Abstract
The PAS-LOV domain is a signal transducing component found in a large variety of proteins that is responsible for sensing different stimuli such as light, oxygen and voltage. The LOV protein VVD regulates blue-light responses in the filamentous fungi Neurospora crassa. Using photocoupled, time-resolved small angle x-ray scattering, we extract the solution protein structure in both dark-adapted and light-activated states. Two distinct dark-adapted conformations are detected in the wild type protein: a compact structure that corresponds to the crystal structure of the dark-state monomer as well as an extended structure that is well modeled by introducing conformational disorder at the N-terminus of the protein. These conformations are accentuated in carefully selected variants, in which a key residue for propagating structural transitions, Cys71, has been mutated or oxidized. Despite different dark state conformations, all proteins form a common dimer in response to illumination. Taken together, these data support a reaction scheme that describes the mechanism for light-induced dimerization of VVD. Envelope reconstructions of the transient light-state dimer reveal structures that are best described by a parallel arrangement of subunits that have significantly changed conformation compared to the crystal structure.
Keywords: LOV domain proteins, blue light activated proteins, VVD, photocoupled small angle x-ray scattering
Introduction
Biological signal transduction systems rely on the ability of intracellular proteins to alter their structures and patterns of association in response to external stimuli. The PAS (Per Arnt Sim) domain is a broadly distributed protein module often found as a component of signaling networks1. PAS domains, either in isolation, or as part of a multidomain protein regulate many different cellular processes, including kinase activity, gene transcription, and channel function2.
LOV domain proteins (for light, oxygen and voltage sensing), which form a subclass of the PAS superfamily, bind cofactors that react chemically to sense changes in ligand concentration, illumination, and redox state. Structural dynamics in LOV proteins have been well characterized in plant phototropins and related phototropin-like photoreceptors3-6, where blue-light triggers the formation of a cysteinyl flavin C4a adduct, which then relays conformational changes to variable N- or C-terminal extensions of the PAS core7,8. The consequences of such processes have been explored in several systems. For example, plant phototropins undergo autophosphorylation to regulate growth9,10, fungal white collar 1 (WC1) activates gene expression11,12, and bacterial YtvA modulates stress responses13. In all of these cases, the mechanisms that link photo-adduct formation to downstream changes in molecular reactivity are not well understood.
Here, we investigate the initial steps in the light-activated response of the fungal PAS-LOV photosensor Vivid (VVD) by monitoring global changes in protein structure triggered by the absorption of a single photon. Vivid is involved in adaptation to blue light responses such as carotenoid production and gating of the circadian clock in the filamentous fungus Neurospora crassa14,15. VVD may also sense intracellular oxidants, such as superoxide, however, the underlying molecular processes have yet to be explored16. Spectroscopic and crystallographic studies15,17 of VVD revealed that cysteine 108 forms an adduct in response to blue light. The crystallographic structure of the light-activated state (Fig. 1) further showed how adduct formation couples to flavin N5 protonation. N5 protonation in turn drives hydrogen bonding rearrangements that alter the conformation of the Ncap, which is composed of N-terminal structural elements that pack against the PAS β-core. In VVD, the Ncap consists of an extended stretch of polypeptide that begins near the adenosine moiety of the flavin, an α helix (aα), a short β-strand (bβ), and a short hinge that connects the rest of the Ncap to the PAS core through the key switching residue Cys7117. In the crystal, light causes a shift in bβ, but in solution the current data suggests that a more dramatic structural response in the Ncap ultimately leads to dimerization17-19. It is currently not clear how the conformational changes observed in the crystal structure of the light state facilitate dimerization.
Fig. 1. Crystal structure of VVD (2PDR.pdb).

The Ncap is shown in green, the β core is shown in purple and the adduct forming flavin is shown in blue. All relevant Cys residues are shown in yellow.
PAS:PAS dimerization is not unique to VVD and plays a significant role in signaling by other PAS domains10,11. Whereas stable PAS dimers have been structurally characterized20-23, their solution dynamics and in vivo relevance remain to be established. Recently, transient gradient methods have demonstrated that phototropin LOV domains can also form light-induced dimers24, but again their role in the context of the full-length proteins is largely unexplored. Solution small angle x-ray scattering (SAXS) reports global structural characteristics of macromolecules and is ideal for studying large conformational changes, including association processes25.
We have previously employed photocoupled SAXS to demonstrate dimerization of the VVD light-state on a rapid time scale. The biologically inactive C71S variant was also shown to be incapable of dimerization17. Here we further apply SAXS to resolve important structural characteristics of the light- and dark-adapted states of two VVD proteins: VVD-36 (an N-terminal truncation of wt VVD with increased stability and solubility17,18) and VVD-36: C71V:C183S, which forms a higher-affinity light-state dimer due to the residue substitution at position 7118. We employ a flow cell which enables time resolved measurements as rapidly as 20 ms after photoexcitation, and furthermore, mitigates radiation damage from x-rays which can confound the interpretation of SAXS data. Two distinct dark-state structures that both lead to a single dimer were discovered and analyzed. By reconstructing the SAXS data into low resolution molecular envelopes, we show that dimerization indeed requires substantial conformational change in the VVD monomer18. Analysis of these structures leads us to propose a mechanism for dimerization of VVD. A similar conformational gate may regulate transient dimerization in other LOV/PAS systems.
Results
The long term goal of this work is to identify the mechanism underlying light induced conformational changes of VVD. To achieve this goal, we measured the structure of VVD in both dark and light-adapted states, using a SAXS compatible flow cell. Previous studies indicate that VVD dimerizes after exposure to light, yet did not reveal the solution structure of or the mechanism for dimer formation18,19. Here, we extract details about the solution protein structure in both dark and light states, and use this information to propose a mechanism that drives this transition.
To enable such biophysical characterization we employ variants of full-length VVD that increase protein solubility, stability and resistance to oxidation. Removal of 36 residues from the N-terminus of VVD that are variable compared to related LOV domains increases the proteins stability, and ability to undergo multiple photocycles17,18. Otherwise, VVD-36 shows similar light-dependent dimerization as the full-length protein17. A point mutation near the C-terminus (C183S) greatly reduces the tendency to form intermolecular disulfide bonds at high concentrations and otherwise deactivate in the presence of oxidants. C183S alone has no known effects on VVD spectral characteristics or dimerization kinetics.
