Abstract
Background
Malarial anaemia is characterized by destruction of malaria infected red blood cells and suppression of erythropoiesis. Interleukin 12 (IL12) significantly boosts erythropoietic responses in murine models of malarial anaemia and decreased IL12 levels are associated with severe malarial anaemia (SMA) in children. Based on the biological relevance of IL12 in malaria anaemia, the relationship between genetic polymorphisms of IL12 and its receptors and SMA was examined.
Methods
Fifty-five tagging single nucleotide polymorphisms covering genes encoding two IL12 subunits, IL12A and IL12B, and its receptors, IL12RB1 and IL12RB2, were examined in a cohort of 913 children residing in Asembo Bay region of western Kenya.
Results
An increasing copy number of minor variant (C) in IL12A (rs2243140) was significantly associated with a decreased risk of SMA (P = 0.006; risk ratio, 0.52 for carrying one copy of allele C and 0.28 for two copies). Individuals possessing two copies of a rare variant (C) in IL12RB1 (rs429774) also appeared to be strongly protective against SMA (P = 0.00005; risk ratio, 0.18). In addition, children homozygous for another rare allele (T) in IL12A (rs22431348) were associated with reduced risk of severe anaemia (SA) (P = 0.004; risk ratio, 0.69) and of severe anaemia with any parasitaemia (SAP) (P = 0.004; risk ratio, 0.66). In contrast, AG genotype for another variant in IL12RB1 (rs383483) was associated with susceptibility to high-density parasitaemia (HDP) (P = 0.003; risk ratio, 1.21).
Conclusions
This study has shown strong associations between polymorphisms in the genes of IL12A and IL12RB1 and protection from SMA in Kenyan children, suggesting that human genetic variants of IL12 related genes may significantly contribute to the development of anaemia in malaria patients.
Background
Malaria continues to be a major global public health burden, causing 250 million clinical cases and nearly one million deaths annually, of which 85% are children under the age of five. About 86% of malaria cases and over 91% of malaria deaths worldwide occur in sub-Saharan Africa [1,2]. Cerebral malaria and severe malarial anaemia (SMA) are two major syndromes causing malaria-related mortality in children. The pattern of these two severe forms varies depending on the intensity of transmission: cerebral malaria is more common in older children in areas with lower intensity of transmission, whereas SMA is often seen in children below two years of age in areas with intense transmission [3], such as western Kenya, where Plasmodium falciparum infection is most common [4].
The human host response to P. falciparum can vary. While some infected individuals die of severe malaria, others survive, and still others are infected without becoming severely ill. It has been estimated that host genetic factors account for ~25% of the risk of severe malaria [5]. Malaria has exerted significant pressure on the human genome to select for mutations conferring refractoriness to malaria [6]. This is illustrated by several well studied mutations that protect against different forms of malaria, including sickle cell allele, Duffy mutations, and alpha thalassaemia [7]. An increasing amount of published information has documented a number of genetic associations with malaria susceptibility. The majority of these concern genes coding for molecules involved in physiology of red blood cells or in the host immune response[8]. It is only beginning to understand the human genetic factors that determine resistance to malaria.
Malarial anaemia is characterized by destruction of malaria infected red blood cells and suppression of erythropoiesis [9-11]. Although the mechanisms of erythropoietic suppression associated with pathogenesis of malarial anaemia are poorly understood, studies in murine models of malarial anaemia have demonstrated that interleukin 12 (IL12) significantly boosts erythropoietic responses by enhancement of erythroid progenitor expansion and immune regulation [12-15], and administration of IL12 corrects malarial anaemia [12,16,17]. Moreover, several immuno-epidemiological studies conducted in malaria endemic areas have shown that decreased IL12 levels are associated with an increased risk of SMA in children [18-20]. Taken together, these findings suggest that reductions of IL12 in malaria infection could lead to SMA by suppression of erythropoiesis.
IL12 is a pro-inflammatory cytokine with a pleiotropic effect as a potent immunoregulatory molecule and haematopoietic growth factor in infections with Plasmodium parasites [14]. It is a disulfide-linked heterodimer composed of a 35 kD subunit encoded by IL12A and 40 kD subunit encoded by IL12B and exerts its biological effects through binding to heterodimeric receptors encoded by IL12RB1 and IL12RB2. The region containing IL12B, chromosome 5q31-q33, is linked to P. falciparum parasite density in linkage analysis studies [21,22]. A functional promoter variant in IL12B, IL12Bpro (rs17860508) is associated with protection against cerebral malaria in children [23]. These results suggest that IL12 is an important candidate gene in host resistance to severe malaria.
Given the biological relevance of IL12 in malarial anaemia and the genetic linkage of IL12B in malaria infection and severe disease (cerebral malaria), the potential associations of differences in genes encoding two human IL12 subunits, IL12A and IL12B, and receptors, IL12RB1 and IL12RB2, in the risk of developing SMA has been investigated by retrospectively genotyping samples collected from an infant cohort study, Asembo Bay Cohort Project (ABCP) [24]. This study presents findings of the first significant genetic association within the IL12A and IL12RB1, which are associated with protection against SMA among young children in western Kenya.
Methods
Participants
This study was part of Asembo Bay Cohort Project, an immuno-epidemiologic birth cohort project conducted from June 1992 through September 1996. Characteristics of this study have been detailed elsewhere [24]. Briefly, the study was carried out in a rural area of western Kenya where more than 95% of the residents belong to the Luo ethnic group, malaria transmission is holoendemic, and severe anaemia is a major cause of malaria-related mortality in children less than two years of age. Pregnant women were enrolled in their second or last trimester; their newborn babies were enrolled soon after birth and followed up to three to five years of life. Village monitors visited each mother-child pair every two weeks for clinical observation. At monthly intervals (and between intervals when children were sick), blood samples were obtained for measurement of parasitaemia and haemoglobin concentrations. Resulting data, along with time to first documented malaria infection, parasite density of first infection, and cumulative treatment with sulphadoxine/pyrimethamine were available as part of routine data collection. For the current genetic association study, longitudinal clinical information up to two-year follow-up from the time of enrollment was used since this age group accounted for most of the observed malarial anaemia [25].
