Synopsis
Dietary isothiocyanates (ITCs) prevent cancer and show other bioactivities in vivo. As electrophiles, ITCs may covalently modify cellular proteins. Using a novel proteomics screen, we identified the Macrophage Migration Inhibitory Factor (MIF) as the principal target of nutrient ITCs in intact cells. ITCs covalently modify the amino-terminal proline residue of MIF and extinguish its catalytic tautomerase activity. MIF deficiency does not prevent induction of Phase 2 gene expression, a hallmark of many cancer chemopreventives including ITCs. Due to the emerging role of MIF in control of malignant cell growth and its clear involvement in inflammation, inhibition of MIF by nutrient ITCs suggests therapeutic strategies for inflammatory diseases and cancer.
Keywords: Sulforaphane, PEITC, Chemoprevention, Cancer, Inflammation, Tautomerase
Introduction
Dietary isothiocyanates, such as phenethyl ITC (PEITC) and sulforaphane (SFN), are prevalent in cruciferous vegetables. Cancer prevention by ITCs is widely attributed to their ability to induce expression of detoxifying enzymes referred to as Phase 2 enzymes, including several of the Glutathione S-transferases [1]. These enzymes are believed to promote the neutralization and excretion of carcinogens. However, ITCs inhibit tumor growth even when administered after the carcinogen insult [2, 3] and in xenograft tumor models [4], when protection from mutagenic damage cannot explain cancer prevention. Other mechanisms such as induction of apoptosis [5], or host-tumor interactions present viable alternate mechanisms of cancer prevention.
Since ITCs are reactive, electrophilic compounds, we hypothesized that they would covalently modify cellular target proteins. We undertook a proteomics screen to purify proteins covalently modified by ITCs. We identified the Macrophage Migration Inhibitory Factor (MIF) as the predominant cellular target of ITC modification. As the first identified cytokine [6, 7], MIF has been extensively characterized for its critical role in inflammation. Studies using neutralizing antibodies or MIF deficient animals show that inhibition of MIF can ameliorate disease processes in animal models of arthritis and inflammatory bowel disease, among many others [8, 9]. Moreover, MIF is overexpressed in cancers (e.g. [10, 11]), and MIF expression correlates with tumor aggressiveness [12]. Recent in vitro evidence has suggested that MIF is required for cellular transformation [13], and that loss of MIF expression reduces growth of tumors in animal models [14, 15].
MIF is a 12kD protein with an enzymatic keto-enol tautomerase activity [16]. While the natural tautomerase substrate remains unknown, tautomerase activity contributes to biological activities associated with MIF [17], leading to the conclusion that pharmacologic agents that block tautomerase activity may have broad applicability. We demonstrate that dietary nutrient ITCs covalently modify MIF and inhibit its tautomerase activity, suggesting that they may represent valuable natural product inhibitors of MIF function in human disease.
Experimental
Cell Culture, Plasmids and Reagents
HeLa cells were cultured in DMEM/10% calf serum and penicillin/streptomycin. MEFs and HepG2 cells were cultured in DMEM/10% fetal calf serum plus antibiotics. Expression plasmids for MIF were prepared in pcDNA3 (Stratagene) by PCR amplifying the MIF coding sequence. In some cases, the MIF coding region was fused to a chitin binding domain (CBD) tag at the C terminus for purification on chitin beads. Recombinant MIF with a similar CBD tag removed using intervening thrombin cleavage site was produced in BL21(DE3) bacteria a using pET11b vector (Novagen). All PCR amplified coding regions were fully sequenced. MIF directed Smartpool siRNAs along with a control non-targeting siRNA pool were purchased from Dharmacon. Anti-MIF antibody was from Santa Cruz Biotechnology and anti-biotin from Cell Signaling. HeLa cells were transfected with Lipofectamine Plus for protein expression or Lipofectamine 2000 for siRNA experiments, HepG2 were transfected with Lipofectamine 2000 (all from Invitrogen).
ITC Proteomics
Biotin-ITC was synthesized as previously described [18] based on modification of a published protocol [19]. HeLa cells were left untreated or pretreated for 30 minutes with 250μM PEITC to promote complete modification of potential target proteins, followed by Dounce homogenization in 25mM HEPES containing aprotinin and leupeptin. Clarified lysates were labeled in vitro with Bio-ITC (500μM for 1hour at room temperature) and then stopped by addition of Laemmli sample buffer. Lysates were separated on SDS-PAGE gels and labeled proteins detected using HRP conjugated streptavidin (Zymed) or anti-biotin (Cell Signaling). For purification, lysates were dialyzed against 25mM HEPES to remove free Bio-ITC, and labeled proteins purified using streptavidin Sepharose (Amersham), separated by SDS-PAGE and detected by silver stain. The small molecular weight band was excised for analysis.
Mass determination of MIF and PEITC-MIF Protein
Recombinant MIF protein was treated with PEITC in vitro until tautomerase activity was completely quenched. Intact rMIF (1μg/μL) and PEITC-labeled rMIF were separately infused into an LTQ mass spectrometer (Thermo Fisher Scientific) at 1-3μL per minute. MS spectra were collected and the protein charge-state envelope for each was de-convoluted to determine the mass with an accuracy of about ±5 Da.