Two VVD Dark State Conformations: Extended and Compact
SAXS data were collected on samples from numerous preparations of protein. A given batch of dark-adapted VVD-36 displays one of two distinct conformational ensembles, shown in Fig. 2. The scattering profile of one of these ensembles exactly matches profiles generated from the crystal structure17, while the other suggests a structure with a larger spatial extent. Comparison of radius of gyration reveals little difference, with the former data set yielding a value of Rg=18.1 +/- 1.1 Å and the latter Rg=18.8 +/- 0.6 Å. To elucidate differences between the two states, we compared distance distribution functions (P(r)) computed from the latter structure with those computed from the former structure, using the 2PD7 coordinates to minimize the noise (Fig. 2). This analysis emphasizes the spatial dimensions of the protein, and requires input of the maximum molecular dimension, Dmax to accurately reproduce the scattering data. The Dmax value required to reproduce scattering from the larger structure exceeds that required for the crystal structure. Because of its larger spatial extent, we refer to the first, extended state as DarkEXT, while the more compact state that agrees with the crystal structure will be referred to as DarkCOMP.
Fig. 2. Comparison of scattering profiles from two dark states, DarkCOMP and DarkEXT.

The left panel compares scattering profiles from the two distinct dark states of VVD-36: DarkCOMP and DarkEXT. The right panel shows P(r) profiles computed from the DarkEXT scattering data and from scattering profiles generated from (a monomer within) the 2PD7 crystal structure. The DarkEXT conformation has a larger apparent maximum dimension and the corresponding P(r) curve peaks at a slightly higher value of r, indicating more extended molecules within the ensemble.
To associate a structural feature with this extension, we turn to results from other biochemical studies of VVD-36. Recent crystal structures display an alternate conformation of the first eight N-terminal residues18. In addition, crosslinking studies of the light-activated conformation of VVD indicate the N-terminal α-helix may reorient to form the light-state dimer18. The Ncap region is the site of the largest rearrangements upon light excitation of the crystallized protein. Taken together, this evidence suggests that reorganization of the N-terminus accounts for the difference between the DarkEXT state and the conformation in the crystal.
In order to assess the length of the N-terminus polypeptide that must rearrange to describe the SAXS data, we conducted computational studies with EOM26. This program generates an ensemble of unfolded or partly unfolded structures from an amino acid sequence and, with a genetic algorithm, searches for a subset of these that combine to match the experimental data. This analysis was performed six times, including selectively longer portions of the Ncap in the unfolded ensemble (Fig. 3). Notably, extension of only the terminal eight peptides, which form alternate conformations in some crystal structures18, is insufficient to describe the data. At a minimum, the N-terminal peptide chain and the N-terminal α-helix must be displaced to obtain good agreement between the calculated and measured SAXS profiles. Flexibility in other regions of the molecule may contribute to the extended structure. For example, there are indications from thermal factors in the crystal structures that the loop regions surrounding the adenosine moiety of FAD, as well as the adenosine itself are quite dynamic17,18. Thus, we must allow that the conformational disorder required to describe the SAXS data does not necessarily involve the entire Ncap if other regions of the protein are also involved. Nonetheless, a substantial number of residues change their conformational state in going from the compact to the extended form of the protein and these changes likely involve the N-terminus.
Fig. 3. Results of the EOM analysis.

The smooth curves are scattering profiles computed from an ensemble of partially extended molecules, beginning with the 3D72 crystal structure (inset). In these models, varying lengths of the amino acid chain, denoted by different colors, are allowed to be flexible. To generate the curves shown here, 10, 17, 22, and 28 residues are displaced; agreement with the measured curve improves as more residues become flexible. Scattering curves generated from the first two ensembles (10 or 17 residues flexible, blue and red curves) do not fit the data well. In contrast, extending residues 36-58, which includes the entire N-terminal α-helix, produces a reasonable fit. The match to the data is further improved if the loop that connects the α-helix with the LOV domain is made flexible.
Conversion between the dark states
Each state appeared uniquely represented within a given batch of protein and no interconversion was measured on a time scale of 24 hours, suggesting that both correspond to stable conformational states of the protein. After extensive experimentation we have found that the emergence of DarkCOMP correlates with the age and treatment of the protein. The DarkEXT state was consistently found with either freshly prepared protein or protein that was treated with the reducing agent DTT. In fact, a sample of DarkCOMP could be converted to DarkEXT on incubation with DTT. Protein that was left at 4°C overnight or repurified in the absence of DTT consistently showed the DarkCOMP conformation. Thus, we suspect that oxidation plays an important role in formation of DarkCOMP. It should also be noted that degradation at the N-terminus of VVD-36 can prevent dimerization18, and in some long experiments minor amounts of N-terminal degradation were observed. However, samples clearly in the DarkCOMP state showed no evidence of proteolysis.
Oxidation at Cys71 alters protein structure
We have observed in many contexts that both passive and promoted oxidation affect the extent to which VVD undergoes light-induced dimerization (Supplementary Fig. 1, online). Passive oxidation correlates with the length of time the protein resides in oxygenated, aqueous buffer. To investigate whether oxidation may underlie the difference between the two forms of dark state VVD, crystal structures were obtained from VVD protein that had been aged several days in oxygenated buffer. These structures show additional electron density at Cys71 indicative of oxidation of this key residue to sulfenic acid (–S-OH) (Fig. 4). The 2.3 Å resolution electron density is fit well by addition of a single oxygen atom and when modeled as such generates no difference electron density peaks. Furthermore, the CH2-S-O(H) bond angle agrees well with that expected for sulfenic acid. The data presented above and in previous studies demonstrate that Cys71 participates in a conformational switch that is essential for dimerization and protein function17. Thus, we believe oxidation of this important residue to be at least partly responsible for generating dark-state samples with different conformational properties.
Fig 4. Oxidation of VVD at Cys71 to a sulfenic acid.