Inclusion criteria for children in this genetic association study included having at least 1 monthly follow-up visit up to two years of age and availability of sample. Mean numbers of visits including both monthly follow-ups and sick visits during first two years of life were 19.8 (SD = 11.5) for the included children. Additionally, for genetic association analyses, it is important that all participants are genetically unrelated individuals. Therefore, only children from singleton births or one of the twins were included in the study. If more than one family member (siblings) was eligible based on the above criteria, only the first child in birth order was included in the study.
Informed written consent was obtained from parents/guardians of all study participants. This study was approved by the Institutional Review Board of the Kenya Medical Research Institute and Centers for Disease Control and Prevention.
Phenotypic definitions
A classification scheme [26], based on previous epidemiological analysis of the same cohort [25], was used in this study. SMA was defined as a haemoglobin concentration of less than 6 g/dL and the presence of P. falciparum greater than 10,000/μl. Three other subphenotypes used in this study were: severe anaemia (SA) (Hb < 6 g/dL), high-density parasitaemia (HDP) (> 10,000 P. falciparum parasites/μl), and severe anaemia with any level of parasitaemia (SAP) (Hb < 6 g/dL with any detectable P. falciparum parasitaemia). All data points collected monthly and at sick visits for two years in children were used and the clinical phenotypes are presented as episode crude incidence/1,000 person-months.
Single nucleotide polymorphism (SNP) selection
For tagging SNP (tagSNP) selection, LDSelect software at default settings (r2 > 0.65) was used to select tagSNPs from all common variation within four IL12 related genes from a public database of SeattleSNPs [27]. LDSelect uses an efficient selection algorithm to select tagging SNPs based on linkage disequilibrium (LD) statistic r2 and does not require direct haplotype inference [28]. At each round of selection, the binning algorithm identifies single SNP, which exceeds threshold r2 with the maximum number of other SNPs, and sets this group of SNPs as a bin. Then each SNP within the bin is analysed to determine whether it exceeds the threshold r2 with all other SNPs in the bin. All SNPs in a bin that meet this criterion are designated as tagSNPs. Only one tagSNP needs to be typed per bin. Binning criteria for tagSNP selection used in this study were minor allele frequency (MAF) cut-off of 10% and an r2 threshold of 0.65. LD varies across different populations [29,30]; therefore, only genotype data for the African-American samples present in the database was used, as this population is most similar to the samples in this study from western Kenya. Fifty five tagSNPs, 7 in IL12 A, 10 in IL12B, 22 in IL12RB1, and 16 in IL12RB2 were selected. Many more SNPs were needed for IL12RB1 gene than for other genes tested due to its larger size and the genetic diversity.
Genotyping
DNA was extracted from frozen blood pellets using a QiaAMP DNA purification kit (Qiagen). For each sample, 10 ng DNA was spotted into 384-well plates (ABgene), dried and frozen until just before use in genotyping assays. SNPs were genotyped using the MassARRAY® iPLEX assay (Sequenom)[31]. Multiplex PCR primers (up to 29-plex) were designed in a region of approximately 100 base pairs around the SNP of interest and an extension primer was designed immediately adjacent to the SNP using MassArray Assay Design software (version 3.1). The single base primer extension step was performed using a standardized cycling programme. Primer extension products were dispensed on 384-well SpectroChip®s that were automatically read by a Compact™ MALDI-TOF mass spectrometer (Sequenom). Data analysis was performed on MassArray Typer software (version 3.4).
Data quality control and statistical analysis
Genotypes were individually examined via a detailed quality control process involving duplicate calling of genotypes, control samples, evaluation of missing calls, and Hardy--Weinberg Equilibrium (HWE) testing. The iPLEX genotype clusters were manually checked. Only SNPs with > 90% calling rate and MAF > 2% were including in the final analysis. Each SNP was analysed using univariate methods and then multivariate Poisson regression analysis to ascertain the association between genotypes and malaria-associated morbidity. Generalized estimating equations and an independent working correlation structure to adjust for correlation between multiple visits from the same individual were used. In the multivariate Poisson regression, SNPs were independently evaluated as covariates using one of four genetic models: dominant ((AA + Aa) vs aa), recessive (AA vs (Aa + aa)), additive (aa vs Aa vs AA, which treat the marker as a continuous variable), and heterozygous advantage (Aa vs (AA + aa)) with three levels based on the most common allele frequency. Here, "A" refers the common allele. The results of the Poisson regression were reported as rate ratios (or risk ratios, RR). Multivariate models were adjusted for confounders, sickle cell type and treatment with anti-malarial drugs since these two factors were significantly associated with all four clinical phenotypes in univariate analysis (Table 1). Data was analysed using SAS software package (version 9.1). Bonferroni adjusted critical alpha level of 0.007 for IL12A, 0.006 for IL12B, 0.005 for IL12RB1, and 0.004 for IL12RB2 were used. Haploview was used to obtain values for LD and perform haplotype block analysis of SNPs in the same gene [32,33]. D prime values (D') were calculated as statistical values for pairwise LD analysis between SNPs.
Table 1.
