SUMMARY
Random effects are often used in generalized linear models to explain the serial dependence for longitudinal categorical data. Marginalized random effects models (MREMs) for the analysis of longitudinal binary data have been proposed to permit likelihood-based estimation of marginal regression parameters. In this paper, we introduce an extension of the MREM to accommodate longitudinal ordinal data. Maximum marginal likelihood estimation is implemented utilizing quasi-Newton algorithms with Monte Carlo integration of the random effects. Our approach is applied to analyze the quality of life data from a recent colorectal cancer clinical trial. Dropout occurs at a high rate and is often due to tumor progression or death. To deal with progression/death, we use a mixture model for the joint distribution of longitudinal measures and progression/death times and principal stratification to draw causal inferences about survivors.
Keywords: marginalized likelihood-based models, ordinal data models, dropout
1. INTRODUCTION
Longitudinal data are repeated measurements from the same subject observed over time. The within-subject measurements (over time) are typically not independent. Although serial correlation may not be of primary interest but it must be taken into account to make proper inferences. In marginal models, the population-averaged effect of covariates on the longitudinal response is directly specified [1, 2] and the regression coefficients have interpretation for the population rather than for any individual [3]. In conditional models, the effect of covariates on responses is specified conditional on random effects or previous history of responses. Hence, the population-averaged effect of covariates is indirectly specified [4, 5]. In this paper, we consider marginal model approaches.
Properly specified probability models lead to efficient estimation even under missing at random (MAR) [6] and nested models can be compared using likelihood ratio tests and non-nested models by penalized criteria such as Akaike information criterion (AIC) [7] or Bayesian information criterion [8]. Recently, marginalized likelihood-based models have been developed for the analysis of longitudinal categorical data [9–13]. Heagerty [9, 10] proposed marginally specified logistic-normal models and marginalized transition models (MTM) for longitudinal binary data. In both models, a marginal logistic-regression model was used for explaining the average response. The models were specified by introducing random effects in the logistic-normal models and Markov dependence for MTM to explain the within-subject dependence. Miglioretti and Heagerty [12] developed marginalized multilevel models for longitudinal binary data in the presence of time-varying covariates. Lee and Daniels [13] extended Heagerty's work to accommodate longitudinal ordinal data using Markov dependence. Marginalized models have advantages over conditional models. First, the interpretation of regression coefficients does not depend on specification of the dependence in the model unlike in conditional models. In addition, estimation of covariate effects has been shown to be more robust to mis-specification of dependence [10, 11, 13].
Models for correlated ordinal data typically fall into two classes based on how the dependence is modeled: via the global odds ratio [14–17] or via random effects [18–20]. A general overview of models for ordinal categorical data can be found in Liu and Agresti [21]. The main contribution of this paper will be to introduce a new marginalized model for longitudinal ordinal data.
A common issue in inference from longitudinal studies is potential biases introduced by missing data. Classes of models to accommodate longitudinal data with dropout are summarized in Hogan et al. [22]. Standard approaches to handle missing data implicitly ‘impute’ values of response after dropout. For quality of life (QOL) data, if a subject drops out due to death, the QOL will not be defined after the dropout time. One way to address the type of dropout is to model the joint distribution of the longitudinal responses and progression/death times [23–25]. Hogan and Laird [23] used a mixture model for the joint distribution of longitudinal measures and progression/death times. Pauler et al. [24] proposed a pattern mixture model (PMM) for longitudinal QOL data with non-ignorable missingness due to dropout and censorship by death. Recently, Kurland and Heagerty [25] explored regression models conditioning on being alive as a valid target of inference. They used regression models that condition on survival status rather than a specific survival time. We will use the ideas in Hogan and Laird [23] similar to the previous work by Pauler et al. [24].
We implement a principal stratification approach [26, 27] here to make inference on the causal effect of the treatment on QOL among (potential) survivors on both treatment arms. Frangakis and Rubin [26] discussed causal effects in studies where the outcome was recorded and unobserved due to death. Rubin [28] and Hayden et al. [29] referred to the estimand in Frangakis and Rubin [26] as ‘the survivors average causal effect (SACE)’. Egleston et al. 27 proposed assumptions to identify the SACE and implemented a sensitivity analysis for some of those assumptions. Rubin [30] introduced the causal effect of a treatment on a outcome that is censored by death in QOL studies. In this paper, we describe an approach that can be used to obtain the causal effect of treatment in the presence of death based on principal stratification for longitudinal ordinal outcomes.
This paper is arranged as follows. In Section 2, we describe the motivating example. We briefly review marginalized random effects models (MREMs) for longitudinal binary data [9] in Section 3. In Section 4, we propose an ordinal MREM (OMREM) for the longitudinal data with ignorable dropout. In Section 5, we conduct a simulation study to examine bias and efficiency in estimation of marginal mean parameters. In the context of QOL data collected in a recent colorectal cancer clinical trial [31], we propose models for the OMREM under dropout due to progression/death and illustrate them on this data in Section 6.
2. MOTIVATING EXAMPLE
We analyzed QOL data from a recent colorectal cancer clinical trial [31]. A total of 795 patients with colorectal cancer were randomly assigned to one of the three treatments (FOLFOX, IFL (control) and IROX) between May 1999 and April 2001. The main objective of this trial was to find a better treatment for colorectal cancer. The median survival for patients receiving IFL was 15.0 months compared with 19.5 months for those receiving FOLFOX and 17.4 months for those receiving IROX. Survival for patients receiving FOLFOX did not differ from those receiving IROX (see [31]).
However, given that the toxicity profiles were quite different on the three treatment arms, it was of interest to see if there was a negative impact of ‘better’ treatments on patients QOL. We focus on one QOL measure, fatigue. Fatigue is measured on a 5-point ordinal scale. Additional complications for analyzing QOL are posed by patients dying during the trial.
The models and analysis to follow focus on two treatments, FOLFOX and IFL (control) and address appropriate analysis of QOL data in the presence of death.
3. REVIEW OF MARGINALLY SPECIFIED LOGISTIC-NORMAL MODELS FOR LONGITUDINAL BINARY DATA
Now we review MREMs for longitudinal binary data [9]. Define . The marginalized logistic-normal model (MLNM) is specified using the following regressions: mean model:
| (1) |
dependence model:
| (2) |
where β is the p × 1 vector of regression coefficients and . We assume that the response vector Yi is conditionally independent given bi = (bi1,..., bini)T and that
| (3) |
The covariance matrix A is assumed to have a simple structure. Conditional on xi and bi, the responses Yit are assumed to be conditionally independent. Parameters in A provide measures of random variation both across individuals and over time.