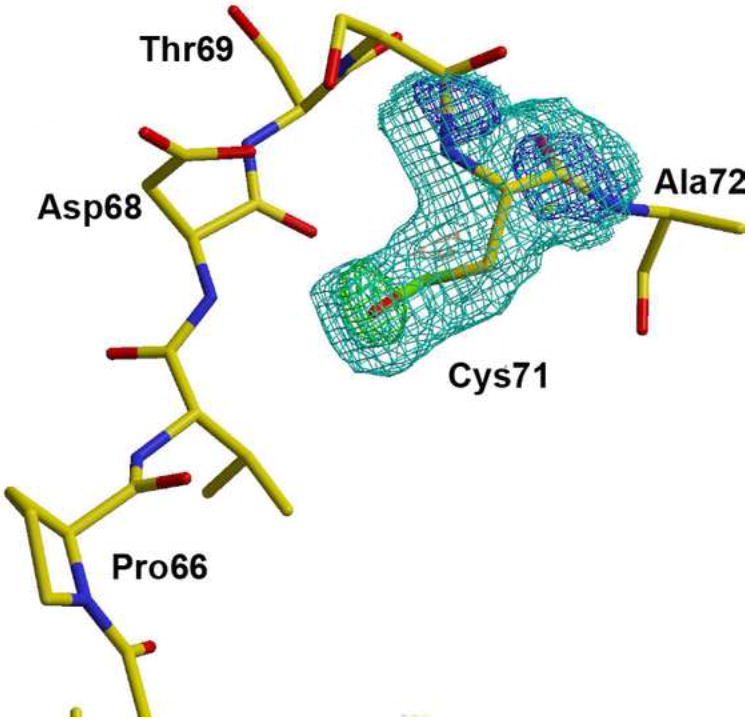
2.3 Å resolution Fo-Fc maps calculated from structures containing a Cys at position 71 instead of the sulfenic acid clearly show the presence of the additional oxygen atom (contoured at 3σ, green). 2Fo-Fc density is contoured at 1σ (cyan) and 2σ (blue).
Light activation of VVD
Within 300 ms of light excitation, scattering profiles from VVD-36 in both compact and extended forms indicate dimerization. Fig. 5 shows the dramatic increase in scattering intensity at low q for both molecules that reflects dimerization. Following standard Guinier analysis, the zero-angle intensity is measured and found to increase by factors of 1.76 ± 0.07 and 1.64 ± 0.04 with DarkCOMP and DarkEXT as respective initial states, less than the factor of 2 expected for complete conversion to dimers. Spectroscopic measurements reveal only ~70 % of the population forms the photo-adduct under these solution and excitation conditions. Thus, the scattering collected from the light-excited states is a mixture of at least two states for each curve: light-activated dimers and dark-state monomers. The light-excited SAXS profiles preserve the shape observed in the high q range of the dark-state monomer profiles, either reflecting different light-state dimers originating from the distinct initial states, or resulting from residual monomers in the ensembles. The more detailed mathematical analysis of the data, presented later, clearly favors the latter scenario.
Fig. 5. Comparison of dark and light-excited scattering data.

The left panel compares scattering profiles from the DarkCOMP state of VVD-36 in the dark adapted (solid curve) and light activated (dashed curve) states. The increase at the lowest scattering angles indicates dimerization of the protein following light exposure. The right panel shows a similar effect in scattering profiles of VVD-36 in the DarkEXT state (dark adapted state: darker solid curve, light activated state: lighter solid curve). Scattering profiles of the VVD variant C71V:C183S are shown as dashed curves in the right panel. The scattering profiles of this variant match those of the DarkEXT state at mid to high q. The enhanced low angle scattering of this variant relative to the DarkEXT conformation suggests the presence of dimers in the dark adapted state of the variant (dark, dashed line). Light excitation further increases the dimer fraction of the variant (light, dashed line).
An activated VVD variant: extended and pretriggered in the dark state
The importance of the Cys71 residue to VVD conformation has already been discussed. Variation of this residue can profoundly impact the protein’s response to light, e.g. the C71S variant forms a cysteinyl-flavin adduct but is biologically inactive17. Here, we studied a different variant, C71V, which appears biased towards the light state conformation, even in the dark-adapted state18. Previous crystallographic studies show that the side chain of Cys71 rotates out from a buried position upon VVD light excitation. This movement leads to functionally relevant conformational change at the N-terminus of the protein17. Crystal structures of VVD C71V show that Val 71 can simultaneously occupy both Cys71 rotamer positions observed in native light- and dark-state structures. Thus, Val71 predisposes VVD to the light-state conformation even in the absence of light.
Following light excitation, data acquired on the C71V variant display a similar change in I(0), although the dark-adapted ensemble of this variant appears heterogeneous. Interestingly, the shape of the high q scattering from the DarkEXT and the C71V variant are in good agreement. When the intensities of these scattering profiles are scaled to match at high q, the low q scattering from the variant exceeds that of the wildtype, and approaches that from the light-state (Fig. 5). This increase suggests that the C71V variant dimerizes to some extent without light excitation (see Supplementary Fig. 2 online). We deduce that the variant monomer is predisposed to a conformation competent for dimerization because the Val side chain partly mimics the light-state conformation of Cys71.
Although dimerization is initiated in the dark state for the C71V variant, it is incomplete. Following illumination, I(0) increases by an additional factor of 1.38 0.03 indicating further dimerization. This is consistent with previous work showing that light induced dimers of C71V variants have dissociation constants in the nm range, as opposed to ~2-15 μM for VVD-3618. Thus while the mutation of Cys71 to Val appears crucial for predisposing VVD to the dimer state, it does not fully represent the changes that cause light-excited VVD to associate as a dimer.
The observed spontaneous dimerization of the C71V variant is interesting given that its scattering profile most closely resembles that of the DarkEXT ensemble. A minimization analysis confirms these states to be a combination of DarkEXT and dimer (see Supplementary Fig. 3 online) whereas DarkCOMP cannot adequately reproduce the data measured for the variant. This result suggests that extension of the Ncap region precedes dimer formation.
The structure of the VVD light-activated dimer
To elucidate the form of the VVD light-activated dimer, SAXS data were collected 8.8 s after light activation (see methods) of VVD-36 in either the DarkCOMP or DarkEXT conformational ensembles. On this long time scale, equilibrium between monomers and dimers has been reached. Assuming that the light-activated dimer takes the same form regardless of the initial protein dark state, we can extract its unique scattering profile by minimizing
Here, Ix(q) are scattering profiles from the different VVD measurements, distinguished by the characteristic scattering of the dark state. For this computation, the scattering profile derived from the 2PD7 structure was employed in place of DarkCOMP to minimize the overall noise. The terms in the denominator represent the errors associated with each measurement. The three parameters r1, r2 and c, which account for fractions of unphotolyzed protein present in each sample and variations in protein concentration, were varied to minimize χ2. The values of r1 and r2, are 0.62 and 0.83 respectively, indicating a mixture of monomer and dimer in each state. Since the dimer fraction varied considerably in the absence of DTT, we hesitate to assign much significance to its absolute value. However, this analysis is valuable because it enables extraction of the scattering profile of the pure light-excited dimer from the data, as ILightCOMP(q)-r1I2PD7(q) or c(ILightEXT(q)-r2IDarkEXT(q)). Reconstruction methods were applied (see below) and yield structural information about the dimer. The close agreement of the dimer scattering profiles obtained from either ensemble (see Supplementary Fig. 4 online), and the χν2 of 0.68 demonstrate the validity of the original assumption. Thus these four scattering curves can be well described by a model containing only three states. At this resolution photoactivated VVD forms the same structure regardless of the conformation of the dark state.