Characteristics of 913 infants selected from Asembo Bay Cohort Project
| Subjects, no. of any incidence (%)a | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Total | SAb | P | HDPc | P | SAPd | P | SMAe | Pf | |
| Sex | |||||||||
| Male | 441 | 213 (48) | .03 | 357 (81) | .33 | 192 (44) | .03 | 77 (18) | .33 |
| Female | 472 | 194 (41) | 370 (78) | 172 (36) | 71 (15) | ||||
| Sickle cell type | |||||||||
| AS | 147 | 51 (35) | .01 | 112 (77) | .02 | 46 (31) | .03 | 13 (9) | .008 |
| AA | 656 | 316 (48) | 547 (83) | 280 (43) | 122 (17) | ||||
| SS | 25 | 13 (52) | 17 (68) | 12 (48) | 2 (8) | ||||
| Anti-malarial use | |||||||||
| Yes | 786 | 371 (47) | < .0001 | 661 (84) | < .0001 | 331 (42) | .0006 | 137 (17) | .01 |
| No | 127 | 36 (28) | 66 (52) | 33 (26) | 11 (9) | ||||
| Placental blood parasitaemia | |||||||||
| Yes | 210 | 84 (40) | .05 | 161 (77) | .11 | 76 (36) | .09 | 35 (17) | .93 |
| No | 481 | 232 (48) | 394 (82) | 207 (43) | 79 (16) | ||||
| Maternal peripheral parasitaemia | |||||||||
| Yes | 296 | 121 (41) | 0.1 | 230 (78) | .25 | 109 (37) | .17 | 57 (19) | .11 |
| No | 598 | 279 (47) | 484 (81) | 249 (42) | 90 (15) | ||||
| Birth weight < 2500 g or gestation age < 37 weeks | |||||||||
| Yes | 82 | 35 (43) | .58 | 59 (72) | .03 | 31 (38) | .54 | 15 (18) | .58 |
| No | 636 | 292 (46) | 522 (82) | 263 (41) | 101 (16) | ||||
| Low birth weight (< 2500 g) | |||||||||
| Yes | 71 | 29 (41) | .41 | 51 (72) | .05 | 25 (35) | .31 | 13 (18) | .61 |
| No | 651 | 299 (46) | 532 (82) | 270 (41) | 104 (89) | ||||
| Pre-term birth (< 37 weeks gestation) | |||||||||
| Yes | 29 | 11 (38) | .44 | 18 (62) | .01 | 10 (35) | .51 | 3 (10) | .37 |
| No | 846 | 382 (45) | 685 (81) | 343 (41) | 141 (17) | ||||
NOTE
a Any incidence: at least once; displayed are frequency counts (percentage).
b SA: Severe anaemia (Hb < 6 g/dL).
c HDP: High density parasitaemia (> 10,000 P. falciparum parasites/μl).
d SAP: Severe anaemia with any density parasitaemia (Hb < 6 g/dL with any detectable P. falciparum parasitaemia).
e SMA: Severe malaria anaemia Hb < 6 g/d Land > 10,000 P. falciparum parasites/μl)
f Chi-square p-values.
Results
Characteristics of study participants
Among 1,459 children enrolled in ABCP, 913 meeting inclusion criteria (62.6%) were selected for genotyping. In this study, incidence of at least one SMA episode in the same individual was 16.5% during the first two years of life, 44.5% for SA, 79.5% for HDP, and 40.0% for SAP. Only sickle cell type and history of anti-malarial use were significantly associated with all four clinical phenotypes of interest in the unadjusted analysis (Table 1). These two covariates were therefore included in the final model of multivariate Poisson regression analysis. Other covariates, such as bed net use, amount of rain in previous 120 days, parasitaemia in the mothers at the time of delivery, and low birth weight (< 2500 g) or pre-term birth (< 37 weeks gestation), were not significantly associated with all four clinical phenotypes although parasitaemia in mother was associated with SA only and low birth weight and pre-term birth were associated with HDP.
Genotyping
Fifty-five tagSNPs were selected for genotyping from genes encoding IL12 A, IL12 B, IL12RB1, and IL12RB2. Eight SNPs were not polymorphic in this population, and nine did not perform sufficiently in the multiplexed assay or pass quality control process (Table 2). Thirty-eight SNPs were included in the final analysis (Additional file 1).
Table 2.
List of SNPs of IL12A, IL12B, IL12RB1 and IL12BR2 tested in this study
| SNP rs# | Gene(s) | Alleles | SNP position | MAF* | Role | AA change |
|---|---|---|---|---|---|---|
| rs2243113 | IL12A | A/G | chr3:161188557 | 0.08 | Promoter | - |
| rs582054 | IL12A | T/A | chr3:161192695 | 0.21 | Intron | - |
| rs583911 | IL12A | A/G | chr3:161193084 | 0.11 | Intron | - |
| rs2243128 | IL12A | G/A | chr3:161193205 | 0.17 | Intron | - |
| rs2243138 | IL12A | A/T | chr3:161197044 | 0.47 | Downstream | - |
| rs2243140 | IL12A | T/C | chr3:161197241 | 0.11 | Downstream | - |
| rs2243143 | IL12A | A/G | chr3:161197496 | 0.45 | Downstream | - |
| rs3212227 | IL12B | A/C | chr5:158675537 | 0.39 | 3' UTR | - |
| rs3213119 | IL12B | C/A | chr5:158676366 | 0.00 | Coding exon | 298 V/F |
| rs2195940 | IL12B | C/T | chr5:158676930 | 0.25 | Intron | - |
| rs3213103 | IL12B | G/A | chr5:158679014 | 0.07 | Intron | - |
| rs3213099 | IL12B | C/G | chr5:158680114 | 0.02 | Intron (boundary) | - |
| rs919766 | IL12B | A/C | chr5:158680142 | 0.28 | Intron (boundary) | - |
| rs2853694 | IL12B | T/G | chr5:158681666 | 0.17 | Intron | - |
| rs3213096 | IL12B | C/T | chr5:158682907 | 0.00 | Coding exon | 33 V/I |
| rs2569253 | IL12B | T/C | chr5:158683571 | 0.17 | Intron | - |
| rs2569254 | IL12B | C/T | chr5:158683827 | 0.06 | Intron | - |
| rs3746190 | IL12RB1 | G/A | chr19:18031384 | 0.07 | 3' UTR | - |
| rs383483 | IL12RB1 | A/G | chr19:18032886 | 0.44 | Intron (boundary) | - |
| rs438421 | IL12RB1 | C/T | chr19:18037086 | 0.30 | Intron | - |