The parameters Δit in (2) are functions of both the marginal mean parameters and the random effects variance and can be obtained using the following identity:
| (4) |
where f is the univariate normal density function. This model has several desirable features. First, the mean model is specified separately from the dependence model. As a result, the interpretation of the regression parameter β does not change as we modify assumptions regarding the dependence in equation (2). This is not true for classical generalized random effects models, which parameterize directly as a function of covariates. In addition, parameters in cov(bi ) provides measures of random variation both across individuals and over time. Second, the MLNM can be used with data where subjects have variable lengths of follow-up, permitting the likelihood analysis in settings where data may be MAR. Further details on MLNM are given in [9].
4. MREMS FOR LONGITUDINAL ORDINAL DATA
In this section, we extend Heagerty's MLNM to accommodate longitudinal ordinal data.
4.1. Proposed models
Let Yi = (Yi1,..., Yini) be a vector of longitudinal K-category ordinal responses on subject i = 1,..., N at times t = 1,..., ni, ni ≤T. We assume that associated exogenous but possibly time-varying covariates, xit = (xit1,..., xitp), are recorded for each subject at each time, and that the regression model properly specifies the full covariate conditional probability such that P(Yit = yit|Xit) = P(Yit = yit|Xi1,..., Xini). The MREM for longitudinal ordinal data, also called OMREM, is specified using the following two regressions: mean model:
| (5) |
dependence model:
| (6) |
where for i = 1,..., N and k = 1,..., K – 1. We assume that the variance–covariance matrix Σi of bi has an autoregressive covariance structure
| (7) |
where log is a c × 1 vector, and λ is a c × 1 coefficient vector of zi. The regression model for σi allows heterogeneity to depend on subject-level covariates such as treatment or gender.
In the dependence model, (6), we need to ensure the monotonicity of Δ in k to guarantee the validity of the mean model in (5).
Theorem 1
For the OMREM given by (5) and (6), if β01 < ··· <β0K–1, then Δit1 <··· <ΔitK–1.
Proof 1
See Appendix.
The marginal mean model is a proportional odds model [32]. Serial dependence is captured by the random effects. The regression parameters in (5) have a marginal interpretations unlike generalized linear mixed models [5]. An advantage of this model is the ability to use conditional models for association (6) while still structuring the marginal mean as a function of covariates directly (5). As a result, the interpretation of the regression coefficients, (β0, β) does not depend on the specification of the dependence model.
For longitudinal data with random effects bi, the marginal probability captures the systematic variation in the marginal probability that is due to xit, whereas parameters in cov(bi) provide a measure of random variation both across individuals and over time. Heagerty and Kurland [33] investigated the impact on the estimates of regression coefficients of incorrect assumptions regarding the random effects in generalized linear mixed models and marginalized models and found that marginalized regression models are much less susceptible to bias resulting from random effects model misspecification.
The marginal and conditional probabilities in (5) and (6) are related as follows:
| (8) |
where and f (·) is a univariate normal distribution with mean 0 and variance var(bit). We use (8) to solve for Δitk given β0k, β and λ.
Reparametrization of the random effects and their covariance matrix: From a computational perspective, it is convenient to orthogonalize the random effects by setting bi = σiΣ*1/2ai where Σ*1/2, a lower triangular matrix with positive diagonal elements, is the Cholesky factor of the ni × ni matrix Σ* [34] and ai is a ni × 1 vector of independent standard normals. The reparameterized conditional model is then given by
where s(t) is the tth row of Σ*1/2 and I is the identity matrix of order ni. This transformation allows us to estimate the Cholesky factor Σ*1/2 instead of the covariance matrix Σ*. Since the Cholesky factor is the square root of the covariance matrix, it allows more stable estimation of near-zero variance terms [35].
4.2. Maximum likelihood estimation
The likelihood function is the integral over random effects of a product of multinominals,
| (9) |
where is the set of K indicators with yitk = 1 if yit = k; yitk = 0 otherwise, for k = 1,..., K, ϕ(·) is a multivariate normal density with mean vector 0 and variance–covariance matrix I, and θ = (β0, β, λ, α). The marginalized likelihood in (9) is not available in the closed form. There are several approaches to (numerically) integrate out the random effects. Gauss–Hermite quadrature is popular for low-dimensional random effects models such as random intercept models [35]; adaptive versions [36, 37] can increase efficiency. Monte Carlo methods are often used for the models with higher-dimensional integrals and use randomly sampled points to approximate the integrals. In our model, we use Monte Carlo methods to evaluate the integral in (9) as the dimension of ai is high.
Maximizing the log likelihood with respect to θ yields the likelihood equation
where
| (10) |
The (K–1+p+c+1)-dimensional likelihood equations are given in the Appendix.
The matrix of second derivatives of the observed data log likelihood has a very complex form. Fortunately, the sample empirical covariance matrix of the individual scores in any correctly specified model is a consistent estimator of the information and involves only the first derivatives. Therefore, the quasi-Newton method can be used to solve the likelihood equations, using
where Ie(θ), an empirical and consistent estimator of the information matrix at step m, is given by
At convergence, the large-sample variance–covariance matrix of the parameter estimates is then obtained as the inverse of .
For the explicit forms of the terms in the quasi-Newton algorithm including the derivatives ∂Δitk/∂β0, ∂Δitk/∂β and ∂Δitg/∂λ calculated using (8), see the Appendix.
The intercepts Δitk are a function of β0k, β, α and λ and must be obtained within the quasi-Newton algorithm. Let . Estimates of Δitk can be obtained using the Newton–Raphson algorithm as follows:
where
| (11) |
Note that the integral in (11) is one dimensional and we use Gauss–Hermite quadrature to evaluate this integral.
5. SIMULATION STUDY
We conducted a simulation to examine the bias and efficiency for estimation of the marginal mean parameters in the setting of misspecification of the dependence model under no missing data and under MAR missingness (common in longitudinal data). We also compare the efficiency of the OMREM to the independence proportional odds model (IPOM), given in (5).
We simulated the longitudinal ordinal data under an OMREM. Covariates were time and group (two levels). The marginal probability for the OMREM was specified as
where t = 1,..., 6, timeit = (t–1)/6, and groupi equals 0 or 1 with an approximately equal sample size per group. The conditional probabilities were specified from (6) and (7) with (σi, α) = (1.1, 0.2) if groupi =0; (σi, α) = (1.5, 0.2) if groupi =1. Note that α = 0.2 corresponds to a lag one correlation of exp(–0.2)=0.819. We simulated 500 data sets each with a sample size of 300. We then fit the IPOM and the OMREM.
