Abstract
Engineered zinc finger nucleases (ZFNs) form the basis of a broadly applicable method for targeted, efficient modification of eukaryotic genomes. In recent work, we described OPEN (Oligomerized Pool ENgineering), an “open-source,” combinatorial selection-based method for engineering zinc finger arrays that function well as ZFNs. We have also shown in direct comparisons that the OPEN method has a higher success rate than previously described “modular assembly” methods for engineering ZFNs. OPEN selections are performed in E. coli using a bacterial two-hybrid system and do not require specialized equipment. Here we provide a detailed protocol for performing OPEN to engineer zinc finger arrays that have a high probability of functioning as ZFNs. Using OPEN, researchers can generate multiple customized ZFNs in approximately 8 weeks.
Introduction
Engineered zinc finger nucleases (ZFNs) can be used as an important and broadly applicable tool for inducing highly efficient, targeted genome modification.1-6 ZFNs consist of a custom-made DNA-binding zinc-finger array fused to a non-specific nuclease domain7, 8 (Fig. 1a). These artificial nucleases bind to DNA as dimers, with ZFN monomers binding to 9 bp half-sites separated by a spacer sequence of variable length into which a double-stranded DNA break (DSB) is introduced (Fig. 1b).9, 10 Repair of a ZFN-induced DSB by non-homologous end-joining can lead to the introduction of mutagenic insertions or deletions (indels) with high frequency.11-22 In addition, DSBs created by ZFNs also stimulate homologous recombination-mediated repair;12, 22, 23 therefore, ZFNs can be used to induce high frequency gene targeting by introducing a homologous “donor DNA template” harboring investigator-specified mutations or insertions into cells.11, 16, 24, 25 ZFN-induced modifications of endogenous genes have been reported to be as high as 50% and to work in a variety of cell types including Drosophila,12, 13, 26, 27 somatic C. elegans,28 zebrafish,18-20 plants,11, 29, 30 and mammalian cells.11, 14-17, 24, 25
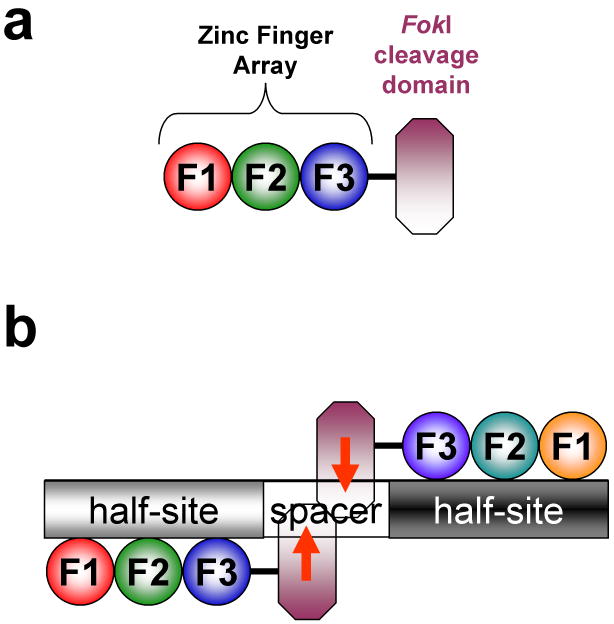
Figure 1. Zinc finger nucleases.
Schematics of (a) a zinc finger nuclease and (b) a pair of zinc finger nucleases bound to their target site. Zinc finger domains are depicted as colored spheres and the FokI nuclease domain is represented by a purple octagon. “F1” represents the amino-terminal finger, “F2” the middle finger, and “F3” the carboxy-terminal finger. Each three-finger array binds to a 9 bp “half-site.” Note that a zinc finger nuclease pair cleaves (red arrows) its target site within the variable length spacer sequence between the half-sites. Figure and legend adapted from Wright et al., 2006.76
Implementing ZFN technology requires the capability to generate custom zinc finger arrays needed to direct DSBs to specific genomic targets. We recently described a highly effective and “open-source” method for engineering zinc finger arrays which we termed OPEN (for Oligomerized Pool ENgineering).11 OPEN utilizes an archive of zinc finger pools, each consisting of a small number (95 or fewer) of different fingers designed to bind to a particular 3 bp “subsite” (Fig. 2a). To perform an OPEN selection, a combinatorial library of multi-finger arrays from these pools is generated for a target 9 bp site of interest (Fig. 2a). Members of this library that bind efficiently to the target site are then isolated using a bacterial two-hybrid (B2H) selection method (Fig. 2c), which has been shown to identify multi-finger arrays that possess high affinities and high specificities31 and that function efficiently as ZFNs in cells.11, 20, 32, 33 Thus, OPEN identifies combinations of fingers that work well together, thereby accounting for the context-dependent DNA-binding activities of zinc fingers in an array. Because ZFNs function as dimers, OPEN selections must be performed for two 9 bp target sites in order to generate a ZFN pair.
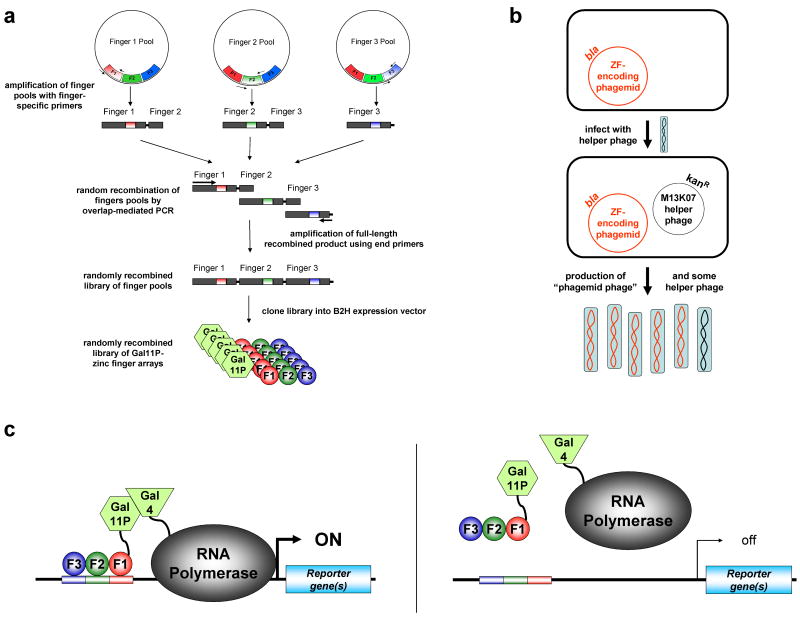
Figure 2. OPEN selection method for engineering multi-finger arrays.
(a) Construction of combinatorial zinc finger libraries. DNA sequences encoding fingers from pre-selected “pools” are amplified by PCR and then fused together to create a library of DNA sequences encoding random combinations of fingers. These DNA fragments are then cloned into B2H vectors which express the zinc fingers as fusions to a fragment of the yeast Gal11P protein.
(b) Schematic illustrating the conversion of zinc finger phagemid DNA library into infectious phage particles. E. coli cells harboring zinc finger-encoding phagemids are infected with M13K07 helper phage (blue ovals with black colored DNA). Infection results in production of infectious phage particles (blue ovals) containing single-stranded copies of the zinc finger-encoding phagemid (red DNA) or helper phage genome (black DNA). Zinc finger-encoding phagemids confer resistance to beta-lactam antibiotics through expression of the beta-lactamase (bla) gene. M13K07 helper phage confer kanamycine resistance (KanR).
(c) Schematic illustrating the bacterial two-hybrid selection system. Binding of a Gal11P-zinc finger array hybrid protein to a target binding site (represented as color-matched rectangles) leads to recruitment of RNA polymerase complexes which have incorporated an RNA polymerase α-subunit-Gal4 hybrid protein to a proximal promoter (left panel). This recruitment occurs as a result of interaction between the Gal11P and Gal4 proteins and results in increased transcription of reporter gene(s) downstream of the promoter. Failure of a Gal11P-zinc finger array hybrid protein to bind a target site results in no activated expression of the reporter gene(s) (right panel).
To date, we have used OPEN to engineer, sequence, and characterize ∼500 zinc-finger arrays targeted to 42 different nine bp target sequences.11, 20, 30, 34 The sequences, activities, and cognate target sites of all these multi-finger arrays have been deposited into the freely available web-based Zinc Finger Database34, 35 (ZiFDB; available at http://www.zincfingers.org/software-tools.htm). Using a subset of these arrays, we have successfully constructed and validated three-finger ZFN pairs for 17 different full target sites from an integrated EGFP reporter gene (four sites) in human cells, three endogenous human genes (six sites), five endogenous zebrafish genes (five sites), and one endogenous plant gene (two sites).11, 20, 30 At present, zinc finger pools have been described for all GNN subsites and for a subset of TNN subsites.
Other zinc finger engineering methods have also been used to create customized ZFNs. Various groups have made ZFNs using “modular assembly”,12, 13, 26-28, 36-38 an approach for engineering multi-finger arrays which treats individual fingers as independent units.39-45 The success rate of modular assembly for making three-finger arrays has been reported to be low46 and in direct comparisons we have demonstrated that OPEN is more robust and efficient than modular assembly for constructing three-finger ZFNs.11 The low success rate of modular assembly may result from its failure to account for the context-dependent activity of zinc finger domains in an array.47-51 The company Sangamo BioSciences, Inc. has also made four-finger ZFNs using their proprietary zinc finger engineering method.16-18, 25 ZFNs made using this approach are now commercially available through Sigma-Aldrich under the brand-name CompoZr™.18, 52 We have performed an indirect comparison of ZFN pairs made by OPEN with one ZFN pair made by the proprietary Sangamo BioSciences approach (designed to different target sites) and found that the activities and toxicities of these ZFNs made by the two approaches were comparable.11
Despite the fact that we have successfully used OPEN to identify zinc finger arrays for a large number of ZFN target half-sites,11, 20 the efficacy of the method is certainly not 100%. Our overall success rate to date is ∼70-80% for obtaining zinc finger arrays that can activate transcription in the B2H system. However, we have also only focused on target half-sites that have one or more GNN “subsite.” Thus, we do not know how well the method will work for sequences that do not contain any GNN subsites. We have deposited the results of both successful and failed selections in the publicly available Zinc Finger Consortium Database34 at http://bindr.gdcb.iastate.edu/ZiFDB/ (or through the Zinc Finger Consortium website at: http://www.zincfingers.org/software-tools.htm) and we encourage all future users of OPEN to do the same. Nonetheless, the less-than-perfect success rate of OPEN suggests that users should target more than one full ZFN site for their gene or locus of interest to improve the chances of successfully obtaining functional zinc finger arrays for pairs of ZFN half-sites.
Although the emphasis of this protocol is on using OPEN zinc finger arrays to construct ZFNs, we note that engineered zinc finger arrays have also been fused to other functional domains to create custom targeted transcription factors and recombinases. Both the modular assembly and proprietary Sangamo BioSciences approaches have been used successfully to generate three, four-, five-, and six-finger arrays for these various fusion proteins.53-70 To date, zinc finger arrays made by OPEN have only been used to make ZFNs, although in principle the method could also be used to construct zinc finger transcription factors and recombinases. One potential limitation of OPEN for these other applications is that it has only been used successfully to create three-finger arrays. It remains unknown whether it, like the modular assembly and proprietary Sangamo approaches, can be successfully adapted and used to create four-, five-, or six-finger arrays.
Here we describe a detailed protocol for using publicly available software and reagents to plan and perform OPEN selections. All experimental steps are performed using E. coli and do not require specialized equipment. The OPEN platform was developed and validated by the Zinc Finger Consortium, a group of academic scientists committed to continued research, use, and application of engineered zinc finger technology (see http://www.zincfingers.org). Software programs used in the OPEN protocol are freely available on the web and do not require registration (http://www.zincfingers.org/software-tools.htm). Protocol-specific reagents required to practice OPEN are available to academic researchers through the non-profit plasmid distribution service Addgene (http://www.addgene.org/zfc) and by request from the Joung lab. All other required materials and reagents are available through standard commercial vendors.
Overview of the Procedure
The process of engineering zinc finger arrays using OPEN can be divided into five parts: (1) identifying potential full ZFN target sites using web-based software, (2) constructing B2H selection strains harboring ZFN target half-sites, (3) creating combinatorial zinc finger array libraries for the ZFN target half-sites, (4) selecting zinc finger arrays using OPEN, and (5) quantifying zinc finger array binding activity in B2H reporter strains harboring the ZFN target half-sites.
Identifying potential target sites using web-based software
In this initial step, genomic sequence of the gene of interest is entered into the ZiFiT program11, 35 which will identify potential full ZFN sites. This step is required because, as noted in the Introduction, at present OPEN can not target all possible sequences due to the availability of “pools” for only a subset of all possible three base pair “subsites” at each finger position (currently 66 of the potential 192 pools are available). ZiFiT will also exclude sites that will be methylated by Dam and Dcm methylases in E. coli. Note that each full ZFN site consists of two 9 bp target “half-sites” separated by a user-defined spacer of 5, 6, or 7 bps and that an OPEN selection must be performed for each of these half-sites. ZFNs with appropriate linkers (between the zinc finger array and the nuclease domain) can recognize full ZFN sites with variable length spacers.71, 72
Constructing B2H selection strains harboring target sites
B2H selection strains harbor a single copy F′ episome with a ZFN target “half-site” positioned upstream of a B2H promoter which drives expression of the HIS3 and aadA selectable marker genes11, 31, 73 (Fig. 2c). The ZFN target half-site used actually consists of the 9 bp target sequence identified by ZiFiT plus an additional upstream and downstream base from the genomic sequence (i.e.—an 11 bp sequence); we use these additional bases because previous studies have shown that the identities of these external bases can influence binding of a zinc finger array.48 To construct a B2H selection strain, a target 11 bp site is first cloned into a reporter plasmid. The selection reporter plasmid contains lacIq and lacZ sequences which flank the binding site-promoter-HIS3-aadA cassette and which can recombine with sequences present on an F′ episome present in the CSH100 strain74, 75 (Fig. 3a). As shown in Fig. 3b, a double cross-over event between these two homologous regions leads to the introduction of the binding site-promoter-HIS3-aadA cassette onto the F′ (although this event will be more infrequent than a single cross-over event or no recombination). Mating of CSH100 cells harboring these various recombinant and non-recombinant F′ episomes with the F- strain KJ1C will enable transfer of the F′ episomes from the former cells to the latter. As shown in Fig. 3b, KJ1C cells harboring the desired double-recombinant F′ can be identified by their growth on plates containing tetracycline, kanamycin, and sucrose (LB/TKS plates). Construction of a B2H selection strain is completed by transforming KJ1C cells bearing a recombinant F′ reporter plasmid with an additional low-copy plasmid which expresses the hybrid “alpha-Gal4” protein consisting of the amino-terminal domain and inter-domain linker of the E. coli RNA polymerase α-subunit fused to a fragment of the yeast Gal4 protein.
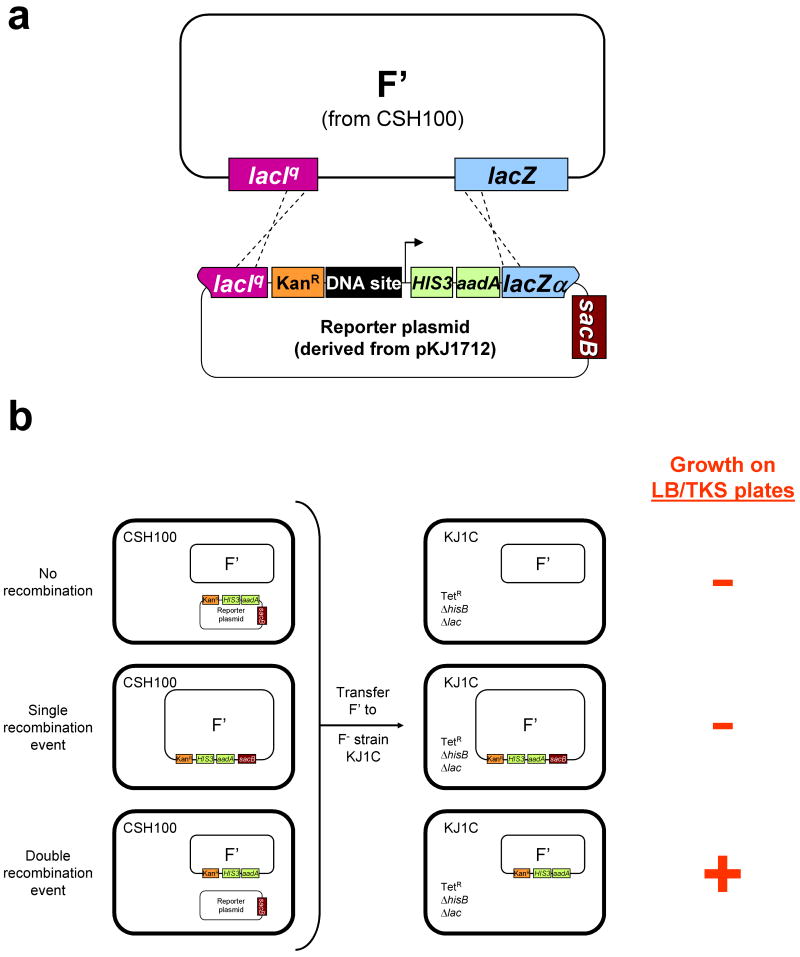
Figure 3. Construction of F′-based reporter episomes required for B2H selection strains (figure and legend adapted from Thibodeau-Beganny & Joung, 200775).
(a) Identical lacIq and lacZ sequences present on both the F′ episome from strain CSH100 and the pKJ1712-derived reporter plasmid permit transfer of a cassette harboring a kanamycin resistance gene (KanR; orange box), target DNA site (black box), B2H promoter (arrow), and the co-cistronic HIS3 and aadA selectable markers (green boxes) from the plasmid to the F′ by a double recombination event (depicted by dashed lines). Note that the desired double-recombinant F′ would not harbor the counter-selectable sacB marker gene (red box) present on the reporter plasmid.
(b) Schematic depicting the various types of cells described in the bacterial mating of step 24. The left side of the figure depicts CSH100 cells in which a double, single, or no recombination event has occurred between the F′ and the reporter plasmid. The right side of the figure depicts KJ1C cells that have received a double, single, or non-recombinant F′ from the CSH100 cells and indicates whether or not these various cells will grow on LB/TKS plates containing tetracycline, kanamycin, and sucrose.
Constructing combinatorial zinc finger array libraries
Zinc finger array libraries are built by using PCR to stitch together random combinations of the three OPEN finger pools corresponding to the three subsites in a given target (Fig. 2a). The DNA encoding these randomly recombined finger arrays is then cloned into a low-copy expression phagemid which expresses them as fusions to a fragment of the yeast Gal11P protein (Fig. 2a). Expression of these Gal11P-zinc finger array hybrid proteins is controlled by the lac repressor and is therefore inducible by the addition of IPTG to the medium.
To facilitate highly efficient transduction of a B2H selection strain by various Gal11P-zinc finger array hybrids, the library of phagemids is converted into infectious phage particles (Fig. 2b). We have found that efficient selection on histidine-deficient NM medium works only with B2H selection cells that have not been grown in rich medium (J.K. Joung, unpublished observations). Use of phage-based transduction enables us to achieve high transformation efficiencies with the B2H selection strain without the need to use electroporation, a method that requires cells to recover in rich SOC medium. In addition, the phage-based approach helps to ensure that only one zinc finger array is introduced into most cells; this can be simply accomplished by using an excess of selection strain cells relative to phage. Finally, the use of phage enables rescue of the zinc finger array-encoding phagemid from selection strain cells. This capability is important for the two-stage selection procedure (see below).