| rs2045386 | IL12RB1 | G/A | chr19:18043266 | 0.28 | Intron | - |
| rs447171 | IL12RB1 | C/T | chr19:18045647 | 0.27 | Intron | - |
| rs11575934 | IL12RB1 | T/C | chr19:18047618 | 0.14 | Coding exon | 214 Q/R |
| rs429774 | IL12RB1 | T/C | chr19:18047752 | 0.24 | Intron (boundary) | - |
| rs17882031 | IL12RB1 | C/T | chr19:18048349 | 0.00 | Intron | - |
| rs11575926 | IL12RB1 | C/T | chr19:18049408 | 0.00 | Coding exon | 156 R/H |
| rs11575925 | IL12RB1 | G/C | chr19:18053977 | 0.00 | Coding exon | 74 S/R |
| rs897751 | IL12RB1 | A/G | chr19:18054349 | 0.30 | Intron | - |
| rs439409 | IL12RB1 | G/A | chr19:18054613 | 0.32 | Intron | - |
| rs17884651 | IL12RB1 | C/A | chr19:18058626 | 0.00 | Coding exon | 3 P/Q |
| rs393548 | IL12RB1 | T/A | chr19:18058744 | 0.27 | Promoter | -111 |
| rs7544381 | IL12RB2 | C/T | chr1:67546881 | 0.49 | Intron | - |
| rs17129749 | IL12RB2 | C/T | chr1:67552339 | 0.30 | Intron | - |
| rs17129751 | IL12RB2 | G/A | chr1:67552503 | 0.04 | Intron | - |
| rs2307147 | IL12RB2 | T/C | chr1:67559874 | 0.24 | Coding exon | 26 D/D |
| rs17129778 | IL12RB2 | A/T | chr1:67560279 | 0.12 | Intron | - |
| rs17129792 | IL12RB2 | G/A | chr1:67565087 | 0.04 | Coding exon | 149 R/Q |
| rs1495963 | IL12RB2 | C/T | chr1:67567907 | 0.32 | Coding exon | 238 S/S |
| rs2307149 | IL12RB2 | A/T | chr1:67568038 | 0.06 | Intron (boundary) | - |
| rs17838054 | IL12RB2 | G/A | chr1:67589869 | 0.00 | Intron | - |
| rs2307145 | IL12RB2 | G/C | chr1:67606115 | 0.11 | Coding exon | 426 Q/H |
| rs2307153 | IL12RB2 | G/A | chr1:67606231 | 0.004 | Coding exon | 465 G/D |
| rs6685568 | IL12RB2 | A/G | chr1:67627885 | 0.45 | Intron | - |
| rs2270614 | IL12RB2 | A/G | chr1:67628609 | 0.17 | Intron | - |
| rs2229546 | IL12RB2 | C/A | chr1:67634108 | 0.46 | Coding exon | 779 P/P |
| rs17838066 | IL12RB2 | T/G | chr1:67634194 | 0.00 | Coding exon | 808 L/R |
*MAF observed in this study.
Association of gene polymorphisms in IL12 and its receptor with clinical phenotypes
This study identified an association with SMA for two SNPs, rs2243140 of IL12A and rs429774 of IL12RB1, both of which were associated with protection from SMA (Table 3). Specifically, for rs2243140 of IL12A, an increasing copy number of minor allele C (TT vs TC vs CC) was significantly associated with a reduced risk of SMA (P = 0.006). For rs429774 of IL12RB1, individuals possessing two copies of minor variant C appeared to be associated with a reduced risk of SMA (P = 5 × 10-5). The risk for developing SMA over time based on risk ratios calculated from multivariate Poisson regression was reduced by 82% (95% CI: 31%-95%) if carrying two copies of minor allele (CC) of rs429774 compared with those carrying one or no copies of the C allele. No statistically significant interaction between these two SNPs (P = 0.38) was found. These two minor alleles are common in the population studied here, 11% and 24%, for rs2243140 and rs429774, respectively (Table 2).
Table 3.
Protective effect of polymorphisms in the genes of IL12A and its receptor, IL12RB1, against severe malaria anaemia
| Severe malaria anaemia episodes crude incidence/1000 person-months | Adjusted risk ratio (95% CI)a | |||||
|---|---|---|---|---|---|---|
| IL12 A-rs2243140b | TT | TC | CC | CC vs TT | TC vs TT | P |
| 12.2 | 7.7 | 0 | 0.28 (0.10-0.79) | 0.52 (0.31-0.89) | .006 | |
| IL12RB1-rs429774c | TT | TC | CC | CC vs (TT+TC) | P | |
| 12.2 | 11.2 | 2.8 | 0.18 (0.05-0.69) | .00005 | ||
NOTE.
a Adjusted by sickle cell type and treatment with anti-malarial drugs.
b An additive genetic model used for IL12A-rs2243140, which treats the marker as a continuous variable and is extrapolated based on Poisson model.
c A dominant model used for the second marker, IL12RB1-rs429774.
Another SNP in IL12A, rs2243138, was associated with protection from SA (P = 0.004) and SAP (P = 0.004) (Table 4). The children homozygous for the rare allele (TT), of which the prevalence is 21.8% in this population, had significantly lower risk for developing SA (RR = 0.69, 95% CI: 0.53-0.89) and SAP (RR = 0.66, 95% CI: 0.49-0.89). A similar trend of association was seen with SMA, but was not statistically significant after adjusting for multiple comparisons (Additional file 1).
Table 4.
Protective effect of polymorphisms in the genes of IL12A and its receptor, IL12RB1, against severe anaemia, severe anaemia with any parasitaemia and high-density parasitaemia
| Crude incidence/1000 person-months | Adjusted risk ratio (95% CI)a | ||||
|---|---|---|---|---|---|
| IL12 A-rs2243138b | AA | AT | TT | TT vs (AA + AT) | P |
| Severe anaemia episodes | 38.3 | 41.3 | 30.3 | 0.69 (0.53-0.89) | .004 |
| Severe anemia with any parasitaemia episodes | 32.8 | 35.8 | 24.5 | 0.66 (0.49-0.89) | .004 |
| IL12RB1-rs383483c | AA | AG | GG | AG vs (AA+GG) | P |
| High-density parasitaemia episodes | 155.2 | 172.8 | 146.8 | 1.21 (1.07-1.36) | .003 |
NOTE.
a Adjusted by sickle cell type and treatment with anti-malarial drugs.
b A dominant model used for IL12 A-rs2243138.
c A heterozygous advantage model used for IL12RB1-rs383483.