For the MAR missingness, we specified the following MAR dropout model:
| (12) |
where Yit–1 ∈{0,1,2,3}.
Table I presents the point estimates, root mean-squared error (√MSE), and 95 per cent Monte Carlo error intervals of the marginal mean parameters. When there were no missing data, the estimates were essentially unbiased for both the OMREM and the IPOM. The root MSEs in the IPOM were also similar to those in the OMREM but sometimes slightly larger (e.g. for β1 and β2). In the presence of MAR missingness, the estimates in the OMREM were still essentially unbiased. However, for the IPOM, we saw considerable biases; for example, the relative bias for the coefficient of time, β1 was 34 per cent ((–0.33+0.5)/(–0.5)). In addition, the root MSEs were larger in the IPOM; for β1 and β2, the root MSEs were eight and three times as large as the OMREM, respectively.
Table I.
Bias of maximum likelihood estimators.
| Complete data |
MAR dropout |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| OMREM |
IPOM |
OMREM |
IPOM |
||||||
| Parameter | Truth | Mean | Mean | Mean | Mean | ||||
| β 01 | –1.00 | –0.99 | 0.01 | –0.99 | 0.01 | –1.01 | 0.01 | –0.98 | 0.02 |
| (–1.00, –0.98) | (–1.00, –0.98) | (–1.02, –0.99) | (–0.99, –0.97) | ||||||
| β 02 | 0.50 | 0.51 | 0.01 | 0.51 | 0.01 | 0.49 | 0.01 | 0.51 | 0.01 |
| (0.50, 0.51) | (0.50, 0.52) | (0.48, 0.50) | (0.50, 0.52) | ||||||
| β 03 | 1.00 | 1.00 | 0.00 | 1.00 | 0.00 | 0.99 | 0.01 | 1.01 | 0.01 |
| (0.99, 1.01) | (1.00, 1.01) | (0.98, 1.00) | (1.00, 1.02) | ||||||
| β1 (Time) | –0.50 | –0.51 | 0.01 | –0.52 | 0.02 | –0.48 | 0.02 | –0.33 | 0.17 |
| (–0.53, –0.49) | (–0.54, –0.50) | (–0.52, –0.45) | (–0.36, –0.30) | ||||||
| β 2 | 0.50 | 0.50 | 0.00 | 0.51 | 0.01 | 0.51 | 0.01 | 0.47 | 0.03 |
| (0.49, 0.51) | (0.49, 0.52) | (0.49, 0.52) | (0.46, 0.48) | ||||||
Displayed are the average regression coefficient estimates, the and 95 per cent Monte Carlo error interval . The true conditional probabilities were specified with σi=1.1 and α=0.2 if groupi=0; σi=1.5 and α=0.2 if groupi=1. The fitted conditional probabilities were specified with random effects, with logσi=λ1+groupiλ2 for the OMREM.
Overall, the simulation shows the increased efficiency of the OMREM over the independence model (IPOM) in complete data and the large biases that can occur in the marginal mean parameters when the dependence is mis-modeled in the presence of MAR missingness.
6. QOL EXAMPLE WITH DROPOUT DUE TO PROGRESSION/DEATH
We focus on one QOL measure, fatigue, measured on a 5-point ordinal scale (1, I am usually not tired at all; 2, I am occasionally rather tired; 3, there are frequently periods when I am quite tired; 4, I am usually very tired; 5, I feel exhausted most of the time). Because very few patients reported category 5, we combined categories 4 and 5 into one category. We used 707 subjects without missing data at baseline and focused on two treatments, FOLFOX and IFL (control).
To examine treatment differences in fatigue levels, we included the type of treatment, Tx,
and visit number(TIME=0.0,0.1,...,0.5) re-scaled. The patient's visit corresponds to the time period during which the survey was filled out (0, baseline; 1, 1–84 days after going on study; 2, 85–168 days after going on study;...,; 5, 337–420 days after going on study). We will analyze the data from the first five windows (up to about 1 year). We assumed that the missing responses (mostly due to dropout) were MAR in our initial analysis. In Section 6.2, we more carefully handled the missingness related to the reason for dropping out (including death).
The quasi-Newton algorithm is not trivial computationally due to the need to obtain estimates and derivatives of Δit using the Gauss–Hermite quadrature for all subjects and at all times, within each quasi-Newton step. Each quasi-Newton step (in which all the Δit need to be computed) on a Pentium with a 1.6 GHz processor took about 3 min for the OMREM with MC sample size of 10 000 and 40 point Gauss–Hermite quadrature. Using good initial values based on fitting an IPOM in standard software results in a minimal number of iterations until convergence. For example, in our analysis below, we obtained convergence in 20 iterations using a fairly strict convergence criterion, where and are current and previous estimates of the parameters, respectively.
6.1. Model fit
We first fitted three OMREMs and one IPOM under an assumption of ignorable dropout. OMREM-1 allowed the random effects variance to depend on treatment, logσi = λ0 + λ1 × Txi. OMREM-2 was a simpler model, with a constant variance, logσi = λ0. Both had autoregressive variance–covariance structures. OMREM-3 was the OMREM-2 with bit = bi0 ~ N(0, σ2).Table II gives the maximum likelihood estimates (MLEs) for all four models.
Table II.
Maximum likelihood estimates for marginalized random effects models under ignorable missingness.
| OMREM-1 | OMREM-2 | OMREM-3 | IPOM | |
|---|---|---|---|---|
| Mean parameters | ||||
| Int1 | –1.099* (0.126) | –1.093* (0.126) | –1.119* (0.124) | –1.163 (0.636) |
| Int2 | 0.839* (0.126) | 0.839* (0.127) | 0.816* (0.123) | 0.796* (0.149) |
| Int3 | 2.280* (0.160) | 2.277* (0.160) | 2.261* (0.156) | 2.277* (0.982) |
| Visit | –0.334 (0.620) | –0.367 (0.609) | –0.166 (0.472) | –0.424* (0.113) |
| Tx | –0.073 (0.168) | –0.072 (0.167) | –0.069 (0.161) | –0.003 (0.039) |
| Visit × Tx | –0.607 (0.922) | –0.469 (0.917) | –0.541 (0.725) | 1.059* (0.063) |
| Dependence parameters | ||||
| Int | 1 .086* (0.634) | 1.251 (0.816) | 0.664* (0.144) | |
| Tx | 0.280 (0.897) | |||
| α | 0.243* (0.076) | 0.264* (0.081) | ||
| Max. logL | –1266.112 | –1267.623 | –1273.577 | –1366.558 |
| AIC | 2550.224 | 2551.246 | 2561.154 | 2745.116 |
OMREM-1 and OMREM-2 are OMREMs with random effects variance logσi=λ0+λ1×Txi and logσi=λ0, respectively. Both had autoregressive covariance structures. OMREM-3 is the OMREM-2 with bit=bi0~ N(0, σ2). IPOM is an independent proportional odds model. Tx is an indicator for FOLFOX. Visit is the patient's visit corresponding to the time period.
