Summary
The Gene Balance Hypothesis states that the stoichoimetry of members of multi-subunit complexes affects the function of the whole due to the kinetics and mode of assembly. Gene regulatory mechanisms would be governed by these principles. Here, we review the impact of this concept with regard to the effects on the genetics of quantitative traits, the fate of duplication of genes following polyploidization events or segmental duplication, the basis of aneuploid syndromes, the constraints on cis and trans variation in gene regulation and the potential involvement in hybrid incompatibilities.
Keywords: aneuploidy, cis-trans regulation, Gene Balance Hypothesis, hybrid incompatibilites, interactome, quantitative traits, polyploidy
Introduction
The Gene Balance Hypothesis posits that altering the stoichiometry of members of multi-subunit complexes will affect the function of the whole as a result of the kinetics and mode of assembly (Birchler et al., 2005; Birchler & Veitia, 2007) (Fig. 1). The idea of balance has a long history in classical genetics on the phenotypic level (Blakeslee et al., 1920) but only recently have parallels been found on the gene expression and evolutionary levels to congeal into a synthesis. The concept of gene balance raises several questions with regard to genetic variation and how it affects evolutionary processes that deserve investigation in this context. In this review, we propose an integration of the phenotypic data with gene regulatory mechanisms and their impact on quantitative trait control.
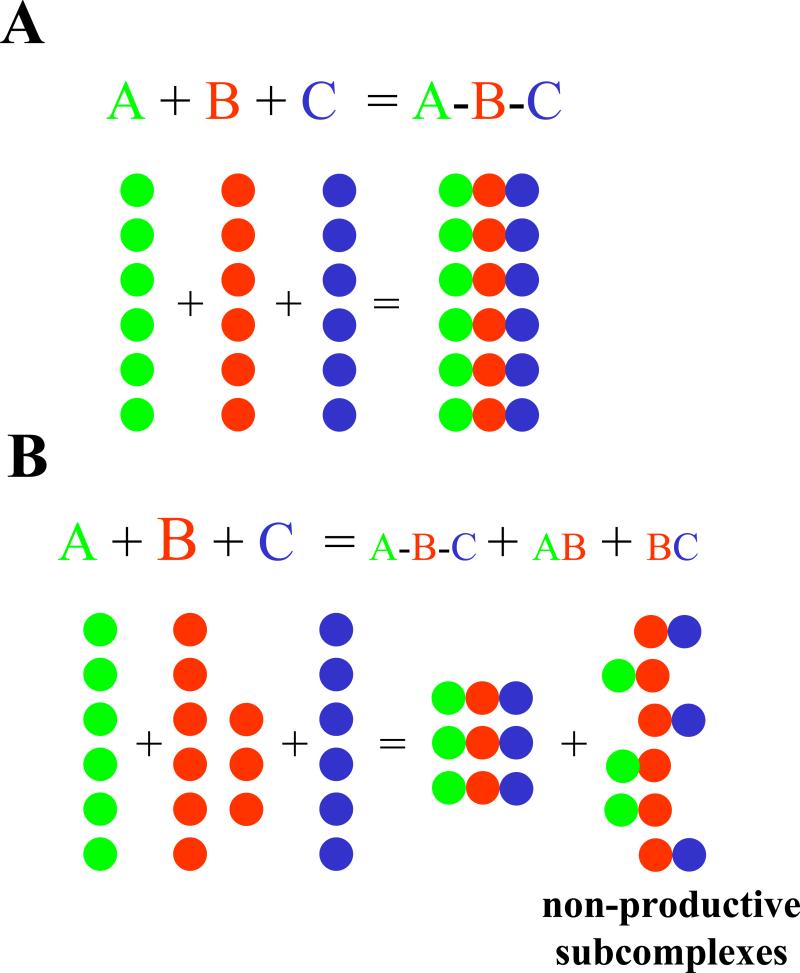
Fig. 1.
Effects of over-expression of a dosage sensitive subunit involved in a macromolecular complex. (a) Irreversible assembly of trimer A-B-C allowing intermediate dimers AB and BC. Although not represented in the figure the reactions involved are A+B=AB, B+C=BC, AB+C=ABC and BC+A=ABC. (b) An excess of the molecular bridge B (1.5 X) leads to a decrease in yield of ABC due to the production of intermediates that cannot be completed because of the lack of enough A and C monomers (notice how incomplete dimeric complexes outnumber the normal expected trimers).
While the Gene Balance Hypothesis applies to any complex of subunits or interaction hierarchy of genes, we will focus on the issue of gene regulation. Clearly, regulatory processes are mediated by multi-subunit complexes and thus would fall into this general concept. Thus, regulatory genes are dosage dependent to some degree--an empirical observation that was one piece of evidence that led to the gene balance hypothesis.
Beginning with the work of Blakeslee on Datura (Blakeslee et al., 1920; Blakeslee, 1934; Satina et al., 1937) a dichotomy was observed between the impact of aneuploidy and whole genome ploidy changes on plant stature. Indeed, extra copies of individual chromosomes or chromosome arms had more detrimental effects on the phenotype than varying the complete set of chromosomes. These observations have been recapitulated in many other plant species and similar results were noted in the animal kingdom particularly with work on Drosophila (Bridges, 1925).