Another SNP in IL12RB1, rs383483, was associated with HDP (P = 0.003) (Table 4). Having an AG genotype, which is common (45.8%) in this population, was significantly associated with susceptibility to HDP at 1.21 (95% CI: 1.07-1.36) times greater risk compared with those with AA or TT genotypes. No association between SNPs from IL12B and IL12RB2 and malaria-related morbidity was identified.
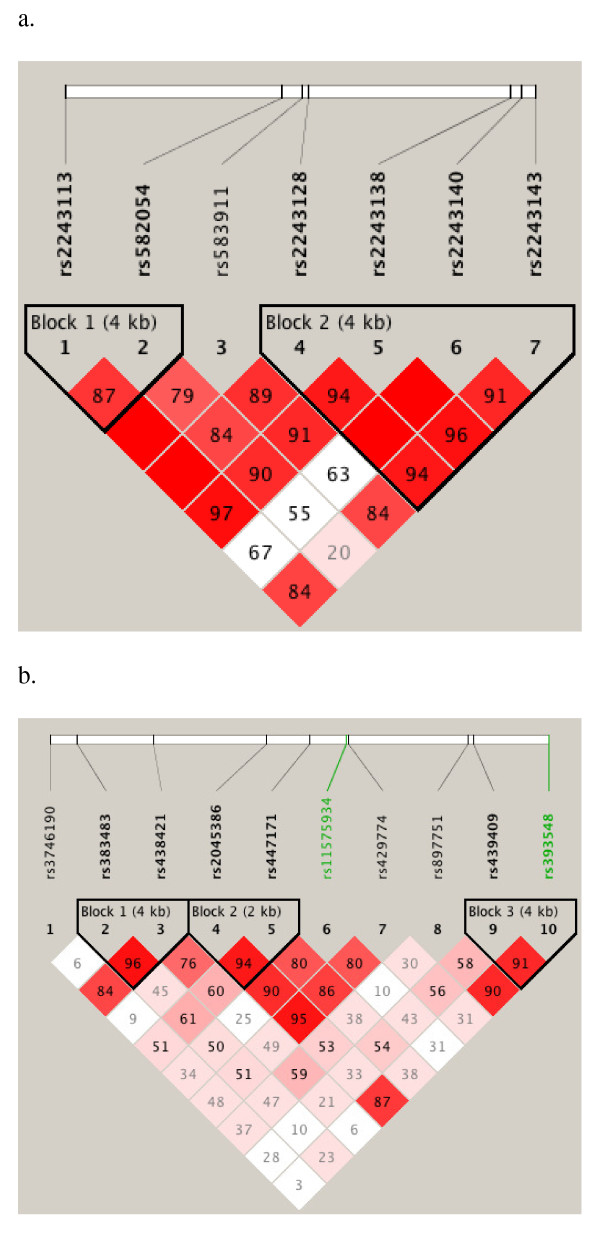
Because the single SNP regression analysis demonstrated that multiple sites within IL12A and IL12RB1 genes are significantly associated with protection from severe malaria anaemia (Additional file 1), analysis of LD in these two genes (Figure 1) was performed respectively. The two hit SNPs of IL12A, rs2243140 and rs2243138, were in high LD (D' = 0.98), but SNPs of IL12RB1, rs383483 and rs429774, were not (D' = 0.47). This suggests that the associations shown by SNPs rs2243140 and rs2243138 of IL12A may be related whereas the rs383483 and rs429774 of IL12RB1 are independent.
Figure 1.
Pairwise linkage disequilibrium (LD) plots of IL12A (a) and IL12RB1 (b) polymorphisms. Data from all 913 participants are included. If LOD ≥ 2, red denotes D primer (D') = 1; shades of pink/red, 0 < D' < 1. If LOD < 2, white indicates 0 < D' < 1. The number in each box = 100 × D' value, except D' = 1 (the box is empty). The plots were generated using Haploview software.
Discussion
In this paper, significant associations between polymorphisms within genes encoding the IL12A and IL12RB1 and protection against SMA are identified. The data shows that a SNP located within the 7th intron boundary of the IL12RB1 gene (rs429774) is associated with protection against developing SMA. In particular, children possessing two minor alleles (C) had an 82% decreased risk of SMA compared to those with one or two copies of the major allele (T); this remained significant after correction for multiple testing. The decreased risk for developing SMA is also associated with increasing minor allele (C) copy number in a SNP located within the downstream of the IL12A gene (rs2243140). Another two SNPs in IL12A (rs22431348) and IL12RB1 (rs383483) were associated with reduced risk of SA and SAP and with increased risk HDP, respectively. There is no evidence to support any significant association between IL12B genetic variants and SMA or the 3 other clinical phenotypes tested in this study. This suggests that common genetic variations within IL12B did not influence the severity of malaria in this Kenyan population, despite previous reported associations of IL12Bpro (rs17860508) with cerebral malaria in Tanzanian and Malian, but not Kenyan children [23,34], and of several other diseases with variation in IL12Bpro (rs17860508) and IL12B 3' UTR (rs3212227) [35]. A lack of significance in IL12B in this study may be the result of lack of power or small sample size; however, a recent study conducted in western African population also showed that there was no association between these two known cis-regulatory element polymophisms in IL12B and malaria parasitaemia [36].