Significance under 95 per cent confidence level.
The inferences for some of the coefficients under the IPOM were very different from those for the dependence models. For example, the interaction of treatment and visit was highly significant under the IPOM, indicating an increase in fatigue over time for FOLFOX relative to IFL, whereas the dependence models indicated a non-significant decrease in fatigue over time.
Comparison of AIC for IPOM and OMREM-3 indicated that the OMREM-3 fit much better than the IPOM (2561.154 for OMREM-3, 2745.116 for IPOM). Point estimates and standard errors for marginal mean parameters for the OMREM-1 and OMREM-2 were similar. To compare the fit of the two models under ignorability, we computed the likelihood ratio test. Comparison of deviances for OMREM-2 and OMREM-1, which were nested yielded ΔD12 = 2 × (1267.623 – 1266.112) = 3.022, p-value=0.082 on 1 d.f. This comparison indicated that the OMREM-1 did not provide a significantly better fit than the OMREM-2. We also computed the likelihood ratio test to compare OMREM-2 and OMREM-3. The deviance difference was ΔD23 = 2 × (1273.577 – 1267.623) = 11.908 p-value<0.01 on 1 d.f. This indicated that the OMREM-2 fit better than the OMREM-3.
The estimate of the correlation parameter ρ in OMREM-2 was significant and corresponded to an estimated correlation of . The MLE for σ (exp(1.251)=3.49) indicated large subject-to-subject variation in the odds of the cumulative probability of fatigue. The coefficients of treatment and the interaction of treatment and visit in the marginal mean model were not significant indicating that fatigue (and its trajectory over time) did not differ between the two treatments.
6.2. Dropout due to progression/death
QOL responses not measured due to participant death do not exist whereas scheduled measurements due to dropout for other reasons can be viewed as existing but unobserved. If we fit models using methods in Section 4 (as we did in Section 6.1), the missing data due to death are implicitly imputed (under MAR). We outline a PMM approach to address this next.
6.2.1. PMM approach
Define Si to be death time for subject i. To address dropout due to death, we can specify the OMREMs conditional on death time, S. The models are given by
| (13) |
where xit is a vector of covariates including treatments, j = 1,..., J, and J is the number of patterns (J≤T). This approach implicitly assumes that for a given death time (pattern) that missing data before the death time is MAR (conditional on pattern).
Now, suppress i without loss of clarity. One target of inference is P(Yt>k|S> j, x) [25], which is recovered from the PMM by summing over the survival distribution [38],
| (14) |
where P(Yt>k|S = g, x) is estimated from (13) and is only defined for t < g. Unfortunately, this approach only uses the ‘survivors’ under each treatment and the corresponding inference is not the causal effect of the treatment.
6.2.2. PMM approach applied
A large number of subjects dropped out of this study due to tumor progression or death (41 per cent). Based on consultation with Mayo investigators for assessing QOL, we grouped tumor progression and death together. When the dropout rates due to progression/death are broken down by treatment arms, the rates were marginally higher in the IFL arm. For subjects randomized to the IFL arm, 50 per cent dropped out due to progression/death, whereas 29 per cent dropped out due to progression/death for subjects with the FOLFOX arm (see Table III).
Table III.
Break down of completers and dropouts by treatment groups for QOL data.
| Dropouts by reason |
||||
|---|---|---|---|---|
| Treatment | Completer | Progression/death | Other | Total |
| IFL | 10 (0.04) | 117 (0.50) | 108 (0.46) | 235 |
| FOLFOX | 4 (0.02) | 68 (0.29) | 161 (0.69) | 233 |
| Total | 14 (0.03) | 185 (0.41) | 269 (0.57) | 468 |
Proportions are given in parentheses.
The number of subjects by progression/death windows by treatment groups is given in Table IV. For a subject who dropped out for reason unrelated to death/progression, we still know when the subject died or progressed if it happened before the study ended. For example, if a subject dropped out for reason unrelated to death/progression after the second visit and was alive until the study was finished, the subject belonged to progression/Death window 6.
Table IV.
Break down of progression/death windows by treatment groups for QOL data.
| Progression/death window |
|||||||
|---|---|---|---|---|---|---|---|
| Treatment | 1 | 2 | 3 | 4 | 5 | 6 | Total |
| IFL | 35 (0.15) | 32 (0.14) | 38 (0.16) | 26 (0.11) | 18 (0.08) | 86 (0.37) | 235 |
| FOLFOX | 16 (0.07) | 28 (0.12) | 24 (0.10) | 17 (0.07) | 19 (0.08) | 129 (0.55) | 233 |
| Total | 51 (0.11) | 60 (0.13) | 62 (0.13) | 43 (0.09) | 37 (0.08) | 215 (0.46) | 468 |
Window 6 means ‘after study.’ Proportions are given in parentheses.
We specify the OMREMs conditional on progression/death time, S as outlined in Section 6.2.1. Due to the small sample sizes when conditioning on individual times, we assumed that the parameters were the same for S=1,...,5 (those who progressed/died before the end of the study) but different for those who did not, S=6.