Subsequently, parallel results were found with gene expression studies. Aneuploid samples of maize or Drosophila exhibited more modulations of gene expression than did changes in ploidy (Birchler et al., 2001). The modulations of enzyme activities or protein levels that change in aneuploids usually fall within the range of either direct or inverse correlations with the dosage of the varied chromosome (Birchler, 1979; Birchler & Newton, 1981; Devlin et al., 1988; Sabl & Birchler, 1993; Guo & Birchler, 1994) to produce direct or inverse dosage effects. Many fewer changes were observed with changes in ploidy (Guo et al., 1996). Similar types of effects were observed at the RNA level for selected genes analyzed (Guo & Birchler, 1994). It was postulated that alterations of the stoichiometry of regulatory genes might be responsible for this spectrum of effects (Birchler & Newton, 1981).
Whenever a regulatory gene producing an inverse dosage effect and a subset of its target loci are varied on the same chromosomal segment, dosage compensation is observed. In other words, increasing the dosage of a structural gene increases its expression in proportion to copy number. However, increasing the dosage of an inverse regulator at the same time would restore the original diploid expression level (Birchler, 1979, 1981; Devlin et al., 1982; Birchler et al., 1989) because the two opposing effects cancel each other. The first described example involved the alcohol dehydrogenase (adh1) gene on the long arm of chromosome 1 in maize (Birchler, 1981). Indeed, varying the dosage of the gene itself produced a gene dosage effect for the amount of product observed. On the other hand, varying another portion of 1L produced an inverse dosage effect on the levels of ADH. When both regions are varied together in the whole of chromosome arm 1L, dosage compensation of ADH levels was observed. Compensation also operates on the RNA level (Birchler et al., 1990; Guo & Birchler, 1994).
The inverse and direct effects on gene expression observed in aneuploids could be mimicked by the recovery of single gene mutations that would increase in the range of two fold or reduce in the range of one half the expression of a reporter gene in Drosophila (Rabinow et al., 1991). A leaky allele of the white eye color gene was used to find modifier mutations that would reduce or increase its expression as a heterozygote. A collection of 47 such mutations in independent genes was recovered over two decades of screening (Birchler et al., 2001). These genes show a dosage effect on the white eye color reporter gene and thus are reasonable candidates for being responsible for the aneuploid effects. The majority showed an increased effect on white while others reduced the expression (Fig. 2).
Fig. 2.
Heuristic example for the explanation of inverse dosage effects produced by a regulatory trimer ABC with variation of subunit B. Consider that trimer assembly is random and irreversible (the reactions are represented within the frame of the graph and are the same as in Fig. 1). The reaction conditions that can lead to the observed inverse dosage effects mentioned in the text are considered. For each mole of monomer of A and C, the trimerization reaction involves three moles of monomer B and that all kinetic constants are the same (unitary for simplicity). This scenario will yield 0.33 moles of ABC (and incomplete subcomplexes). This reaction will produce 100% of ABC. For the same (unitary) amounts of A and C, halving the amount of B (1.5 moles of B) leads to 200% of trimer yield and increasing the relative amount of B (to 4.5 mole) leads to 67% of trimer. Thus, if the trimer acts positively on target genes and B is varied in an aneuploid series, the inverse effect on target loci would result. This simplified view considers that the reacting amounts of A, B and C in steady state. Notice that A, B and C can also be multi-subunit subcomplexes. Such an example does not exclude the possibility that an inverse dosage effect is produced by negatively acting regulators.
The connection between dosage effects and QTL
It is an interesting question of how there can be such a large number of modifiers of the white eye color gene. The answer likely resides in the fact that gene regulatory systems operate in a hierarchy with one regulator controlling some downstream, which in turn control other regulators (Birchler et al., 2001). Moreover, regulators are often multisubunit complexes and thus each subunit-encoding gene can be a modifier of the phenotype. The developmental progression of the Drosophila embryo has been studied in great detail and illustrates the cascade of dosage dependent regulatory molecules that trigger off each subsequent step in development (Driever et al., 1988; Struhl et al., 1989; Warrior & Levine, 1990; Sauer & Jackle, 1991; Schulz & Tautz, 1994; Cribb et al., 1995). If the amount of one modifier is changed and it modulates another dosage dependent modifier, the change in gene expression is transmitted through the hierarchy. The combination of several gene products contributing to any particular regulatory complex and the fact that regulation often operates through a hierarchy would contribute to the multigenic control of the ultimate phenotype.
Such a multigenic control of a single phenotypic trait has parallels from quantitative traits. Thus, a particular trait is affected by multiple loci when genetic variation for the trait is analyzed (Tanksley, 1993). When lines are crossed that show phenotypic extremes, there is some measure of additivity in the hybrid. The first quantitative trait locus cloned, fruit weight 2.2 (fw2.2), shows a negative dosage effect on the size of tomato fruit (Frary et al., 2000). In other words the controlling gene shows allelic dosage effects. Other QTL are associated with regulatory changes that are additive (Liu et al., 2002; Cong et al., 2002, 2008). A comprehensive analysis of quantitative trait differences between wild and domesticated sunflower found 78 QTL for 18 characters, which were mostly of small effect and not completely dominant or recessive, i.e. demonstrated a dosage effect to some degree (Burke et al., 2002). In an exhaustive study of QTL affecting flowering time in maize, a large number were identified largely of very small effect (Buckler et al., 2009). In the artificial selection experiment for different oil content in maize, greater than 50 QTL appear to be involved (Laurie et al., 2004). These observations from the field of quantitative genetics fit together with the fact that multiple aneuploidies have small effects on the same traits and suggest that multiple dosage sensitive modifiers can control one phenotype (Guo & Birchler, 1994; Birchler et al., 2001).