This study was conducted in a large and clinically well-characterized cohort of children using a tagSNP method based on the following considerations. First, although methods have been developed to predict the functional impact of variants such as nonsynonymous coding SNPs, which introduce amino acid changes in their corresponding proteins [35], these SNPs represent a small subset of all common variants. Second, several genome wide association studies have indicated that variations in non-coding regions rather than nonsynonymous coding SNPs are likely to be causative to disease susceptibility or resistance in most instances [37]. Regulation of the protein products, rather than differences in the structure or function of the protein may be most important for disease risk [38]. Third, many SNPs show correlated genotypes, or LD, suggesting that a subset of all SNPs (known as tagSNPs) can be genotyped for disease association studies [39]. Lastly, several tagSNP methods have been developed to select a subset of SNPs that efficiently describes existing patterns of variation in candidate gene regions for subsequent genotyping and association analysis, without making assumption about the potential functional impact of each SNP [40]. An efficient set of tagSNPs to cover each candidate gene region, rather than simply focusing on a few putatively interesting SNPs have been selected, such that genotype at untyped SNPs can be accurately inferred from genotype at a tagSNP [28].
Previous immunological studies have shown that IL12 exerts its biological function via binding to its receptors. This study has identified associations of polymorphisms not only in IL12 related gene, IL12A, but also in its receptor, IL12RB1 with SMA and HDP with different effects. Two SNPs in the IL12A (rs2243140 and rs2243138) and one SNP in the IL12RB1 (rs429774) were associated with protection against SMA or SA whereas one SNP in the IL12RB1 (rs383483) was associated with susceptibility to HDP. There are a number of studies that examined the effect of IL12 on erythropoiesis and malarial anaemia in mouse models, in which IL12 was found to be extremely effective in correcting malarial anaemia [12-14]. The dramatic effect of IL12 treatment is mediated by enhancement of erythroid progenitor expansion in bone marrow and by immune regulation of IFN-γ, TNF-α and NO [16]. In humans, a decrease in IL12 production is also clearly associated with SMA [18-20]. Although it remains unknown how the noncoding polymorphic SNPs in IL12A and IL12RB1 identified in this study control or regulate the expression of IL12 and its receptors, the results from this study support the assertion that non-coding regions rather than nonsynonymous coding SNPs potentially have effects on disease outcomes [39]. The results from this study also suggest that the effects of IL12A and IL12RB1 on malaria disease outcomes most likely result from the long evolutionary history of P falciparum parasite within the human population.
The IL23 pathway may offer further insights on the genetic results from this study since IL23 and IL12 share the IL12p40 subunit encoded by IL12B and IL23 also binds to the IL12 receptor subunit encoded by IL12RB1. A recent study has shown that children with malarial anaemia had increased circulating IL23 and IL10 and decreased IL12 relative to healthy controls in Kenyan children, suggesting that IL23 and IL12 are counter-regulators in malaria anaemia [20]. The findings from this study that polymorphisms (3 SNPs) in IL12A and IL12RB1, but not in IL12B, have protective effects on SMA and one SNP in IL12RB1 has an opposite effect on HDP are interesting. It is possible that the genetic effects on disease outcomes observed in this study may occur at the phenotypic level of IL12 and IL23 systems, which may be influenced by the interaction of genetic variants in IL12A, IL12RB1 and IL12B. This hypothesis requires testing. Currently, whether the polymorphisms in IL12A and IL12RB1 observed in this study influence the level and expression of IL12, IL12 receptor and IL23 in vitro in relation to severity of malaria anaemia have being examined in Kenyan children.
This study included a large sample size and used longitudinal data for association analysis. However, it will be important to replicate these findings in additional African populations [41]. These efforts are complicated by the patterns of disease seen in regions of differing malaria transmission, but these studies are needed to allow us to further understand the most important pathways in successfully controlling infection by the malaria parasite and to assess the effects of racial differences, transmission intensity, and other environmental factors on the interaction of IL12 related gene and severe malaria anaemia. In addition, studies are needed to examine whether the polymorphisms observed in these genes influence the level and expression of IL12, IL12 receptor and IL23 in vitro in other African populations in relation to malarial anaemia.
Conclusions
This study has shown strong associations between polymorphisms in the genes of IL12A and IL12RB1 and protection from SMA in Kenyan children, suggesting that human genetic variants of IL12 related genes may significantly contribute to the development of anaemia in malaria patients.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
LZ was responsible for tagSNP selection, carried out genotyping work and genetic data analysis, and wrote manuscript. DP conducted genotyping and genetic data analysis. JVE and SC performed statistical analysis. SK, FTK and DT implemented Asembo Bay Cohort Project including collection of clinical and epidemiological data and laboratory diagnosis. BN and AAL were the PIs for Asembo Bay Cohort Project and designed ABCP. LS and VU participated in design of the genetic association study and contributed to data interpretation. YPS was the PI for this genetic association study, designed the study and wrote manuscript. All authors read and approved the final manuscript.
Supplementary Material
Summary of p-value of associations between genotypes tested and malaria-associated morbidity.
Contributor Information
Lyna Zhang, Email: chn6@cdc.gov.
Donald Prather, Email: dwj5@cdc.gov.
Jodi Vanden Eng, Email: jev8@cdc.gov.
Sara Crawford, Email: SaraCrawford@valpo.edu.
Simon Kariuki, Email: SKariuki@ke.cdc.gov.
Feiko ter Kuile, Email: terkuile@liverpool.ac.uk.
Dianne Terlouw, Email: D.J.Terlouw@liverpool.ac.uk.
Bernard Nahlen, Email: bnahlen@usaid.gov.
Altaf A Lal, Email: LalA@state.gov.
Laurence Slutsker, Email: lms5@cdc.gov.
Venkatachalam Udhayakumar, Email: vxu0@cdc.gov.
Ya Ping Shi, Email: yps0@cdc.gov.
Acknowledgements
We sincerely thank all the study subjects who participated in the Asembo Bay Cohort Project and community field workers for supporting the collection of data and samples. DP was supported by the Emerging Infectious Disease Postdoctoral Fellowship. LZ was funded by the Career Development Award from American Association of Teachers and Preventive Medicine. We thank the Director, Kenya Medical Research Institute for permission to publish this paper and Dr William Dotson for editorial assistance. This study was supported by WHO/TDR Collaborative Research Grant #A30322.