MS Identification of Modification site
PEITC labeled MIF was digested with trypsin (Roche) and loaded onto a 100μm i.d. fused silica pre-column self-packed with irregular (5-25μm, 120 Å) reverse phase phenyl beads (Phenomenex). The pre-column was attached to an analytical 75μm i.d. PicoFrit column (New Objective, Woburn, MA) self-packed with regular (5 μm, 120 (x001FA)) reverse-phase phenyl beads (Phenomenex). Peptides were eluted using a gradient of 0 to 70% acetonitrile in 0.1% acetic acid and directly electrosprayed into the LTQ mass spectrometer. The duty cycle was a full scan (300-2000 m/z) followed by MS2 CID fragmentation of the 5 most abundant ions with dynamic exclusion. MS2 scans identified using SEQUEST were manually verified.
MIF Tautomerase Activity
Substrate [16] was prepared by mixing equal volumes of 4mM L-3,4-Dihydroxyphenylalanine methyl ester hydrochloride (dopachrome, Sigma) and 8mM sodium (meta)periodate (Sigma) and incubating at room temperature for five minutes. Cell lysate or recombinant MIF was assayed in 25mM phosphate buffer (pH6.0)/1mM EDTA buffer containing 100μl/ml freshly prepared substrate. Decolorization was monitored over time by OD at 475nM.
ARE luciferase and Phase2/ARE-driven gene induction in MEFs
HepG2 cells seeded in 24well plates were transfected with an ARE driven luciferase plasmid with either control siRNA or MIF targeting siRNAs (50pmoles). After 24 hours, media was replaced with fresh media containing various concentration of sulforaphane or DMSO vehicle. Cells were incubated for an additional 24 hours and then harvested in cell culture lysis reagent (Promega) and assayed for luciferase activity. For ITC-induced gene expression in MEFs, wild type or MIF deficient MEFs were treated with 10μM sulforaphane or vehicle control for 12 hours and RNA prepared using Trizol Reagent (Invitrogen). RNA was quantified and 1ug of RNA reverse transcribed using iScript kit (Bio-Rad). Real-time PCR analysis was performed using IQ SYBR Green Supermix and the iCycler Instrument (Bio-Rad).
Results
Proteomics Screen for Targets of ITC Modification
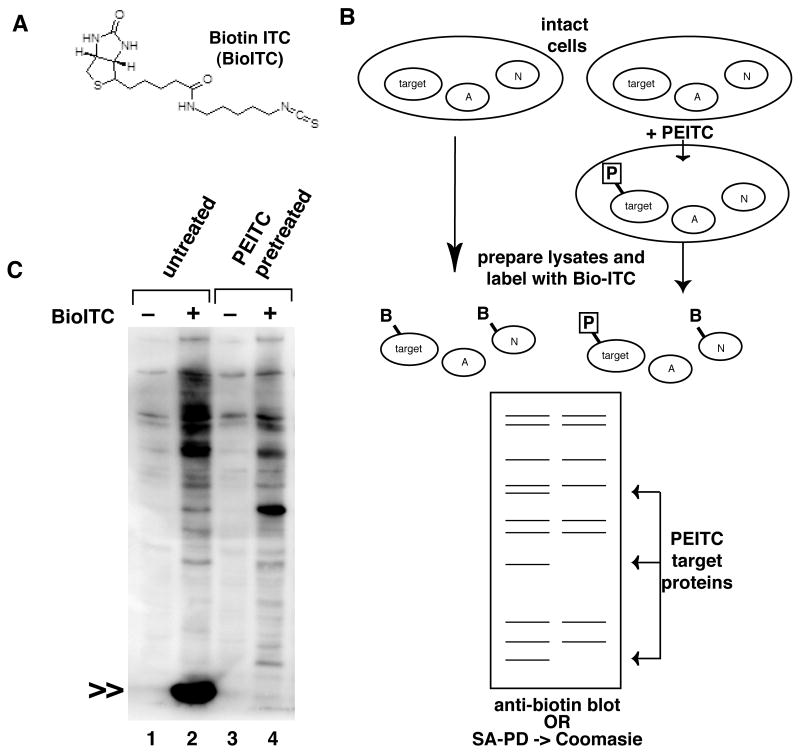
We synthesized a biotin-based ITC affinity probe, Bio-ITC, (Figure 1A), and used a competition assay to identify protein targets of natural ITCs, as outlined in Figure 1B. We first incubated intact cells with an excess of the natural ITC, PEITC, so that covalent protein targets of PEITC would be fully modified. Then, PEITC treated cells and untreated control cells were lysed and protein extracts reacted with Bio-ITC in vitro. Proteins that were fully modified by PEITC in cells were not labeled with Bio-ITC, since the target sites were occupied. Therefore, biotin labeled proteins present in untreated cell lysates, but absent in the PEITC pretreated cell lysates represent targets of PEITC in intact cells.
Figure 1. Proteomics screen for protein targets of covalent ITC modification.