This principle is further illustrated by the mechanistic example of multigenic control of a phenotype by unlinked non-complementation. In this condition, two heterozygous mutations in different loci lead to a phenotype while the single heterozygotes are normal. This phenomenon has been reported in physically interacting products, in proteins that do not interact directly but belong to the same complex, between gene products involved with the same pathway and in proteins apparently not involved in the same pathway but functionally related (Yook et al., 2001). One explanation of unlinked non-complementation is that the combined haplo-insufficiency due to a decrease in the dosage of both loci would be responsible for the abnormal phenotype. Of course, this paradigm can be extended to more loci in which case a greater number of defective alleles would be required to engender an abnormal phenotype. A wider panoply of phenotypes can be expected for measurable characters depending on the number of deleterious alleles at different loci, appearing in the same individual (Fig. 3).
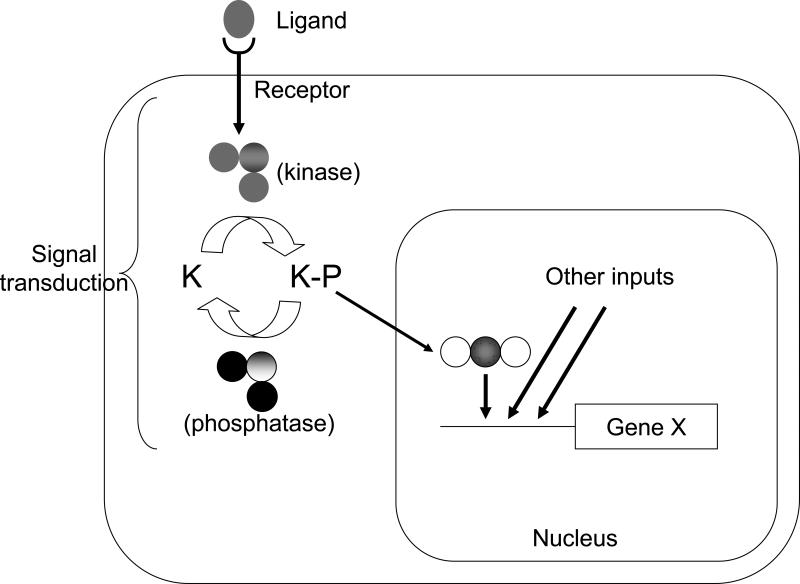
Fig. 3.
Idealized informational pathway showing how many genes can have an impact on a transcriptional (and phenotypic output). The membrane receptor and its ligand must respect some balance to prevent outright receptor saturation or insufficient stimulation. The receptor, upon binding its ligand, is able to activate the trimeric kinase (which can undergo the very same dosage effects outlined in Figs 1 and 2). This kinase is counteracted by a phosphatase and they must respect some stoichiometric balance as proposed (Veitia, 2004). Their action leads to some degree of phosphorylation of kinase K which in turns activates a transcription factor that will promote transcription of gene X, which is under the combined control of other transcription factors, responding to the same or different transduction cascades.
Dosage effects: molecular bases
The molecular nature of the collection of white eye color modifiers has been elucidated to a large extent. The collection of the dosage sensitive modifiers was revealed to contain transcription factors, chromatin proteins or members of signal transduction cascades (Birchler et al., 2001). This fact parallels nicely with the realization that most haplo-insufficient clinical conditions in human are lesions in transcription factors or other proteins involved with interacting complexes (Seidman & Seidman, 2002; Veitia, 2002; Kondrashev & Koonin, 2004).
This realization led to the modeling of haplo-insufficiencies with the idea that the stoichiometry of interacting subunits would generate a semi-dominant effect on the phenotype based on the action on the target loci (Veitia, 2002). An experimental demonstration of this principle was provided by evidence from diploid yeast in which the heterozygote fitness was measured for null mutations in essential genes. As the genes were grouped into bins of different heterozygous fitness, a greater proportion were involved in protein complexes as the fitness declined (Papp et al., 2003). Furthermore, over-expression of genes involved in complexes is also detrimental, which can be corrected by co-expression of inter-actors.
A biophysical property of proteins that is an indication of involvement in protein complexes is the degree of underwrapping, which is a measure of how protein stability relies on interactions with other partners. Interestingly, when the degree of underwrapping is plotted against gene duplicability, there is a negative correlation in several major taxa examined (Liang et al., 2008). This correlation further indicates that genes whose products are involved in complexes have detrimental effects on fitness when changed from the normal stoichiometric relationship.
Recently, it has been realized that polyploidization events have been quite common in the evolution of eukaryotes (Wolfe & Shields, 1997; Simillion et al., 2002; Bowers et al., 2003; Maere et al., 2005; Aury et al., 2006; Blomme et al., 2006). In fact, basically all eukaryotes have had a history of ancient polyploidization events followed by diploidization and then repeated polyploidizations. The yeast genome was found to show evidence of duplicated genes that indicate an ancient whole genome doubling (Wolfe & Shields, 1997). Such events were subsequently shown to be quite common in flowering plants. In the first sequenced angiosperm, Arabidopsis, analysis of gene duplicates provides evidence of at least three whole genome duplications (WGD) stretching back a quarter of a billion years (Blanc & Wolfe, 2004; Freeling & Thomas, 2006). Rice and maize share a WGD event that is independent of the two most recent ones in Arabidopsis and maize has yet another genome doubling layered atop the one in common with rice (Freeling & Thomas, 2006). In Paramecium tetraurelia, there are three tetraploidization events that can be documented from the analysis of the sequenced genome, each followed by a reduction in gene number (Aury et al., 2006). The most recent event allows one to determine the classes of genes that show evidence of deletion or degeneration into pseudogenes.