References
- WHO, UNICEF. World Marlaria Report 2005. Geneva: World Health Organization; 2005. [Google Scholar]
- Hay SI, Guerra CA, Gething PW, Patil AP, Tatem AJ, Noor AM, Kabaria CW, Manh BH, Elyazar IR, Brooker S, Smith DL, Moyeed RA, Snow RW. A world malaria map: Plasmodium falciparum endemicity in 2007. PLoS Med. 2009;6:e1000048. doi: 10.1371/journal.pmed.1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood BM. The epidemiology of malaria. Ann Trop Med and Parasitol. 1997;91:763–769. doi: 10.1080/00034989760518. [DOI] [PubMed] [Google Scholar]
- Bloland PB, Boriga DA, Ruebush TK, McCormick JB, Roberts JM, Oloo AJ, Hawley W, Lal A, Nahlen B, Campbell CC. Longitudinal cohort study of the epidemiology of malaria infections in an area of intense malaria transmission II. Descriptive epidemiology of malaria infection and disease among children. Am J Trop Med Hyg. 1999;60:641–648. doi: 10.4269/ajtmh.1999.60.641. [DOI] [PubMed] [Google Scholar]
- Mackinnon MJ, Mwangi TW, Snow RW, Marsh K, Williams TN. Heritability of malaria in Africa. PLoS Med. 2005;2:e340. doi: 10.1371/journal.pmed.0020340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Williams SM. Genetic analysis of African populations: human evolution and complex disease. Nat Rev Genet. 2002;3:611–621. doi: 10.1038/ni0702-611. [DOI] [PubMed] [Google Scholar]
- Kwiatkowski DP. How malaria has affected the human genome and what human genetics can teach us about malaria. Am J Hum Genet. 2005;77:171–192. doi: 10.1086/432519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirugo G, Hennig BJ, Adeyemo AA, Matimba A, Newport MJ, Ibrahim ME, Ryckman KK, Tacconelli A, Mariani-Costantini R, Novelli G, Soodyall H, Rotimi CN, Ramesar RS, Tishkoff SA, Williams SM. Genetic studies of African populations: an overview on disease susceptibility and response to vaccines and therapeutics. Hum Genet. 2008;123:557–598. doi: 10.1007/s00439-008-0511-y. [DOI] [PubMed] [Google Scholar]
- Chang KH, Stevenson MM. Malarial anaemia: mechanisms and implications of insufficient erythropoiesis during blood-stage malaria. Int J Parasitol. 2004;34:1501–1516. doi: 10.1016/j.ijpara.2004.10.008. [DOI] [PubMed] [Google Scholar]
- Ekvall H. Malaria and anemia. Curr Opin Hematol. 2003;10:108–114. doi: 10.1097/00062752-200303000-00002. [DOI] [PubMed] [Google Scholar]
- Menendez C, Fleming AF, Alonso PL. Malaria-related anaemia. Parasitol Today. 2000;16:469–476. doi: 10.1016/S0169-4758(00)01774-9. [DOI] [PubMed] [Google Scholar]
- Mohan K, Stevenson MM. Interleukin-12 corrects severe anemia during blood-stage Plasmodium chabaudi AS in susceptible A/J mice. Exp Hematol. 1998;26:45–52. [PubMed] [Google Scholar]
- Jacobsen SE. IL12, a direct stimulator and indirect inhibitor of haematopoiesis. Res Immunol. 1995;146:506–514. doi: 10.1016/0923-2494(96)83024-0. [DOI] [PubMed] [Google Scholar]
- Stevenson MM, Su Z, Sam H, Mohan K. Modulation of host responses to blood-stage malaria by interleukin-12: from therapy to adjuvant activity. Microbes Infect. 2001;3:49–59. doi: 10.1016/S1286-4579(00)01354-X. [DOI] [PubMed] [Google Scholar]
- Mohan K, Sam H, Stevenson MM. Therapy with a combination of low doses of interleukin 12 and chloroquine completely cures blood-stage malaria, prevents severe anemia, and induces immunity to reinfection. Infect Immun. 1999;67:513–519. doi: 10.1128/iai.67.2.513-519.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng VM, Car BD, Schnyder B, Lorenz M, Lugli S, Aguet M, Anderson TD, Ryffel B, Quesniaux VF. The stimulatory effects of interleukin (IL)-12 on hematopoiesis are antagonized by IL-12-induced interferon gamma in vivo. J Exp Med. 1995;181:1893–1898. doi: 10.1084/jem.181.5.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman SL, Crutcher JM, Puri SK, Ansari AA, Villinger F, Franke ED, Singh PP, Finkelman F, Gately MK, Dutta GP, Sedegah M. Sterile protection of monkeys against malaria after administration of interleukin-12. Nat Med. 1997;3:80–83. doi: 10.1038/nm0197-80. [DOI] [PubMed] [Google Scholar]
- Perkins DJ, Weinberg JB, Kremsner PG. Reduced interleukin-12 and transforming growth factor-beta1 in severe childhood malaria: relationship of cytokine balance with disease severity. J Infect Dis. 2000;182:988–992. doi: 10.1086/315762. [DOI] [PubMed] [Google Scholar]
- Chaisavaneeyakorn S, Othoro C, Shi YP, Otieno J, Chaiyaroj SC, Lal AA, Udhayakumar V. Relationship between plasma Interleukin-12 (IL-12) and IL-18 levels and severe malarial anemia in an area of holoendemicity in western Kenya. Clin Diagn Lab Immunol. 2003;10:362–366. doi: 10.1128/CDLI.10.3.362-366.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong'echa JM, Remo AM, Kristoff J, Hittner JB, Were T, Ouma C, Otieno RO, Vulule JM, Keller CC, Awandare GA, Perkins DJ. Increased circulating interleukin (IL)-23 in children with malarial anemia: in vivo and in vitro relationship with co-regulatory cytokines IL-12 and IL-10. Clin Immunol. 2008;126:211–221. doi: 10.1016/j.clim.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flori L, Kumulungui B, Aucan C, Esnault C, Traore AS, Fumoux F, Rihet P. Linkage and association between Plasmodium falciparum blood infection levels and chromosome 5q31-q33. Genes Immun. 2003;4:265–268. doi: 10.1038/sj.gene.6363960. [DOI] [PubMed] [Google Scholar]