A. Bio-ITC structure. B. Schematic representation of competition based screen. In cells pretreated with PEITC, ITC target sites are occupied by the natural isothiocyanate (P). After lysis and labeling with Bio-ITC, these specific target sites are unavailable, though nonspecific proteins (N) may be labeled. Detection of Bio-ITC will show specific and non-specific proteins. Proteins detected in the samples prepared from the untreated cells, but absent from the PEITC treated cells represent specific targets of the natural isothiocyanate in intact cells. C. Representative labeling experiment. HeLa cells were left untreated or pretreated with 250μM PEITC to promote complete modification of potential target proteins. Lysates were labeled in vitro with Bio-ITC, and labeled proteins detected on SDS-PAGE with HRP conjugated streptavidin. The predominant target of Bio-ITC modification was a small molecular weight protein of less than 29kD, indicated by double arrows; labeling was completely inhibited by pretreatment of the intact cells with PEITC.
Using this screen, a single small molecular weight protein of less than 29kDa accounted for the majority of the total Bio-ITC labeling of cell proteins (Figure 1C). This labeled protein was completely absent from samples pretreated with PEITC, suggesting that it was quantitatively modified under these conditions. Dose response experiments showed that Bio-ITC labeling of this small protein was nearly completely inhibited by 30 minute incubation with as little as 25μM PEITC, and was partially inhibited at even lower doses (Supplementary Figures 1 and 4). Similar results were obtained with other ITCs, including sulforaphane (e.g. Supplementary Figure 4).
Bio-ITC labeled proteins purified from untreated or PEITC pretreated lysates by streptavidin affinity chromatography were separated on an SDS-PAGE gel (not shown), and a small molecular weight band present in the untreated sample but absent from the PEITC pretreated sample was excised and subjected to tryptic peptide fingerprinting using LC-mass spectrometry. We identified a peptide in the sample (LLCGLLAER), unique in the human genome and identical to a predicted peptide of the Macrophage Migration Inhibitory Factor, MIF.
ITCs Covalently Modify MIF on proline 2
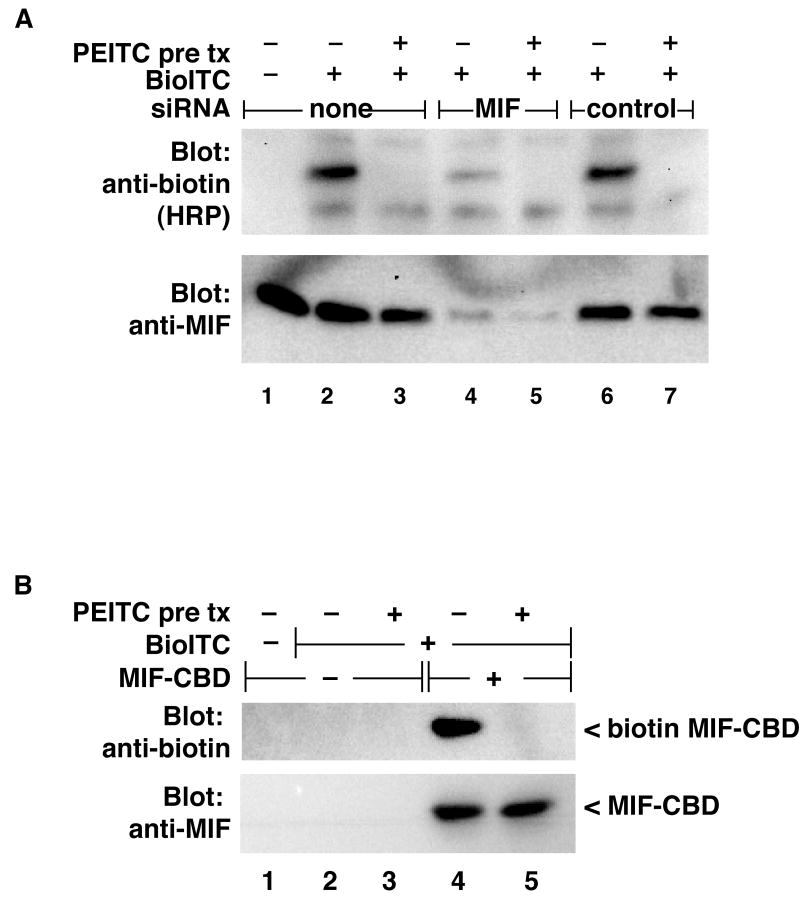
To confirm the identification of MIF as the PEITC target protein, we depleted MIF from HeLa cells using siRNA and measured the effect on Bio-ITC labeling of the small protein (Figure2A). Depletion of MIF protein resulted in a concomitant loss of the biotin-labeled band. We made expression plasmids for MIF that fuse a chitin binding domain (CBD) tag to the C-terminus of MIF. MIF-CBD expressed in transfected cells was efficiently labeled by Bio-ITC treatment of cell lysates, and this labeling was completely inhibited by pretreatment of intact cells with PEITC (Figure 2B). These results confirm that MIF is a covalent target of dietary ITCs.
Figure 2. MIF is a target for ITC modification.