Selecting zinc finger arrays using OPEN
OPEN selections are performed in two stages: an initial lower stringency selection enriches for zinc finger arrays that bind to the ZFN target half-site; a second higher stringency selection identifies the final candidates. In the first selection stage, IPTG is added to induce higher levels of Gal11P-zinc finger array and alpha-Gal4 hybrid protein expression. In the second selection step, IPTG is omitted so that the two hybrid proteins are expressed at lower levels. Both selection steps are performed by introducing a combinatorial zinc finger array phagemid library into a B2H selection strain and by plating on media which selects for cells that exhibit increased expression of the HIS3 and aadA marker genes. If a Gal11P-zinc finger array hybrid protein can bind to the 11 bp ZFN target half-site on the reporter, then transcription of the selectable marker genes is activated because the DNA-bound Gal11P-zinc finger hybrid protein recruits RNA polymerase complexes harboring the alpha-Gal4 protein to the promoter via a protein-protein interaction between the Gal11P and Gal4 protein fragments (Fig. 2c).
The ability to rescue the zinc finger-encoding phagemids from selection strain cells is a critical capability utilized during the two-stage selection. Following the initial stage of selection, zinc finger-encoding phagemids are rescued from these cells by infection with M13K07 helper phage and subsequent packaging of single-stranded phagemids in infectious phage particles. This “enriched phagemid library” is then harvested and used to transduce fresh B2H selection strain cells for the second round of selection.
Quantifying zinc finger array DNA-binding activity using B2H reporter strains
To confirm that zinc finger arrays identified in the selection process bind to the ZFN target half-site, the phagemids encoding these candidates are isolated from colonies on the selection plates and then introduced by transformation into a B2H reporter strain.76 The B2H reporter strain is similar to the B2H selection strain in that it harbors: (1) a single-copy plasmid (in this case a mini-BAC plasmid instead of a full F′ episome) with the ZFN target half-site positioned upstream of a weak promoter which, in turn, controls expression of a lacZ reporter gene; (2) a low-copy number plasmid expressing the alpha-Gal4 hybrid protein. If a Gal11P-zinc finger array hybrid that can bind to the ZFN target half-site is expressed in the B2H reporter strain, then transcription of lacZ will be activated. Because the lacZ gene encodes β-galactosidase, its expression level can be measured by a simple quantitative assay.75, 76 By comparing β-galactosidase expression in the presence and absence of a given zinc finger array, a “fold-activation” value can be calculated which can guide the choice of which arrays to carry forward for testing as ZFNs.
Experimental Design
Initial trial selections
Before attempting to perform selections for new target sites of interest, we recommend that users first complete the entire OPEN protocol start-to-finish with at least one target site that has worked successfully in previous experiments. We define a “successful” selection as one that has previously yielded one or more zinc finger arrays that activate transcription more than three-fold in the B2H system. The choice of this positive control target site may be influenced by the pools that the investigator has requested and therefore has on hand. The Zinc Finger Database (ZiFDB; available at http://www.zincfingers.org/software-tools.htm) contains information on target DNA sites for which OPEN selections have been successful as well as the sequences and B2H activities of finger arrays obtained from those experiments with which investigators can compare their own results.
For any initial selection, we strongly recommend that investigators follow the protocol outlined here precisely. We have noted throughout the protocol certain steps that are particularly critical to success. Although some of these suggestions may not appear to be important or significant to the first-time user, we have learned that these parameters can be critical to success of the protocol. Examples of such recommendations include the use of a specific thermostable polymerase for amplification of the finger pools, the use of glucose from a specific vendor for the NM medium, and the method for inoculating colonies into NM medium.
Time required to complete the protocol
Although an experienced lab can complete the entire OPEN protocol in eight weeks or less, investigators should anticipate that the protocol will require significantly more time the first few times they perform it due to the inevitable need to repeat certain steps. In addition, completing OPEN in the optimal timeframe requires significant planning and coordination. We anticipate which plasmids, PCR products, restriction and modification enzymes, plates, medium, and cells will be required at least several days in advance to ensure that these reagents do not become a rate limiting step. An optimal timeline for performing each part of the protocol is given below in the TIMING section. Once the protocol has been mastered, in our experience it is possible for a single individual to perform 12 or more selections in parallel in eight weeks time or less.
Materials
Reagents
OPEN zinc finger pools (available by request from the Joung lab)
Plasmids and expression vectors (see REAGENT SET UP)
Bacterial strain CSH100 (F′ lacproA+,B+(lacIq lacPL8)/araD(gpt-lac)5; available by request from the Joung lab); this strain is used to construct B2H selection strains.
Bacterial strain KJ1C (F- ΔhisB463 Δ(gpt-proAB-arg-lac)XIII zaj∷Tn10); available by request from the Joung lab); this strain is required to construct B2H selection strains.
Bacterial strain XL-1 Blue (recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIq lacZDM15 Tn10 (TetR)]; Stratagene cat. no. 200249); this strain is used for routine subcloning of plasmids and for building the OPEN zinc finger libraries.
Bacterial strain Transformax EPI300 (F− mcrA Δ (mrr-hsdRMS-mcrBC) Φ80dlacZDM15 ΔlacX74 recA1 endA1 araD139 Δ(ara, leu)7697 galU galK λ− rpsL nupG trfA dhfr; Epicentre, cat. no. C300C105); this strain is used to perform subcloning steps with the pBAC-lacZ-derived B2H reporter plasmids.
Bacterial strain KJBAC1 (F- lacIq ΔhisB463 Δ(gpt-proAB-arg-lac)XIII zaj∷Tn10; strain available through Addgene; http://www.addgene.org/zfc); this strain is required for propagation and subcloning of pBAC-lacZ-derived B2H reporter plasmids.
-
M13K07 helper phage (New England Biolabs, Cat #N0315S; store indefinitely at -20°C)
<!>CAUTION Care should be taken to avoid contaminating laboratory equipment and benches with bacteriophage
Restriction enzymes (all from New England Biolabs): BamHI (cat. no. R0136S), BbsI (cat. no R0539L), BsaI (cat. no. R0533L), EcoRI (cat. no. R0101L), HindIII (cat. no. R0104L), PstI (cat. no. R0140S), SapI (cat. no. R0569L), XbaI (cat. no. R0145L)
10× restriction enzyme buffers (New England Biolabs, included with enzymes)
T4 DNA Ligase and associated standard T4 DNA Ligase reaction buffer (New England Biolabs, cat. no. M0202S)
Quick Ligation Kit (New England Biolabs, cat. no. M2200S)
T4 Polynucleotide Kinase (New England Biolabs, cat. no. M0201S)
Cloned Pfu polymerase and associated 10× reaction buffer (Stratagene, cat. no. 600159-81)
-
Expand High-Fidelity thermostable polymerase and associated 10× Expand buffer with MgCl2 (Roche, cat. no. 11732641001)
<!>CRITICAL We have found the use of Expand enzyme to be critical for successful amplification of the zinc finger pools
-
AccuGel 29:1 acrylamide:bis-acrylamide solution (National Diagnostics, cat. no. EC-852)
<!>CAUTION Acrylamide is a neurotoxin and should be handled with gloves.
10% (w/v) ammonium persulfate in ddH2O (Fisher, cat. no. 7727-54-0; store indefinitely at -20°C)
TEMED (Fisher, cat. no. BP150-100; store indefinitely at 4°C)
100% ethanol (Pharmco, cat. no. 111ACS200)
70% (v/v) ethanol in ddH2O
QIAprep Spin Miniprep kit (Qiagen, cat. no. 27106)
QIAquick PCR Purification kit (Qiagen, cat. no. 28106)
MinElute PCR Purification kit (Qiagen, cat. no. 28006)
LB medium (Difco, cat. No. 244620)
LB agar medium (Difco, cat. No 244520)
2xYT medium (Difco, cat. No. 244020)
SOB medium (Difco, cat. No. 244310)
SOC medium (SOB medium with 0.4% (v/v) glucose)
Bacto-Agar (Difco, cat. no. 214010)
Bacto-Tryptone (Difco, cat. no. 211705)
Sterile 10% (v/v) glyercol and sterile 50% (v/v) glycerol both in ddH2O (100% glycerol – Fisher, cat. no. BP229-1)
10× M9 salts (Difco, cat. no. 248510)
-
20% (w/v) Glucose in ddH2O (Mallinckrodt Baker, cat. No. 4912-06)
<!>CRITICAL We have found the use of glucose from this specific vendor to be critical for high density growth of our B2H selection strains in NM medium
20 mM Adenine HCl in ddH2O (Sigma, cat. no A8751; store indefinitely at room temperature (20-25°C)
Amino Acid Mixture (see REAGENT SET UP)
Individual amino acid powders (all available through Sigma): Phenylalanine (#P5482), Lysine (#L5626), Arginine (#A6969), Glycine, Valine (#V0500), Alanine (#A7469), Tryptophan (#T8941), Threonine (#T8625), Serine (#S4311), Proline (#P0380), Asparagine(#A4159), Aspartic Acid (#A4534), Glutamine (#G3126), Isoleucine (#I2752), Leucine (#L8000), L-Glutamic acid Potassium salt monohydrate (#G1501), and Tyrosine (#T3754).
1 M MgSO4 in ddH2O (Fisher, cat. no. BP213-1)
Thiamine (10 mg ml-1 stock solution in ddH2O; filter sterilize and store indefinitely at -20°C)
10 mM ZnSO4 in ddH2O
100 mM CaCl2 in ddH2O
Carbenicillin (Sigma, cat. no. T4625; 50 mg ml-1 stock solution in ddH2O; filter sterilize and store indefinitely as 1 ml aliquots at -20°C)
Chloramphenicol (Sigma, cat. no. C0378; 30 mg ml-1 stock solution in 100% ethanol; store indefinitely at -20°C)
Kanamycin (Sigma, cat. no. K4000-5G; 30 mg ml-1 stock solution in ddH2O; filter sterilize and store indefinitely at 4°C)
Tetracycline (Sigma, cat. no. T3383; 12.5 mg ml-1 stock solution in 80% (v/v) ethanol; filter sterilize and store indefinitely wrapped in foil at -20°C)
Streptomycin (Sigma, cat. no. S6501; 100 mg ml-1 stock solution in ddH2O; filter sterilize and store indefinitely at 4°C)
3-aminotriazole (3-AT; US Biochemical, cat. no. 11245; 1 M stock solution in ddH2O; filter sterilize and store indefinitely at -20°C)
Sucrose (Fisher, cat. no. 8360; 50% (w/v) stock solution in ddH2O; store indefinitely at room temperature)
Glycogen (Sigma, cat. no. G1767; 10 mg ml-1 stock solution; store indefinitely at -20°C)
IPTG (isopropyl-beta-D-thiogalactopranoside; Sigma, cat. no. 16758; 1 M stock solution in ddH2O; filter sterilize and store indefinitely as 1 ml aliquots at -20°C)
Sterile 100 mM ZnSO4 in ddH2O (Fisher, cat. no. Z68-500)
Ice-cold Solution A for preparing competent cells (10 mM MnCl2 (Fisher, cat. no. BP541-100), 50 mM CaCl2 (Fisher, cat. no. BP510-250), 10 mM MES, pH 6.3 (Fisher, cat. no. BP300-11) in ddH2O; filter sterilize and store wrapped in foil at 4 °C; solution is usable until it acquires brown discoloration; Note: to make Solution A, use a 100 mM MES stock solution prepared in ddH2O which has been brought to a pH of 6.3 using KOH)
Ice-cold Solution A with 15% (v/v) glycerol for preparing competent cells (Solution A with 15% (v/v) glycerol; filter sterilize and store wrapped in foil at 4°C; solution is usable until it acquires brown discoloration)
ONPG (Sigma, cat. no. N1127; o-nitrophenyl-beta-D-galactopryanoside, 4 mg ml-1 in ddH2O; solution can be stored indefinitely as 10 ml aliquots wrapped in foil at -20°C.
Z-buffer (1 liter: 16.1 g of Na2HPO4-7H2O, 5.5 g of NaH2PO4-H2O, 0.75 g of KCl, 0.246 g of MgSO4-7H2O dissolved in ddH2O; filter sterilize and store at room temperature)
Z-buffer with β-mercaptoethanol (prepare fresh by adding 2.7 μl of β-mercaptoethanol to every 1 ml of Z-buffer)
Popculture reagent (Novagen, cat. no. 71092)
-
R-Lysozyme (30,000 units μl-1) and associated dilution buffer (Novagen, cat. no. 71110)
<!>CRITICAL Once diluted in dilution buffer, R-Lysozyme can not be re-frozen. R-Lysozyme can be stored indefinitely at -20°C when diluted to 400 units μl-1 in dilution buffer containing 50% glycerol.
Lysis Master Mix (10:1 mixture of Popculture reagent to diluted R-Lysozyme [4 units μl-1])
-
7.5 M NH4OAc in ddH2O (Fisher, cat. no. A637-500; store indefinitely at room temperature)
<!>CRITICAL Due to the volatility of NH4OAc, we seal storage containers by capping tightly and wrapping with Parafilm.
10% SDS (Fisher, cat. no. BP2436-200)
1 M MgOAc in ddH2O (Fisher, cat. no. M13-500)
0.5 M EDTA, pH 8.0 (Fisher, cat. no. S311-500)
1M Tris, pH 8 in ddH2O (Fisher, cat. no. BP152-1)
1M MgCl2 in ddH2O (Sigma, cat. no. M2393)
5M NaCl in ddH2O (Fisher, cat. no. BP358-212)
Ammonium acetate elution buffer (see REAGENT SET UP)
5× PEG/NaCl solution (see REAGENT SET UP)
dNTP nucleotide set (Roche, cat. no. 11969064001; use this to make a 10 mM stock solution of dCTP, a 10 mM stock solution of dTTP and a 10 mM stock solution of dNTP mixture; store indefinitely in 200 μl aliquots at -20°C)
10× BSA (bovine serum albumin; 1 mg ml-1 solution; New England Biolabs, cat. no. B9001S; store indefinitely at -20°C)
PEG8000 (Fisher, cat. no. BP233-1)
1 M Arabinose in ddH2O(Sigma, cat. no. A-3256)
10× Annealing Buffer (see REAGENT SET UP)
Sequencing and PCR primers (see Table 1)
Table 1. Sequences of primers required for OPEN.
| Primer name | Sequence | Purpose |
|---|---|---|
| OK61 | 5′-GGGTAGTACGATGACGGAACCTGTC-3′ | Sequencing of plasmid pBR-UV-GP-FD2-dervatives |
| OK.163 | 5′-CGCCAGGGTTTTCCCAGTCACGAC-3′ | Sequencing of plasmid pBAC-lacZ-derivatives |
| OK181 | 5′-CCAGAGCATGTATCATATGGTCCAGAAACCC-3′ | Sequencing of plasmid pKJ1712-derivatives |
| HIS3 2F | 5′-CGTATCACGAGGCCCTTTC-3′ | PCR |
| HIS3 2R | 5′-GCAAATCCTGATCCAAACCT-3′ | PCR |
| OK1424 | 5′-GAGCGCCCCTTCCAGTGTCGC-3′ | PCR |
| OK1425 | 5′-CGCATACAGATCCGACACTGAAACGG-3′ | PCR |
| OK1426 | 5′-GTGTCGGATCTGTATGCGAAATTTCTCC-3′ | PCR |
| OK1427 | 5′-TCGGCATTGGAATGGCTTCTCG-3′ | PCR |
| OK1428 | 5′-GCCATTCCAATGCCGAATATGCA-3′ | PCR |
| OK1429 | 5′-CCCTCAGGTGGGTTTTTAGGTG-3′ | PCR |
| OK1430 | 5′-GGGGAGCGCCCCTTCCAGTGTCGC-3′ | PCR |
| OK1432 | 5′-GTGCAGAGGATCCCCTCAGGTGGGTTTTTAGGTG-3′ | PCR |
Equipment
96-well thermocycler
Wooden sticks (Fisher, cat. no. 23-400-102; sterilize by autoclaving)
200 μl filtered pipet tips (CLP, cat. no. BT200)
25 mm glass culture tubes (Fisher, cat. no. 14-961-34)
18 mm glass culture tubes (Fisher, cat. no. 14-961-32)
Glass beads, 3mm (Fisher, cat. no. 11-312A)
Rotating wheel drum (for bacterial cultures)
LabQuake shaker/rotisserie (Barnstead Thermolyne, cat. no. 415110)
1 mm gap electroporation cuvettes (BTX, cat. no. 45-0124, model no. 610)
Electroporation device with adjustable settings
Programmable, multi-channel (8 or 12) pipet
250 ml, 1 liter, and 2 liter glass flasks
Orbital platform shaker with adjustable speed
2 ml cryogenic storage vials (Corning, cat. no. 430659)
0.22 μm PES (polyethersulfone) filter units (Millipore, cat. no. SLGP033RS)
245 mm square plates (Corning, cat. no. 431301)
100 × 15 mm round Petri plates (Fisher, cat. no. 08-757-13)
96-well flat-bottom, microtiter plates (Corning-Costar, cat. no. 3596)
Deep-well 96-well blocks (optional; Corning, cat. no. 3960)
Microtiter plate reader with temperature control option
Sterile 250-ml centrifuge bottles
Sterile 1 liter centrifuge bottles
Sterile 50 ml conical tubes (Corning, cat. no. 430290)
Autoclave
Polyacrylamide gel running apparatus with gel casting system (Thermo Scientific, cat. no. P9DS-1)
Biorad Model 680 Microplate Reader (or other microtiter plate reader with temperature control)
Microtitertron orbital shaker for 96-well blocks (Appropriate Technical Resources; optional)
Reagent Set Up
-
Plasmids and expression vectors: The following are available by request through the Joung lab: pKJ1712 reporter plasmid: KANR, p15A origin of replication, full sequence and plasmid features described in Supplementary Fig. 1; pBR-UV5-GP-FD2 zinc finger B2H expression plasmid: AMPR, ColEI origin of replication, full sequence and plasmid features described in Supplementary Fig. 2; pAC-alphaGal4 expression plasmid: CAMR, p15A origin of replication, full sequence and plasmid features described in Supplementary Fig. 3. The following plasmids (together with their full sequences and maps) are available through Addgene (http://www.addgene.org/zfc): pBAC-lacZ reporter plasmid: CAMR, primary F′ and secondary oriV origins of replication; pAC-KAN-alphaGal4 expression plasmid: KANR, p15A origin of replication.
<!>CRITICAL Plasmids pBR-UV5-GP-FD2, pAC-alphaGal4, and pAC-KAN-alphaGal4 must be propagated in a lacIq strain (e.g.--XL-1 Blue) in order to avoid toxicity due to unregulated expression of fusion proteins encoded on these plasmids.
<!>CRITICAL pBAC-lacZ is a single-copy plasmid that gives low yields when propagated in standard E. coli strains. However, it also harbors a second, higher copy origin (oriV) that requires for activity a protein encoded by the trfA gene. For routine sub-cloning, pBAC-lacZ and its derivatives should therefore be propagated in Transformax EPI300 cells. These cells express trfA from a promoter that can be induced with arabinose (S.T.B. and J.K.J., unpublished data).
-
Ammonium acetate elution buffer: To prepare 25 ml, add the following to 22.8 ml ddH2O: 1.65 ml of 7.5 M NH4OAc, 250 μl of 10% (w/v) SDS, 250 μl of 1 M MgOAc, and 50 μ1 of 0.5 M EDTA. Store at room temperature for no more than one month.
<!>CRITICAL To avoid irreversible precipitation of SDS, do not store elution buffer at temperatures below room temperature. Also, seal storage container tightly with Parafilm to avoid loss of ammonium acetate through volatilization.
10× Annealing Buffer: To prepare 1 ml, combine the following: 400 μl 1 M Tris (pH 8), 200 μl 1 M MgCl2, 100 μl 5 M NaCl, 20 μl 0.5 M EDTA (pH 8), and 280 μl ddH2O. Buffer can be stored indefinitely at -20°C.
M9 minimal medium agar plates: To prepare 500 ml, autoclave 439 ml H2O with 7.5 g Bacto-agar and a stir bar. When agar has cooled to approximately 65°C, add 50 ml 10× M9 salts, 1 ml 1 M MgSO4, 10 ml 20% (w/v) glucose and 0.5 ml 100mM CaCl2 and then pour plates. Plates can be stored indefinitely at 4°C in sealed plastic bags.