As the genomes lose genes following polyploidization events, the spectrum of retained genes is not random. Indeed, in the independent diploidizations in yeast, Arabidopsis, rice, composites and Paramecium, the classes of genes retained are preferentially represented by those that are involved in protein complexes or that interact with other proteins (Seoighe & Gehring, 2004; Blanc & Wolfe, 2004; Freeling & Thomas, 2006; Aury et al., 2006; Hakes et al., 2007; Barker et al., 2008). Duplicates of transcription factors and members of signal transduction pathways, which are included among those involved in complexes and interacting hierarchies, are typically maintained. Thus, the same classes of genes are held in duplicate as those that show dosage effects as regulatory modifiers and quantitative trait determinants (Birchler et al., 2001). It seems likely that the stoichiometry of these gene products is important and that deletion of one member of a duplicate pair might act like an aneuploid effect via an influence on target gene expression and be selected against due to reduced fitness. These genes would therefore be retained in the genome over longer periods of evolutionary time than other classes. Indeed, in the Paramecium study, the retained duplicates showed evidence of purifying selection indicating that there is strong selection to maintain the function of these genes and in the proper balance (Aury et al., 2006).
The finding of retention of duplicate pairs in such a strict manner implies that dosage is important, as is the relationship of the gene products to other protein quantities. This fact also illustrates the narrow range of modulation that appears to be tolerated. Following a WGD, there would be four copies of a particular transcription factor gene, for example, instead of the two present in the progenitor diploid species. The mere reduction to only three functional copies is apparently not usually tolerated. All of these findings are in agreement with the Gene Balance Hypothesis, which makes different predictions on the fate of duplicate genes in evolution depending on whether they are duplicated via polyploidization or via a segmental duplication. Indeed, WGD events do not perturb gene dosage balance and allow retention of multiple interacting components of a complex during the diploidizaiton process. On the contrary, isolated duplication of genes encoding subunits of a complex will be forbidden due to dosage effects. Accordingly, such duplicated genes are underrepresented in genomic analyses in several organisms (Dopman & Hartl, 2007; Freeling et al., 2009).
Consistent with the results of preferential retention of genes in regulatory balance following ancient polyploidization events are the results of a comparison of mutation accumulation lines compared to natural isolates in C. elegans (Denver et al., 2005). These authors propagated a set of lines of nematodes that would accumulate mutations and compared them with natural variants at the transcriptomic level. The results suggested that very strong purifying selection was operating on the expression patterns that could not vary over a great magnitude and that this observation could not be explained by a neutral hypothesis. There appears to be a very limited tolerance for modulation of many regulatory genes, which is consistent with the balance concept and the evolutionary retention following polyploidization events.
There are several possible explanations for the retention of specific classes of genes following WGD. Insight into the reason can be gained from studies of classes of genes found in segmental duplications, of which there is also a nonrandom distribution. In general, genes involved with molecular complexes and interactions are underrepresented (Yang et al., 2003; David & Petrov, 2005; Maere et al., 2005; Freeling & Thomas, 2006; Freeling et al., 2009). In this case, the duplication of only a single member of an interacting complex would mimic an aneuploid effect via an altered stoichiometry. Thus, there is a generalized complementary pattern of gene ontologies present in segmental duplications compared to those that are retained following polyploidization events.
This principle extends to copy number polymorphisms (CNP) in populations as studied in Drosophila and human (Dopman & Hartl, 2007; Kim et al., 2007; Nguyen et al., 2006, 2009). CNP for genes with network centrality are underrepresented for deletions. The proportion of CNPs involved with at least one interaction was not reduced for duplications. However, for genes with greater network centrality, duplications were underrepresented. CNPs in humans are depleted for genes with network centrality but tolerated for those on the periphery (Korbel et al., 2008; Ionita-Laza et al., 2009). Thus, the more intricately interacting a particular gene product is, the more likely that quantitative variation for it will be constrained.
Implications
The above-mentioned studies have several implications that are deserving of study. First of all, the fact that most regulatory processes involve multi-subunit complexes when considered together with the evolutionary and quantitative genetic studies suggests that new mutations that arise in genes encoding one member of a complex will exhibit a dosage effect on the phenotype, however subtle. If this new mutation is detrimental, it will be selected against quickly. If it is adaptive, then the heterozygous mutation can spread readily through the population. Whether this new mutation becomes fixed will depend on the nature of the effect of the homozygous mutation. If the new mutation is neutral, then its fate will depend on the vagaries of genetic drift if the effective population size is small or to founder effects, to bottleneck effects or to hitchhiking effects for spread in a population. The important aspect to realize is that any new regulatory mutation that provides a reproductive advantage, while rare in relation to detrimental mutations, has the capability to spread in a population as a heterozygote due to its subtle semi-dominant dosage effect. Thus, there is potentially a greater availability for adaptive selection for regulatory genes than for other classes of genes that do not exhibit tight dosage stoichiometries.
A second implication is that the balanced nature of regulatory systems can maintain the status quo (Williams, 1966) in the absence of adaptive selection. The retention of duplicates following tetraploidization events for long evolutionary time periods (Aury et al., 2006; Freeling & Thomas, 2006) indicates the resistance to change that the balance relationship maintains. Selection pressure is needed to alter the impact of changes in the stoichiometric relationship of molecular complexes. Examples are known from the fossil record of the maintenance of morphological form for some species over extended periods (e. g. ginkgo trees, horseshoe crabs) while other species can change rapidly. The balance concept suggests that the default mode is the maintenance and the changes will be selected under altered environmental conditions.