A: HeLa cells were seeded in 12 well plates and transfected with a pool of siRNAs directed against MIF or the control non-targeting siRNA pool (100pmoles/well) as indicated. Thirty minutes prior to harvest, indicated wells were pretreated with 250μM PEITC (pre tx). Cells were lysed and labeled with Bio-ITC as described in Figure 1. Effective depletion of MIF from the lysates (lower panel) was accompanied by loss of Bio-ITC reactivity of the Bio-ITC target protein (upper panel). Panel B: CBD-tagged MIF was expressed in HeLa cells, pretreated with PEITC or not, and cell lysates labeled with Bio-ITC as above. CBD tagged proteins were purified on chitin beads, and Bio-ITC labeling detected by immunoblot. CBD-tagged MIF was efficiently labeled by Bio-ITC, and this label was completely inhibited by pretreatment of the cells with PEITC.
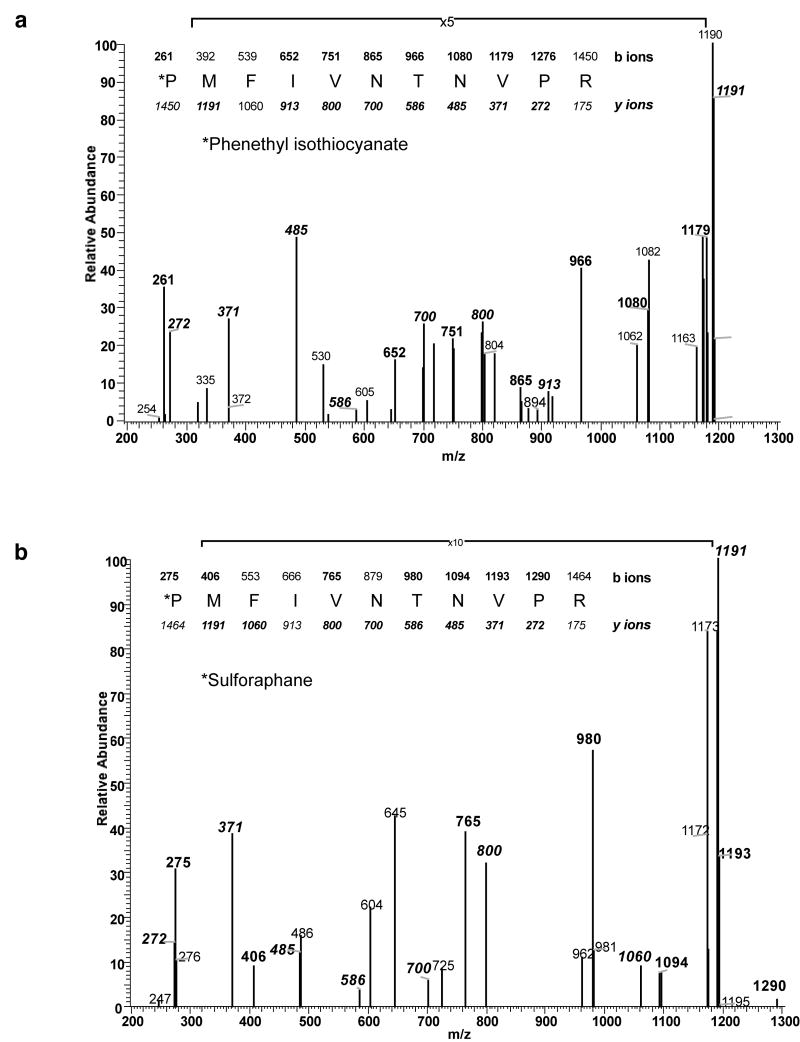
Mass spectrometry showed a mass of the intact ITC-modified recombinant MIF (rMIF) protein consistent with one ITC moiety per polypeptide (Supplementary Figure 2). To determine the site of ITC modification, rMIF protein was incubated with PEITC, trypsinized and then analyzed by LC/MS/MS. We found spectra from a single tryptic peptide containing the predicted 163 Da addition; this was on a peptide containing the PEITC label on the amino terminal proline at position 2 (Figure 3 A), following cleavage of the initiator methionine. Separately, rMIF was treated with other isothiocyanates including sulforaphane (Figure 3B). In each case, ITC modification of the expected mass was found on proline 2. Therefore, rMIF is singly modified on proline 2 by ITCs in vitro.
Figure 3. Mass Spectrometry analysis of ITC treated MIF reveals modification of Proline 2.
Recombinant MIF modified with PEITC (Panel A) or sulforaphane (Panel B) was trypsinized and analyzed using LC-MS. Spectra from the N terminal peptide of MIF was found modified by the predicted 163Da (PEITC) or 177Da (sulforaphane) addition with analysis of the MS2 scans localizing the modification to the N terminal proline.