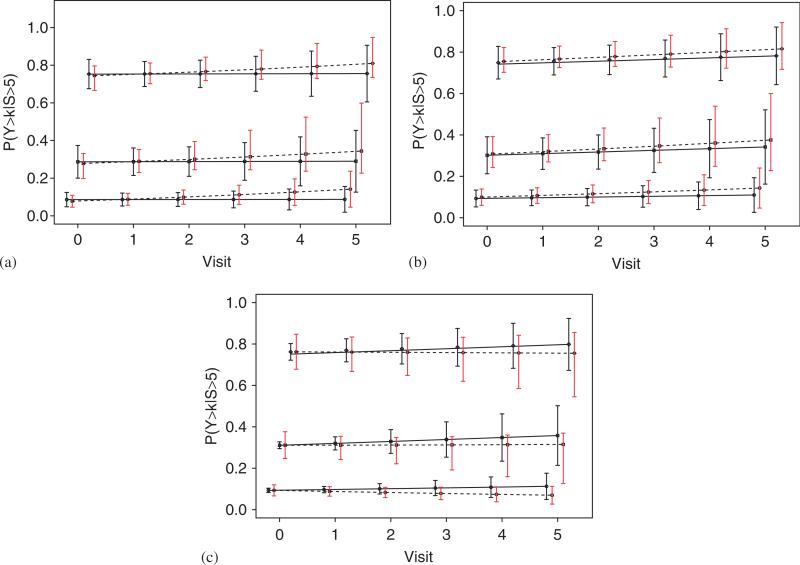
In our analysis here, we compared three models, an OMREM (OMREM-2), an independent proportional odds model (IPOM) under ignorable missingness and a mixture model (PMM) as outlined in Section 6.2.1. In the PMM, the target probabilities given in (14) evaluated at j = 5 correspond to those who did not progress/die before the end of the study. Table V presents the estimated target probabilities on the two treatment arms. Figure 1 indicates MLEs of P(Yt>k|S>5, Tx) for the PMM and those of P(Yt>k|Tx) for the MREM (OMREM) and the independent proportional odds model (IPOM) under ignorable missingness, respectively. In the PMM and OMREM, P(Yt>k|S>5, Tx) and P(Yt>k|Tx), which, respectively, evaluate the probability of fatigue for those who did not progress/die before the end of study and that for all patients were calculated. In the PMM, the target probabilities for the FOLFOX arm increased over time, whereas that for the IFL arm did not change. However, there were no significant differences over time between the treatment arms. In the OMREM, we have a pattern similar to that in the PMM. In the IPOM, the target probabilities for the FOLFOX were higher than those for the IFL unlike the other models as time increases because the estimate of coefficient of interaction between visit and arm was positive and large compared with those in the other models (1.059 for the IPOM and 0.469 for the OMREM). Because the PMM handled the missingness due to progression/death properly, we focus on the PMM and conclude that patients’ fatigue was not affected by treatment like in the inappropriate ignorable models. However, because we restrict the analyses for the QOL data to subgroup of patients who survived, the resulting treatment comparisons will no longer have a causal interpretation. In the following section, we present a principal stratification approach to address this.
Table V.
Maximum likelihood estimates of P(Yt>k|S>5, Tx) (standard errors) where S is progression/death time.
| Visit(t) |
|||||||
|---|---|---|---|---|---|---|---|
| Trt | k | 1 | 2 | 3 | 4 | 5 | 6 |
| IFL | 1 | 0.753 (0.039) |
0.754 (0.034) |
0.754 (0.037) |
0.755 (0.047) |
0.755 (0.061) |
0.756 (0.077) |
| 2 | 0.287 (0.044) |
0.288 (0.037) |
0.288 (0.040) |
0.288 (0.051) |
0.289 (0.066) |
0.290 (0.084) |
|
| 3 | 0.086 (0.019) |
0.086 (0.017) |
0.087 (0.019) |
0.087 (0.023) |
0.087 (0.028) |
0.087 (0.035) |
|
| FOLFOX | 1 | 0.731 (0.033) |
0.757 (0.028) |
0.781 (0.032) |
0.803 (0.039) |
0.823 (0.047) |
0.842 (0.054) |
| 2 | 0.264 (0.034) |
0.291 (0.031) |
0.320 (0.038) |
0.350 (0.054) |
0.381 (0.073) |
0.413 (0.095) |
|
| 3 | 0.078 (0.016) |
0.088 (0.016) |
0.099 (0.019) |
0.112 (0.026) |
0.126 (0.036) |
0.141 (0.049) |
|
P(Yt>k|S>5, Tx) evaluates probability of fatigue stress for those who did not progress/die before the end of the study.
Figure 1.
Maximum likelihood estimates of P(Yt>k|S>5, Tx) for the pattern mixture model (PMM) and those of P(Yt>k|Tx) for the marginalized random effects model (OMREM) and the independent proportional odds model (IPOM) under ignorable missingness, respectively, and 95 per cent confidence intervals, where S is progression/death time. P(Yt>k|S>5, Tx) evaluates the probability of fatigue for those who did not progress/die before the end of study. Solid line is for IFL; dashed line is for FOLFOX. ○, □, and ◇ are for k = 3, 2, 1, respectively. (a) PMM for k = 1, 2, 3; (b) OMREM for k = 1, 2, 3; and (c) IPOM for k = 1, 2, 3.
6.2.3. Principal stratification
Before defining the causal effect of treatment, we introduce potential outcomes. Potential outcomes are all the outcomes that would be observed if both treatments had been applied to each of the subjects [39–41]. Frangakis and Rubin [26] used the concept of potential outcomes in an approach called ‘principal stratification’. Principal stratification partitions subjects into sets with respect to post-treatment variables. The principal strata are not affected by treatment assignment and therefore can be used as any pre-treatment covariate. Causal effects are defined within these principal strata. In the following, the post-treatment variable of interest is progression/death.
Let Tx be the treatment indicator as defined earlier. Let Yt(Tx), the potential outcome at time t, be an ordinal QOL response. We only observe either Yt(1) or Yt(0) for each subject. Now, let Dt(Tx) be tumor progression/death indicator at visit t on treatment Tx,
Obviously, if Dt(Tx) = 1, then Ds(Tx) = 1 for s>t. In this case, there are four principal strata defined by the pairs of potential values of Dt(Tx):
At(0)={i|(Dt(0), Dt(1))=(0,0)}: the subjects who would be alive under both arms at visit t;
At(1)={i|(Dt(0), Dt(1)) = (1,0)}: the subjects who would be alive under the treatment arm but not alive under the control arm at visit t;
At(2)={i|(Dt(0), Dt(1)) = (0,1)}: the subjects who would be alive under control arm but not alive under the treatment arm at visit t;
At(3)={i|(Dt(0), Dt(1)) = (1,1)}: the subjects who would not be alive under either treatment at visit t.