These considerations raise the issue of how selection can cause rapid change under certain circumstances. Dogs were domesticated from wolves (Gray et al., 2009), which probably still look similar to the original. However, domesticated dogs under intense artificial selection have diverged into many forms. In another example, Darwin was fascinated by the diversity of pigeons that could be selected by fanciers (Darwin, 1859). The famous Illinois high oil selection experiment in corn has been in progress for over 100 yr with no plateau (Laurie et al., 2004). The basis of why these selection experiments seem to respond so well is still a matter of debate as to whether they do so from standing variation, new mutations, epigenetic changes (Gibson & Dworkin, 2004; Johannes et al., 2008) or a combination of the above. A possibility that should be explored in the context of balance is that many loci would be predicted to affect any particular characteristic. Any standing variation, be it genetic or epigenetic, at any of the potential loci affecting specific characteristics is likely to be very subtle, much less than two-fold effects surrounding the population mean. This estimate is based on the evolutionary results of retention from WGD and depletion in segmental duplications for regulatory factors, as noted above. However, if standing variation with subtle changes does exist, the fact that so many dosage dependent factors can modify any one characteristic has the potential to explain the progression to dramatically new phenotypes in the event of strong selection.
The available data from evolutionary studies suggests that, for regulatory genes, variation in expression at or beyond the two-fold range is largely selected against. Consistent with this observation is that monosomics in most organisms are either lethal or highly detrimental indicating two-fold changes for developmental regulation is not tolerated (Lindsley & Sandler et al., 1972; Bond & Chandley, 1983). However, because selection experiments work and because examination of individuals in a population reveal morphological variation, some level of subtle variation is standing in populations but is probably neutral under most circumstances and much below a two-fold effect (a room of men show height variation. A nearly 2-m male would be in the normal range but there are no nearly 4 m males!). The implication for quantitative traits is thus that changes will occur by very small changes accumulated from many genes. It is also reasonable to suppose that selection for a particular characteristic in one direction from the norm can occur in thousands of ways given the possibility for selecting different alleles at multiple dosage sensitive loci. Lastly, the quantitative relationship of interacting gene products would also likely contribute to the outcome.
These considerations are consistent with studies on the cis versus trans regulatory variation found in global studies of gene expression. For interacting regulatory genes that produce trans-acting variation, there is likely to be a restriction to very small changes. As noted above, the evolutionary results from whole genome or segmental duplications suggest a very narrow window of magnitude for the variation to remain neutral. By contrast, cis variation in genes that are not dosage sensitive or detrimental can be much greater without any fitness consequences. Studies of cis versus trans allele specific effects in hybrid conditions and from eQTL mapping of gene expression profiles across various genomes is generally consistent with a large number of global trans-acting genes with effects of relatively small magnitude (or in some cases below detection) while the cis effects on individual genes are of greater magnitude (Schadt et al., 2003; Yvert et al., 2003; Morley et al., 2004; Wayne et al., 2004; Wittkopp et al., 2004; Hughes et al., 2006; Petretto et al., 2006; Wang et al., 2007; West et al., 2007; Grieve et al., 2008; Tirosh et al., 2009).
The realization that two-fold effects are not well tolerated for many regulatory processes raises the interesting question of parental gene imprinting in which there is monoallelic expression at a locus depending on the history of the two alleles. Most imprinted genes appear to be dosage sensitive and thus the phenomenon is a non-mutational mechanism to alter the amount of gene expression (Beaudet & Jiang, 2002). When biparental inheritance for imprinted genes does not occur, highly detrimental phenotypes result. If two inactive alleles are present, one can easily rationalize these results as being due to the absence of the encoded gene product. However, there are also detrimental effects when two active alleles are present. This observation illustrates the sensitivity of the amount of gene product present with regard to the stoichiometry of interacting factors, whether physically or not.
Hybrid incompatibilities
Speciation can occur by many mechanisms that cause the lack of gene flow and thus establish separate genetic lineages. These effects can be prezygotic or postzygotic. The latter can involve sterility or lethality of hybrids. The genetics of such hybrid reactions has been the subject of much conjecture for decades (Orr & Turelli, 2001). Muller (Muller, 1942) and Dobzhansky (Dobzhansky, 1937) noted that a minimum of two interacting loci must be involved and suggested co-evolving gene complexes that can operate among themselves within a species but that produce detrimental interactions in hybrids. Many candidates for Muller–Dobzhansky (M–D) incompatibilities have been suggested but will not be reiterated here. Indeed, there are many probable sources of hybrid incompatibilities. Here we note that balanced regulatory complexes that might diverge in different lineages are worthy candidates to consider in future thought about M–D incompatibilities. The evolutionary evidence that balance is critically important might suggest that if an interacting complex were to be selected to differ from a progenitor state, then when two diverged set of interactors are brought together in a hybrid, detrimental effects similar to aneuploid syndromes might be predicted to occur. It is also likely that stochastic changes in target gene expression that might ultimately shift the optimum balance of the regulators could cause divergence of regulatory networks in a completely neutral fashion that would produce incompatibilities when diverged genomes are combined in a hybrid. Indeed, evidence has been presented that coevolution of cis and trans effects for genes will lead to misregulation in Drosophila (Landry et al., 2005) and yeast (Tirosh et al., 2009) hybrids as well as in interspecific recombinant congenic mice strains (Lhote et al., 2008). Although not extensively studies, the available evidence suggests that there is a greater nonadditive gene expression in hybrids with increasing evolutionary distance between the parents (Gibson et al., 2004; Ranz et al., 2003). At the phenotypic level, the morphological differences between D. simulans and D. mauritiana can be attributed to multiple additive (dosage sensitive) QTL (Liu et al., 1996).
