: 10.1016/j.ejca.2004.07.023. [DOI] [PubMed] [Google Scholar]
- 91.Druker BJ. Imatinib as a paradigm of targeted therapies. Adv Cancer Res. 2004;91:1–30. doi: 10.1016/S0065-230X(04)91001-9. [DOI] [PubMed] [Google Scholar]
- 92.Pietras K, Sjoblom T, Rubin K, Heldin CH, Ostman A. PDGF receptors as cancer drug targets. Cancer Cell. 2003;3(5):439–443. doi: 10.1016/s1535-6108(03)00089-8. [DOI] [PubMed] [Google Scholar]
- 93.Graff JR, McNulty AM, Hanna KR, et al. The protein kinase Cbeta-selective inhibitor, Enzastaurin ( LY317615.HCl), suppresses signaling through the AKT pathway, induces apoptosis, and suppresses growth of human colon cancer and glioblastoma xenografts. Cancer Res. 2005;65(16):7462–7469. doi: 10.1158/0008-5472.CAN-05-0071. [DOI] [PubMed] [Google Scholar]
- 94.Mendel DB, Laird AD, Xin X, et al. In vivo antitumor activity of SU11248, a novel tyrosine kinase inhibitor targeting vascular endothelial growth factor and platelet-derived growth factor receptors: determination of a pharmacokinetic/pharmacodynamic relationship. Clin Cancer Res. 2003;9(1):327–337. [PubMed] [Google Scholar]
- 95.Pietras K, Hanahan D. A multitargeted, metronomic, and maximum-tolerated dose “chemo-switch” regimen is antiangiogenic, producing objective responses and survival benefit in a mouse model of cancer. J Clin Oncol. 2005;23(5):939–952. doi: 10.1200/JCO.2005.07.093. [DOI] [PubMed] [Google Scholar]
- 96.Ferrara N, Hillan KJ, Gerber HP, Novotny W. Discovery and development of bevacizumab, an anti-VEGF antibody for treating cancer. Nat Rev Drug Discov. 2004;3(5):391–400. doi: 10.1038/nrd1381. [DOI] [PubMed] [Google Scholar]
- 97.Hurwitz H, Fehrenbacher L, Novotny W, et al. Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N Engl J Med. 2004;350(23):2335–2342. doi: 10.1056/NEJMoa032691. [DOI] [PubMed] [Google Scholar]
- 98.Holash J, Davis S, Papadopoulos N, et al. VEGF-Trap: a VEGF blocker with potent antitumor effects. Proc Natl Acad Sci USA. 2002;99(17):11393–11398. doi: 10.1073/pnas.172398299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Manley PW, Bold G, Bruggen J, et al. Advances in the structural biology, design and clinical development of VEGF-R kinase inhibitors for the treatment of angiogenesis. Biochim Biophys Acta. 2004;1697(1–2):17–27. doi: 10.1016/j.bbapap.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 100.Morgan B, Thomas AL, Drevs J, et al. Dynamic contrast-enhanced magnetic resonance imaging as a biomarker for the pharmacological response of PTK787/ZK 222584, an inhibitor of the vascular endothelial growth factor receptor tyrosine kinases, in patients with advanced colorectal cancer and liver metastases: results from two phase I studies. J Clin Oncol. 2003;21(21):3955–3964. doi: 10.1200/JCO.2003.08.092. [DOI] [PubMed] [Google Scholar]
- 101.Wedge SR, Ogilvie DJ, Dukes M, et al. ZD6474 inhibits vascular endothelial growth factor signaling, angiogenesis, and tumor growth following oral administration. Cancer Res. 2002;62(16):4645–4655. [PubMed] [Google Scholar]