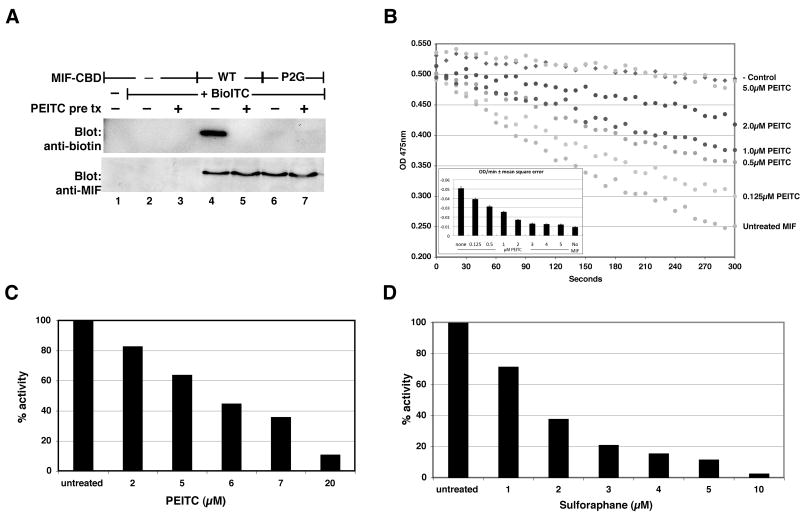
To confirm the specificity of labeling in intact mammalian cells, we made an expression plasmid encoding MIF with the proline 2 replaced by glycine (P2G MIF). The P2G MIF was not labeled with Bio-ITC (Figure 4A), demonstrating that proline 2 is required for the modification of MIF in mammalian cells.
Figure 4. ITC modification of MIF requires proline 2 and inhibits tautomerase activity.
Panel A: CBD tagged wild type or a P2G mutant MIF expressed in HeLa cells was labeled and analyzed as in Figure 2. WT MIF, but not the P2G MIF, was labeled by Bio-ITC. Pretreatment with PEITC (pre tx) completely inhibited Bio-ITC labeling. This figure is representative of three independent experiments that confirmed the absence of Bio-ITC labeling of the P2G mutant. Panel B: Tautomerase activity of rMIF was measured using a D-dopachrome decolorization assay. PEITC dose responsively inhibited the tautomerase activity with an IC50 less than 1μM. The slope of the reaction line, representing tautomerase activity, was calculated by determining the least squares regression of each data set (inset). Error bars represent the mean squared error of the regression. Panel C: HeLa cells were treated with PEITC for 30 minutes at 37°C, then tautomerase assayed in cell lysates. PEITC inhibited MIF in intact HeLa cells with an IC50 between 5-6μM. Panel D: A similar dose response assay with sulforaphane revealed an IC50 for MIF inhibition between 1-2μM.
ITCs inhibit MIF tautomerase activity
The MIF protein encodes an enzymatic tautomerase activity that has previously been shown to require proline 2. Incubation of rMIF with PEITC inhibited the tautomerase activity in a dose dependent manner (Figure 4B). In vitro, rMIF is exquisitely sensitive to inhibition by PEITC, with an IC50 for tautomerase inhibition of less than 1μM, as might be expected for a covalent inhibitor. Similar results were obtained with sulforaphane (not shown).
In intact HeLa cells, PEITC treatment resulted in dose dependent inhibition of tautomerase activity (Figure 4C), with an IC50 for inhibition between 5 and 6μM. Inhibition of tautomerase in HeLa cells by sulforaphane was even more efficient, with an IC50 between 1 and 2μM (Figure 4D). Since MIF is the only protein contributing to the tautomerase activity measured under our assay conditions in HeLa cell lysates (Supplementary Figure 3), these results demonstrate that ITCs modify MIF in intact cells and efficiently inhibit the tautomerase activity. Furthermore, they demonstrate differential inhibition of MIF by distinct ITC compounds. Importantly, inhibition of MIF tautomerase activity correlated closely with the covalent modification by ITCs as reflected by inhibition of BioITC labeling (Supplementary Figure 4).
MIF deficiency does not alter the induction of Phase 2/ARE-dependent genes by ITCs
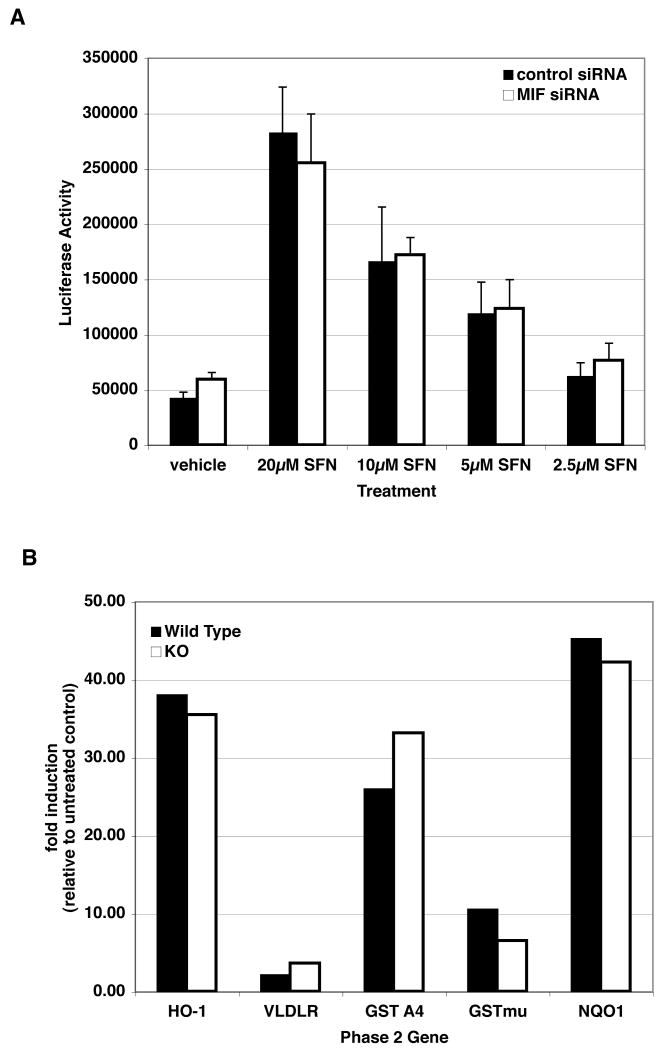
MIF has been proposed to play a role in regulation of several transcription factors, including AP1 [20]. Therefore, ITC modification of MIF could explain at a molecular level the induction of Phase 2 genes and other ARE-driven gene expression in ITC exposed cells. However, depletion of MIF with siRNA did not alter the activation of ARE dependent transcription in response to sulforaphane treatment (Figure 5A). Moreover, MEFs prepared from MIF -/- embryos were not impaired in their ability to induce transcription of Phase 2 or other ARE-dependent genes in response to sulforaphane treatment (Figure 5B). Together, these results suggest that modification and inhibition of MIF is not involved in the induction of Phase 2 gene expression in response to isothiocyanate treatment.