In the plant kingdom, polyploidy serves as an isolating mechanism, at least in part, because of failure of the endosperm development with crosses between ploidies (Birchler, 1993). The basis of this endosperm failure likely involves an incompatible multigenic interaction of the dosage of maternal regulators with zygotic factors (Birchler, 1980, 1993; Birchler & Hart, 1987; Dilkes & Comai, 2004; Walia et al., 2009). Natural variation for cross incompatibilities involving the endosperm that can be corrected by changing the ploidy are referred to as endosperm balance number and are predictive of whether endosperm development will be successful in crosses (Johnston & Hanneman, 1982). Imprinting of dosage sensitive regulators might play a role in establishing the proper balance. This phenomenon is consistent with the involvement of dosage sensitive interactions in initiating endosperm development and might fit into the context of regulatory balance.
Perspectives
The divergence of balanced complexes presents a problem that has yet to be fully explored experimentally or theoretically. Namely, if a balanced set of interacting genes is resistant to changes in stoichiometry, as the broad evolutionary evidence suggests, how is it possible for them to diverge? One possibility is that selection operating on one member of a complex will present conflict with the interacting members and place a selection pressure on the interaction properties (Birchler et al., 2007). An example of selection on one member of a complex affecting interacting partners has been described in the case of the Male Specific Lethal complex in Drosophila (Levine et al., 2007; Rodriquez et al., 2007). Another potential means for a new balance to evolve is if there are changes among the target loci that occur in cis that alter the level of expression (Birchler et al., 2007). There is some evidence for increasing cis regulatory divergence with increasing genetic distance (Lemos et al., 2008; Wittkopp et al., 2008). Over time such changes could accumulate among the critical target genes controlled by a regulatory complex and allow subtle variation in the balanced interactors to shift the stoichiometric relationships. We are not aware of whether any evidence for such a scenario has been sought. Further, a completely unexplored concept is the role of microRNA modulation of translation of proteins that are involved in balanced complexes. MicroRNAs could potentially modulate the expression of genes subtly to either maintain a protein-protein balance or cause it to diverge. Lastly, retained duplicates during diploidization may find themselves duplicated again in turn during a subsequent tetraploidizaiton event. With now eight copies of a gene present, changes in the amount of the gene product might be tolerated and a copy of the gene can be lost or diverge.
One of the future directions that deserves attention is experimental work on the basis of how changing the stoichiometry of subunits of multi-molecular complexes affects the action of the complex. One possibility involves the ordered assembly of the complex in such a manner that subcomplexes form regularly and diminish the amount of the full complex that can be assembled (Veitia, 2002; Veitia et al., 2003, 2008). Another possibility is that with varied amounts of subunits, the multisubunit complex will form and the unassociated subunits are degraded (Veitia et al., 2008).
Another issue of interest is how the regulatory balance is manifested. The genetic and evolutionary evidence indicates that regulatory systems that operate in complexes are tightly conserved whereas the target loci can be deleted back to a diploid level following tetraploidization or can be varied regularly in copy number variants. However, the effect of the regulatory balance must ultimately be realized through the expression of target loci that perform cellular metabolism and developmental processes. The dynamics of the regulators and their target loci has not been explored in the context of balance.
Another topic worthy of theoretical exploration in the context of regulatory balance is the fate of duplicate genes and their evolution of new functions. Duplicate genes have the potential to divide their activities via a process referred to as subfunctionalization. Alternatively, one of the copies could evolve a new property, which is referred to as neofunctionalization (Lynch & Conery, 2000). These processes would hold the duplicate pair in the evolutionary lineage because the original function has been separated into two genes. Clearly, we know that neofunctionalization has given rise to new regulatory and housekeeping genes. However, we now know from repeated tetraploidization events followed by diploidization in plants and Paramecium that duplicate genes can be held in a lineage because of stoichiometric constraints involved with molecular complexes. Other classes of genes are regularly deleted back to the diploid level and are thus rarely maintained by subfunctionalization or neofunctionalization. However, exceptions exist such as disease resistance genes that are constantly in an arms race with pathogens. Nevertheless, it is important to note that the maintenance of duplicate regulatory genes via balance constraints will hold them in an evolutionary lineage for a longer time frame than other classes of genes. This scenario would provide a longer time frame for cases of neofunctionalization to occur. Also, the degree to which these factors hold duplicate genes in a lineage deserves further study. In plants and Paramecium, as the tetraploidization events recede into the distant past, fewer of the duplicate pairs persist, suggesting that subfunctionalization and neofunctionalization, while important for evolutionary novelty, are not pervasive (Freeling, 2008).
The Gene Balance Hypothesis suggests that quantitative traits will be controlled by a large number of genes that can contribute variation. This variation is constrained due to the interacting balance of regulators to a very narrow window around the population norm under most conditions. However, because of the large number of loci involved with any one trait, subtle progressive changes to large extremes are possible under intense selection. Subtle regulatory variation of small magnitude, but present at many loci, can be neutral for long periods of evolutionary time providing the opportunity for novelty to evolve with a change of environment. Thus, the Gene Balance Hypothesis has the potential to explain the apparently contradictory observations that the status quo can be maintained for eons but extreme novelty can evolve within a few generations.