Significantly, the robust scattering profile obtained for the VVD dimer does not match the scattering curve calculated from the dimer found in the asymmetric unit of the crystal structure (see Supplementary Fig. 5 online). This disagreement is not surprising; the crystallized dimer is unlikely to be functionally relevant18. Even for photoexcited crystals17, the crystal packing interactions likely prevent the full conformational changes needed to gate functional dimerization and surely preclude rearrangement to a different dimer. To investigate the structure of the solution, light-induced dimer we reconstructed low-resolution molecular envelopes from the scattering profiles of the pure dimer, derived as described above. These methods produced a shape with 2-fold symmetry (Fig. 6), regardless of whether or not the reconstruction program was seeded with this information. In contrast to the cylindrical outline of the crystallographic dimer, this shape has a large bulge along the dyad axis, indicating an expansion of the molecule at the dimer interface and a contraction of the molecule along the long axis perpendicular to the molecular interface. Importantly, we could find no orientation of the crystallographic VVD-36 monomer that upon application of two-fold symmetry could fit the reconstructed envelope. Thus, the subunit in the light-induced dimer must have a conformation substantially different from that held by the dark-state, crystallographic monomer.
Fig. 6. Molecular envelope of the VVD light-activated dimer.
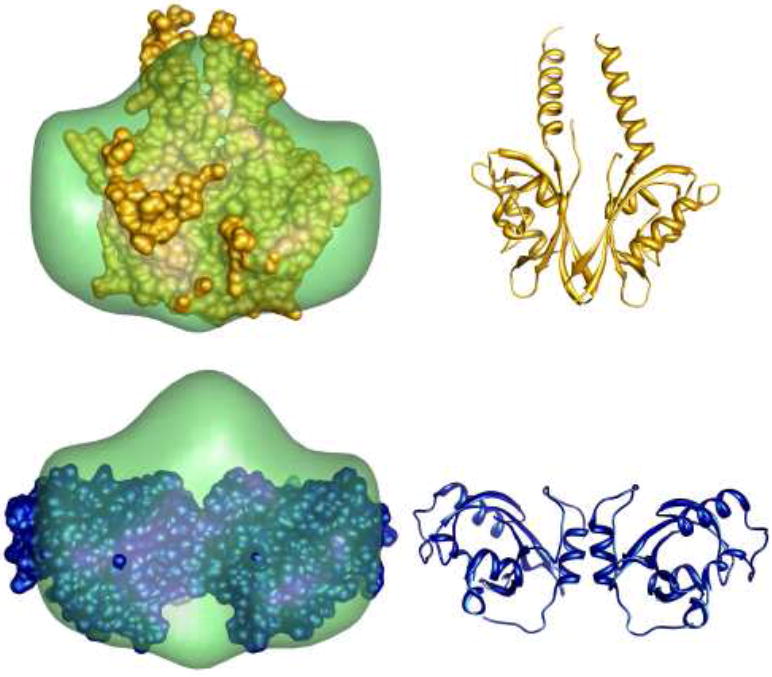
The smooth envelope represents a shape reconstruction based on the scattering profile of the dimer, extracted by minimization analysis. The mean normalized spatial discrepancy of the reconstruction is 0.55, indicating a unique shape envelope. The top image also shows the structure of YtvA, a dimeric LOV domain with a very different dimer interface compared to that found in the VVD crystal structure. At left, the two structures are superimposed; the ribbon diagram of YtvA is shown at right. The bottom panel illustrates the superposition of the crystallized VVD dimer on the reconstructed shape. The VVD ribbon diagram is shown at the bottom, right. The reconstructed shape suggests that the VVD solution dimer has extended helices, similar to YtvA.
Discussion
SAXS profiles of light- and dark-adapted states of VVD-36 and a C71V SAXS data are effectively described by linear combinations of scattering profiles from three states: the monomer reported in the dark-state crystal structure, the same monomer with an extended N-terminus, and a light-state dimer composed of subunits with structures that are distorted compared to the crystal structure. Furthermore, VVD-36 molecules can become oxidized to a form which chemically resembles the nonfunctional C71S mutant. This evidence raises further questions such as: What distinguishes DarkEXT from DarkCOMP? What is the relationship between DarkEXT and the C71V mutant dark state? How does a unique dimer structure emerge from two distinct starting conformations? Consideration of these points leads us to propose a model for the conformational transitions involved in VVD activation.
As stated above, the C71V variant of VVD partially dimerizes without light, suggesting its monomer form closely mimics the light-induced conformation that is competent for dimerization. Light excitation results in additional dimerization. In contrast, DarkEXT has the same scattering profile as the variant monomer but does not dimerize until exposure to light. These functional distinctions indicate conformational differences between C71V and DarkEXT beyond the resolution of these ensemble measurements. Modeling demonstrates good, albeit not necessarily unique, agreement between DarkEXT and a system where the ~ 28 N-terminal amino acids are treated as a flexible ensemble. Involvement of Ncap conformational change in light-activation is also supported by (a) the structure of the molecule, which shows the Ncap near the surface and thus extendable, (b) crystal structures which demonstrate that alternate conformations of the N-terminus are possible, (c) movement of structural elements that stabilize the Ncap against the PAS core in light-excited crystals, (d) residues buried beneath the Ncap that when altered affect the dimerization equilibrium (e) involvement of N-terminal regions in the constitutive dimerization of other PAS domains21,27 and (f) subunit cross-linking patterns in the light-induced dimer that are best explained if the N-terminus assumes a different position than it does in the dark-state crystal structure. Taken together, the current data support a structural model in which the Ncap packs weakly against the PAS-scaffold, in a manner that allows transient undocking from the PAS core. We propose that this undocking exposes and stabilizes the interaction surface.