- Rihet P, Traore Y, Abel L, Aucan C, Traore-Leroux T, Fumoux F. Malaria in humans: Plasmodium falciparum blood infection levels are linked to chromosome 5q31-q33. Am J Hum Genet. 1998;63:498–505. doi: 10.1086/301967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquet S, Doumbo O, Cabantous S, Poudiougou B, Argiro L, Safeukui I, Konate S, Sissoko S, Chevereau E, Traore A, Keita MM, Chevillard C, Abel L, Dessein AJ. A functional promoter variant in IL12B predisposes to cerebral malaria. Human Molecular Genetics. 2008;17:2190–2195. doi: 10.1093/hmg/ddn118. [DOI] [PubMed] [Google Scholar]
- Bloland PB, Ruebush TK, McCormick JB, Ayisi J, Boriga DA, Oloo AJ, Beach R, Hawley W, Lal A, Nahlen B, Udhayakumar V, Campbell CC. Longitudinal cohort study of the epidemiology of malaria infections in an area of intense malaria transmission I. Description of study site, general methodology, and study population. Am J Trop Med Hyg. 1999;60:635–640. doi: 10.4269/ajtmh.1999.60.635. [DOI] [PubMed] [Google Scholar]
- McElroy PD, Lal AA, Hawley WA, Bloland PB, Kuile FO, Oloo AJ, Harlow SD, Lin X, Nahlen BL. Analysis of repeated hemoglobin measures in full-term, normal birth weight Kenyan children between birth and four years of age. III. The Asemobo Bay Cohort Project. Am J Trop Med Hyg. 1999;61:932–940. doi: 10.4269/ajtmh.1999.61.932. [DOI] [PubMed] [Google Scholar]
- Aidoo M, Terlouw DJ, Kolczak MS, McElroy PD, ter Kuile FO, Kariuki S, Nahlen BL, Lal AA, Udhayakumar V. Protective effects of the sickle cell gene against malaria morbidity and mortality. Lancet. 2002;359:1311–1312. doi: 10.1016/S0140-6736(02)08273-9. [DOI] [PubMed] [Google Scholar]
- SeattleSNPs Variation Discovery Resource. http://pga.mbt.washington.edu/education.html
- Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DM, Cardon LR. A comparison of linkage disequilibrium patterns and estimated population recombination rates across multiple populations. Am J Hum Genet. 2005;76:681–687. doi: 10.1086/429274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall JD, Pritchard JK. Haplotype blocks and linkage disequilibrium in the human genome. Nat Rev Genet. 2003;4:587–597. doi: 10.1038/nrg1123. [DOI] [PubMed] [Google Scholar]
- Schaeffeler E, Zanger UM, Eichelbaum M, Asante-Poku S, Shin JG, Schwab M. Highly multiplexed genotyping of thiopurine s-methyltransferase variants using MALD-TOF mass spectrometry: reliable genotyping in different ethnic groups. Clin Chem. 2008;54:1637–1647. doi: 10.1373/clinchem.2008.103457. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Wigginton JE, Cutler DJ, Abecasis GR. A note on exact tests of Hardy-Weinberg equilibrium. Am J Hum Genet. 2005;76:887–893. doi: 10.1086/429864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morahan G, Boutlis CS, Huang D, Pain A, Saunders JR, Hobbs MR, Granger DL, Weinberg JB, Peshu N, Mwaikambo ED, Marsh K, Roberts DJ, Anstey NM. A promoter polymorphism in the gene encoding interleukin-12 p40 (IL12B) is associated with mortality from cerebral malaria and with reduced nitric oxide production. Genes Immun. 2002;3:414–418. doi: 10.1038/sj.gene.6363909. [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. Predicting the effects of amino acid substitutions on protein function. Annu Rev Genomics Hum Genet. 2006;7:61–80. doi: 10.1146/annurev.genom.7.080505.115630. [DOI] [PubMed] [Google Scholar]
- Barbier M, Atkinson A, Fumoux F, Rihet P. IL12B polymorphisms are linked but not associated with Plasmodium falciparum parasitemia: a familial study in Burkina Faso. Genes Immun. 2008;9:405–411. doi: 10.1038/gene.2008.31. [DOI] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J Clin Invest. 2008;118:1590–1605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardlie KG, Kruglyak L, Seielstad M. Patterns of linkage disequilibrium in the human genome. Nat Rev Genet. 2002;3:299–309. doi: 10.1038/nrg777. [DOI] [PubMed] [Google Scholar]
- Halldorsson BV, Istrail S, De La Vega FM. Optimal selection of SNP markers for disease association studies. Hum Hered. 2004;58:190–202. doi: 10.1159/000083546. [DOI] [PubMed] [Google Scholar]
- Chanock SJ, Manolio T, Boehnke M, Boerwinkle E, Hunter DJ, Thomas G, Hirschhorn JN, Abecasis G, Altshuler D, Bailey-Wilson JE, Brooks LD, Cardon LR, Daly M, Donnelly P, Fraumeni JF Jr, Freimer NB, Gerhard DS, Gunter C, Guttmacher AE, Guyer MS, Harris EL, Hoh J, Hoover R, Kong CA, Merikangas KR, Morton CC, Palmer LJ, Phimister EG, Rice JP, Roberts J, Rotimi C, Tucker MA, Vogan KJ, Wacholder S, Wijsman EM, Winn DM, Collins FS. Replicating genotype-phenotype associations. Nature. 2007;447:655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Summary of p-value of associations between genotypes tested and malaria-associated morbidity.