LB/CK plates (LB agar supplemented with 30 μg ml-1 chloramphenicol and 30 μg ml-1 kanamycin). Plates can be stored for up to two months at 4°C in sealed plastic bags.
LB/CCK plates (LB agar supplemented with 100 μg ml-1 carbenicillin, 30 μg ml-1 chloramphenicol and 30 μg ml-1 kanamycin). Plates can be stored for up to two months at 4°C in sealed plastic bags.
LB/TK plates (LB agar supplemented with 12.5 μg ml-1 tetracycline and 30 μg ml-1 kanamycin). Plates can be stored for up to two months at 4°C in sealed plastic bags wrapped in aluminum foil to protect the tetracycline from light.
LB/TKS plates (LB agar supplemented with 12.5 μg ml-1 tetracycline, 30 μg ml-1 kanamycin, and 5% (w/v) sucrose). Plates can be stored for up to two months at 4°C in sealed plastic bags wrapped in aluminum foil to protect the tetracycline from light.
LB/TC plates (LB agar supplemented with 12.5 μg ml-1 tetracycline and 100 μg ml-1 carbenicillin). Plates can be stored for up to two months at 4°C in sealed plastic bags wrapped in aluminum foil to protect the tetracycline from light.
LB/KCarb plates (LB agar supplemented with 100 μg ml-1 carbenicillin and 70 μg ml-1 kanamycin). Plates can be stored for up to two months at 4°C in sealed plastic bags.
NM medium: To prepare 500 ml, combine the following components in the order listed: 418 ml ddH2O, 50 ml 10× M9 salts, 10 ml 20% (w/v) glucose, 5 ml 20 mM Adenine HCl, 15 ml Amino Acid Mixture, 500 μl 1M MgSO4, 500 μl thiamine (10 mg/ml), 500 μl 10 mM ZnSO4 and 500 μl 100 mM CaCl2. Filter sterilize and store at 4°C. Wrap container in alumnimum foil to protect from light.
NM plates: To prepare 500 ml, autoclave 7.5g Bacto-agar, 418 ml H2O and a stir bar in a 1 or 2 liter Erlenmyer flask. In a separate sterile container, mix together the following components in the order listed: 50 ml 10× M9 salts, 10 ml 20% (w/v) glucose, 5 ml 20mM Adenine HCl, 15 ml Amino Acid Mixture, 500 μl 1M MgSO4, 500 μl thiamine (10mg/ml), 500 μl 10 mM ZnSO4, 500 μl 100 mM CaCl2 and carbenicillin, chloramphenicol, kanamycin, IPTG, 3-AT, and streptomycin as needed. When agar has cooled to ∼65-70°C, add the separate mixture, mix well and pour plates. Plates can be stored for up to two months at 4°C in sealed plastic bags.
NM/CCK plates (NM agar supplemented with 100 μg ml-1 carbenicillin, 30 μg ml-1 chloramphenicol and 30 μg ml-1 kanamycin). Plates can be stored for up to two months at 4°C in sealed plastic bags.
NM/CCKI plates (NM agar supplemented with 100 μg ml-1 carbenicillin, 30 μg ml-1 chloramphenicol, 30 μg ml-1 kanamycin, and 50 μM IPTG). Plates can be stored for up to two months at 4°C in sealed plastic bags.
Z-buffer: To prepare 1 liter, dissolve 16.1 g of Na2HPO4-7H2O, 5.5 g of NaH2PO4-H2O, 0.75 g of KCl and 0.246 g of MgSO4-7H2O in ddH2O; filter sterilize. Buffer can be stored indefinitely at room temperature.
Z-buffer with β-mercaptoethanol: Prepare fresh on day of use by adding 2.7 μl of β-mercaptoethanol to every 1 ml of Z-buffer.
H Top Agar: To prepare 100 ml, autoclave 0.8 g Bacto-agar, 0.8 g NaCl, and 1 g Bacto-Tryptone in 100 ml ddH2O. Media can be stored indefinitely in a sealed container at room temperature.
-
5× PEG/NaCl Solution: To prepare 500 ml, dissolve 87.5 g PEG 8000 and 62.5 g NaCl in ddH2O to a final volume of 500 ml. Filter sterilize. Solution can be stored indefinitely at room temperature.
<!>CRITICAL Dissolving the PEG and NaCl into solution may require stirring for many hours. We typically leave the mixture stirring overnight.
-
Custom synthesized oligonucleotides required to create Selection Reporter Plasmids bearing ZFN half-site target sequences
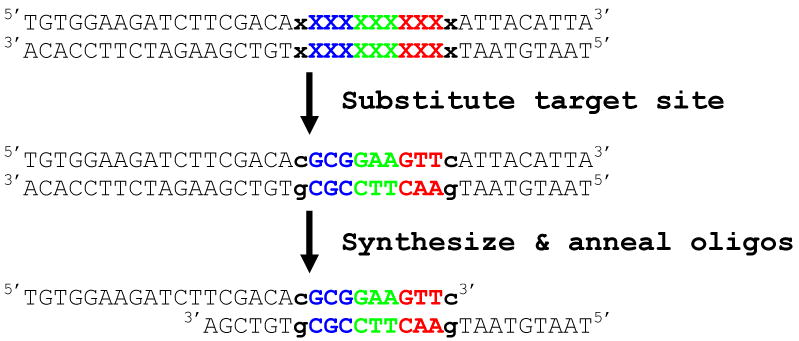
<!>CRITICAL Two oligonucleotides must be synthesized for each ZFN half-site target sequence. Details regarding design of these oligonucleotides are provided in Box 1.
-
Amino Acid Mixture: Prepare six separate amino acid solutions by dissolving the components listed below in ddH2O to a final volume of 100 ml each. After preparing all six solutions, combine them together and filter sterilize. Store wrapped in foil at 4°C. Solution is good for at least 30 days.
Solution 1: 0.99 g Phenylalanine, 1.1 g Lysine, 2.5 g Arginine
Solution 2: 0.2 g Glycine, 0.7 g Valine, 0.84 g Alanine, 0.41 g Tryptophan
Solution 3: 0.71 g Threonine, 8.4 g Serine, 4.6 g Proline, 0.96 g Asparagine
Solution 4: 9.1 ml HCl, 1.04 g Aspartic Acid, 14.6 g Glutamine
Solution 5: 18.7 g L-Glutamic acid Potassium salt monohydrate, 0.36 g Tyrosine, 4 g NaOH
Solution 6: 0.79 g Isoleucine, 0.79 g Leucine
<!>CRITICAL The six solutions must first be made up individually and then combined together. For each of the six solutions, add each amino acid component in the order listed and make sure that each component is completely dissolved before adding the next.
Procedure
Identifying potential OPEN ZFN sites using the web-based Zinc Finger Targeter (ZiFiT) program
1| Open a web browser to the ZiFiT v3.0 program webpage (address: http://binder.gdcb.iastate.edu/ZiFiT; a link to the website is also permanently available at: http://www.zincfingers.org/software-tools.html).
2| Click on the ZiFiT option on the left hand menu and then click on the “Design Zinc Finger Nucleases” option under the “OPEN (Oligomerized Pool Engineering)” menu. Note that on the sequence entry page, the DNA triplets for which OPEN pools are currently publicly available are already automatically checked. Additional pools for other triplets can also be selected by the user.
3| In the “Sequence” box, type or paste in the DNA sequence that will be searched for potential OPEN sites. Note that any spaces or numbers are ignored and that the DNA sequence to be analyzed can be pasted in as raw sequence or in FASTA format.
4| Select the length of the target site spacer sequence desired from the Spacer drop down box. Various ZFN expression vectors encoding different length linkers between the zinc finger arrays and the FokI nuclease domain have been described that enable cleavage of ZFN target sites harboring spacer sequences of five, six, or seven base pairs.71, 72 The ZFN linker encoded in expression vectors available from the Zinc Finger Consortium76 works best on target sites with spacers of five or six base pairs.71
5| Click the “Advanced” button to set upper limits on the number of GNN, ANN, CNN, or TNN subsites that will be allowed for a target ZFN site. This setting is useful especially if a large number of potential target sites are returned. The number of GNN subsites selected can be a useful parameter to alter because to date all target ZFN half-sites for which OPEN has worked have harbored one or more GNN subsites.
6| Click the “Submit” button to retrieve a list of ZFN target sites. For each potential target site, ZiFiT returns the DNA sequence of the full site with the 3 bp subsites of each half-site highlighted in different colors and a number indicating the nucleotide position of the site within the submitted sequence. The subsites are highlighted on the so-called “primary strand” which is the one that is predominantly contacted by amino acid residues in the zinc finger recognition helix. Because the nucleotide just 3′ to a given triplet subsite can influence binding of a zinc finger, some F1 subsites have multiple pools that were each generated in the context of a different 3′ nucleotide. If for a given F1 target subsite, a pool is available that was generated in the context of the specific 3′ nucleotide adjacent to the target site, then only that pool is returned in the output. However, if no pool was specifically generated in the context of the 3′ nucleotide present adjacent to the target site, then all of the available pools for that F1 target subsite (selected in the context of various other 3′ nucleotides) are returned. Note that each pool is assigned a unique “Reference Number” which can be used when requesting pools from the Joung lab.
? TROUBLESHOOTING
7| Use the nucleotide position information in the ZiFiT output to locate the target within your region of interest. Determine if the target location is compatible with your application. Ideally, for homologous recombination, ZFN cleavage sites should fall very close to the alteration or insertion to be introduced. Targets for gene knockout via NHEJ should preferably be located in the beginning or in a critical region of the coding sequence.
8| Determine whether highly similar off-target sites exist in your cell type of interest. To do this, use the BLAST button and organism list that is present in each target's ZiFiT output window. The BLAST button queries the NCBI BLAST servers for highly similar ZFN target sequences within the host genome. ZiFiT substitutes N's for the nucleotides within the spacer. This prevents matches to sequence within the spacer from positively influencing the BLAST results. ZiFiT implements the following the BLAST parameters to locate similar ZFN target matches: Database=NCBI Genomes, Expect=100000, word size=7, Match/Mismatch=1/-1. Filtering, masking, and automatic parameter adjustment by the BLAST program are disabled. The query may take up to a minute and results are returned to the ZiFiT window.
9| Check whether individual three-finger zinc finger arrays have already been successfully (or unsuccessfully) identified for your ZFN half-sites. To do this, use the Zinc Finger Database (ZiFDB) at http://bindr.gdcb.iastate.edu/ZiFDB/ (or through the Zinc Finger Consortium website at: http://www.zincfingers.org/software-tools.htm), a repository which describes sequences and activities of engineered zinc finger arrays previously described in the literature.34 The color-coded arrays in the ZiFiT output are hyperlinks which automatically query ZiFDB for zinc finger arrays generated for exact and similar sites. In addition, the color coded DNA triplets in the table are set to query ZiFDB for all known recognition helices that are believed to bind specifically to the given triplet.
10| To determine whether a potential ZFN target site occurs within a repeat sequence (e.g. a transposon) or a low complexity region, use the RepeatMasker Web Server at http://www.repeatmasker.org/. Follow the RepeatMasking link from the services section and paste the entire sequence for the region of interest. (Note: Be sure to use at least several hundred base pairs on both sides of the target because pasting just the ZFN site alone is not sufficient for the program to recognize longer repeats.) Select your target organism from the DNA source drop down list and click the submit button. RepeatMasker will return a summary page identifying known repeat and low complexity regions. Select one of the annotation files under the Results section to verify your target is not in one of these regions.
Construction of B2H Selection Strains
11| To construct a Selection Reporter Plasmid bearing a ZFN half-site target sequence, digest plasmid pKJ1712 with SapI (as tabulated below) at 37°C for 2 hours and purify the digested vector backbone on a 5% non-denaturing polyacrylamide gel. The full sequence of pKJ1712 is given in Supplementary Fig. 1. Elute and ethanol-precipitate the digested vector DNA as described in Box 2.
| Component | Amount | Final Concentration |
| Plasmid pKJ1712 | 1 μl (1 μg) | 20 ng μl-1 |
| 10× Buffer (NEBuffer 4) | 5 μl | 1× |
| SapI (2 U μl-1) | 5 μl | 0.04 U μl-1 |
| Nuclease-free water | 39 μl | |
| Total | 50 μl | |
<!>CAUTION Acrylamide is a neurotoxin and therefore polyacrylamide gels should be prepared wearing gloves.
12| Create extended overhangs by treating the SapI-digested pKJ1712 vector backbone of step 11 with Pfu polymerase in the presence of dCTP nucleotide (as tabulated below). Incubate this reaction for 15 minutes at 72°C and then place immediately at 4°C.
| Component | Amount | Final Concentration |
| SapI-digested pKJ1712 vector backbone | 10 μl | ∼10 ng μl-1 |
| 10× Pfu Buffer | 2 μl | 1× |
| dCTP nucleotide (10 mM) | 2 μl | 1 mM |
| Cloned Pfu polymerase (2.5 U μl-1) | 1.2 μl | 1.25 U μl-1 |
| Nuclease-free water | 4.8 μl | |
| Total | 20 μl | |
13| For each target ZFN half-site, design a pair of synthetic oligonucleotides as described in Box 1. Anneal each pair of oligos together by mixing components as tabulated below. Incubate oligo mixture at 95°C for 2 min, slowly cool to 35°C, and then immediately place on ice. This incubation can be performed either in a programmed thermocycler or in a heat block with a thermometer.
| Component | Amount | Final Concentration |
| Binding site oligo #1 (10 μM) | 1 μl | 50 nM |
| Binding site oligo #2 (10 μM) | 1 μl | 50 nM |
| 10× Annealing Buffer | 20 μl | 1× |
| Nuclease-free water | 178 μl | |
| Total | 200 μl | |
14| Ligate the annealed binding site oligonucleotides (from step 13) to the gel purified, SapI-digested, Pfu-treated pKJ1712 vector backbone (from step 12) as tabulated below. As a control, also set up a ligation that lacks the annealed oligonucleotides. Incubate these reactions at room temperature for 15 minutes and then halt the reactions by placing them on ice.
| Component | Amount | Final Concentration |
| SapI- and Pfu-treated pKJ1712 backbone | 2 μl | ∼1 ng μl-1 |
| Annealed binding site oligos (or water for control) | 8 μl | 19 nM |
| 2× Quick Ligase Buffer | 10 μl | 1× |
| T4 DNA Ligase (400 U μl-1) | 1 μl | 19 U μl-1 |
| Total | 21 μl | |
15| Transform chemically competent XL-1 Blue cells with the actual and control ligations from step 14. To do this, add 200 μl of chemically competent XL-1 Blue cells (prepared as described in Box 3) to each ligation. Leave on ice for 5 minutes, heat shock at 42°C for 2 minutes, return immediately to ice for 2 minutes, add 700 μl LB media, and incubate on a LabQuake shaker/rotisserie at 37°C for 40 minutes. Plate 300 μl of each transformation on an LB plate supplemented with 30 μg ml-1 kanamycin and incubate overnight at 37°C.
16| Verify that the desired ligation(s) yields at least three-fold more transformants than the control ligation. If this is the case, for each ligation inoculate two or more candidates into 10 ml LB cultures supplemented with 30 mg ml-1 kanamycin. Grow these cultures overnight at 37°C for 14 to 18 hours for performing plasmid miniprep isolation.
? TROUBLESHOOTING
17| Isolate plasmid DNA from overnight cultures using a QIAprep Spin Miniprep kit following the manufacturer's instructions including the optional PB buffer wash step.
18| Confirm the reporter plasmid candidates by digesting them with EcoRI and HindIII for 1 hour at 37°C under the conditions described below. Digestion products can be visualized on a 5% non-denaturing polyacrylamide gel (a typical gel is shown in Supplementary Fig. 4). Reporter plasmids that possess the binding sites should yield fragments of sizes 6108, 1006, 963, 431, and 190 bp, compared with the control parental pKJ1712 plasmid, which will yield fragments of sizes 6108, 1006, 963, 456, and 190 bp.
| Component | Amount | Final Concentration |
| Plasmid DNA | 5 μl (∼0.75 μg) | ∼25 ng /μl-1 |
| 10× EcoRI Buffer | 3 μl | 1× |
| EcoRI (20 U μl-1) | 1 μl | 0.66 U μl-1 |
| HindIII (20 U μl-1) | 1 μl | 0.66 U μl-1 |
| Nuclease-free water | 20 μl | |
| Total | 30 μl | |
<!>CAUTION Acrylamide is a neurotoxin and therefore polyacrylamide gels should be prepared wearing gloves.
19| Confirm the sequence of selection reporter plasmids between the EcoRI and SalI sites that flank the target ZFN half-site using sequencing primer OK181 (this primer is an anti-sense primer that anneals ∼270 bp downstream of and points back toward the binding site). Candidate reporter plasmids can be aligned with the sequence in Supplementary Fig. 5 (“EcoRI-SalI binding site reporter”). Note that the target 11 bp ZFN half-site is shown as a series of Xs in this file.
Recombination-based Transfer of Selection Reporter Plasmid Sequences to a Single-Copy Episome
20| Transform sequence-confirmed selection reporter plasmids (one for each ZFN target half-site) into bacterial strain CSH100. Add 1 μl of mini prep plasmid DNA (∼0.1 μg) to 50 μl of ice-cold chemically competent CSH100 cells (prepared as described in Box 4), incubate at 42°C for 2 minutes, return immediately to ice for 2 minutes, add 250 μl of LB medium, incubate at 37°C for 40 minutes on a LabQuake shaker/rotisserie, plate entire transformation on a LB plate containing 30 μg ml-1 kanamycin, and incubate overnight at 37°C.
21| On the same day that step 20 is performed, inoculate a fresh colony of bacterial strain KJ1C into 10 ml of LB medium supplemented with 12.5 μg ml-1 tetracycline and grow with agitation on a roller drum (∼60 rpm) overnight at 37°C.
22| Examine plates with transformants of CSH100 (from step 20 above) and confirm the presence of thousands of confluent colonies. Harvest these transformants by scraping all of the colonies from a single plate using a sterile wooden stick and resuspend these cells into 10 ml of LB medium using gentle vortexing in a sterile 25 mm glass tube.
CRITICAL STEP: Only very gentle vortexing should be used to resuspend the CSH100 transformants to minimize damage to the F pili expressed on the surface of these cells.
23| Subculture ∼200 μl of the resuspended CSH100 transformants (prepared in step 22) into a sterile 25 mm glass tube containing 5 ml of LB (without antibiotics). Also subculture ∼200 μl of the overnight culture of strain KJ1C (from step 21 above) into a sterile 25 mm glass tube containing 5 ml of LB (without antibiotics). The density of cells in each of these subcultures should initially resemble a prelog phase culture by visual inspection (i.e.—an OD600 of ∼0.1). As a control, also add 10 ml of LB to a sterile 25 mm glass tube. Incubate all of these tubes for 2 hours at 37°C without agitation.
24| Perform matings by setting up the following mixtures of subcultures prepared in step 23 above in sterile 18 mm glass tubes:
| Actual Mating: | 1 ml of CSH100 transformants + 1 ml of KJ1C |
| Controls: | 1 ml of CSH100 transformants + 1 ml of LB |
| 1 ml of KJ1C + 1 ml of LB | |
| 2 ml of LB |
Incubate matings at 37°C for 1 hr without agitation and then transfer tubes to a roller drum (∼60 rpm) at 37°C for 90 minutes.
25| To identify desired double recombinant F's that have been successfully transferred to strain KJ1C (Fig. 3), plate 300 μl of the actual desired mating from step 24 above on a LB/TKS plate (as a control, also plate 300 μl of the actual mating on a LB/TK plate). Spot 5 μl of each of the control matings from step 24 on LB/TKS and LB/TK plates. Incubate all plates overnight at 37°C.