Acknowledgements
Research on this topic was supported by NSF DBI 0733857 and NIH RO1GM068042 to J.B. The research of R.A.V. is supported by the CNRS, the Universite Paris-Diderot and the Institut Universitaire de France.
References
- Aury J-M, Jaillon O, Duret L, Jubin C, Porcel BM, Segurens B, Daubin V, Anthouard V, Aiach N, Arnaiz O, et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006;444:171–178. doi: 10.1038/nature05230. [DOI] [PubMed] [Google Scholar]
- Barker MS, Kane NC, Matvienko M, Kozik A, Michelmore RW, Knapp SJ, Rieseberg LH. Multiple paleopolyploidizations during the evolution of the compositae reveal parallel patterns of duplicate gene retention after millions of years. Molecular Biology and Evolution. 2008;25:2445–2455. doi: 10.1093/molbev/msn187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudet AL, Jiang Y-H. A rheostat model for a rapid and reversible form of imprinting-dependent evolution. Am J Hum Genet. 2002;70:1389–1397. doi: 10.1086/340969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA. A study of enzyme activities in a dosage series of the long arm of chromosome one in maize. Genetics. 1979;92:1211–1229. doi: 10.1093/genetics/92.4.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA. On the nonautonomy of the small kernel phenotype produced by B-A translocations in maize. Genetical Research. 1980;36:111–116. [Google Scholar]
- Birchler JA. The genetic basis of dosage compensation of alcohol dehydrogenase-1 in maize. Genetics. 1981;97:625–637. doi: 10.1093/genetics/97.3-4.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA. Dosage analysis of maize endosperm development. Ann. Rev. Genetics. 1993;27:181–204. doi: 10.1146/annurev.ge.27.120193.001145. [DOI] [PubMed] [Google Scholar]
- Birchler JA, Bhadra U, Pal Bhadra M, Auger DL. Dosage dependent gene regulation in multicellular eukaryotes: Implications for dosage compensation, aneuploid syndromes and quantitative traits. Developmental Biology. 2001;234:275–288. doi: 10.1006/dbio.2001.0262. [DOI] [PubMed] [Google Scholar]
- Birchler JA, Hart JR. Interaction of endosperm size factors in maize. Genetics. 1987;117:309–317. doi: 10.1093/genetics/117.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Hiebert JC, Paigen K. Analysis of autosomal dosage compensation involving the Alcohol dehydrogenase locus in Drosophila melanogaster. Genetics. 1990;124:677–686. [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Newton KJ. Modulation of protein levels in chromosomal dosage series of maize: The biochemical basis of aneuploid syndromes. Genetics. 1981;99:247–266. doi: 10.1093/genetics/99.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Riddle NC, Auger DL, Veitia RA. Dosage balance in gene regulation: biological implications. Trends in Genetics. 2005;21:219–226. doi: 10.1016/j.tig.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Birchler JA, Veitia RA. The gene balance hypothesis: From classical genetics to modern genomics. The Plant Cell. 2007;19:395–402. doi: 10.1105/tpc.106.049338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Yao H, Chudalayandi S. Biological consequences of dosage dependent gene regulatory systems. Biochemica Biophysica Acta-Gene Structure and Expression. 2007;1769:422–428. doi: 10.1016/j.bbaexp.2006.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakeslee AF. New Jimson weeds from old chromosomes. J. Hered. 1934;24:80–108. [Google Scholar]
- Blakeslee AF, Belling J, Farnham ME. Chromosomal duplication and Mendelian phenomena in Datura mutants. Science. 1920;52:388–390. doi: 10.1126/science.52.1347.388. [DOI] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. The Plant Cell. 2004;16:1679–1691. doi: 10.1105/tpc.021410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomme T, Vandepoele K, De Bodt S, Simillion C, Maere S, Van de Peer Y. The gain and loss of genes during 600 million years of vertebrate evolution. Genome Biology. 2006;7:R43. doi: 10.1186/gb-2006-7-5-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond DJ, Chandley AC. Aneuploidy. Oxford University Press; Oxford, UK: 1983. [Google Scholar]
- Bowers JE, Chapman BA, Rong J, Paterson AH. Unraveling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature. 2003;422:433–438. doi: 10.1038/nature01521. [DOI] [PubMed] [Google Scholar]
- Bridges CB. Sex in relation to chromosomes and genes. Am. Naturalist. 1925;59:127–137. [Google Scholar]
- Buckler ES, Holland JB, Bradbury PJ, Acharya CB, Brown PJ, Browne C, Ersoz E, Flint-Garcia S, Garcia A, Glaubitz JC, et al. The genetic architecture of maize flowering time. Science. 2009;325:714–718. doi: 10.1126/science.1174276. [Author, please check inserted names to make up to 10 authors before et al.] [DOI] [PubMed] [Google Scholar]
- Chapman BA, Bowers JE, Feltus FA, Paterson AH. Buffering of crucial functions by paleologous duplicated genes may contribute cyclicality to angiosperm genome duplication. Proc. Natl. Acad. Sci. USA. 2006;103:2730–2735. doi: 10.1073/pnas.0507782103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong B, Barrero LS, Tanksley SD. Regulatory change in YABBY-like transcription factor led to evolution of extreme fruit size during tomato domestication. Nature Genetics. 2008;40:800–804. doi: 10.1038/ng.144. [DOI] [PubMed] [Google Scholar]