Differences in the extent of dimerization in DarkEXT and C71V dark and light states, despite their common scattering states, must result from subtle changes in molecular structure. SAXS is a low resolution technique; small changes in molecular conformation or shifts in ensemble populations may be imperceptible. While our data are consistent with extensive undocking of the N-terminus, they do not indicate the exact size or orientation of the undocked region, nor do they favor a homogeneous versus heterogeneous population of states. Additional conformational or chemical changes to the extended state might shift the orientation or stabilize a subpopulation and alter the dimerization potential, explaining why DarkEXT does not dimerize but C71V (dark) does. The more extensive dimerization of C71V in the light state implies some degree of continuity between these extended states. Mutation of a cysteine to a valine at a key site could limit possible conformations of an otherwise flexible extended state, facilitating formation of a small fraction of dimer in the dark state, while light activation further stabilizes the interface involved in dimer formation. Alternatively or additionally, Cys71 could participate directly in the dimer interface and substitution to Val could stabilize subunit contacts. Either case could account for the dimerization patterns we report here.
Either the extended monomer (DarkEXT) or the compact monomer (DarkCOMP) can dimerize on light excitation, and the structure of the dimer formed is the same in each case (see Fig. 7). Unlike the C71V variant, there is no change in protein sequence that explains the difference between these two populations. However, Cys71 also turns out to be a prominent site of oxidation on the protein in that it forms a sulfenic acid after incubation in aerated buffer. Because oxidation inhibits light-induced dimerization, we must conclude that this modification (S-OH) stabilizes the compact monomer to a degree that disfavors progression to the extended precursor state. This is consistent with the observations that the C71S substitution will also not undergo any light-induced dimerization and that protein oxidation also inhibits dimerization (Supplementary Fig. 1 online). A Ser substitution at residue 71, like a sulfenic acid, buries a hydrogen-bonding hydroxyl group into the turn that separates the Ncap from the PAS core. In fact, the sulfenic acid or Ser at position 71 both hydrogen bond to the same back-bone amide on bβ which shifts in crystal-structures of light-activated VVD-36. We suspect that the hydroxyl-to-amide hydrogen bond present in both C71S and oxidized VVD-36 locks the hinge region in the inactive conformation.
Fig. 7. Scheme illustrating the proposed dimerization mechanism.

All of the dark states discussed in the text are shown on the left, in a continuum from less (top) to more (bottom) competent for dimerization. In the case of extensive oxidation or mutation of Cys71 to Ser, shown at the top of the left column, the protein does not dimerize. The DarkCOMP conformation dimerizes less readily than the more extended DarkEXT conformation, presumably due to oxidation which affects the placement of the N terminal helix. Despite small differences in the structures of the DarkCOMP and DarkEXT states, a common dimer forms following illumination. In agreement with reconstructions, the Ncaps detach from the core and extend upwards along the dimer interface in the orientation shown in this scheme. Differences in conformation resulting from mutation of Cys71 to Val appear to be below the resolution of these SAXS experiments, but are evidenced by partial VVD dimerization in the absence of light, suggested by the dark state dimer at the bottom of the left column. Upon light excitation, additional conformational changes enhance the dimer fraction of these proteins as well.
The presence of the two distinct dark states in the VVD-36 population may then be related to oxidation at Cys71, identified above as a key switch point that leads to VVD dimerization. Given that the protein preparations that lead to DarkEXT had minimal oxygen exposure or could be generated by treatment with reductant, DarkEXT likely represents the unoxidized form of the protein. However, since crystals structures of VVD that do not have Cys71 oxidized are composed of compact molecules, we must conclude that crystallization itself stabilizes the N-terminus against the PAS core. (This is not surprising given that the Ncap forms extensive crystal contacts in the lattice.) The increases in I(0) and the fitting constants r1 and r2 give very rough estimates of dimerization and indicate that both populations of molecules dimerize extensively on exposure to light. Mass-spectrometry analysis and some high-resolution crystal structures indicate that Met residues also undergo partial oxidation in VVD-36. The extent of oxidation at key residues may provide a biologically relevant mechanism to tune the structure and the extent of dimerization in the same way that mutation of the residues has been observed to affect both (above and17,18). In fact, it has been demonstrated that under conditions of oxidative stress Neurospora shows behavior consistent with VVD inactivation16. Specifically, a sod mutant exhibits increased production of carotenoids following exposure to blue-light, which is analogous to the vvd null phenotype16. Moreover, the response to reactive oxygen species is enhanced in the sod:vvd double mutant. Upregulation of carotenoids through vvd inactivation maybe an effective strategy for scavenging radicals and preventing damage under conditions of oxidative stress.
Whether or not the two VVD dark states are represented in vivo, they provide insight into the conformational processes that gate and mediate light-induced dimerization of VVD. Adoption of the extended state is necessary, but not sufficient, for subunit association, and factors that favor the compact state, e.g. oxidation, inhibit subunit association. The compact state is well represented by the previously determined crystal structure, and it would be difficult to imagine that the specific contacts made by the Ncap were not relevant for the fold of the protein in some context. These properties then provide physicochemical constraints upon which biological function can be based. In the context of full-length VVD within the cell and surrounded by potential partners, the protein may primarily assume a compact form. Regardless, upon light activation, the protein must transition through an extended state prior to dimerization. The expanded dark state is newly characterized here as a feature of the conformational switches that ultimately produce light-induced dimerization.
In spite of the distinct states present in dark-adapted VVD, a common structure for the light-activated dimer is measured. Reconstruction of the dimer curve supports conclusions from previous work that the solution dimer takes a different form from the crystallized dimer18. This is not surprising, given that the crystal structure was obtained by light exposure of a crystal containing dark-adapted VVD that has already formed a dimer. The reconstructed envelope depicts a dimer with a single bulge on the dyad axis, which we speculate is due to the rearranged Ncaps of both molecules. The molecular envelope suggests that the dimer is parallel with respect to the orientation of the central β-scaffold, an orientation consistently present in homodimeric PAS dimers21,27-29. Such a model is suggestive of the related bacterial LOV domain YtvA20, which displays a dimer interface between β-sheets, with no appreciable intervening Ncap. Instead a C-terminal helix extends up along the dimer plane, producing a shape similar to our reconstructed envelope. Parallel orientations are also found in homodimers of Arnt and HIF30 and those of the LOV1 and 2 subdomains from phototropins24,28,31. However, we note that the ARNT:HIF heterodimers are anti-parallel with respect to the β-strand directions. Adopting a parallel arrangement in VVD would place the loops containing residue 171 in close contact, consistent with observed light-induced solution cross-linking from residues at that position18. A number of different arrangements are known for PAS dimers associated through the subunit beta-sheets, although the same residue positions often participate in the respective interfaces32. In VVD, these conserved contact sites generally hold hydrophobic residues that could also mediate dimerization.