26| Inspect all plates from step 25 for bacterial colony growth. KJ1C cells that have successfully received the desired double-recombinant F′ should be able to grow on LB/TKS plates (see Fig. 3). For the actual mating, we will typically see hundreds of colonies on the LB/TK control plate and about a ∼10-fold decrease in colony number observed on the matched LB/TKS plate. This reduction indicates that the sacB gene on the selection reporter plasmid is expressed, a critical requirement for successful identification of double recombinant F's (see Fig. 3). All plates on which control matings were spotted should be free of colonies or bacterial growth (although occasionally, we will observe a few colonies on spots from the controls on LB/TK plates).
? TROUBLESHOOTING
27| For each mating, pick two independent colonies (designated “A” and “B”) to carry forward for confirmation. To purify clonal isolates, serially re-streak each colony to a LB/TKS plate (one plate can be divided in two and a candidate streaked on each side) and grow for 12-18 hours at 37°C. Colonies for each candidate are then re-streaked again to LB/TKS plates and grown at 37°C for 12-18 hours.
28| As an additional check for successful transfer of the F′ to strain KJ1C, test the re-streaked colonies of candidates “A” and “B” from step 27 for their abilities to grow in the absence of proline by resuspending a colony for each candidate in 100 μl of 1× M9 salts (this is conveniently done in the wells of a sterile 96-well plate) and spotting on a M9 minimal medium plate. Incubate this plate overnight at 37°C. (Strain KJ1C lacks the proAB genes for proline biosynthesis and the F′ from CSH100 contains the proAB genes. Thus, KJ1C cells that have successfully acquired the F′ from CSH100 cells should be proline prototrophs (i.e.—they should be able to grow in the absence of exogenously supplied proline) and therefore should be able to grow on M9 minimal medium plates.)
29| On the same day that candidates are spotted on M9 plates (as described in step 28), use 20 μl of each cell resuspension (from step 28) to inoculate 4 ml of LB medium containing 30 μg ml-1 kanamycin and grow these cultures overnight at 37°C on a roller drum (∼60 rpm).
30| After overnight incubation, verify the growth of cells spotted on the M9 minimal medium plate (from step 28) and discard any candidates that fail to grow.
31| For all candidates that do show growth on M9 minimal medium plates, isolate F′ DNA from the overnight cultures inoculated in step 29 by centrifuging 100 μl of culture in a 1.5 ml microcentrifuge tube at highest speed for 1 minute in a microfuge, removing the media, resuspending the cells in 100 μl of nuclease-free water, heating the resuspension at 95°C for 10 minutes, pelleting the cellular debris by centrifuging for 1 minute at maximum speed, and removing 50 μl of supernatant to a fresh microcentrifuge tube.
32| To verify the sequence of the F′ reporter, amplify the region of the target binding site using the crude preparation of DNA isolated in step 31 as template for a PCR reaction as tabulated below:
| Component | Amount | Final Concentration |
| Crude DNA preparation (from step 31) | 5 μl | Unknown |
| 10× Expand Buffer with MgCl2 | 5 μl | 1× |
| 10 mM dNTPs | 4 μl | 0.8 Mm |
| Primer HIS3-2F (10 μM) | 1 μl | 0.2 μM |
| Primer HIS3-2R (10 μM) | 1 μl | 0.2 μM |
| Expand thermostable polymerase (3.5 U μl-1) | 0.375 μl | 0.02625 U μl-1 |
| Nuclease-free water | 33.625 μl | |
| Total | 50 μl | |
33| Perform PCR of reactions set up in step 32 using the following cycling conditions:
| Step Number | Denature | Anneal | Extend |
| 1 | 94°C, 5 min | ||
| 2-36 | 94°C, 30 s | 55°C, 30 s | 72°C, 1 min |
| 37 | 72°C, 7 min | ||
34| Purify PCR products from step 33 using the QIAQuick PCR Purification Kit following the manufacturer's directions with the following exception: at the final step, elute the DNA from each column using 50 μl 0.1× EB. Re-sequence the DNA between the EcoRI and HindIII sites in the fragment using primer OK181 as described in step 19 above.
35| Make glycerol stocks and chemically competent cells (using the protocol in Box 4) from the overnight cultures inoculated in step 29.
36| For each target site, choose one strain that passes all of the checks described above and transform the competent cells made in step 35 with the plasmid pAC-alphaGal4. Add 10 ng of pAC-alphaGal4 plasmid to 100 μl of competent cells made in step 35, incubate on ice for 10 minutes, heat shock at 42°C for two minutes, return to ice for two minutes, add 900 μl of LB, incubate on a LabQuake shaker/rotisserie at 37°C for 45 minutes, and plate 100 μl of the transformation on an LB/CK plate. Incubate the plate overnight at 37°C.
? TROUBLESHOOTING
37| Colonies from the transformations performed in step 36 can be inoculated into LB cultures containing chloramphenicol (30 μg ml-1) and kanamycin (30 μg ml-1), grown overnight at 37°C, and stored as glycerol stocks. These transformants are referred to as “B2H selection strains” in the remainder of this protocol.
Construction of Combinatorial Zinc Finger Libraries
38| Digest the pBR-UV5-GP-FD2 plasmid vector with BbsI at 37°C overnight as tabulated below. The full sequence of pBR-UV5-GP-FD2 is given in Supplementary Fig. 2.
| Component | Amount | Final Concentration |
| pBR-UV5-GP-FD2 plasmid DNA | 40 μg | 100 ng μl-1 |
| 10× NEBuffer 2 | 40 μl | 1× |
| BbsI (5 U μl-1) | 40 μl | 0.5 U μl-1 |
| Nuclease-free water | to 400 μl | |
CRITICAL STEP: We use fresh 10× NEBuffer 2 (so that the DTT component is at optimal concentration) and BbsI enzyme to maximize digestion efficiency.
39| The following morning, perform an additional BamHI digestion of the reaction from step 38 as tabulated below. Incubate this reaction for 2.5 hours at 37°C.
| Component | Amount | Final Concentration |
| BbsI-digest from step 38 | 400 μl | |
| 10× BSA (1 mg ml-1) | 50 μl | 1× |
| 10× BamHI buffer | 47.5 μl | 1× |
| BamHI (20 U μl-1) | 2.5 μl | 0.1 Uμl-1 |
| Total | 500 μl | |
CRITICAL STEP: Complete digestion with BamHI is critical to cleave the BbsI fragment liberated from the pBR-UV5-GP-FD2 plasmid in step 38 (Fig. 5) into two smaller pieces which can then be efficiently removed from the reaction by purification with a QIAQuick PCR purification kit column in the next step.
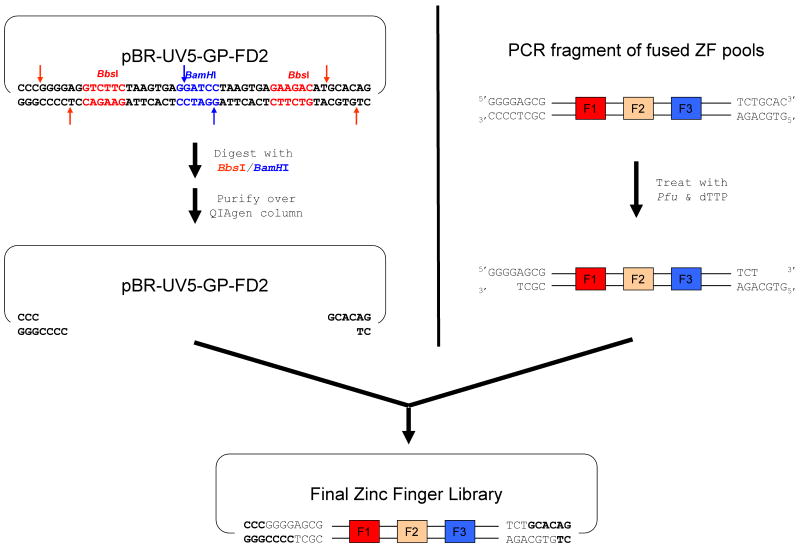
Figure 5. Schematic overview of zinc finger library construction.
The left side illustrates how digestion of B2H expression plasmid pBR-UV5-GP-FD2 with restriction enzymes BbsI and BamHI results in the liberation of two small DNA fragments which are eliminated by purifying the reaction with a QIAgen PCR purification column. The right side illustrates how treatment of a PCR fragment encoding fused ZF pools with Pfu polymerase and dTTP nucleotide results in creation of 5′ overhangs that are 4 bases long. Ligation of the digested plasmid and Pfu polymerase-treated PCR fragment leads to the desired zinc finger library.
40| Purify the vector backbone by dividing the reaction from step 39 into four equal portions of 125 μl each and purify each part using a QIAQuick PCR purification kit column following the manufacturer's directions with the following exception: at the final step, elute the DNA by adding 50 μl of 0.1× EB (pre-warmed to 60°C) to the column, allow to stand for 1 minute, cap the column, centrifuge for 1 minute at highest speed, and then re-elute DNA from each column using the flow-through. Following the second elution step, pool all four eluates together (final total volume of ∼200 μl). Quantify the purified DNA by measuring A260 on a spectrophotometer. The DNA concentration should be ∼200 ng μl-1. To determine the concentration of DNA, use the following equation:
Concentration (μg ml-1) = A260 × dilution factor × 50
PAUSE POINT: Purified vector DNA can be stored indefinitely at -20°C.
Assembly of DNA Fragments Encoding Multi-finger Arrays
41| For each target site, set up PCR reactions using the three required pools (identified from the ZiFiT output obtained in steps 1-10 and obtained by request from the Joung lab) as template as tabulated below. Note that in the following steps the naming convention F1, F2 and F3 will specify finger positions where F1 is the amino-terminal finger, F2 is the middle finger and F3 is the carboxy-terminal finger.
| Component | Amount | Final Concentration |
| Zinc Finger Pool DNA (∼0.15-0.2 μg) | 1 μl | 3-4 ng μl-1 |
| 10× Expand Buffer with MgCl2 | 5 μl | 1× |
| 10mM dNTPs | 4 μl | 0.8 mM |
| Forward primer (10 μM) | 3 μl | 0.6 μM |
| for F1 use OK1424 | ||
| for F2 use OK1426 | ||
| for F3 use OK1428 | ||
| Reverse primer (10 μM) | 3 μl | 0.6 μM |
| for F1 use OK1425 | ||
| for F2 use OK1427 | ||
| for F3 use OK1429 | ||
| Nuclease-free H2O | 33.625 μl | |
| Expand Enzyme (3.5 U μl-1) | 0.375 μl | 0.02625 U μl-1 |
| Total | 50 μl | |
42| Perform PCR using the finger position-specific cycling conditions listed in Tables 2, 3 and 4.
Table 2. PCR conditions for amplifying Finger 1 pools (steps 41 and 42).
| Step number | Denature | Anneal | Extend |
|---|---|---|---|
| 1 | 94°C, 5 min | ||
| 2-6 | 94°C, 30 s | 55°C, 30 s | 72°C, 30 s |
| 7-26 | 94°C, 30 s | 59°C, 30 s | 72°C, 30 s |
| 27 | 72°C, 2 min |
Table 3. PCR conditions for amplifying Finger 2 pools (steps 41 and 42).
| Step number | Denature | Anneal | Extend |
|---|---|---|---|
| 1 | 94°C, 5 min | ||
| 2-6 | 94°C, 30 s | 52°C, 30 s | 72°C, 30 s |
| 7-26 | 94°C, 30 s | 57°C, 30 s | 72°C, 30 s |
| 27 | 72°C, 2 min |
Table 4. PCR conditions for amplifying Finger 3 pools (steps 41 and 42).
| Step number | Denature | Anneal | Extend |
|---|---|---|---|
| 1 | 94°C, 5 min | ||
| 2-6 | 94°C, 30 s | 41°C, 30 s | 72°C, 30 s |
| 7-26 | 94°C, 30 s | 56°C, 30 s | 72°C, 30 s |
| 27 | 72°C, 2 min |
PAUSE POINT: PCR products can be stored indefinitely at -20°C.
43| Purify PCR products from step 42 by electrophoresis on a 10% non-denaturing polyacrylamide gel run at ∼100 volts. Stain the gel with ethidium bromide and use long-wave UV light to visualize the desired ∼100 bp products (note that these products will typically run as a smeared, rather than a distinct, band and that a significant “primer dimer” product is always seen with the F1 primers). Excise the PCR products in a gel slice, crush the gel piece, and elute DNA overnight at 37°C in 700 μl of ammonium acetate elution buffer.
<!>CAUTION Acrylamide is a neurotoxin and therefore polyacrylamide gels should be prepared wearing gloves.
? TROUBLESHOOTING
44| Purify the DNA fragments from the overnight elution of step 43 by spinning at highest speed in a benchtop microfuge for 2 minutes and removing as much of the supernatant as possible (typically ∼600 μl) being very careful to avoid the acrylamide pellet and transfer to a fresh microfuge tube.
45| Spin the recovered supernatant of step 44 at highest speed in the microfuge for 2 minutes, remove 475 μl of supernatant (again taking care to avoid the residual acrylamide pelleted at the bottom of the tube) and transfer to a fresh microfuge tube.
46| Add 2 μl of 10 mg ml-1 glycogen to the recovered supernatant of step 45, mix well, add 1 ml of 100% ethanol, mix well, place on dry ice for >15 minutes, and spin the tube at highest speed in a microfuge for 15 minutes at 4°C. Remove and discard the supernatant (carefully avoiding the pellet), add 500 μl of 70% (v/v) ethanol, spin at highest speed in a microfuge for 5 minutes, remove all residual liquid (again avoiding the pellet), air dry the pellet for 5-10 minutes, and resuspend the pellet in 40 μl of nuclease-free water.
PAUSE POINT: Purified DNA can be stored indefinitely at -20°C.
47| Fuse together the purified F1, F2 and F3 PCR fragments purified in step 46 by setting up a PCR reaction as tabulated below.
| Component | Amount | Final Concentration |
| Purified F1 PCR product (from step 46) | 3 μl | unknown |
| Purified F2 PCR product (from step 46) | 3 μl | unknown |
| Purified F3 PCR product (from step 46) | 3 μl | unknown |
| 10× Expand Buffer with MgCl2 | 5 μl | 1× |
| 10mM dNTPs | 4 μl | 0.8 mM |
| Nuclease-free water | 31.625 μl | |
| Expand Enzyme (3.5 U μl-1) | 0.375 μl | 0.02625 U μl-1 |
| Total | 50 μl | |
48| Perform PCR of reaction set up in step 47 using the following cycling conditions:
| Step number | Denature | Anneal | Extend |
| 1 | 94°C, 5 min | ||
| 2-11 | 94°C, 30 s | 50°C, 30 s | 72°C, 1 min |
| 12 | 72°C, 7 min | ||
PAUSE POINT: PCR product can be stored indefinitely at -20°C.
49| Purify the DNA from the PCR reaction of step 48 using a QIAQuick PCR purification kit column following the manufacturer's directions with the following exception: at the final step, elute the DNA from each column using 50 μl 0.1× EB.
PAUSE POINT: Purified DNA can be stored indefinitely at -20°C.
50| Amplify the fused PCR product encoding various combinations of three-finger arrays by setting up the PCR reaction tabulated below.
| Component | Amount | Final Concentration |
| Purified PCR product of step 49 | 24 μl | unknown |
| Forward primer OK1430 (10 μM) | 3 μl | 0.6 μM |
| Reverse primer OK 1432 (10 μM) | 3 μl | 0.6 μM |
| 10× Expand Buffer with MgCl2 | 5 μl | 1× |
| 10mM dNTPs | 4 μl | 0.8 mM |
| Nuclease-free water | 10.625 μl | |
| Expand Enzyme (3.5 U μl-1) | 0.375 μl | 0.02625 U μl-1 |
| Total | 50 μl | |
51| Perform PCR of reaction set up in step 50 using the following cycling conditions:
| Step number | Denature | Anneal | Extend |
| 1 | 94°C, 5 min | ||
| 2-11 | 94°C, 30 s | 56°C, 30 s | 72°C, 1 min |
| 12-31 | 94°C, 30 s | 64°C, 30 s | 72°C, 1 min |
| 32 | 72°C, 7 min | ||
PAUSE POINT: PCR product can be stored indefinitely at -20°C.
52| Purify the PCR product from step 51 on a 5% non-denaturing polyacrylamide gel run at ∼200 volts with a 100 bp marker ladder. Stain the gel with ethidium bromide, visualize DNA products using a hand-held long-wave UV light source, and excise the portion of the lane that corresponds to fragments of size ∼200 to ∼400 bps. Elute and ethanol precipitate the DNA fragments as described in Box 2.
<!>CAUTION Acrylamide is a neurotoxin and therefore polyacrylamide gels should be prepared wearing gloves.
PAUSE POINT: Purified DNA can be stored indefinitely at -20°C.
53| Create 4 bp overhangs on the DNA fragment purified in step 52 by treating with thermostable Pfu polymerase and the single nucleotide triphosphate dTTP (Fig. 5) as tabulated below. Incubate this reaction for exactly 15 minutes at 72°C and then place on ice or keep at <4°C.
| Component | Amount | Final Concentration |
| Purified PCR fragment (from step 52) | 10 μl | Unknown |
| 10mM dTTP | 3 μl | 1 mM |
| 10× Cloned Pfu Buffer | 3 μl | 1× |
| Cloned Pfu polymerase (2.5 U μl-1) | 3 μl | 0.25 U μl-1 |
| Nuclease-free water | 11 μl | |
| Total | 30 μl | |
CRITICAL STEP: Incubation of the Pfu reaction for 15 minutes is critical for creating the required 4 bp overhangs. However, this reaction should also not be allowed to proceed for longer than 15 minutes to avoid the risk of depleting the dTTP nucleotide and the resulting degradation of the DNA fragments.
54| Purify the DNA from the Pfu reaction of step 53 using a QIAQuick PCR purification kit column following the manufacturer's directions with the following exception: at the final step, elute the DNA from each column using 45 μl 0.1× EB.
PAUSE POINT: Purified DNA can be stored indefinitely at -20°C.
55| Phosphorylate the 5′-OH groups of the purified DNA of step 54 by treating with T4 polynucleotide kinase as tabulated below. Incubate this reaction for 30 minutes at 37°C and then immediately purify the DNA using a MinElute PCR Purification kit column following the manufacturer's directions with the following exception: at the final step, elute the DNA from each column using 22 μl 0.1× EB.
| Component | Amount | Final Concentration |
| Purified DNA (from step 54) | 44 μl | Unknown |
| 10× T4 DNA ligase buffer | 5 μl | 1× |
| T4 polynucleotide kinase (10 U ul-1) | 1 μl | 0.2 U μl-1 |
| Total | 50 μl | |
CRITICAL STEP: We have found that phosphorylation of the PCR product is critical for the subsequent ligation step. In our experience, failure to perform this step results in very few ligation events.
PAUSE POINT: Purified DNA can be stored indefinitely at -20°C.
Ligation of Digested Vector to DNA Fragments Encoding Multi-finger Arrays
56| Ligate the purified BbsI-digested pBR-UV5-GP-FD2 vector (from step 40) to the purified, Pfu-treated, phosphorylated PCR fragment encoding various combinations of three fingers (from step 55) as tabulated below. Incubate ligations overnight at 16°C. In parallel, also set up a control ligation reaction where 10 μl of nuclease-free water is substituted for the PCR fragment encoding the three-finger arrays.
| Component | Amount | Final Concentration |
| pBR-UV5-GP-FD2 backbone | 1 μg | 20 ng μl-1 |
| PCR fragment encoding three-finger arrays | 10 μl | Unknown |
| 10× standard T4 DNA Ligase buffer (NEB) | 5 μl | 1× |
| T4 DNA ligase (400U μl-1) | 2 μl | 16 U μl-1 |
| Nuclease-free water | to 50 μl | |
PAUSE POINT: Ligations can be stored indefinitely at -20°C.