Figure 5. MIF is not required for induction of Phase 2/ARE dependent gene expression.
Panel A: HepG2 cells cotransfected with an ARE driven luciferase reporter and siRNAs directed against MIF (as in Figure 2) were treated with sulforaphane followed by measurement of luciferase. Depletion of MIF did not alter the induction of ARE dependent transcription in response to sulforaphane. Panel B: Mouse embryo fibroblasts (MEFs) from wild type and MIF knockout embryos were treated with 10μM sulforaphane for 12hrs. Expression of Phase 2 genes was quantified relative to the untreated control. Sulforaphane induced transcription of all of the tested genes, ranging from 2 fold to greater than 40 fold, in both wild type and MIF deficient MEFs.
Discussion
The role of MIF as an essential mediator of disease, particularly inflammatory disease, has been long appreciated (e.g. [8, 9]). In animal models, neutralization of MIF with antibodies and/or experiments in MIF deficient mice have supported the conclusion that maintenance of functional MIF is required for disease progression. Thus, inhibition of MIF using ITCs as nutritional or pharmacologic agents may provide therapeutic intervention in a broad range of disease processes, and different inhibitory efficiencies for distinct ITC agents may enable optimization of therapeutic modalities. Further, biochemical inhibition of the MIF tautomerase should serve as a biomarker of ITC therapeutic action in target organs or for pharmacokinetic measurements in biological fluids.
Importantly, the low micromolar concentration of ITCs necessary for inhibition of MIF in cells is similar to levels found in the circulation of human subjects after oral consumption of broccoli sprouts, a rich source of dietary ITCs [21]. While ITCs are metabolized by conjugation to glutathione in vivo, glutathione conjugated ITCs, and the terminal metabolite, N-acetylcysteine conjugated ITCs are as effective as unconjugated ITCs for MIF tautomerase inhibition (Supplementary Figure 5 and not shown). Thus, metabolism does not limit the potential utility of ITCs as anti-MIF human therapeutics.
The tautomerase activity of MIF has been correlated with its biological activities. However, the natural substrate of the MIF tautomerase remains unknown. This has led at least one group to suggest that the tautomerase activity cannot be relevant to the biological activities [22], though these same workers simultaneously reported the merit of a synthetic competitive tautomerase inhibitor. This inhibitor has proven effective in several animal models of MIF-dependent disease (e.g. [22, 23]). The ITCs offer an advantage over this competitive inhibitor owing to irreversibility and potentially lower concentrations required for therapeutic effect.
Isothiocyanates are among the most appreciated nutraceutical components of what has become known as ‘functional foods’. The cancer preventive activities of ITCs are well known, though identification of a primary mechanism of action has been elusive. Phase 2 gene induction by ITCs is a feature common to nearly all cancer preventives. Our data show clearly that MIF does not contribute to induction of Phase 2 genes or other ARE dependent gene expression. It is not certain if covalent targets of ITCs other than MIF may be responsible for Phase 2 gene expression, and ongoing proteomics work is addressing this possibility.
Previous reports have shown that MIF expression in cells is required for malignant transformation by combinations of dominant oncogenes [13], and that overexpression of MIF can suppress cell death induced by p53 expression [24]. Because of these observations, it is likely that suppression of part of the cancer phenotype by ITCs may be a direct result of inhibition of MIF function in tumor cells. Alternatively, the role for MIF in cancer prevention may reflect disruption of the stimulatory interactions between cancer cells and the host, for example through inhibition of the inflammatory cells that contribute to the tumor microenvironment. This possibility is supported by the numerous reports of cancer prevention in animals, and the relative paucity of reports of tumor cell growth interruption by the same agents excepting at extra-physiologic doses.