VVD has been shown to work in concert with WC-1 to regulate circadian rhythms in Neurospora crassa12. Indeed, VVD antagonizes the ability of WC-1 to activate gene transcription in response to light and thus generates an adaptive response. The mechanism for conformational dynamics in PAS domains has been observed in phototropins24,33-36, and HIF:ARNT heterodimers23,37. In phototropins, N-terminal and C-terminal elements similar to the VVD N-terminal α-helix have been shown to undergo large scale conformational changes following photoexcitation23,37. The HIF:ARNT system is intriguing in its similarities. In this case, the HIF:ARNT heterodimer competes with the ARNT homodimer on the signaling pathway23,37. Notably, WC-1 has a very similar LOV domain to VVD and perhaps both homo and hetero dimerization of these domains are important elements in their regulation18.
In conclusion, the combined data present a model in which light driven flavin adduct chemistry in VVD reorganizes structural elements adjacent to the β-scaffold. The reorganized surface is then conducive to subunit association. Such a system is highly adaptive because evolution can remodel the mobile elements to incorporate different effector modules and the interaction surface to target different partners. For example, phototropin LOV domains display a similar mechanism, but recruit a C-terminal helix as the mobile elements instead of the Ncap38. The degree to which the signaling processes of LOV/PAS domains are similar or divergent likely forms a continuum, which is only beginning to be described. Visualization of transient states via SAXS reconstructions has the potential to elucidate key aspects of these mechanisms and thereby broaden our understanding of how proteins transduce environmental stimuli into cellular responses.
Materials and Methods
Sample Preparation
Preparation of VVD Variants
The C71V variant of VVD-36 was produced and characterized in a previous study18. However, to help ward against the complicating issues associated with oxidation and protein instability, we produced this variant in the background of C183S, which has a known site of oxidation (Cys183) removed. The C71V:C183S variant was constructed according to the QuickChange protocol (Stratagene). Resultant mutants were sequenced in their entirety at the Biotechnology Resource Center at Cornell University.
Protein expression and Purification
VVD-36 and C71V:C183S variants were overexpressed in E. coli BL21(DE3) cells. Two-liter cultures of the variants were grown to an OD600 of 0.6-0.8 at 37°C. When the cell density reached 0.6, the cultures were cooled to 18°C and induced with 100 M IPTG. Protein was then expressed for 22 hours prior to harvesting the cells.
24-liters of both VVD-36 and C71V:C183S were prepared via the above protocol and lysed in buffer containing 13 % glycerol, 300 mM NaCl, 50 mM Hepes pH 8.0 and 5 mM imidazole pH 8, as the sonicated and soluble cell lysate was factionated by centrifugation. The supernatant was then collected and protein purified via Ni:NTA affinity chromatography. Eluted VVD was subsequently treated with 1 unit of thrombin/mg of protein for 6 hours in buffer containing 2 mM DTT, 13% glycerol, 150 mM NaCl, 50 mM Hepes pH 8.0 and 100 mM imidazole. The protein was then purified on a Superdex 75 26/60 Hi-load column and concentrated to 5 mg/mL. Final protein samples contained 5 mM DTT.
Crystallization of oxidized VVD
VVD-36 was cocrystallized in the presence of 10 mM imidazole via the hanging drop method. Crystals grew from droplets containing 2 μl each of 5 mg/ml VVD-36 and 2 μl of the reservoir solution containing 20 mM imidazole, 100 mM NaCl, 26% PEG 4k and 100 mM trisodium citrate pH 5.6. Crystals were obtained overnight at 22° C and diffracted to 2.3 Å resolution at the F3 beamline at the Cornell High Energy Synchrotron Source (CHESS). Diffraction data for 30-2.3 Å was processed with HKL2000 and the structure determined using molecular replacement (AMORE) using 2PD7 as a search model. The model was rebuilt using Xfit followed by positional and thermal refinement in CNS. Residual density in 2fo-fc (2σ) and fo-fc (3σ) connecting to the Cys71 side chain was consistent with oxidation to cysteinic acid (Cys-OH).
SAXS Data Collection
Small angle x-ray scattering data was collected at the G1 beamline at the CHESS at an energy of 8 keV. A continuous flow cell made of a 1 mm polyester tube (Advanced Polymers, Inc., VT)39 was employed to collect time resolved data, using a method described in ref. 18. A 473 nm laser from Holograms and Laser, International (Houston, TX) was aligned with the x-ray beam using a 20 μm slit to confirm their coincidence. The x-ray beam was focused with a glass capillary40,41 for better position definition. The laser focal point was then moved against the direction of fluid flow to introduce delay. Eight 30 s x-ray exposures were collected on the protein for each sample, to improve signal to noise and ensure reproducibility. A PIN diode was mounted onto the x-ray beamstop to measure changes in beam intensity.
Although this method allows for data to be collected at several points after light excitation, minimal changes were observed after the first data point (Supplementary Figs. 6 and 7 online and Supplementary Discussion). Previous time-resolved SAXS measurements on VVD reflected additional conformational changes after several seconds, in contrast to current observations19. The main difference between these measurements was the current inclusion of DTT in the protein buffer. This may indicate that oxidation is associated with conformational changes in the light excited state. Static SAXS experiments confirm that synchrotron X-ray exposure reduce the VVD flavin and thereby inhibit dimerization17. However, the flow cell configuration greatly diminishes X-ray exposure and thereby mitigates problems associated with flavin reduction. In flow cell experiments the x-ray exposure time was varied simply by changing the flow speed (while carefully ensuring identical light exposure of the sample). No effect on the yield of VVD dimerization with X-ray exposure was found. UV/Vis absorption spectra collected on samples after irradiation and X-ray exposure confirmed a substantial conversion of VVD to the light-state adduct.
Data analysis
Images were converted to scattering profiles of intensity (I) as a function of q, where, , with θ being half the scattering angle and λ the x-ray wavelength. An image of a silver stearate42 scattering ring collected under the same beam conditions was used to find the beam center and determine the radial calibration. Each scattering profile was normalized for changes in beam intensity and checked for reproducibility. Final scattering profiles were obtained using MATLAB (The Mathworks, Natick, MA) by first averaging images, then converting the data to intensity versus q, and subtracting the buffer background.
Scattering data plotted as a Guinier plot, log(I(q)) versus q2, is approximately linear at low angles43. Typically, this approximation is considered valid for qRg <1.3. Fitting to this line allows extrapolation to I(0) and provides the slope, which is proportional to Rg. Errors on these quantities were determined by propagating the 95% confidence intervals from the slope and y-intercept of the line fit with MATLAB.