57| Check the efficiency of the ligation from step 56 by transforming the actual and control ligations into chemically competent E. coli XL-1 Blue cells. Add 1 μl of actual ligation or 10 μl of control each to 200 μl of chemically competent XL-1 Blue cells (prepared as described in Box 3) in a sterile microfuge tube. Incubate transformations on ice for 10 minutes, heat shock by placing in a 42°C water bath for 2 minutes, return immediately to ice for 2 minutes, add 900 μl of LB, and incubate on a roller drum (∼60 rpm) at 37°C for 40 minutes. Make 10-1 and 10-2 dilutions of the transformations by serial dilution as described in Box 5. Spot 5 μl of the undiluted and diluted transformations in triplicate (i.e.—15 μl total) on LB/TC plates and incubate overnight at 37°C.
58| The next morning, count colonies from the highest dilution spots for which distinct countable colonies are visible. Calculate the number of transformants μl-1 of ligation reaction by using the equations below.
Equation for transformants μl-1 of the actual ligation:
(# of colonies in 3 spots × 1100)/(15 × dilution factor)
Equation for transformants μl-1 of the control ligation:
(# of colonies in 3 spots × 1100)/(15 × dilution factor × 10)
CRITICAL STEP: The number of transformants μl-1 from the actual ligation should be >10-fold the number of transformants from the control ligation. Note that Steps 57 and 58 only need to be performed once for each preparation of purified, digested vector backbone.
? TROUBLESHOOTING
Introduction of Combinatorial Zinc Finger Library into E. coli Cells
59| Purify 25 μl of the actual ligation from step 56 using a QIAgen MinElute PCR Purification Kit column following the manufacturer's instructions but eluting at the final step into 10 μl of nuclease-free water.
PAUSE POINT: Purified ligation DNA can be stored indefinitely at -20°C.
60| Transform 70 μl of electrocompetent XL-1 Blue cells (prepared as described in Box 6) with the entire purified ligation from step 59 (∼8-9 μl volume). Perform electroporation using a 1 mm gap cuvette and with the following settings: 1.75 kV, 200 Ohms, and 25 μF. Immediately following application of current, add 1 ml of SOC medium to the cuvette and then transfer cells to 9 ml of 2XYT media in a sterile 25 mm glass culture tube. Allow the cells to recover by placing them on a roller drum (∼60 rpm) at 37°C for 1 hour. Also perform positive and negative control electroporations with 1 μg of pBR-UV5-GP-FD2 plasmid DNA and no DNA, respectively.
CRITICAL STEP: Use of electroporation is critical at this step in order to achieve sufficient oversampling of the theoretical library size. Chemically competent cells do not yield enough transformants to achieve this.
61| Determine the number of transformants for each electroporation performed in step 60 by performing a “pre-amplification quantification.” To do this, prepare dilutions of each electroporation in triplicate from 10-1 to 10-6 using the method described in Box 5. Spot 5 μl of each serial dilution in triplicate (15 μl total) on LB/TC plates. A set of six dilutions can be conveniently spotted in triplicate on half of a plate (Fig. 6). Incubate plates overnight at 37°C.
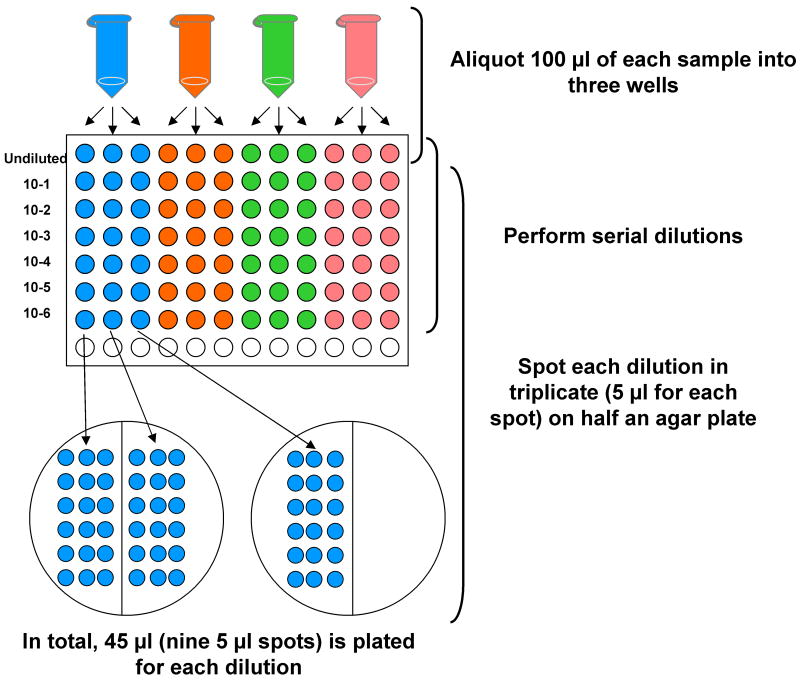
Figure 6. Schematic depicting triplicate serial dilution and triplicate spotting of dilutions on agar plates.
100 μl of each sample to be diluted is placed in triplicate into three wells in the top row of a 96-well plate. 10-fold serial dilutions are performed of each sample and then 5 μl of each dilution is plated in replicate on half of an agar plate.
62| Amplify the zinc finger library transformation from step 60 (not the controls) by transferring the entire remaining volume to 90 ml of 2xYT supplemented with 12.5 μg ml-1 of tetracycline and 50 μg ml-1 of carbenicillin in a sterile Erlenmeyer flask. Record the volume of the transformation transferred (this will be needed later to calculate the “pre-amplification quantification” value). Incubate the culture for 2 hours at 37°C with agitation on an orbital platform shaker (250 rpm).
63| Determine the number of transformed cells in the amplified culture of step 62 by performing a “post-amplification quantification.” To do this, prepare dilutions of each electroporation in triplicate from 10-1 to 10-6 using the method described in Box 5. Spot 5 μl of each serial dilution in triplicate (15 μl total) on LB/TC plates. Incubate plates overnight at 37°C.
64| Pellet the cells from the amplified culture of step 62 by transferring 90 ml into two sterile 50 ml conical tubes (i.e.—45 ml in each tube) and spinning at 2000-3000 × g for 40 minutes at 4°C in a tabletop Sorvall centrifuge. Drain the media supernatant and resuspend each cell pellet in 1 ml of 2XYT media containing 15% (v/v) glycerol. The cell pellet will typically be difficult to resuspend and requires repeated pipeting to adequately break apart the cells. Combine the two 1 ml resuspensions together and then transfer four equal volume aliquots (∼0.5 ml each) into four sterile 2 ml cryogenic vials. Freeze on dry ice or in a dry ice/ethanol bath. Store aliquots at -80°C.
PAUSE POINT: Frozen cell libraries can be stored indefinitely at -80°C.
65| Using the equations below, calculate the “pre-amplification quantification” and “post-amplification quantification” numbers from the plates of steps 61 and 63, respectively. Count colonies from the highest dilution spots for which distinct countable colonies are visible. The pre-amplification quantification number must be >2.5 × 106 to ensure three-fold over-sampling of the theoretical library size of 953=∼8.6 × 105). The positive control (using 1 μg of pBR-UV5-GP-FD2 plasmid DNA should yield >108 transformants. The post-amplification quantification number should be >1 × 107 if adequate amplification (4-fold minimum) of the transformed cells has occurred.
PRE-AMPLIFICATION QUANTIFICATION = (Total # of colonies from 45 μl of transformation dilution for which distinct countable colonies are visible × volume in μl transferred in step 62)/(45 × dilution factor)
POST-AMPLIFICATION QUANTIFICATION = (Total # of colonies from 45 μl of transformation dilution for which distinct countable colonies are visible × 90,000)/(45 × dilution factor)
? TROUBLESHOOTING
Conversion of Combinatorial Zinc Finger Library into Phage
66| Based on the post-amplification quantification number obtained in step 65, thaw enough vials (typically 1 or 2) of the frozen cell library of step 64 to inoculate >2.5 × 106 transformed cells into 9 ml of 2XYT medium containing 12.5 μg ml-1 of tetracycline and 50 μg ml-1 of carbenicillin in a sterile 25 mm glass tube. Incubate on a roller drum (∼60 rpm) at 37°C for 1.5 hours.
67| Determine the number of transformed cells in the subculture of step 66 by performing a “pre-infection titer.” To do this, prepare dilutions in triplicate from a small amount of the subculture from 10-1 to 10-6 using the method described in Box 5. Spot 5 μl of each serial dilution in triplicate (15 μl total) on LB/TC and on LB/KCarb plates. Cells transformed with zinc finger plasmid will grow on LB/TC plates but should fail to grow on LB/KCarb plates (the latter is a control to check that cells are not already infected with M13K07 helper phage which confers resistance to kanamycin). Incubate plates overnight at 37°C.
68| Infect the subculture of step 66 with helper phage by adding 140 μl of high titer M13K07 stock (109 KTU μl-1) and mixing well to ensure even distribution of the phage throughout the culture medium. The high-titer M13K07 stock is prepared as described in Box 7. Allow the infected culture to sit at room temperature for 15 minutes without agitation and then incubate on a roller drum (∼60 rpm) for 1.5 hours at 37°C.
69| Determine the number of helper phage-infected cells in the culture of step 68 by performing a “post-infection titer.” To do this, prepare dilutions in triplicate from a small amount of the subculture from 10-1 to 10-6 using the method described in Box 5. Spot 5 μl of each serial dilution in triplicate (15 μl total) on LB/TC and on LB/KCarb plates. Cells transformed with zinc finger plasmid will grow on LB/TC plates and helper phage-infected cells will also grow on LB/KCarb plates. Incubate plates overnight at 37°C.
70| Add the remainder of the culture from step 68 to a sterile flask containing 90 ml of 2XYT medium containing 50 μg ml-1 carbenicillin and 70 μg ml-1 kanamycin. Record the volume of culture transferred to this flask (it will be important for calculations performed later). Shake at 250 rpm, 37°C on an orbital platform shaker overnight.
71| The next day, verify that the pre-infection titer plates of step 67 show that none of the cells were initially infected with helper phage (i.e.—that there are no colonies on the LB/KCarb plates). In addition, confirm that the library cells have been successfully super-infected by helper phage by verifying that the post-infection titer plates of step 69 show that >50% of the cells were infected by helper phage (i.e.—that the titers from the LB/KCarb plates are >50% those from the LB/TC plates). If these titers look appropriate, harvest the phage from the culture of step 70. To do this, transfer ∼30 ml of the culture to a sterile 50 ml conical tube, pellet the cells by centrifuging at ∼1500 × g in a tabletop Sorvall centrifuge for 30 minutes, and filter the supernatant through a 0.22 μm PES filter.
PAUSE POINT: Phage libraries can be stored at 4°C for up to a month. For long-term storage, freeze the phage libraries at -80°C.
? TROUBLESHOOTING
Performing OPEN Selections
72| To begin the first stage of OPEN selection, streak the selection strain harboring the target DNA site from the glycerol stock made in step 37 onto a LB/CK plate and incubate at 37°C overnight.
73| Inoculate two or three colonies from the plate streaked in step 72 into a sterile 125 ml Erlenmeyer flask containing 20 ml NM medium supplemented with 30 μg ml-1 kanamycin, 30 μg ml-1 chloramphenicol, 50 μM IPTG. Grow the culture overnight for 16-24 hours at 37°C on an orbital platform shaker at 125 rpm.
CRITICAL STEP: Do not shake cells at speeds higher than 125 rpm. For reasons that we do not as yet understand, the cells will grow suboptimally if shaken at higher speeds in NM medium.
CRITICAL STEP: When inoculating colonies into NM medium, it is important to make sure that the colony is well dispersed into the medium. Failure to adequately disperse the colony will result in failure of the cells to grow. A reliable way to disperse a colony is to take up the entire colony into the base of a CLP 200 μl filtered pipet tip and then to force air through the tip several times as it is pressed against the bottom of the flask. To do this, push air forcefully through while the tip is pressed against the bottom of the flask (bubbles should be generated if this is done correctly), lift the tip out of the medium, draw air back into the tip gently by slowly releasing the pipet plunger, and then place the tip against the bottom of the flask and repeat several times.
74| Measure the density of the selection strain culture inoculated in step 73 using a spectrophotometer. This culture should have an OD600 ∼1.0.
? TROUBLESHOOTING
75| In a sterile 125 ml Erlenmeyer flask, infect 5 ml of the selection strain culture inoculated in step 73 with 50 μl of the phage library from step 71 and mix well. Let infection proceed at room temperature for 30 minutes without agitation.
76| Add 20 ml of pre-warmed NM medium containing 30 μg ml-1 chloramphenicol, 30 μg ml-1 kanamycin, and 50 μM IPTG to the infected cells of step 75 and slowly shake at 125 rpm, 37°C, for 1.5 hours.
77| Harvest infected cells by transferring the culture of step 76 to a sterile 50 ml conical tube, centrifuging at 1300 × g in a tabletop Sorvall centrifuge for 25 minutes at room temperature, draining the supernatant media, and resuspending the cell pellet in 2.5 ml of pre-warmed NM media containing 30 μg ml-1 chloramphenicol, 30 μg ml-1 kanamycin, and 50 μM IPTG.
78| To determine the number of total and infected cells in the resuspended cells of step 77, prepare dilutions of these cells in triplicate ranging from 10-1 to 10-6 using the method described in Box 5. Spot 5 μl of each serial dilution in triplicate (15 μl total) on LB/CK, LB/CCK/ and NM/CCKI plates. Incubate the LB/CK and LB/CCK plates for 14-18 hours at 37°C and the NM/CCKI plates for 24 hours at 37°C.
79| Plate 250 μl of the resuspended cells of step 77 onto two different NM/CCKI plates: one supplemented with 10 mM 3-AT/20 μg ml-1 streptomycin and the other with 25 mM 3-AT/40 μg ml-1 streptomycin. Spread resuspended cells on these plates using sterile glass beads (∼10-12 per plate), allow plates to dry, cover, invert, and tap the beads down onto the lid. Incubate plates (do not remove the beads) for 40-48 hours at 37°C.
80| After the plates from step 78 have completed their incubation, perform calculations using the equations below by counting from the highest dilution spots for which distinct countable colonies are visible. The total number of infected cells in 250 μl of resuspended cells from step 77 should be >2.5 × 106 (to ensure three-fold oversampling of the theoretical zinc finger library size of 953=∼8.6 × 105). The total number of cells in 250 μl of resuspended cells from step 77 should be at least three-fold more than the total number of infected cells to ensure that the vast majority of infected cells have been infected by only a single phage particle).
Total number of cells in 250 μl of resuspended cells from step 77 = (Total # of colonies from 45 μl of dilution for which distinct countable colonies are visible on LB/CK plates × 250)/(45 × dilution factor)
Total number of infected cells in 250 μl of resuspended cells from step 77 = (Total # of colonies from 45 μl of dilution for which distinct countable colonies are visible on LB/CCK plates × 250)/(45 × dilution factor)
Total number of infected cells capable of growing on NM medium in 250 μl of resuspended cells from step 77 = (Total # of colonies from 45 μl of dilution for which distinct countable colonies are visible on NM/CCKI plates × 250)/(45 × dilution factor)
? TROUBLESHOOTING
81| After the plates from step 79 have completed their incubation, harvest the highest stringency plate that has >1000 colonies. To do this, invert the plate, tap the glass beads back onto the agar, add 1.5 ml of pre-warmed NM media to the plate, gently swirl to resuspend the colonies in the medium, and transfer the cell suspension to a sterile glass tube.
82| Rescue the phagemids encoding zinc finger arrays from the cell suspension harvested in step 81 by infecting with helper phage and inducing the production of infectious phage harboring these phagemids. To do this, add enough cell suspension from step 81 (typically 25 to 150 μl) to a sterile 25 mm glass tube containing 9 ml of 2xYT media supplemented with 50 μg ml-1 carbencillin and 30 μg ml-1 kanamycin to generate a subculture with an OD600 of ∼0.1. Place tube on a roller drum (∼60 rpm) at 37°C for 1 hour. Add 1011 KTU of M13K07 helper phage (100 μl of a 109 KTU μl-1 M13K07 stock), mix well, let sit without agitation at room temperature for 30 minutes, and then place on the roller drum (∼60 rpm) at 37°C for 6 hours.
83| While the infected culture of cells from step 82 are incubating, add glycerol to a final (v/v) concentration of 15% to the remaining cell suspension of step 81 and freeze at -80°C. These cells can be used to repeat step 82 if necessary.
84| Harvest the phage supernatant from the infected culture of step 82. To do this, transfer the culture to a sterile 50 ml conical tube, pellet the cells by centrifuging at 1300 × g in a tabletop Sorvall centrifuge for 25 minutes, and pass the supernatant through a 0.22 μm PES filter. This filtered supernatant is the enriched zinc finger phage library.
PAUSE POINT: The enriched zinc finger phage library stock can be stored at 4°C for up to 1 month. After this, phage can be stored indefinitely at -80°C.
85| To perform the second stage of OPEN selection, determine the titer of the enriched zinc finger phage library stock of step 84. To prepare cells for doing this, inoculate two or three colonies of the selection strain from a freshly streaked plate (<1 week old) into a sterile 125 ml Erlenmeyer flask containing 20 ml NM medium supplemented with 30 μg ml-1 kanamycin and 30 μg ml-1 chloramphenicol. Grow the culture overnight for 16-24 hours at 37°C on an orbital platform shaker at 125 rpm.
86| Measure the density of the selection strain culture inoculated in step 85 using a spectrophotometer. This culture should have an OD600 >1.0.
87| In a sterile 18 mm glass tube, infect 500 μl of the selection strain culture inoculated in step 85 with 200 μl of the enriched zinc finger phage library from step 84 and mix well by gently swirling. Let infection proceed at room temperature for 30 minutes without agitation.
88| Add 2 ml of pre-warmed NM medium containing 30 μg ml-1 chloramphenicol and 30 μg ml-1 kanamycin to the infected cells of step 87 and incubate on a roller drum (∼60 rpm) for 1.5 hours at 37°C.
89| To determine the number of total and infected cells in the culture of step 88, prepare dilutions of these cells ranging from 10-1 to 10-6 using the method described in Box 5. Spot 5 μl of each serial dilution in triplicate (15 μl total) on LB/CK, LB/CCK and NM/CCK plates. Incubate the LB/CK and LB/CCK plates for 14-18 hours at 37°C and the NM/CCK plates for 24 hours at 37°C.
90| After the plates from step 89 have completed their incubation, perform calculations using the equations below by counting from the highest dilution spots for which distinct countable colonies are visible.
Total number of cells in the culture of step 88 = (Total # of colonies from 45 μl of dilution for which distinct countable colonies are visible on LB/CK plates × 2700)/(45 × dilution factor)
Total number of infected cells in the culture of step 88 = (Total # of colonies from 45 μl of dilution for which distinct countable colonies are visible on LB/CCK plates × 2700)/(45 × dilution factor)
Titer of the enriched zinc finger library phage stock (in carbenicillin-transducing units [CTU]/μl) = (Total # of colonies from 45 μl of dilution for which distinct countable colonies are visible on NM/CCK plates × 2700)/(45 × dilution factor × 200)
The total number of cells in the culture of step 88 should be at least three-fold the number of infected cells to ensure that the vast majority of infected cells harbor only a single zinc finger-encoding phagemid and therefore that the titer is accurate. The titer of the enriched zinc finger library phage stock must be >1×104 CTU μl-1 in order to successfully perform the next step. (Note that phage particles are measured in CTUs because the zinc finger array-encoding phagemid encodes resistance to carbenicillin.) If phage titer is not sufficiently high, it may be necessary to concentrate the phage library by PEG precipitation (Box 8).
? TROUBLESHOOTING
91| Perform the second stage of OPEN selection by infecting selection strain cells with the enriched zinc finger phage library. To do this, inoculate a colony of the selection strain from a freshly streaked plate (<1 week old) into a sterile 125 ml Erlenmeyer flask containing 20 ml NM medium supplemented with 30 μg ml-1 kanamycin and 30 μg ml-1 chloramphenicol. Grow the culture overnight for 16-24 hours at 37°C on an orbital platform shaker at 125 rpm.