As we are interested in subjects who were alive until the end of the study, At (3) is not of direct interest. The full set of potential outcomes at visit t is
= where for treatment tx, Yt(tx) is the potential response if a subject is alive at visit t (Dt(tx) = 0). If the patient is alive (Dit(Txi)=0), define Rit to be the indicator that Yit = Yit(Txi) is observed. In the following, we assume monotone dropout (Rit = 1 ⇒ Rit–1 = 1). Hence, the observed data for individual i is
We formally define the causal effects of interest and then state some additional assumptions that are necessary to estimate them.
Suppress i here for clarity and let T be the last visit in the study. We define the causal effect of interest [27] as
SACEk(1,0) is the odds ratios based on the probability that response at time T is larger than k for subjects who would be alive under both treatments between the treatment arm (FOLFOX) and the control arm (IFL).
Now, define to be the history of potential outcomes up to and including the outcomes at visit t. We first make the Stable Unit Treatment Value Assumption (SUTVA, [40]) which states that a patient's potential outcomes are unrelated to the treatment status of other patients. Next, we list the additional assumptions necessary to identify the causal effect of interest:
Assumption 1 (Monotonicity)
If Dt(1) = 1, then Dt(0) = 1; if Dt(0) = 0, then Dt(1) = 0.
Assumption 1 assumes that the active treatment is effective. As such, if a patient is alive under the control arm, then the patient will also be alive under the active arm. Note that At(2) is empty from Assumption 1.
Assumption 2 (Ignorability)
.
Note that is all the potential outcome up to and including time t. Assumption 2 states that the treatment arm is unrelated to the set of potential outcomes.
Assumption 3
.
This assumption states that missingness of the outcome is independent of the value of the outcome given all the potential outcome up to and including time t–1. It is similar to an assumption of sequential MAR [42].
Assumption 4 (Proportional odds)
where and
This assumption can be re-written as
| (15) |
We use this to reduce the number of sensitivity parameters. This will become apparent in what follows.
In general, the goal here is to use the observed data, , along with these assumptions to draw inference about SACE. To identify SACE, we need to identify P(YT(0)>k|AT(0)) and P(YT(1)>k|AT(0)). We provide the details of this identification using Assumptions 1–4 in the Appendix.
6.2.4. Principal stratification applied
In our study, even though there is a known survival difference between the groups, differential toxicity between the treatment arms led us to examine QOL in a manner not influenced by the survival difference.
The causal estimand of interest, SACE is a function of gT(Tx) and hT,Tx(k) (see below and Appendix for more details). For Tx=0, 1,
where N(tx) is the number of subjects who had the treatment Tx=tx. The estimate of gT(tx) is given by
| (16) |
where NT(tx) is the number of subjects who were alive after the study was complete under Tx=tx.
To define hT,Tx(k), we define S to be the progression/death time and specify the OMREM conditional on progression/death time as in (13). Within this framework, hT,Tx(k) is given by
| (17) |
where P(S = j|Tx) is a multinomial mass function for S, and P(YT>k|S=j, Tx) is calculated from (14) given Tx and is only defined for t < j. To calculate standard errors for SACE, we use the delta method.
The identification of the SACE relies on untestable assumptions. We implemented a sensitivity analysis procedure to draw inference about SACE by varying the sensitivity parameter τ in (15) (see Assumption 4). This is similar to the approach in [27]. The sensitivity parameter τ is the odds ratio of the probability of fatigue under the FOLFOX arm among the group that is alive under the FOLFOX arm but not alive under the control treatment at visit T when compared with the group that is alive under both arms. A large value of τ means that the probability of fatigue under the FOLFOX arm among patients in stratum AT(1) is higher than that among patients in stratum AT(0). The Mayo investigator told us it was very unlikely that τ was outside the range (0.5,2.0), corresponding to a two-fold or less change in either direction. Thus, we used this as the range for our sensitivity analysis.
The causal effect of treatment may be estimated for principal stratum AT(0), patients who would survive to the end of the study period regardless of the treatment. Table IV shows the proportions of patients who were alive after the study was finished (progression/Death window 6). Using the data in Table IV and (16), the MLEs of gT(·) were and .
To estimate SACEK–1(1,0) we also need to estimate P(DT(0)=0|DT(1)=0), P(YT(0)>K–1|AT(0)) and P(YT(1)>K–1|AT(0)). All these probabilities are estimable given Assumptions 1–4. The estimate of P(DT(0)=0|DT(1)=0), the probability of surviving under the control arm given survival under the FOLFOX arm, was 0.661. The estimate of P(YT(0)>K–1|AT(0)) was and P(YT(1)>K–1|AT(0)) was .
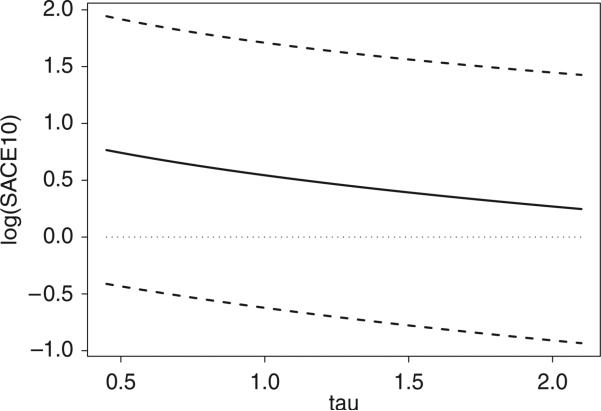
The estimated values of SACEK–1(1,0) and associated confidence intervals over τ ranging from 0.5 to 2.0 are given in Figure 2. As τ increased, the estimated values of SACEK–1(1,0) decreased. The estimated values of SACEK–1(1,0) were always larger than 1. This indicated that the probability of a patient's fatigue on the FOLFOX treatment was larger than the probability of a patient's fatigue on the IFL treatment. However, there was no significant difference between the two arms (at 95 per cent level). Thus, we conclude that patients’ fatigue was not affected by the treatment.
Figure 2.
SACEK–1(1,0) as a function of τ (Solid line). Dashed lines are 95 per cent confidence intervals.
7. CONCLUSION
We have proposed MREMs for longitudinal ordinal data that directly model marginal probabilities as a function of covariates while accounting for the longitudinal correlation via random effects. To evaluate the marginalized likelihood, we used Monte Carlo integration. Parameter estimation was based on ML using a quasi-Newton algorithm.