GNOM, DAMMIN, DAMAVER, CRYSOL and EOM are all analysis tools for scattering data made available by the Biological Small Angle Scattering group at the European Molecular Biology Laboratory44. CRYSOL was used to calculate scattering from crystal structures. Scaling was determined by CRYSOL through comparison to appropriate experimental data. Conversion of scattering data to P(r) was done with GNOM45. In general Dmax was found by testing input values to GNOM based on the Rg determined from Guinier analysis and choosing the one which best maximized the default regularization parameters used by GNOM. We determined Dmax to within 5 Å with this method.
All minimizations described in the text were performed using the fmin function in MATLAB. Statistical errors for the analysis were determined using GNOM based on the noise in the data. Degrees of freedom, ν, were calculated by subtracting the number of fitting parameters from the number of points in a single scattering curve. Reconstructions were carried out by running DAMMIN46 ten times on the output from GNOM, and then using DAMAVER to average the results and Situs47 to generate a shape envelope. Crystal structures were fitted in the molecular envelopes also using Situs. Analysis with EOM was carried out using the 3D72 crystal structure.
Supplementary Material
Acknowledgments
Funding for this work was provided by the Cornell Nanobiotechnology Center, which is supported by the STC Program of the National Science Foundation (NSF) under Agreement No. ECS-9876771. This work was also supported by National Institutes of Health (NIH) grant R01-GM079679. CHESS is supported by the NSF and the NIH/National Institute of General Medical Sciences under award DMR-0225180. The authors would like to thank Sterling Cornaby and Arthur Woll for their assistance at the G1 station and Steve Meisburger for valuable comments. Computations were carried out at the Cornell Center for Materials Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pellequer JL, Wager-Smith KA, Kay SA, Getzoff ED. Photoactive yellow protein: a structural prototype for the three-dimensional fold of the PAS domain superfamily. Proc Natl Acad Sci USA. 1998;95:5884–90. doi: 10.1073/pnas.95.11.5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taylor BL, Zhulin IB. PAS Domains: Internal Sensors of Oxygen, Redox Potential, and Light. Microbiol Mol Biol Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swartz TE, et al. The photocycle of a flavin-binding domain of the blue light photoreceptor phototropin. J Biol Chem. 2001;276:36493–500. doi: 10.1074/jbc.M103114200. [DOI] [PubMed] [Google Scholar]
- 4.Briggs WR, Christie JM, Salomon M. Phototropins: A new family of flavin-binding blue light receptors in plants. Antioxid Redox Signaling. 2001;3:775–788. doi: 10.1089/15230860152664975. [DOI] [PubMed] [Google Scholar]
- 5.Crosson S, Rajagopal S, Moffat K. The LOV domain family: Photoresponsive signaling modules coupled to diverse output domains. Biochemistry. 2003;42:2–10. doi: 10.1021/bi026978l. [DOI] [PubMed] [Google Scholar]
- 6.Losi A. The bacterial counterparts of plant phototropins. Photochem Photobiol Sci. 2004;3:566–574. doi: 10.1039/b400728j. [DOI] [PubMed] [Google Scholar]
- 7.Salomon M, et al. An optomechanical transducer in the blue light receptor phototropin from Avena sativa. Proc Natl Acad Sci USA. 2001;98:12357–61. doi: 10.1073/pnas.221455298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crosson S, Moffat K. Photoexcited Structure of a Plant Photoreceptor Domain Reveals a Light-Driven Molecular Switch. Plant Cell. 2002;14:1067–1075. doi: 10.1105/tpc.010475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huala E. Arabidopsis NPH1: A Protein Kinase with a Putative Redox-Sensing Domain. Science. 1997;278:2120–2123. doi: 10.1126/science.278.5346.2120. [DOI] [PubMed] [Google Scholar]
- 10.Crosson S, Moffat K. Structure of a flavin-binding plant photoreceptor domain: insights into light-mediated signal transduction. Proc Natl Acad Sci USA. 2001;98:2995–3000. doi: 10.1073/pnas.051520298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loros JJ, Dunlap JC. Genetic and molecular analysis of circadian rhythms in Neurospora. Annu Rev Physiol. 2001;63:757–94. doi: 10.1146/annurev.physiol.63.1.757. [DOI] [PubMed] [Google Scholar]
- 12.Brunner M, Kaldi K. Interlocked feedback loops of the circadian clock of Neurospora crassa. Mol Microbiol. 2008;68:255–262. doi: 10.1111/j.1365-2958.2008.06148.x. [DOI] [PubMed] [Google Scholar]
- 13.Ávila-Pérez M, Hellingwerf KJ, Kort R. Blue Light Activates the σB-Dependent Stress Response of Bacillus subtilis via YtvA. J Bacteriol. 2006;188:6411–6414. doi: 10.1128/JB.00716-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heintzen C, Loros JJ, Dunlap JC. The PAS protein VIVID defines a clock-associated feedback loop that represses light input, modulates gating, and regulates clock resetting. Cell. 2001;104:453–464. doi: 10.1016/s0092-8674(01)00232-x. [DOI] [PubMed] [Google Scholar]
- 15.Schwerdtfeger C, Linden H. Blue light adaptation and desensitization of light signal transduction in Neurospora crassa. Mol Microbiol. 2001;39:1080–7. doi: 10.1046/j.1365-2958.2001.02306.x. [DOI] [PubMed] [Google Scholar]
- 16.Yoshida Y, Hasunuma K. Reactive oxygen species affect photomorphogenesis in Neurospora crassa. J Biol Chem. 2004;279:6986–6993. doi: 10.1074/jbc.M310060200. [DOI] [PubMed] [Google Scholar]