92| Measure the density of the selection strain culture inoculated in step 91 using a spectrophotometer. This culture should have an OD600 >1.0.
93| In a sterile 18 mm glass tube, infect 500 μl of the selection strain culture inoculated in step 91 with 2.5 × 106 CTU of the enriched zinc finger phage library from step 84 and mix well by gently swirling. Let infection proceed at room temperature for 30 minutes without agitation.
94| Add 2 ml of pre-warmed NM medium containing 30 μg ml-1 chloramphenicol and 30 μg ml-1 kanamycin to the infected cells of step 93 and incubate on the roller drum (∼60 rpm) for 1.5 hours at 37°C.
95| To determine the number of total and infected cells in the culture of step 94, prepare dilutions of these cells ranging from 10-1 to 10-6 using the method described in Box 5. Spot 5 μl of each serial dilution in triplicate (15 μl total) on LB/CK, LB/CCK and NM/CCK plates. Incubate the LB/CK and LB/CCK plates for 14-18 hours at 37°C and the NM/CCK plates for 24 hours at 37°C.
96| Plate the remainder of the culture of step 94 on a 245 mm square NM/CCK plate containing gradients of 3-AT (0 to 80 mM) and streptomycin (0 to 100 μg ml-1). Gradient plates are poured as described in Box 9. Record the volume of culture plated (this will be required for calculations performed in step 97). Incubate the gradient plate for 4 days at 37°C.
97| After the titer plates from step 95 have completed their incubation, perform calculations using the equations below by counting from the highest dilution spots for which distinct countable colonies are visible.
Total number of cells on the gradient plate = (Total # of colonies from 45 μl of dilution for which distinct countable colonies are visible on LB/CK plates × volume in μl of culture plated in step 96)/(45 × dilution factor)
Total number of infected cells on the gradient plate = (Total # of colonies from 45 μl of dilution for which distinct countable colonies are visible on LB/CCK plates × volume in μl of culture plated in step 96)/(45 × dilution factor)
Total number of infected cells on the gradient plate that are able to grow on NM plates = (Total # of colonies from 45 μl of dilution for which distinct countable colonies are visible on NM/CCK plates × volume in μl of culture plated in step 96)/(45 × dilution factor)
The total number of cells in the culture of step 94 should be at least three-fold the number of infected cells to ensure that the vast majority of infected cells harbor only a single zinc finger-encoding phagemid. The total number of infected cells plated on the gradient plate (and calculated based on the titers from the NM/CCK plate) should be >2.5 × 106.
? TROUBLESHOOTING
98| After the gradient plate of step 96 has completed its 4 day incubation, pick 6-12 colonies from the most selective/stringent region of the plate (Fig. 8) and inoculate into 4 ml LB medium containing 50 μg ml-1 carbenicillin. Grow cultures overnight and then isolate plasmids with the QIAprep Spin Miniprep Kit using the manufacturer's instructions but with the following modifications: perform two serial washes with PB buffer instead of one and elute with 0.1× EB that is pre-warmed to 65°C.
Figure 8. Appearance of a typical gradient selection plate with a successful OPEN selection.
The density of bacterial colonies (beige circles) on the gradient plate (grey square) ranges from confluent (on the end of the plate containing the lowest concentrations of selective agents) to sparse (on the end of the plate with maximal concentrations of selective agents). We typically pick colonies from the end of the plate where the colony density is most sparse.
PAUSE POINT: Plasmid DNA can be stored indefinitely at -20°C.
99| Sequence the plasmids isolated in step 98 using primer OK61 to determine the amino acid residues selected in the recognition helices of the finger arrays. OK61 anneals just upstream of and fires toward the sequence encoding the zinc finger array. The resulting sequences can be aligned with the sequence in Supplementary Fig. 6 entitled “XbaI-BamHI three-finger array” – note that the triplet codons encoding randomized positions of the recognition helices are shown as a series of Ns in this file. The sequences of the recognition helices in the selected clones will typically resemble one another closely and one or more consensus sequences can be observed at each position.
Constructing B2H reporter strains (steps 100-111 below are reproduced with minor changes from steps 31-41 of Wright et al., 200676)
100| Digest the single-copy vector pBAC-lacZ with BsaI restriction enzyme using the following conditions:
| Component | Amount | Final Concentration |
| pBAC-lacZ plasmid DNA | 1 μg | 50 ng μl-1 |
| 10× NEBuffer 3 | 2 μl | 1× |
| BsaI (10 U μl-1) | 0.5 μl | 0.5 U μl-1 |
| Nuclease-free water | to 20 μl | |
101| Purify the ∼11,100 bp vector backbone by electrophoresis through a 5% polyacrylamide gel. Elute and ethanol precipitate this purified vector as described in Box 2.
CRITICAL STEP: BsaI digests should be incubated at 50° C.
PAUSE POINT: Purified DNA can be stored indefinitely at -20° C.
102| Incubate purified, BsaI-digested pBAC-lacZ vector backbone with Pfu polymerase in the presence of the dCTP only (i.e.--omit dATP, dGTP, and dTTP) at 72° C for 10 minutes using the conditions listed below. The combined 5′ to 3′ polymerase activity and 3′ to 5′ exonuclease activity of Pfu will generate the extended overhangs as previously described.76
| Component | Amount | Final Concentration |
| BsaI-digested pBAC-lacZ vector backbone | 10 μl | ∼25 ng μl-1 |
| 10× Pfu Buffer | 2 μl | 1× |
| dCTP nucleotide (10 mM) | 2 μl | 1 mM |
| Cloned Pfu polymerase (2.5 U μl-1) | 1.2 μl | 1.25 Uμl-1 |
| Nuclease-free water | 4.8 μl | |
| Total | 20 μl | |
103| For each target binding site, ligate the pBAC-lacZ vector backbone from step 102 to annealed oligonucleotides from step 13. As a control, perform a ligation with the pBAC-lacZ vector backbone only. Detailed conditions are provided below. Ligations are incubated for 15 minutes at room temperature.
| Component | Amount | Final Concentration |
| BsaI- and Pfu-treated pBAC-lacZ vector | 2 μl | ∼2.5 ng μl-1 |
| Annealed binding site oligos (or water for control) | 8 μl | 19 nM |
| 2× Quick Ligase Buffer | 10 μl | 1× |
| T4 DNA Ligase (400 U μl-1) | 1 μl | 19 U μl-1 |
| Total | 21 μl | |
104| Transform ligations into E. coli strain Transformax EPI300 using standard chemical transformation. To do this, add 200 μl of chemically competent cells (prepared as described in Box 3) to each ligation. Leave on ice for 5 minutes, heat shock at 42°C for 2 minutes, return immediately to ice for 2 minutes, add 700 μl LB media, and incubate on a LabQuake shaker/rotisserie at 37°C for 40 minutes. Plate transformations on LB plates supplemented with 12.5 μg ml-1 chloramphenicol and incubate overnight (14-18 hours) at 37° C.
105| Inspect and count colonies on the transformation plates from step 104. If the transformation of a ligation yields at least three-fold more colonies than that of the control ligation, we consider that ligation to be successful. From each actual successful ligation/transformation plate, inoculate two 3 ml cultures of LB containing 12.5 μg ml-1 chloramphenicol and grow overnight (14-18 hours) at 37° C for isolating plasmid DNA.
106| Inoculate 1 ml of each overnight culture into 9 ml of LB containing 14 μg ml-1 chloramphenicol and 1.1 mM arabinose. Grow cultures for 6 hours at 37° C and then harvest cells and isolate plasmid DNA using a QIAprep Spin Miniprep Kit.
CRITICAL STEP: Subculturing the cells containing the plasmids in arabinose is critical to induce expression of the trfA gene product, which in turn increases the copy number of the pBAC-lacZ plasmid.
PAUSE POINT: Plasmids can be stored indefinitely at -20° C.
107| Digest the candidate reporter plasmids (and the original pBAC-lacZ plasmid as a control) with EcoRI and HindIII using the conditions detailed below and visualize the digestions on a 5% polyacrylamide gel. Candidates that have taken up the annealed oligonucleotides should yield fragments of sizes 159 and 10,970 base pairs whereas the original pBAC-lacZ plasmid should yield fragments of sizes 182 and 10,973 base pairs.
| Component | Amount | Final Concentration |
| Plasmid DNA | 5 μl (∼0.75 μg) | ∼25 ng /μl-1 |
| 10× EcoRI Buffer | 3 μl | 1× |
| EcoRI (20 U μl-1) | 1 μl | 0.66 U μl-1 |
| HindIII (20 U μl-1) | 1 μl | 0.66 U μl-1 |
| Nuclease-free water | 20 μl | |
| Total | 30 μl | |
108| Confirm the sequence of the target DNA binding site in the candidate reporter plasmid by sequencing with primer OK163 (this primer is an anti-sense primer that anneals ∼150 bp downstream of the binding site). We will refer to the sequence-confirmed reporter plasmid as the pBAC-ZFBS-lacZ plasmid (ZFBS = Zinc Finger Binding Site) in subsequent steps of the protocol.
109| Transform strain KJBAC1 (which constitutively expresses high levels of lac repressor) with each pBAC-ZFBS-lacZ reporter plasmid. Plate transformations on LB plates containing 12.5 μg ml-1 chloramphenicol and incubate overnight at 37° C.
110| From each successful transformation of step 109, inoculate a single colony into a 3 ml LB culture supplemented with 12.5 μg ml-1 chloramphenicol. Grow cultures overnight (12-16 hours) with agitation at 37° C.
111| Make chemically competent cells from each of the overnight cultures of step 110 as described in Box 4. Also prepare glycerol stocks of each of the B2H reporter strains for long-term storage at -80° C.
PAUSE POINT: Competent B2H reporter strain cells can be stored and frozen indefinitely at -80° C.
Quantitative B2H assay of OPEN-selected zinc finger arrays
112| Prepare the zinc finger-encoding plasmid minipreps isolated in step 98 for use in transformation of their cognate B2H reporter strain by digestion with PstI overnight at 37°C as tabulated below. This digestion removes the pAC-alphaGal4 plasmid that is isolated together with the desired zinc finger-encoding plasmid.
| Component | Amount | Final Concentration |
| Miniprep DNA (of step 98) | 3 μl | ∼15 ng μl-1 |
| 10× NEBuffer 3 | 3 μl | 1× |
| 10× BSA (1 mg ml-1) | 3 μl | 1× |
| Nuclease-free water | 19 μl | |
| PstI (20 U μl-1) | 2 μl | 1.3 Uμl-1 |
| Total | 30 μl | |
113| Inactivate the PstI enzyme in the reactions of step 112 by heating at 80°C for 20 minutes.
PAUSE POINT: PstI-digested DNA can be stored indefinitely at -20°C.
114| Perform a double plasmid transformation of each B2H reporter strain prepared in step 111. To do this, add 5 μl (∼75 ng) of the PstI-digested DNA of step 101 and 150 ng of the pAC-KAN-alphaGal4 plasmid to 50 μl of chemically competent B2H reporter strain prepared in step 111, keep on ice for 10 minutes, heat shock at 42°C for 2 minutes, return immediately to ice for 2 minutes, add 175 μl of LB medium and shake or agitate at 37°C for 75 minutes. To obtain well-isolated colonies, make serial 10-1 and 10-2 dilutions and spot 5 μl of the undiluted and 10-1 and 10-2 diluted transformations in triplicate on a LB agar plate supplemented with 100 μg ml-1 carbenicillin, 12.5 μg ml-1 chloramphenicol and 30 μg ml-1 kanamycin. Also perform an additional transformation of each B2H reporter strain in parallel using 10 ng of the pBR-UV5-GP-FD2 plasmid and 150 ng of the pAC-KAN-alphaGal4 plasmid (this serves as a control lacking expression of a zinc finger array). Incubate these plates overnight at 37°C.
115| Check the transformation plates of step 114 for growth of colonies.
PAUSE POINT: These plates may be stored for up to a week at 4°C.
? TROUBLESHOOTING
116| Using the colonies obtained from the plates of step 115, perform β-galactosidase assays as described in Box 10. Zinc finger arrays that mediate three-fold activation of lacZ expression relative to control cells that do not express a zinc finger array have a high probability of functioning as ZFNs in human and zebrafish cells.11, 20, 32, 33 Occasionally, we will observe zinc finger arrays that cause toxicity in E. coli cells. Typically, this will manifest itself as the inability to sub-culture cells for β-galactosidase assay.
? TROUBLESHOOTING
117| Clone candidate zinc finger arrays selected by OPEN into ZFN expression vectors. DNA fragments encoding zinc finger arrays can be conveniently excised by digestion with XbaI and BamHI and cloned into various Zinc Finger Consortium ZFN expression vectors for human, zebrafish, and plant cells (Table 5).
Table 5. Zinc Finger Consortium vectors for expression of zinc finger arrays as ZFNs.
| Plasmid name | For expression in… | FokI cleavage domain type |
|---|---|---|
| pST1374a | mammalian cells, zebrafish (RNA) | wild-type |
| pMLM290b | mammalian cells, zebrafish (RNA) | “+” obligate heterodimer15 |
| pMLM292b | mammalian cells, zebrafish (RNA) | “−” obligate heterodimer15 |
| pDW1775a | plants | wild-type |
Zinc finger arrays selected by OPEN can be directly cloned into the ZFC expression plasmids listed using unique XbaI and BamHI sites. The resulting plasmids express a ZFN with a linker of sequence LRGS between the last conserved histidine of the carboxy-terminal zinc finger and the first amino acid of the FokI nuclease domain.
Available through Addgene (http://www.addgene.org/zfc)
Available by request through the Joung lab
● TIMING
Steps 1-10, Identification of ZFN target sites using ZiFiT software: <1 d
Steps 11-37, Construction of B2H Selection Strains: ∼12-14 d
Steps 38-71, Construction of Combinatorial Zinc Finger Libraries: ∼6-8 d
Note: Steps 11-37 and 38-71 can be performed in parallel
Steps 72-99, Performing OPEN Selections: ∼11-13 d
Steps 100-111, Construction of B2H Reporter Strains: ∼7-9 d
Note: Steps 72-99 and 100-111 can be performed in parallel
Steps 112-117, Quantitative B2H assay of OPEN-selected zinc finger arrays, 3 d
? TROUBLESHOOTING
Troubleshooting advice can be found in Table 6.
Table 6. Troubleshooting table.
| Step(s) | Problem | Possible Reason | Solution |
|---|---|---|---|
| 6 | No ZFN target sites identified | Insufficient length of sequence to identify an OPEN target site | Consider expanding the DNA search length |
| 6 | Too many ZFN target sites identified | Consider using only sites that contain two or more GNN subsites | |
| 16, 58 | No transformants obtained from ligation | Low efficiency of transformation | Consider re-making competent cells or purchasing commercially available competent cells. |
| 16, 58 | No transformants obtained from ligation | Insufficient concentration of vector backbone or insert | Check DNA concentrations using 260 nm absorbance readings on a spectrophotometer. |
| 16, 58 | Too many transformants from vector backbone control ligation | Incomplete digestion of vector backbone | Screen additional colonies from the actual ligation/transformation plate or consider re-digesting vector using higher concentration of enzyme or less DNA |
| 26 | Number of colonies on TK and TKS plates are the same | sacB gene in the reporter plasmid acquired a mutation | Repeat steps 20-26 using another plasmid candidate |
| 36 | Failure to obtain transformants | Low efficiency of transformation | Consider re-making competent cells (step 35) |
| 36 | Failure to obtain transformants | Insufficient amount of pAC-alphaGal4 plasmid used for transformation | Use larger amount of DNA for transformation and/or plate a larger portion of the transformation |
| 43 | Failure to obtain PCR product on gel | PCR reaction was unsuccessful | Verify that you are using Expand PCR enzyme and that PCR cycling conditions are correct |
| 43 | Failure to obtain PCR product on gel | Low amounts of DNA template in PCR reaction | Verify that the pools (obtained from the Joung lab) were fully resuspended by checking DNA concentration on a spectrophotometer. |
| 65 | Number of transformants from pre-amplification quantification is too low | Low efficiency of transformation | Consider re-making electrocompetent cells or purchasing commercially available electrocompetent XL1-blue cells. |
| 65 | Number of transformants from the positive control is adequate but number of library transformants is too low | Insufficient quantity of library DNA | Repeat step 60 with remaining half of ligation and repeat electroporation. If this fails, try repeating the ligation. |
| 71 | Titers show that <50% of cells were infected by helper phage | Titer of helper phage was too low to successfully superinfect the culture | Repeat steps 66-71 using a larger volume of helper phage or consider re-making high-titer helper phage stock. |
| 74 | Failure of selection strain culture to grow to correct density | Colonies on plate were older than 1 week | Re-streak strain and repeat innoculation with fresh colonies |
| 74 | Failure of selection strain culture to grow to correct density | Glassware used for innoculation contained residual soap detergent | Thoroughly rinse glassware with distilled water before autoclaving to remove any residual detergent. |
| 74 | Failure of selection strain culture to grow to correct density | Insufficient colony dispersal during innoculation | Repeat innoculation making sure to completely disperse colony in media |
| 74 | Failure of selection strain culture to grow to correct density | Incorrect composition of NM media | Verify that M9 salt stock is actually 10× (not 5× as described by recipe on the bottle supplied by Difco) |
| 74 | Failure of selection strain culture to grow to correct density | Incorrect composition of NM media | Verify that glucose used was obtained from Mallinckrodt-Baker |
| 74 | Failure of selection strain culture to grow to correct density | Incorrect composition of NM media | Verify composition of all other NM reagents |
| 74 | Failure of selection strain culture to grow to correct density | Inability of strain to grow in NM media | Go back to the “B” candidate from step 35, repeat step 36 and proceed with selections using this strain. |
| 80 | Total number of cells in 250 μl of resuspended cells from step 77 is not at least three-fold more than the total number of infected cells. | Too much phage library was used for the selection | Repeat steps 72-80 using an appropriately decreased number of CTUs (carbenicillin-transducing units) |
| 80 | The total number of infected cells in 250 μl of resuspended cells from step 77 is <2.5 × 106 | Too little phage library was used for the selection | Repeat steps 72-80 using an appropriately increased number of CTUs (carbenicillin-transducing units) |
| 90 | Titer of the enriched zinc finger library phage stock is not >1×104 ATU/μl | Too little phage library was used for the selection | Concentrate the enriched zinc finger library phage stock and re-titer the concentrated stock as described in Box 8. |
| 97 | The total number of cells in the culture of step 94 is less than three-fold the number of infected cells | Too much enriched zinc finger library phage was used for infection | Check calculation of enriched zinc finger library phage titer. Consider repeating steps 85-90 to obtain an accurate titer. |
| 97 | The total number of infected cells plated on the gradient plate is <2.5 × 106 | Too little enriched zinc finger library phage was used for infection | Check calculation of enriched zinc finger library phage titer. Consider repeating steps 85-90 to obtain an accurate titer. |
| 97 | The total number of infected cells plated on the gradient plate is <2.5 × 106 | Too little enriched zinc finger library phage was used for infection | Consider PEG precipitation to concentrate enriched zinc finger library (Box 8). |
| 97 | The total number of infected cells plated on the gradient plate is <2.5 × 106 | Too little enriched zinc finger library phage was used for infection | If colonies appear to be growing on gradient plate, proceed with protocol despite the fact that the numbers were not high enough to three-fold oversample the library. |
| 115 | Failure to obtain colonies | One of the recognition helix sequences selected in the zinc finger expression plasmid contains a PstI site | Isolate zinc finger-encoding plasmid from pAC-alphaGal4 plasmid by an alternative method. Dilute undigested plasmid isolated in step 98 by 100-fold in water and transform into chemically competent XL-1 Blue cells. Patch streak transformants onto LB/TC and LB+30mg/ml chloramphenicol plates. Pick candidates that fail to grow on LB+30mg/ml chloramphenicol plates (as these were transformed only by the zinc finger expression plasmid and not the pAC-alpha-Gal4 expression plasmid) and isolate miniprep DNA. Repeat steps 114-116. |
| 116 | Failure to obtain zinc finger arrays that mediate at least 3-fold activation | Target site may not be suitable for OPEN | Consider other targets |
| 116 | Failure to obtain zinc finger arrays that mediate at least 3-fold activation | High basal level of transcription | If the absolute ß-galactosidase activity units for the Gal11P control are particularly high, consider proceeding with zinc finger arrays that activate >2-fold. |
Boxes
Box 1
Designing oligonucleotides to create the target DNA-binding site for the B2H reporter vector (reproduced from Box 2 of Wright et al., 200676); TIMING: <1 d
-
In the double-stranded DNA fragment shown in Fig. 4, replace the nine bp XXXXXXXXX sequence with the nine bp target site of the three-finger protein to be tested. Replace the single bases (x) that flank the nine bp XXXXXXXXX sequence with the bases that flank the target site in the genomic sequence.