As an efficient, natural product inhibitor of MIF tautomerase, ITCs represent a safe, effective class of inhibitors that could rapidly reach clinical application for a number of diseases in which MIF has been identified as an attractive therapeutic target.
Supplementary Material
Acknowledgments
The authors thank Fang Xu for technical assistance in preparation of the rMIF protein. The ARE luciferase reporter plasmid was generously provided by Mark Hannink, University of Missouri, Columbia, MO. The authors thank Dr. Scott Vande Pol for many helpful discussions and for critical reading of the manuscript.
Funding: This work was supported by NIH grants R01AT004323 (JVC) and R01CA113899 (DJT) and was aided by Grant #IRG 81-001-23 from the American Cancer Center (JVC).
Abbreviations
- Bio-ITC
biotin-ITC
- CBD
chitin binding domain
- IC50
inhibitory concentration 50
- ITC(s)
Isothiocyanate(s)
- MEFs
mouse embryo fibroblasts
- MIF
Macrophage Migration Inhibitory Factor
- PEITC
phenethyl isothiocyanate
- SFN
Sulforaphane
Footnotes
Authors' Contributions: Janet V. Cross co-conceived the project, performed most of the experiments, and wrote the original draft of the manuscript. Joshua M. Rady performed several of the experiments and assisted with preparation of the figures. Frank W. Foss, Jr. and Timothy L Macdonald synthesized the biotin-ITC, Charles E. Lyons performed the mass spectrometry analyses. Dennis J. Templeton coordinated the laboratory, co-conceived the project, and contributed to the preparation of the final manuscript.
References
- 1.Prochaska HJ, Talalay P. Regulatory mechanisms of monofunctional and bifunctional anticarcinogenic enzyme inducers in murine liver. Cancer Res. 1988;48:4776–4782. [PubMed] [Google Scholar]
- 2.Morse MA, Reinhardt JC, Amin SG, Hecht SS, Stoner GD, Chung FL. Effect of dietary aromatic isothiocyanates fed subsequent to the administration of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone on lung tumorigenicity in mice. Cancer Lett. 1990;49:225–230. doi: 10.1016/0304-3835(90)90163-r. [DOI] [PubMed] [Google Scholar]
- 3.Hecht SS, Upadhyaya P, Wang M, Bliss RL, McIntee EJ, Kenney PM. Inhibition of lung tumorigenesis in A/J mice by N-acetyl-S-(N-2-phenethylthiocarbamoyl)-L-cysteine and myo-inositol, individually and in combination. Carcinogenesis. 2002;23:1455–1461. doi: 10.1093/carcin/23.9.1455. [DOI] [PubMed] [Google Scholar]
- 4.Srivastava SK, Xiao D, Lew KL, Hershberger P, Kokkinakis DM, Johnson CS, Trump DL, Singh SV. Allyl isothiocyanate, a constituent of cruciferous vegetables, inhibits growth of PC-3 human prostate cancer xenografts in vivo. Carcinogenesis. 2003;24:1665–1670. doi: 10.1093/carcin/bgg123. [DOI] [PubMed] [Google Scholar]
- 5.Chen YR, Han J, Kori R, Kong AN, Tan TH. Phenylethyl isothiocyanate induces apoptotic signaling via suppressing phosphatase activity against c-Jun N-terminal kinase. J Biol Chem. 2002;277:39334–39342. doi: 10.1074/jbc.M202070200. [DOI] [PubMed] [Google Scholar]
- 6.David JR. Delayed hypersensitivity in vitro: its mediation by cell-free substances formed by lymphoid cell-antigen interaction. Proc Natl Acad Sci U S A. 1966;56:72–77. doi: 10.1073/pnas.56.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bloom BR, Bennett B. Mechanism of a reaction in vitro associated with delayed-type hypersensitivity. Science. 1966;153:80–82. doi: 10.1126/science.153.3731.80. [DOI] [PubMed] [Google Scholar]
- 8.Ichiyama H, Onodera S, Nishihira J, Ishibashi T, Nakayama T, Minami A, Yasuda K, Tohyama H. Inhibition of joint inflammation and destruction induced by anti-type II collagen antibody/lipopolysaccharide (LPS)-induced arthritis in mice due to deletion of macrophage migration inhibitory factor (MIF) Cytokine. 2004;26:187–194. doi: 10.1016/j.cyto.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 9.de Jong YP, Abadia-Molina AC, Satoskar AR, Clarke K, Rietdijk ST, Faubion WA, Mizoguchi E, Metz CN, Alsahli M, ten Hove T, Keates AC, Lubetsky JB, Farrell RJ, Michetti P, van Deventer SJ, Lolis E, David JR, Bhan AK, Terhorst C, Sahli MA. Development of chronic colitis is dependent on the cytokine MIF. Nat Immunol. 2001;2:1061–1066. doi: 10.1038/ni720. [DOI] [PubMed] [Google Scholar]