As discussed in Section 6, about 40 per cent of the patients dropped out due to tumor progression or death. We adjusted for this using a pattern mixture approach with patterns defined based on the observed progression/death times and then using a principal stratification approach to estimate treatment effects. We plan on extending this to a fully Bayesian approach and as such, using informative priors for the sensitivity parameters to obtain a single inference that characterizes our uncertainty about the sensitivity parameters. We also will consider an extension to three treatments (two active and one placebo) and a weakening of the monotonicity assumptions.
We can extend marginalized ordinal models to allow both serial dependence via a Markov structure and random effects [43]. These models are particularly useful in longitudinal analyses with a moderate to large number of repeated measurements per subject. We are also working on extensions to multivariate longitudinal ordinal responses which would be applicable to QOL data [44].
ACKNOWLEDGEMENTS
We would like to thank Dr Daniel Sargent and Ms Erin Green of the Mayo clinic for providing the data, for their help in data collection, and for clarifying some issues with the data. The authors gratefully acknowledge two referees for their helpful comments on this paper. This project was supported by NIH grants CA85295 and HL079457.
Contract/grant sponsor: Publishing Arts Research Council; contract/grant number: 98-1846389
Contract/grant sponsor: NIH; contract/grant numbers: CA85295, HL079457
APPENDIX A
A.1. Proof of Theorem 1
We know that β01<···<β0K–1 implies that
where
Hence,
for all k. We can rewrite this as
| (A1) |
We now claim that Δitk>Δitk–1 for all k. We will show that if it is not true (i.e, Δitk≤Δitk–1 for some k), then condition (A1) cannot be satisfied.
If Δitk = Δitk–1 for some k, then the left-hand side term of (A1) is zero. This is a contradiction to (A1).
- If Δitk < Δitk–1 for some k, then let Δitk–1 = Δitk + ε for some ε>0. Therefore, (A1) is given by
As , this is a contradiction to (A1).
A.2. Detailed calculations of quasi-Newton under ignorability
The contribution of subject i to the log likelihood is given by
where , and .
The forms of the derivatives for quasi-Newton algorithm are
where L(θ,ai|yi) is given by (10) and reexpressed as
for j = 1,..., K–1. The integrals are estimated using Monte Carlo integration. We simply generate 10 000 random vectors from multivariate standard normal distributions to compute the integrals.
To make the derivatives simpler, (8) can be reexpressed as
| (A2) |
where ϕ(·) is the standard normal density function. Note that the integral in (A2) is one dimensional. To compute the score vector and information matrix, we also need derivatives of Δit with respect to β0, β and λ. They can be obtained from relationship (A2),
Similarly, we have
A.3. Identification of the causal effects
Based on observed data and Assumptions 1–4 in Section 6.2.3, we show how to estimate the causal effects SACE. Assumption 1 implies that
for tx=0,1. Assumption 2 implies that
| (A3) |
and
for tx=0,1. From Assumptions 2 and 3, we have that
and
for tx=0,1.
Recall we need to estimate P(YT(1)>k|AT(0)) to estimate SACEk(1,0). To do this, we show that P(YT(1)>k|DT(1)=0) can be expressed as
| (A4) |
The original expressions P(YT(tx)>k|DT(tx)=0) for tx=0,1 are identifiable. However, only some of the factors in (A4) are identified. The mixing probabilities, P(DT(0)=0|DT(1)=0) and P(DT(0)=1|DT(1)=0), are identifiable by (A3) as
However, P(YT(1)>k|AT(0)) and P(YT(1)>k|AT(1)) in (A4) are not identified. Given the identified components, to identify these unidentified quantities, we only need to know their ratios. All three ratios are identified via Assumption 4,
| (A5) |
From (A4) and (15), we can identify P(YT(1)>k|AT(0)) by solving the following quadratic equation:
where x = P(YT(1)>k|AT(0)). When τ=1,
When , we have
where a(τ) = (1–τ)gT(0), b(τ) = (τ–1)(hT,1(k)gT(1)+gT(0))–τgT(1), and c(1) = hT,1(k)gT(1). Note that the solution is a decreasing function of τ.
REFERENCES
- 1.Liang KY, Zeger SL. Longitudinal data analysis using generalized linear models. Biometrika. 1986;73:13–22. [Google Scholar]
- 2.Fitzmaurice GM, Laird NM. A likelihood-based method for analysing longitudinal binary responses. Biometrika. 1993;80:141–151. [Google Scholar]
- 3.Zeger SL, Liang KY, Albert PS. Models for longitudinal data: a generalized estimating equation approach. Biometrics. 1988;44:1049–1060. [PubMed] [Google Scholar]
- 4.Zeger SL, Karin MR. Generalized linear model with random effects: a Gibbs sampling approach. Journal of the American Statistical Association. 1991;86:79–86. [Google Scholar]
- 5.Breslow NE, Clayton DG. Approximate inference in generalized linear mixed models. Journal of the American Statistical Association. 1993;88:125–134. [Google Scholar]
- 6.Laird NM. Missing data in longitudinal studies. Statistics in Medicine. 1988;7:305–315. doi: 10.1002/sim.4780070131. [DOI] [PubMed] [Google Scholar]
- 7.Akaike H. A new look at the statistical model identification. IEEE Transactions on Automatic Control. 1974;19:716–723. [Google Scholar]
- 8.Schwarz G. Estimating the dimension of a model. The Annals of Statistics. 1978;6:461–464. [Google Scholar]
- 9.Heagerty PJ. Marginally specified logistic-normal models for longitudinal binary data. Biometrics. 1999;55:688–698. doi: 10.1111/j.0006-341x.1999.00688.x. [DOI] [PubMed] [Google Scholar]
- 10.Heagerty PJ. Marginalized transition models and likelihood inference for longitudinal categorical data. Biometrics. 2002;58:342–351. doi: 10.1111/j.0006-341x.2002.00342.x. [DOI] [PubMed] [Google Scholar]
- 11.Heagerty PJ, Zeger SL. Marginalized multilevel models and likelihood inference (with Discussion). Statistical Science. 2000;15:1–26. [Google Scholar]
- 12.Miglioretti D, Heagerty P. Marginal modeling of multilevel binary data with time-varying covariates. Biostatistics. 2004;5:381–398. doi: 10.1093/biostatistics/5.3.381. [DOI] [PubMed] [Google Scholar]