CRITICAL STEP: Note that the XXXXXXXXX site is positioned so that the F3 triplet is on the “left” and the F1 triplet is on the “right” in the double-stranded fragment shown in Fig. 4. Replacement of the flanking “x” bases with the bases present in the genomic target sequence is important because the identities of these adjacent bases can influence binding of the multi-finger domain to the target site.
Place an order for oligonucleotides corresponding to the first 29 bases of the top strand of the fragment (starting from the 5′ end) and to the first 26 bases of the bottom strand (starting from the 5′ end) (Fig. 4).
Box 2
Purification of DNA from a polyacrylamide gel slice; TIMING: 1 d
Crush the excised gel slice in a microfuge tube using a thin metal spatula.
Add 700 μl of ammonium acetate elution buffer, seal the tube shut by wrapping Parafilm around the top of the tube, and elute the DNA overnight at 37°C with agitation on a LabQuake shaker/rotisserie.
The next morning, spin the microfuge tube at 16,000 × g in a microcentrifuge for 2 minutes to pellet the acrylamide fragments. Transfer the supernatant (∼600 μl) to a fresh microfuge tube being careful to avoid the acrylamide pellet.
Spin the supernatant at 16,000 × g in a microcentrifuge for 2 minutes. Transfer 475 μl of the supernatant to a fresh microfuge tube. Add 950 μl of 100% ethanol and mix well. Place on dry ice for at least 15 minutes.
Spin the frozen sample at 16,000 × g in a microcentrifuge for 15 minutes at room temperature. Immediately after the spin ends, remove the supernatant being careful not to dislodge or disturb the pellet.
Add 500 μl of 70% (v/v) ethanol to the tube. Invert gently several times. Spin at 16,000 × g in a microcentrifuge for 5 minutes. Remove most of the supernatant with a larger 1000 μl tip being careful to avoid the pellet. Re-spin the tube briefly and then use a 200 μl pipet tip to remove all remaining supernatant.
-
Air dry the pellet at room temperature for 10 minutes. Resuspend the pellet in 20 μl of nuclease-free water by gently pipeting up and down until the pellet is completely dissolved.
PAUSE POINT: Purified DNA fragments may be stored indefinitely at -20°C.
Box 3
Preparation of highly competent bacterial cells for chemical transformation; TIMING: 3 d
Streak out cells (from frozen glycerol stock) on a LB plate with appropriate antibiotic and grow overnight at 37°C.
Autoclave 1 liter of LB medium in a 2 liter baffled Erlenmeyer flask.
The next day, pick a single, well-isolated colony from the plate of step 1 and start a 10ml overnight culture in LB medium containing any appropriate antibiotics or chemical inducers. Incubate the culture overnight at 37°C on a roller drum (∼60 rpm).
Add 15 ml of 1M MgCl2 to the LB prepared in step 2 and shake at 37°C, 250 rpm overnight to check for any undesired contamination.
The next day, add any appropriate antibiotics or inducers to the LB medium prepared in step 4 (if the medium appears to be free of any bacterial contamination). Inoculate 1 ml of the overnight culture of step 3 into the LB medium and incubate at 37°C with agitation on an orbital shaker (250 rpm).
Monitor the density of the culture of step 5 by taking OD600 readings on a spectrophotometer. The first reading should be taken at approximately 2 hours and subsequent readings should be taken every 15-45 minutes depending on the growth rate of the culture.
After the culture has reached an OD600 between 0.4-0.6, pour the culture into a sterile 1 liter centrifuge bottle and pellet the cells by centrifuging at 4°C, 2700 × g for 30 minutes.
Decant the media and resuspend the cell pellet in 300 ml of ice-cold Solution A. Perform this resuspension step on ice and then incubate the resuspended cells on ice for 20 minutes.
Centrifuge resuspended cells at 4°C, 2700 × g for 30 minutes and then decant the supernatant.
Resuspend the remaining cell pellet in 60 ml of ice-cold Solution A with 15% (v/v) glycerol.
Make 500 μl and/or 1 ml aliquots of the cell suspension in sterile 1.5 ml microfuge tubes and freeze on dry ice.
-
Store competent cells at -80°C.
PAUSE POINT: Competent cells may be stored indefinitely at -80° C.
Box 4
Preparing competent bacterial cells for chemical transformation (reproduced with minor alterations from Box 3 of Wright et al., 200676); TIMING: 2 d
Inoculate a single colony of the bacterial strain to be made competent into a 2 ml culture of LB supplemented with any appropriate antibiotics. Grow overnight at 37°C with agitation.
The next morning, subculture 1 ml of the saturated overnight culture from step 1 into 50 ml LB supplemented with any appropriate antibiotics and 15 mM MgCl2 in a 250 ml flask. Grow for 1 hour with agitation at 37° C or until cells reach log phase growth (an OD600 of ∼0.4-0.6).
Pellet cells in a sterile 50 ml conical tube by centrifuging at 3000-6000 × g in a tabletop centrifuge.
Pour off media and resuspend cell pellet in 3 ml of ice-cold Solution A with 15% glycerol.
Leave sitting on ice for 20 minutes and then aliquot into sterile microcentrifuge tubes.
-
Snap freeze the tubes on dry ice
PAUSE POINT: Competent cells may be stored indefinitely at -80° C.
Box 5
Serial dilution of bacterial cultures; TIMING: <1 d
For each sample to be diluted, place 100 μl of undiluted culture in a well in the top row of a sterile, flat-bottom, 96-well microtiter plate (Fig. 6).
In the wells below each sample, add 90 μl of media. For each desired dilution, fill a row of wells with media. For example, if one wishes to create 6 dilutions, fill the six rows below each undiluted sample with media.
Using a multi-channel pipet, transfer 10 μl of the undiluted samples from the top row of the plate to the second row of wells (containing media). Mix thoroughly but gently to avoid splashing and excessive bubble formation. Discard the tips from the multi-channel pipet.
Repeat the actions of step 3 to transfer samples from the second row of the plate to the third row of the plate. Continue this process until the last row of wells is used on the plate.
Box 6
Preparation of competent XL-1 Blue bacterial cells for transformation by electroporation; TIMING: 5-7 d
Streak XL-1 Blue cells from a glycerol stock onto a LB plate supplemented with 12.5 μg ml-1 tetracycline.
The next day, pick a single colony and start a 10 ml overnight culture in LB with 12.5 μg ml-1 tetracycline. Grow the culture overnight on a roller drum (∼60 rpm) or orbital shaker (∼250 rpm) at 37°C.
The following morning, subculture 1 ml of overnight culture into a 2 liter baffled flask containing 1 liter of 2xYT media with 12.5 μg ml-1 tetracycline.
Shake culture at 18°C on an orbital platform shaker at 250 rpm. After 24 hours of shaking, add an additional 1 ml of tetracycline stock (12.5 mg ml-1).
After culture has been shaking for a total of approximately 48 hours, check the density by measuring the OD600 on a spectrophotometer.
Continue to check the density of the culture until the OD600 is between 0.4 and 0.6.
Harvest the cells by transferring to a sterile 1 liter centrifuge bottle and centrifuging for 20 minutes at ∼4,800 × g, 4°C.
Decant the media supernatant and resuspend the cell pellet in 425 ml ice-cold, autoclaved water. Perform this resuspension on ice. In order to ensure that all solutions remain ice-cold, we also perform all resuspension steps in the cold room. Transfer the resuspended cells to a sterile 500 ml centrifuge bottle and centrifuge for 20 minutes at ∼7,500 × g, 4°C.
Decant supernatant immediately after centrifugation is completed (the cell pellet is very loose and disperses quickly following centrifugation). Resuspend the cell pellet in 210 ml ice-cold, autoclaved water. Perform resuspension on ice. Transfer resuspended cells to a sterile 250 ml centrifuge bottle and centrifuge for 20 minutes at ∼8,600 × g, 4°C.
Decant supernatant immediately after centrifugation and resuspend the cell pellet in 105 ml of ice-cold, filter-sterilized 10% (v/v) glycerol. Perform resuspension on ice. Centrifuge resuspension for 20 minutes at ∼10,500 × g, 4°C
Decant supernatant immediately after centrifugation and resuspend the cell pellet in 1 ml of sterile, ice-cold 10% (v/v) glycerol.
Make small aliquots (∼150 μl each) of the resuspension in pre-chilled, sterile microfuge tubes and freeze on dry ice. Store competent cells at -80°C.
Test the competency of cells by electroporating 1 μg of pBR-UV5-GP-FD2 plasmid DNA into 70μl of electrocompetent cells. Also perform a negative control electroporation by using 70 μl of cells and no DNA. For the electroporations, use a 1 mm gap cuvette and with the following settings: 1.75 kV, 200 Ohms, and 25 μF. Pre-chill all cuvettes on ice and perform all electroporation steps in the cold room. Immediately following application of current, add 1 ml of SOC medium to the cuvette and then transfer cells to 9 ml of 2xYT media in a sterile 25 mm glass culture tube. Allow the cells to recover by incubating them on a roller drum (∼60 rpm) at 37°C for 1 hour.
Determine the number of transformants by preparing dilutions of each electroporation in triplicate from 10-1 to 10-6 using the method described in Box 5. Spot 5 μl of each serial dilution in triplicate (15 μl total) on LB/TC plates. A set of six dilutions can be conveniently spotted in triplicate on half of a plate (Fig. 6). Incubate plates overnight at 37°C.
Calculate the total number of transformants from the plates of step 14. Total number of transformants in the 10 ml culture of step 14 should be in the range of 5 × 108 to 5 × 109.
Box 7
Protocol for preparing high-titer stock of M13K07 helper phage; TIMING: ∼4-5 d
<!>CAUTION Care should be taken to avoid contaminating laboratory equipment and benches with bacteriophage. Use filtered pipet tips and pipets, dedicated tube racks, and change bench paper frequently.
From a glycerol stock, streak out XL-1 Blue cells on a LB plate supplemented with 12.5 mg ml-1 tetracycline. Incubate plate overnight at 37°C.
The next day, inoculate a well-isolated colony of XL-1 Blue into a sterile 25 mm glass tube containing LB medium supplemented with 12.5 mg ml-1 tetracycline. Incubate this inoculated culture on a roller drum (∼60 rpm) overnight at 37°C.
The next morning, melt a bottle of H Top Agar in the microwave and equilibrate the temperature of this molten agar to ∼60-65°C.
Dip the tip of a sterile loop or a sterile wooden stick into a commercially available stock of M13K07 and serially streak this tip with a side-to-side motion and down the surface of one LB plate supplemented with 12.5 mg ml-1 tetracycline and then another second plate. This process creates a progressive dilution of the phage across the surfaces of these plates.
Thoroughly mix together 7 ml of molten H Top Agar (∼60°C) with 0.5 ml of the XL-1 Blue culture of step 2 and plate 3.75 ml onto each of the phage-streaked plates of step 4. When depositing the molten agar-bacteria mix, place it on the end of the plate that should contain the more dilute streaks of phage and then tilt the plate slightly to allow the molten agar to cover the entire plate. After the top agar has hardened (typically ∼2 minutes), incubate the plates face-up at 37°C overnight.
In the morning, use a sterile spatula to scoop off a region of the top agar (∼5 to 25 cm2) which contains dozens of well-isolated phage plaques and inoculate this into a baffled 2 liter flask containing 500 ml of 2xYT media supplemented with 12.5 mg ml-1 tetracycline. Shake on an orbital shaker at 250 rpm, 37°C for 2 hours.
Add 1.17 ml of 30 mg ml-1 kanamycin stock to the culture of step 6 to achieve a final concentration of 70 μg ml-1. Continue to shake the culture at 250 rpm, 37°C for an additional 22 hours.
-
Harvest the supernatant of the culture of step 7 by transferring the culture to a sterile 1 liter centrifuge bottle and spinning the cells at ∼2700 × g. Remove residual cells by filtering the culture supernatant through a 0.2 μm PES filter unit.
PAUSE POINT: Filtered phage stock can be stored for up to a month at 4°C.
Concentrate the phage by PEG precipitation. Divide the 500 ml of phage stock from step 8 into four sterile 250 ml centrifuge bottles (i.e.--125 ml of phage in each bottle). Add 31.25 ml of 5× PEG/NaCl solution to each bottle, seal the cap tightly, and mix thoroughly by inverting each bottle several times. Incubate on ice for at least 2.5 hours. Spin the bottles at ∼8600 × g in a Sorvall centrifuge, pour off and discard the supernatant. Resuspend phage pellets from the four bottles in a total of 5 ml of 2xYT with 15% (v/v) glycerol. Make 500 μl aliquots in 2 ml cryovials and store at -80°C.
Titer the phage stock of step 9 by serially diluting the stock in triplicate from 10-1 to 10-8 in 2xYT and using 10 μl of each dilution to infect 50 μl of XL-1 Blue cells from a fresh overnight culture. Allow the infection to proceed at room temperature for 20 minutes without shaking. Add 190 μl of 2xYT to each infection and incubate without shaking at 37°C for 90 minutes. Spot 5 μl of each infection in triplicate on 2xYT plates supplemented with 70 μg ml-1 kanamycin. Incubate plates overnight at 37°C.
Calculate the titer of the phage stock by counting colonies from the most dilute phage infections that yield distinct colonies.
Box 8
PEG precipitation of phage library stocks; TIMING: ∼1-2 d
In a sterile 2 ml microfuge tube, combine 1.44 ml of phage library with 360 μl of 5× PEG/NaCl solution and mix well. Set up four such tubes for each phage library.
Incubate the tubes on ice for a minimum of 2.5 hours.
Spin the tubes at 4°C in a microcentrifuge at 16,000 × g for 30 minutes. Mark tubes so that you know where the pellet should be because the phage precipitate will not be visible.
Immediately following completion of the spin, carefully remove the supernatant from each tube using a 1000 μl tip and resuspend the 4 pellets for each phage library in a total of 500 μl of 2xYT media containing 15% (v/v) glycerol.
-
Titer the concentrated phage library as described in steps 85-90, except use only 100 μl of phage for infection (instead of 200 μl). The equation for calculating the titer of the phage library is:
Titer of the enriched zinc finger library phage stock (in carbenicillin-transducing units [CTU] μl-1) = (Total # of colonies from 45 μl of dilution for which distinct countable colonies are visible on NM/CCK plates × 2600)/(45 × dilution factor × 100)
PAUSE POINT: Concentrated phage stock can be stored for up to a month at 4°C. For longer-term storage, keep the phage stock at -80°C.
Box 9
Pouring gradient selection plates; TIMING: <1 d
Make two batches of NM/CCK agar. For each gradient plate to be poured, ∼125 ml of agar from each batch will be required.
To one of the agar batches, add 3-AT to a final concentration of 80 mM and streptomycin to a final concentration of 100 μg ml-1.
Place 245mm square plates at about a 5° angle from the bench top as depicted in Fig. 7.
Using a 100ml pipet, pipet 120ml of the NM agar containing 3-AT and streptomycin onto the plates, making sure to cover the entire surface (Fig. 7). Keep plates at an angle until agar has completely hardened (approximately 15 minutes).
Lay plates flat and pipet 120ml of the NM agar which lacks 3-AT and streptomycin onto each plate, making sure to completely cover the surface (Fig. 7).
Allow plates to sit at room temperature overnight and then store with lids down at 4°C. Gradient plates should be used within 2 days because the gradient will dissipate over time.
Box 10
Performing beta-galactosidase assays of B2H reporter strain cells (reproduced with minor changes from steps 43-49 of Wright et al., 200676); TIMING: 2 d
For each transformant to be assayed, inoculate three overnight cultures (each from an independent colony) in LB containing 50 μg ml-1 carbenicillin, 30 μg ml-1 kanamycin, 12.5 μg ml-1 chloramphenicol, 10 μM ZnSO4, and 500 μM IPTG. Grow overnight (14-18 hours) with agitation at 37° C. Cultures can be grown either in standard sterile glass or plastic culture tubes or in 96 well blocks. For growth in deep well blocks, we recommend shaking on a platform with a sufficiently small throw radius to ensure uniform and adequate agitation of each well in the block (e.g.--the Microtitertron orbital shaker, Appropriate Technical Resources).77
Subculture saturated overnight cultures from step 1 by diluting them 1:40 into pre-warmed LB containing 50 μg ml-1 carbenicillin, 30 μg ml-1 kanamycin, 12.5 μg ml-1 chloramphenicol, 10 μM ZnSO4, and 500 μM IPTG. Monitor the growth of cultures by measuring OD600 (relative to a media only blank) on a spectrophotometer and harvest them for lysis when they reach log phase (OD600 = 0.3-0.8). Record the OD600 value at which cultures are lysed.
-
Lyse log-phase subcultures from step 2 in a 96-well microtiter plate by adding 100 μl of culture to 11 μl of Lysis Master Mix77 (already in the plate) and mixing well by pipetting up and down. Allow lysis to proceed for a minimum of 15 minutes at room temperature.
PAUSE POINT: The activity of β-galactosidase is stable in the cell lysates for at least 18 hours when stored at room temperature.77 Lysates should be covered to prevent evaporation.
Set up β-galactosidase assay by adding 15 μl of lysate to a microtiter plate well containing 135 μl of Z buffer with β-mercaptoethanol and 30 μl of 4 mg ml-1 ONPG and mixing well. Perform duplicate assays for each lysate.
-
Place microtiter plate containing β-galactosidase assay reactions in a microtiter plate reader with temperature control (e.g.--Biorad Model 680 Microplate Reader). Incubate reactions at 28° C and take timed serial measurements of absorbance at 420 nm. Calculate the velocity of ONPG cleavage (v) by plotting A420 vs. time and calculating the slope of the line.
CRITICAL STEP: Many microtiter plate readers can be programmed to take absorbance measurements at fixed intervals and to calculate the velocity of ONPG cleavage. We typically take measurements every 10-30 seconds. Reactions should not be allowed to proceed for more than 30 minutes as substrate can become limiting.
-
Calculate the units of β-galactosidase for each assay using the following formula:
V × 1000/(OD600)
We include the 1000 multiplier to make all units larger than 1. The final value obtained for each B2H reporter strain transformant is the average of triplicate cultures, which in turn are measured in duplicate.
CRITICAL STEP: If a microtiter plate reader is not available, the β-galactosidase assay can be performed in glass tubes as an endpoint assay using a standard spectrophotometer as previously described by Miller.77
Calculate the fold-activation mediated by each zinc finger protein tested by comparing β-galactosidase units obtained from cells expressing a zinc finger-Gal11P hybrid protein with the units obtained from control cells in which no zinc finger protein is expressed. In our experience, zinc finger domains that mediate three-fold activation or higher will have a high probability of generating active ZFNs in cell-based gene targeting assays.