- 10.Campa MJ, Wang MZ, Howard B, Fitzgerald MC, Patz EF., Jr Protein expression profiling identifies macrophage migration inhibitory factor and cyclophilin a as potential molecular targets in non-small cell lung cancer. Cancer Res. 2003;63:1652–1656. [PubMed] [Google Scholar]
- 11.Kettunen E, Anttila S, Seppanen JK, Karjalainen A, Edgren H, Lindstrom I, Salovaara R, Nissen AM, Salo J, Mattson K, Hollmen J, Knuutila S, Wikman H. Differentially expressed genes in nonsmall cell lung cancer: expression profiling of cancer-related genes in squamous cell lung cancer. Cancer Genet Cytogenet. 2004;149:98–106. doi: 10.1016/S0165-4608(03)00300-5. [DOI] [PubMed] [Google Scholar]
- 12.Bando H, Matsumoto G, Bando M, Muta M, Ogawa T, Funata N, Nishihira J, Koike M, Toi M. Expression of macrophage migration inhibitory factor in human breast cancer: association with nodal spread. Jpn J Cancer Res. 2002;93:389–396. doi: 10.1111/j.1349-7006.2002.tb01269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petrenko O, Fingerle-Rowson G, Peng T, Mitchell RA, Metz CN. Macrophage migration inhibitory factor deficiency is associated with altered cell growth and reduced susceptibility to Ras-mediated transformation. J Biol Chem. 2003;278:11078–11085. doi: 10.1074/jbc.M211985200. [DOI] [PubMed] [Google Scholar]
- 14.Ren Y, Chan HM, Fan J, Xie Y, Chen YX, Li W, Jiang GP, Liu Q, Meinhardt A, Tam PK. Inhibition of tumor growth and metastasis in vitro and in vivo by targeting macrophage migration inhibitory factor in human neuroblastoma. Oncogene. 2006;25:3501–3508. doi: 10.1038/sj.onc.1209395. [DOI] [PubMed] [Google Scholar]
- 15.Hagemann T, Robinson SC, Thompson RG, Charles K, Kulbe H, Balkwill FR. Ovarian cancer cell-derived migration inhibitory factor enhances tumor growth, progression, and angiogenesis. Mol Cancer Ther. 2007;6:1993–2002. doi: 10.1158/1535-7163.MCT-07-0118. [DOI] [PubMed] [Google Scholar]
- 16.Rosengren E, Bucala R, Aman P, Jacobsson L, Odh G, Metz CN, Rorsman H. The immunoregulatory mediator macrophage migration inhibitory factor (MIF) catalyzes a tautomerization reaction. Mol Med. 1996;2:143–149. [PMC free article] [PubMed] [Google Scholar]
- 17.Swope M, Sun HW, Blake PR, Lolis E. Direct link between cytokine activity and a catalytic site for macrophage migration inhibitory factor. Embo J. 1998;17:3534–3541. doi: 10.1093/emboj/17.13.3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cross JV, Foss FW, Rady JM, Macdonald TL, Templeton DJ. The isothiocyanate class of bioactive nutrients covalently inhibit the MEKK1 protein kinase. BMC Cancer. 2007;7:183. doi: 10.1186/1471-2407-7-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szalecki W. Synthesis of norbiotinamine and its derivatives. Bioconjug Chem. 1996;7:271–273. doi: 10.1021/bc950094f. [DOI] [PubMed] [Google Scholar]
- 20.Kleemann R, Hausser A, Geiger G, Mischke R, Burger-Kentischer A, Flieger O, Johannes FJ, Roger T, Calandra T, Kapurniotu A, Grell M, Finkelmeier D, Brunner H, Bernhagen J. Intracellular action of the cytokine MIF to modulate AP-1 activity and the cell cycle through Jab1. Nature. 2000;408:211–216. doi: 10.1038/35041591. [DOI] [PubMed] [Google Scholar]
- 21.Ye L, Dinkova-Kostova AT, Wade KL, Zhang Y, Shapiro TA, Talalay P. Quantitative determination of dithiocarbamates in human plasma, serum, erythrocytes and urine: pharmacokinetics of broccoli sprout isothiocyanates in humans. Clin Chim Acta. 2002;316:43–53. doi: 10.1016/s0009-8981(01)00727-6. [DOI] [PubMed] [Google Scholar]
- 22.Al-Abed Y, Dabideen D, Aljabari B, Valster A, Messmer D, Ochani M, Tanovic M, Ochani K, Bacher M, Nicoletti F, Metz CN, Pavlov VA, Miller EJ, Tracey KJ. ISO-1 binding to the tautomerase active site of MIF inhibits its pro-inflammatory activity and increases survival in severe sepsis. J Biol Chem. 2005;280:36541–36544. doi: 10.1074/jbc.C500243200. [DOI] [PubMed] [Google Scholar]
- 23.Cvetkovic I, Al-Abed Y, Miljkovic D, Maksimovic-Ivanic D, Roth J, Bacher M, Lan HY, Nicoletti F, Stosic-Grujicic S. Critical role of macrophage migration inhibitory factor activity in experimental autoimmune diabetes. Endocrinology. 2005;146:2942–2951. doi: 10.1210/en.2004-1393. [DOI] [PubMed] [Google Scholar]
- 24.Hudson JD, Shoaibi MA, Maestro R, Carnero A, Hannon GJ, Beach DH. A proinflammatory cytokine inhibits p53 tumor suppressor activity. J Exp Med. 1999;190:1375–1382. doi: 10.1084/jem.190.10.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.