- 13.Lee K, Daniels M. A class of Markov models for longitudinal ordinal data. Biometrics. 2007;63:1060–1067. doi: 10.1111/j.1541-0420.2007.00800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dale JR. Global cross-ratio models for bivariate, discrete, ordered responses. Biometrics. 1986;42:909–917. [PubMed] [Google Scholar]
- 15.Molenberghs G, Lesaffre E. Marginal modeling of correlated ordinal data using a multivariate Plackett distribution. Journal of the American Statistical Association. 1994;89:633–644. [Google Scholar]
- 16.Williamson JM, Kim K, Lipsitz SR. Analyzing bivariate ordinal data using a global odds ratio. Journal of the American Statistical Association. 1995;90:1432–1437. [Google Scholar]
- 17.Heagerty PJ, Zeger SL. Marginal regression models for clustered ordinal measurements. Journal of the American Statistical Association. 1996;91:1024–1036. [Google Scholar]
- 18.Gibbons R, Hedeker D. Random effects probit and logistic regression models for three-level data. Biometrics. 1997;53:1527–1537. [PubMed] [Google Scholar]
- 19.Todem D, Kim K, Lesaffre E. Latent-variable models for longitudinal data with bivariate ordinal outcomes. Statistics in Medicine. 2006;26:1034–1054. doi: 10.1002/sim.2599. [DOI] [PubMed] [Google Scholar]
- 20.Liu LC, Hedeker D. A mixed-effects regression model for longitudinal multivariate ordinal data. Biometrics. 2006;62:261–268. doi: 10.1111/j.1541-0420.2005.00408.x. [DOI] [PubMed] [Google Scholar]
- 21.Liu I, Agresti A. The analysis of ordered categorical data: an overview and a survey of recent developments. Test. 2005;14:1–73. [Google Scholar]
- 22.Hogan JW, Roy J, Korkontzelou C. Tutorial in biostatistics handling drop-out in longitudinal studies. Statistics in Medicine. 2004;23:1455–1497. doi: 10.1002/sim.1728. [DOI] [PubMed] [Google Scholar]
- 23.Hogan JW, Laird NM. Mixture models for the joint distribution of repeated measures and event times. Statistics in Medicine. 1997;16:239–257. doi: 10.1002/(sici)1097-0258(19970215)16:3<239::aid-sim483>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 24.Pauler DK, McCoy S, Moinpour C. Pattern mixture models for longitudinal quality of life studies in advanced stage disease. Statistics in Medicine. 2003;22:795–809. doi: 10.1002/sim.1397. [DOI] [PubMed] [Google Scholar]
- 25.Kurland BF, Heagerty PJ. Directly parameterized regression conditioning on being alive: analysis of longitudinal data truncated by deaths. Biostatistics. 2005;6:241–258. doi: 10.1093/biostatistics/kxi006. [DOI] [PubMed] [Google Scholar]
- 26.Frangakis CE, Rubin DB. Principal stratification in causal inference. Biometrics. 2002;58:21–29. doi: 10.1111/j.0006-341x.2002.00021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Egleston B, Scharfstein DO, Freeman ER, West SK. Causal inference for non-mortality outcomes in the presence of death. Biostatistics. 2007;8:526–545. doi: 10.1093/biostatistics/kxl027. [DOI] [PubMed] [Google Scholar]
- 28.Rubin DB. Comment on causal inference without counterfactuals by A. P. Dawid. Journal of the American Statistical Association. 2000;6:34–58. [Google Scholar]
- 29.Hayden D, Pauler DK, Schoenfeld D. An estimator for treatment comparisons among survivors in randomized trials. Biometrics. 2005;61:305–310. doi: 10.1111/j.0006-341X.2005.030227.x. [DOI] [PubMed] [Google Scholar]
- 30.Rubin DB. Causal inference through potential outcomes and principal stratification: application to studies with ‘censoring’ due to death. Statistical Science. 2006;21:299–309. [Google Scholar]
- 31.Goldberg RM, Sargent DJ, Morton RF, Fuchs CS, Ramanathan RK, Williamson SK, Findlay BP, Pitot HC, Albets SR. A randomized controlled trial of fluorouracil plus leucovorin, irinotecan, and oxaliplatin combinations in patients with previously untreated metastatic colorectal cancer. Journal of Clinical Oncology. 2004;22:23–30. doi: 10.1200/JCO.2004.09.046. [DOI] [PubMed] [Google Scholar]
- 32.McCullagh P. Regression models for ordinal data. Journal of the Royal Statistical Society, Series B. 1980;42:109–142. [Google Scholar]
- 33.Heagerty PJ, Kurland BF. Misspecified maximum likelihood estimates and generalised linear mixed models. Biometrika. 2001;88:973–985. [Google Scholar]
- 34.Gibbons R, Bock R. Trend in correlated proportions. Psychometrika. 1987;52:113–124. [Google Scholar]
- 35.Hedeker D, Gibbons RD. A random-effects ordinal regression model for multilevel analysis. Biometrics. 1994;50:933–944. [PubMed] [Google Scholar]
- 36.Liu Q, Pierce DA. A note on Gauss–Hermite quadrature. Biometrika. 1994;81:624–629. [Google Scholar]
- 37.Pinheiro JC, Bates DM. Approximations to the log-likelihood function in the nonlinear mixed-effects model. Journal of Computational and Graphical Statistics. 1995;4:12–35. [Google Scholar]
- 38.Fitzmaurice GM, Laird NM. Generalized linear mixture models for handling nonignorable dropouts in longitudinal studies. Biostatistics. 2000;1:141–156. doi: 10.1093/biostatistics/1.2.141. [DOI] [PubMed] [Google Scholar]
- 39.Rubin DB. Estimating causal effects of treatments in randomized and nonrandomized studies. Journal of Educational Psychology. 1974;66:688–701. [Google Scholar]
- 40.Rubin DB. Bayesian inference for causal effects. Annals of Statistics. 1978;6:34–58. [Google Scholar]
- 41.Holland P. Statistics and causal inference. Journal of the American Statistical Association. 1986;81:945–961. [Google Scholar]
- 42.Robins JM, Rotnitzky A, Zhao LP. Analysis of semiparametric regression models for repeated outcomes in the presence of missing data. Journal of the American Statistical Association. 1995;90:106–121. [Google Scholar]
- 43.Schildcrout J, Heagerty PJ. Marginalized models for moderate to long series of longitudinal binary response data. Biometrics. 2007;63:322–331. doi: 10.1111/j.1541-0420.2006.00680.x. [DOI] [PubMed] [Google Scholar]
- 44.Ilk O, Daniels M. Marginalized transition random effects models for multivariate longitudinal binary data. Canadian Journal of Statistics. 2007;35:105–123. [Google Scholar]