Anticipated Results
A typical successful OPEN selection will yield many different zinc finger arrays that all show activity in the B2H system. Because zinc finger arrays targeted to two half-sites are required to target a full ZFN site, the identification of multiple candidates for each half-site results in a very large number of potential pairs that could be tested as ZFNs. To date, we typically have chosen one or two zinc finger arrays for each half-site and tested the small number of resulting pairs as ZFNs.11, 20 We have chosen zinc finger arrays that exhibit the highest fold-activation levels in the B2H system and that do not show toxicity when assayed for β-galactosidase activity in E. coli cells. With this approach, for any given target site, we have found either that all (or nearly all) of the ZFN pairs we test show activity or that none of the ZFN pairs show activity. We believe that this demonstrates that OPEN yields zinc finger arrays that bind their sites but that other factors can influence whether they are functional at their intended target in cells (see below). Given this experience, we suggest that investigators choose one, or at most two, zinc finger arrays for each half-site (using the criteria described above) and test the combinations of these proteins as ZFNs. If these pairs fail to work, it is unlikely that testing additional zinc finger arrays from the selections will yield a functional pair. (We note that, when expressed as ZFNs, OPEN zinc finger arrays can be fused to either wild-type or optimized heterodimeric15, 19, 78 FokI nuclease domains.)
We do not know why some zinc finger array pairs fail to show activity when expressed as ZFNs. Possible explanations include (but are not limited to): inadequate expression levels of one or more of the ZFNs, inaccessibility of the genomic ZFN target site due to chromatin, or interference with DNA-binding due to methylation of the genomic ZFN target site. Expression levels of ZFNs can be checked by Western blot analysis using epitope tags included on all Zinc Finger Consortium ZFN expression vectors. Various observations from our experience to date suggest that chromatin or methylation may be a factor in the ultimate success of a ZFN at its target gene. For example, we have found that when we have made ZFN pairs to different sequences in the same gene, sometimes some will work while others will not.11 We have also found that some ZFNs will show cell-type dependent activity (i.e.—they will work in one cell line but not in another) (S.T.-B., M.L.M., and J.K.J., unpublished data). All of these observations further strengthen the argument for targeting more than one full ZFN site for a gene or locus of interest to optimize chances of success.
Supplementary Material
Figure 4. Strategy for designing binding site oligonucleotides (figure from Wright et al., 200676).
Schematic illustrating the design of target zinc finger binding site oligonucleotides as described in Box 1.
Figure 7. Schematic illustrating a method for pouring gradient selection plates.
Each plate is first tilted at ∼5 degrees from the horizontal and a first layer of molten agar containing selective agents (dark red) is added. After this initial layer hardens, the plate is laid flat and a second layer of molten agar lacking selective agents (light red) is poured.
Acknowledgments
We thank Magdalena Eichtinger, Tao Jiang, and Shondra Pruett-Miller for technical assistance and Matthew Porteus for helpful discussions during early stages of protocol development. M.L.M., S.T.-B., and J.K.J. are supported by the National Institutes of Health (R01 GM069906, R24 GM078369, R21 RR024189) and the MGH Pathology Service. D.F.V. is supported by the National Science Foundation (DBI 0501678). J.D.S. is supported by a Pioneer-Hi-Bred International, Inc. 2008 Graduate Fellowship, the National Institutes of Health (R33 GM066387), and the Iowa State University CIAG. The authors declare competing financial interests (see the HTML version of this article for details).
References
- 1.Cathomen T, Joung JK. Zinc-finger nucleases: the next generation emerges. Mol Ther. 2008;16:1200–1207. doi: 10.1038/mt.2008.114. [DOI] [PubMed] [Google Scholar]
- 2.Porteus MH, Carroll D. Gene targeting using zinc finger nucleases. Nat Biotechnol. 2005;23:967–973. doi: 10.1038/nbt1125. [DOI] [PubMed] [Google Scholar]
- 3.Durai S, et al. Zinc finger nucleases: custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res. 2005;33:5978–5990. doi: 10.1093/nar/gki912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carroll D. Progress and prospects: Zinc-finger nucleases as gene therapy agents. Gene Ther. 2008 doi: 10.1038/gt.2008.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu J, Kandavelou K, Chandrasegaran S. Custom-designed zinc finger nucleases: what is next? Cell Mol Life Sci. 2007;64:2933–2944. doi: 10.1007/s00018-007-7206-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Camenisch TD, Brilliant MH, Segal DJ. Critical parameters for genome editing using zinc finger nucleases. Mini Rev Med Chem. 2008;8:669–676. doi: 10.2174/138955708784567458. [DOI] [PubMed] [Google Scholar]
- 7.Kim YG, Shi Y, Berg JM, Chandrasegaran S. Site-specific cleavage of DNA-RNA hybrids by zinc finger/FokI cleavage domain fusions. Gene. 1997;203:43–49. doi: 10.1016/s0378-1119(97)00489-7. [DOI] [PubMed] [Google Scholar]
- 8.Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci U S A. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mani M, Smith J, Kandavelou K, Berg JM, Chandrasegaran S. Binding of two zinc finger nuclease monomers to two specific sites is required for effective double-strand DNA cleavage. Biochem Biophys Res Commun. 2005;334:1191–1197. doi: 10.1016/j.bbrc.2005.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith J, et al. Requirements for double-strand cleavage by chimeric restriction enzymes with zinc finger DNA-recognition domains. Nucleic Acids Res. 2000;28:3361–3369. doi: 10.1093/nar/28.17.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maeder ML, et al. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bibikova M, Beumer K, Trautman JK, Carroll D. Enhancing gene targeting with designed zinc finger nucleases. Science. 2003;300:764. doi: 10.1126/science.1079512. [DOI] [PubMed] [Google Scholar]
- 13.Bibikova M, Golic M, Golic KG, Carroll D. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics. 2002;161:1169–1175. doi: 10.1093/genetics/161.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Santiago Y, et al. Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases. Proc Natl Acad Sci U S A. 2008;105:5809–5814. doi: 10.1073/pnas.0800940105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller JC, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 16.Lombardo A, et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat Biotechnol. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- 17.Perez EE, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doyon Y, et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol. 2008;26:702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat Biotechnol. 2008;26:695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foley JE, et al. Rapid Mutation of Endogenous Zebrafish Genes Using Zinc Finger Nucleases Made by Oligomerized Pool ENgineering. PLoS ONE. 2009;4:e4348. doi: 10.1371/journal.pone.0004348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lloyd A, Plaisier CL, Carroll D, Drews GN. Targeted mutagenesis using zinc-finger nucleases in Arabidopsis. Proc Natl Acad Sci U S A. 2005;102:2232–2237. doi: 10.1073/pnas.0409339102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wright DA, et al. High-frequency homologous recombination in plants mediated by zinc-finger nucleases. Plant J. 2005;44:693–705. doi: 10.1111/j.1365-313X.2005.02551.x. [DOI] [PubMed] [Google Scholar]
- 23.Porteus MH, Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- 24.Moehle EA, et al. Targeted gene addition into a specified location in the human genome using designed zinc finger nucleases. Proc Natl Acad Sci U S A. 2007;104:3055–3060. doi: 10.1073/pnas.0611478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Urnov FD, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 26.Beumer K, Bhattacharyya G, Bibikova M, Trautman JK, Carroll D. Efficient gene targeting in Drosophila with zinc-finger nucleases. Genetics. 2006;172:2391–2403. doi: 10.1534/genetics.105.052829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beumer KJ, et al. Efficient gene targeting in Drosophila by direct embryo injection with zinc-finger nucleases. Proc Natl Acad Sci U S A. 2008;105:19821–19826. doi: 10.1073/pnas.0810475105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morton J, Davis MW, Jorgensen EM, Carroll D. Induction and repair of zinc-finger nuclease-targeted double-strand breaks in Caenorhabditis elegans somatic cells. Proc Natl Acad Sci U S A. 2006;103:16370–16375. doi: 10.1073/pnas.0605633103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cai CQ, et al. Targeted transgene integration in plant cells using designed zinc finger nucleases. Plant Mol Biol. 2009;69:699–709. doi: 10.1007/s11103-008-9449-7. [DOI] [PubMed] [Google Scholar]
- 30.Townsend JA, et al. High Frequency Modification of Plant Genes Using Engineered Zinc Finger Nucleases. Nature. 2009 doi: 10.1038/nature07845. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hurt JA, Thibodeau SA, Hirsh AS, Pabo CO, Joung JK. Highly specific zinc finger proteins obtained by directed domain shuffling and cell-based selection. Proc Natl Acad Sci U S A. 2003;100:12271–12276. doi: 10.1073/pnas.2135381100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cornu TI, et al. DNA-binding Specificity Is a Major Determinant of the Activity and Toxicity of Zinc-finger Nucleases. Mol Ther. 2008;16:352–358. doi: 10.1038/sj.mt.6300357. [DOI] [PubMed] [Google Scholar]
- 33.Pruett-Miller SM, Connelly JP, Maeder ML, Joung JK, Porteus MH. Comparison of zinc finger nucleases for use in gene targeting in mammalian cells. Mol Ther. 2008;16:707–717. doi: 10.1038/mt.2008.20. [DOI] [PubMed] [Google Scholar]
- 34.Fu F, et al. Zinc Finger Database (ZiFDB): a repository for information on C2H2 zinc fingers and engineered zinc-finger arrays. Nucleic Acids Res. 2009;37:D279–283. doi: 10.1093/nar/gkn606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sander JD, Zaback P, Joung JK, Voytas DF, Dobbs D. Zinc Finger Targeter (ZiFiT): an engineered zinc finger/target site design tool. Nucleic Acids Res. 2007;35:W599–605. doi: 10.1093/nar/gkm349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Porteus MH. Mammalian gene targeting with designed zinc finger nucleases. Mol Ther. 2006;13:438–446. doi: 10.1016/j.ymthe.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 37.Alwin S, et al. Custom zinc-finger nucleases for use in human cells. Mol Ther. 2005;12:610–617. doi: 10.1016/j.ymthe.2005.06.094. [DOI] [PubMed] [Google Scholar]
- 38.Mani M, Kandavelou K, Dy FJ, Durai S, Chandrasegaran S. Design, engineering, and characterization of zinc finger nucleases. Biochem Biophys Res Commun. 2005;335:447–457. doi: 10.1016/j.bbrc.2005.07.089. [DOI] [PubMed] [Google Scholar]
- 39.Segal DJ, et al. Evaluation of a modular strategy for the construction of novel polydactyl zinc finger DNA-binding proteins. Biochemistry. 2003;42:2137–2148. doi: 10.1021/bi026806o. [DOI] [PubMed] [Google Scholar]
- 40.Segal DJ. The use of zinc finger peptides to study the role of specific factor binding sites in the chromatin environment. Methods. 2002;26:76–83. doi: 10.1016/S1046-2023(02)00009-9. [DOI] [PubMed] [Google Scholar]
- 41.Segal DJ, Barbas CF., 3rd Custom DNA-binding proteins come of age: polydactyl zinc-finger proteins. Curr Opin Biotechnol. 2001;12:632–637. doi: 10.1016/s0958-1669(01)00272-5. [DOI] [PubMed] [Google Scholar]
- 42.Mandell JG, Barbas CF., 3rd Zinc Finger Tools: custom DNA-binding domains for transcription factors and nucleases. Nucleic Acids Res. 2006;34:W516–523. doi: 10.1093/nar/gkl209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beerli RR, Barbas CF., 3rd Engineering polydactyl zinc-finger transcription factors. Nat Biotechnol. 2002;20:135–141. doi: 10.1038/nbt0202-135. [DOI] [PubMed] [Google Scholar]
- 44.Bae KH, et al. Human zinc fingers as building blocks in the construction of artificial transcription factors. Nat Biotechnol. 2003;21:275–280. doi: 10.1038/nbt796. [DOI] [PubMed] [Google Scholar]
- 45.Liu Q, Xia Z, Zhong X, Case CC. Validated zinc finger protein designs for all 16 GNN DNA triplet targets. J Biol Chem. 2002;277:3850–3856. doi: 10.1074/jbc.M110669200. [DOI] [PubMed] [Google Scholar]
- 46.Ramirez CL, et al. Unexpected failure rates for modular assembly of engineered zinc-fingers. Nat Methods. 2008;5:374–375. doi: 10.1038/nmeth0508-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wolfe SA, Grant RA, Elrod-Erickson M, Pabo CO. Beyond the “recognition code”: structures of two Cys2His2 zinc finger/TATA box complexes. Structure (Camb) 2001;9:717–723. doi: 10.1016/s0969-2126(01)00632-3. [DOI] [PubMed] [Google Scholar]
- 48.Wolfe SA, Greisman HA, Ramm EI, Pabo CO. Analysis of zinc fingers optimized via phage display: evaluating the utility of a recognition code. J Mol Biol. 1999;285:1917–1934. doi: 10.1006/jmbi.1998.2421. [DOI] [PubMed] [Google Scholar]
- 49.Elrod-Erickson M, Rould MA, Nekludova L, Pabo CO. Zif268 protein-DNA complex refined at 1.6 A: a model system for understanding zinc finger-DNA interactions. Structure. 1996;4:1171–1180. doi: 10.1016/s0969-2126(96)00125-6. [DOI] [PubMed] [Google Scholar]
- 50.Isalan M, Klug A, Choo Y. Comprehensive DNA recognition through concerted interactions from adjacent zinc fingers. Biochemistry. 1998;37:12026–12033. doi: 10.1021/bi981358z. [DOI] [PubMed] [Google Scholar]
- 51.Isalan M, Choo Y, Klug A. Synergy between adjacent zinc fingers in sequence-specific DNA recognition. Proc Natl Acad Sci U S A. 1997;94:5617–5621. doi: 10.1073/pnas.94.11.5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pearson H. The Fate of Fingers. Nature. 2008;455:160–164. doi: 10.1038/455160a. [DOI] [PubMed] [Google Scholar]
- 53.Tan S, et al. Zinc-finger protein-targeted gene regulation: genomewide single-gene specificity. Proc Natl Acad Sci U S A. 2003;100:11997–12002. doi: 10.1073/pnas.2035056100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blancafort P, et al. Genetic reprogramming of tumor cells by zinc finger transcription factors. Proc Natl Acad Sci U S A. 2005;102:11716–11721. doi: 10.1073/pnas.0501162102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blancafort P, Magnenat L, Barbas CF. Scanning the human genome with combinatorial transcription factor libraries. Nat Biotechnol. 2003;21:269–274. doi: 10.1038/nbt794. [DOI] [PubMed] [Google Scholar]
- 56.Stege JT, Guan X, Ho T, Beachy RN, Barbas CF., 3rd Controlling gene expression in plants using synthetic zinc finger transcription factors. Plant J. 2002;32:1077–1086. doi: 10.1046/j.1365-313x.2002.01492.x. [DOI] [PubMed] [Google Scholar]
- 57.Ordiz MI, Barbas CF, 3rd, Beachy RN. Regulation of transgene expression in plants with polydactyl zinc finger transcription factors. Proc Natl Acad Sci U S A. 2002;99:13290–13295. doi: 10.1073/pnas.202471899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guan X, et al. Heritable endogenous gene regulation in plants with designed polydactyl zinc finger transcription factors. Proc Natl Acad Sci U S A. 2002;99:13296–13301. doi: 10.1073/pnas.192412899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Beerli RR, Dreier B, Barbas CF., 3rd Positive and negative regulation of endogenous genes by designed transcription factors. Proc Natl Acad Sci U S A. 2000;97:1495–1500. doi: 10.1073/pnas.040552697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Beerli RR, Segal DJ, Dreier B, Barbas CF., 3rd Toward controlling gene expression at will: specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc Natl Acad Sci U S A. 1998;95:14628–14633. doi: 10.1073/pnas.95.25.14628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gordley RM, Gersbach CA, Barbas CF., 3rd Synthesis of programmable integrases. Proc Natl Acad Sci U S A. 2009;106:5053–5058. doi: 10.1073/pnas.0812502106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Blancafort P, et al. Modulation of drug resistance by artificial transcription factors. Mol Cancer Ther. 2008;7:688–697. doi: 10.1158/1535-7163.MCT-07-0381. [DOI] [PubMed] [Google Scholar]
- 63.Nomura W, Barbas CF., 3rd In vivo site-specific DNA methylation with a designed sequence-enabled DNA methylase. J Am Chem Soc. 2007;129:8676–8677. doi: 10.1021/ja0705588. [DOI] [PubMed] [Google Scholar]
- 64.Gordley RM, Smith JD, Graslund T, Barbas CF., 3rd Evolution of programmable zinc finger-recombinases with activity in human cells. J Mol Biol. 2007;367:802–813. doi: 10.1016/j.jmb.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 65.Eberhardy SR, et al. Inhibition of human immunodeficiency virus type 1 replication with artificial transcription factors targeting the highly conserved primer-binding site. J Virol. 2006;80:2873–2883. doi: 10.1128/JVI.80.6.2873-2883.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tan W, Dong Z, Wilkinson TA, Barbas CF, 3rd, Chow SA. Human immunodeficiency virus type 1 incorporated with fusion proteins consisting of integrase and the designed polydactyl zinc finger protein E2C can bias integration of viral DNA into a predetermined chromosomal region in human cells. J Virol. 2006;80:1939–1948. doi: 10.1128/JVI.80.4.1939-1948.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rebar EJ, et al. Induction of angiogenesis in a mouse model using engineered transcription factors. Nat Med. 2002;8:1427–1432. doi: 10.1038/nm1202-795. [DOI] [PubMed] [Google Scholar]
- 68.Liang Y, et al. Activation of vascular endothelial growth factor A transcription in tumorigenic glioblastoma cell lines by an enhancer with cell type-specific DNase I accessibility. J Biol Chem. 2002;277:20087–20094. doi: 10.1074/jbc.M201766200. [DOI] [PubMed] [Google Scholar]
- 69.Liu PQ, et al. Regulation of an endogenous locus using a panel of designed zinc finger proteins targeted to accessible chromatin regions. Activation of vascular endothelial growth factor A. J Biol Chem. 2001;276:11323–11334. doi: 10.1074/jbc.M011172200. [DOI] [PubMed] [Google Scholar]
- 70.Zhang L, et al. Synthetic zinc finger transcription factor action at an endogenous chromosomal site. Activation of the human erythropoietin gene. J Biol Chem. 2000;275:33850–33860. doi: 10.1074/jbc.M005341200. [DOI] [PubMed] [Google Scholar]
- 71.Handel EM, Alwin S, Cathomen T. Expanding or restricting the target site repertoire of zinc-finger nucleases: the inter-domain linker as a major determinant of target site selectivity. Mol Ther. 2009;17:104–111. doi: 10.1038/mt.2008.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bibikova M, et al. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol Cell Biol. 2001;21:289–297. doi: 10.1128/MCB.21.1.289-297.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Joung JK, Ramm EI, Pabo CO. A bacterial two-hybrid selection system for studying protein-DNA and protein-protein interactions. Proc Natl Acad Sci U S A. 2000;97:7382–7387. doi: 10.1073/pnas.110149297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Whipple FW. Genetic analysis of prokaryotic and eukaryotic DNA-binding proteins in Escherichia coli. Nucleic Acids Res. 1998;26:3700–3706. doi: 10.1093/nar/26.16.3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Thibodeau-Beganny S, Joung JK. Engineering Cys2His2 Zinc Finger Domains Using a Bacterial Cell-Based Two-Hybrid Selection System. Methods in Molecular Biology. 2007;408:317–334. doi: 10.1007/978-1-59745-547-3_17. [DOI] [PubMed] [Google Scholar]
- 76.Wright DA, et al. Standardized reagents and protocols for engineering zinc finger nucleases by modular assembly. Nat Protoc. 2006;1:1637–1652. doi: 10.1038/nprot.2006.259. [DOI] [PubMed] [Google Scholar]
- 77.Thibodeau SA, Fang R, Joung JK. High-throughput beta-galactosidase assay for bacterial cell-based reporter systems. Biotechniques. 2004;36:410–415. doi: 10.2144/04363BM07. [DOI] [PubMed] [Google Scholar]
- 78.Szczepek M, et al. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat Biotechnol. 2007;25:786–793. doi: 10.1038/nbt1317. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.