- 17.Zoltowski BD, et al. Conformational switching in the fungal light sensor vivid. Science. 2007;316:1054–1057. doi: 10.1126/science.1137128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zoltowski BD, Crane BR. Light activation of the LOV protein Vivid generates a rapidly exchanging dimer. Biochemistry. 2008;47:7012–7019. doi: 10.1021/bi8007017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lamb JS, Zoltowski BD, Pabit SA, Crane BR, Pollack L. Time-resolved dimerization of a PAS-LOV protein measured with photocoupled small angle X-ray scattering. J Am Chem Soc. 2008;130:12226–7. doi: 10.1021/ja804236f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Möglich A, Moffat K. Structural basis for light-dependent signaling in the dimeric LOV domain of the photosensor YtvA. J Mol Biol. 2007;373:112–26. doi: 10.1016/j.jmb.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma XL, Sayed N, Baskaran P, Beuve A, van den Akker F. PAS-mediated dimerization of soluble guanylyl cyclase revealed by signal transduction histidine kinase domain crystal structure. J Biol Chem. 2008;283:1167–1178. doi: 10.1074/jbc.M706218200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strickland D, Moffat K, Sosnick TR. Light-activated DNA binding in a designed allosteric protein. Proc Natl Acad Sci USA. 2008;105:10709–14. doi: 10.1073/pnas.0709610105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Card PB, Erbel PJA, Gardner KH. Structural basis of ARNT PAS-B dimerization: Use of a common beta-sheet interface for hetero- and homodimerization. J Mol Biol. 2005;353:664–677. doi: 10.1016/j.jmb.2005.08.043. [DOI] [PubMed] [Google Scholar]
- 24.Nakasone Y, Eitoku T, Matsuoka D, Tokutomi S, Terazima M. Kinetic measurement of transient dimerization and dissociation reactions of Arabidopsis phototropin 1 LOV2 domain. Biophys J. 2006;91:645–653. doi: 10.1529/biophysj.106.084772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakasako M, Iwata T, Matsuoka D, Tokutomi S. Light-induced structural changes of LOV domain-containing polypeptides from Arabidopsis phototropin 1 and 2 studied by small-angle X-ray scattering. Biochemistry. 2004;43:14881–90. doi: 10.1021/bi0485530. [DOI] [PubMed] [Google Scholar]
- 26.Bernadó P, Pérez Y, Svergun DI, Pons M. Structural characterization of the active and inactive states of Src kinase in solution by small-angle X-ray scattering. J Mol Biol. 2008;376:492–505. doi: 10.1016/j.jmb.2007.11.066. [DOI] [PubMed] [Google Scholar]
- 27.Key J, Hefti M, Purcell EB, Moffat K. Structure of the redox sensor domain of Azotobacter vinelandii NifL at atomic resolution: Signaling, dimerization, and mechanism. Biochemistry. 2007;46:3614–3623. doi: 10.1021/bi0620407. [DOI] [PubMed] [Google Scholar]
- 28.Nakasako M, Zikihara K, Matsuoka D, Katsura H, Tokutomi S. Structural basis of the LOV1 dimerization of Arabidopsis phototropins 1 and 2. J Mol Biol. 2008;381:718–733. doi: 10.1016/j.jmb.2008.06.033. [DOI] [PubMed] [Google Scholar]
- 29.Lee J, et al. Changes at the KinA PAS-A dimerization interface influence histidine kinase function. Biochemistry. 2008;47:4051–4064. doi: 10.1021/bi7021156. [DOI] [PubMed] [Google Scholar]
- 30.Erbel PJA, Card PB, Karakuzu O, Bruick RK, Gardner KH. Structural basis for PAS domain heterodimerization in the basic helix-loop-helix PAS transcription factor hypoxia-inducible factor. 2003;100:15504–15509. doi: 10.1073/pnas.2533374100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salomon M, Lempert U, Rudiger W. Dimerization of the plant photoreceptor phototropin is probably mediated by the LOV1 domain. FEBS Lett. 2004;572:8–10. doi: 10.1016/j.febslet.2004.06.081. [DOI] [PubMed] [Google Scholar]
- 32.Ayers RA, Moffat K. Changes in Quaternary Structure in the Signaling Mechanisms of PAS Domains. Biochemistry. 2008;47:12078–12086. doi: 10.1021/bi801254c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harper SM, Neil LC, Gardner KH. Structural basis of a phototropin light switch. Science. 2003;301:1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- 34.Harper SM, Neil LC, Day IJ, Hore PJ, Gardner KH. Conformational changes in a photosensory LOV domain monitored by time-resolved NMR spectroscopy. J Am Chem Soc. 2004;126:3390–3391. doi: 10.1021/ja038224f. [DOI] [PubMed] [Google Scholar]
- 35.Nakasone Y, Eitoku T, Matsuoka D, Tokutomi S, Terazima M. Dynamics of conformational changes of Arabidopsis phototropin 1 LOV2 with the linker domain. J Mol Biol. 2007;367:432–442. doi: 10.1016/j.jmb.2006.12.074. [DOI] [PubMed] [Google Scholar]
- 36.Nakasone Y, Ono TA, Ishii A, Masuda S, Terazima M. Transient dimerization and conformational change of a BLUF protein: YcgF. J Am Chem Soc. 2007;129:7028–7035. doi: 10.1021/ja065682q. [DOI] [PubMed] [Google Scholar]
- 37.Erbel PJA, Card PB, Karakuzu O, Bruick RK, Gardner KH. Structural basis for PAS domain heterodimerization in the basic helix-loop-helix-PAS transcription factor hypoxia-inducible factor. Proc Natl Acad Sci USA. 2003;100:15504–15509. doi: 10.1073/pnas.2533374100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ko WH, Nash AI, Gardner KH. A LOVely view of blue light photosensing. Nat Chem Biol. 2007;3:372–374. doi: 10.1038/nchembio0707-372. [DOI] [PubMed] [Google Scholar]
- 39.Kalinin Y, et al. A new sample mounting technique for room-temperature macromolecular crystallography. J Appl Crystallogr. 2005;38:333–339. [Google Scholar]
- 40.Engström P, et al. A submicron synchrotron X-ray beam generated by capillary optics. Nucl Instrum Methods Phys Res, Sect A. 1991;302:547–552. [Google Scholar]
- 41.Lamb JS, et al. Focusing capillary optics for use in solution small-angle X-ray scattering. J Appl Crystallogr. 2007;40:193–195. [Google Scholar]
- 42.Vand V, Aitken A, Campbell RK. Crystal structure of silver salts of fatty acids. Acta Crystallogr. 1949;2:398–403. [Google Scholar]
- 43.Guinier A, Fournet G. Small-Angle Scattering of X-Rays. John Wiley and Sons, New York; New York, USA: 1955. [Google Scholar]
- 44.Konarev PV, Petoukhov MV, Volkov VV, Svergun DI. ATSAS 2.1, a program package for small-angle scattering data analysis. J Appl Crystallogr. 2006:277–286. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Svergun DI. Determination of the Regularization Parameter in Indirect-Transform Methods Using Perceptual Criteria. J Appl Crystallogr. 1992;25:495–503. [Google Scholar]
- 46.Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys J. 1999;76:2879–86. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wriggers W, Chacón P. Using Situs for the registration of protein structures with low-resolution bead models from X-ray solution scattering. J Appl Crystallogr. 2001;34:773–776. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