- Cong B, Liu J, Tanksley SD. Natural alleles at a tomato fruit size quantitative trait locus differ by heterochronic regulatory mutations. Proc. Natl. Acad. Sci. USA. 2002;99:13606–13611. doi: 10.1073/pnas.172520999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cribb DL, Benassayag C, Randazzo FM, Kaufman TC. Levels of homeotic protein function can determine developmental identity: evidence from low-level expression of the Drosophila homeotic gene proboscipedia under Hsp70 control. EMBO J. 1995;14:767–778. doi: 10.1002/j.1460-2075.1995.tb07055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C. The origin of species by means of natural selection. John Murray; London, UK: 1859. [Google Scholar]
- Davis JC, Petrov DA. Do disparate mechanisms of duplication add similar genes to the genome? Trends in Genetics. 2005;21:548–551. doi: 10.1016/j.tig.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Denver DR, Morris K, Streelman JT, Kim SK, Lynch M, Thomas WK. The transcriptional consequences of mutation and natural selection in Caenorhabditis elegans. Nature Genetics. 2005;37:544–548. doi: 10.1038/ng1554. [DOI] [PubMed] [Google Scholar]
- Devlin RH, Holm DG, Grigliatti TA. Autosomal dosage compensation in Drosophila melanogaster strains trisomic for the left arm of chromosome 2. Proc Natl Acad Sci USA. 1982;79:1200–1204. doi: 10.1073/pnas.79.4.1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin RH, Holm DG, Grigliatti TA. The influence of whole-arm trisomy on gene expression in Drosophila. Genetics. 1988;118:87–101. doi: 10.1093/genetics/118.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilkes BP, Comai L. A differential dosage hypothesis for parental effects in seed development. Plant Cell. 2004;16:3174–3180. doi: 10.1105/tpc.104.161230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T. Genetics and the origin of species. Columbia University Press; New York, USA: 1937. [Google Scholar]
- Driever W, Nusslein-Volhard C. The bicoid protein determines position in the Drosophila embryo in a concentration-dependent manner. Cell. 1988;54:95–104. doi: 10.1016/0092-8674(88)90183-3. [DOI] [PubMed] [Google Scholar]
- Dopman EB, Hartl DL. A portrait of copy-number polymorphism in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 2007;104:19920–19925. doi: 10.1073/pnas.0709888104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frary A, Nesbitt RC, Frary A, Grandillo S, van der Knaap E, Cong B, Liu J, Meller J, Elber R, Alpert KB, et al. fw2.2: A quantitative trait locus key to the evolution of tomato fruit size. Science. 2000;289:85–88. doi: 10.1126/science.289.5476.85. [DOI] [PubMed] [Google Scholar]
- Freeling M. The evolutionary position of subfunctionalization, downgraded. Genome Dynamics. 2008;4:25–40. doi: 10.1159/000126004. [DOI] [PubMed] [Google Scholar]
- Freeling M, Lyons E, Pedersen B, Alam M, Ming R, Lisch D. Many or most genes in Arabidopsis transposed after the origin of the order Brassicales. Genome Research. 2008;18:1924–1937. doi: 10.1101/gr.081026.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M, Thomas BC. Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity. Genome Research. 2006;16:805–14. doi: 10.1101/gr.3681406. [DOI] [PubMed] [Google Scholar]
- Gibson G, Dworkin I. Uncovering cryptic genetic variation. Nat Rev Genetics. 2004;5:681–690. doi: 10.1038/nrg1426. [DOI] [PubMed] [Google Scholar]
- Gibson G, Riley-Berger R, Harshman L, Kopp A, Vacha S, Nuzhdin S, Wayne M. Extensive sex-specific nonadditivity of gene expression in Drosophila melanogaster. Genetics. 2004;167:1791–1799. doi: 10.1534/genetics.104.026583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MM, Granka JM, Bustamante CD, Sutter NB, Boyko Ar, Zhu L, Ostrander EA, Wayne RK. Linkage disequilibrium and demographic history of wild and domestic canids. Genetics. 2009;181:1493–1505. doi: 10.1534/genetics.108.098830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieve IC, Dickens NJ, Pravenec M, Kren V, Hubner N, Cook SA, Ailtman TJ, Petretto E, Mangion J. Genome-wide co-expression analysis in multiple tissues. PLoS one. 2008;3:e4033. doi: 10.1371/journal.pone.0004033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Birchler JA. Trans-acting dosage effects on the expression of model gene systems in maize aneuploids. Science. 1994;266:1999–2002. doi: 10.1126/science.266.5193.1999. [DOI] [PubMed] [Google Scholar]
- Guo M, Davis D, Birchler JA. Dosage effects on gene expression in a maize ploidy series. Genetics. 1996;142:1349–1355. doi: 10.1093/genetics/142.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakes L, Pinney JW, Lovell Sc, Oliver SG, Robertson DL. All duplicates are not equal: the difference between small-scale and genome duplications. Genome Biology. 2007;8:R209. doi: 10.1186/gb-2007-8-10-r209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes KA, Ayroles JF, Reedy MM, Drnevich JM, Rowe KC, Ruedi EA, Caceres CE, Paige KN. Segregating variation in the transcriptome: Cis regulation and additivity of effects. Genetics. 2006;173:1347–1355. doi: 10.1534/genetics.105.051474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionita-Laza I, Rogers AJ, Lange C, Raby BA, Lee C. Genetic association analysis of copy-number variation (CNV) in human disease pathogenesis. Genomics. 2009;93:22–26. doi: 10.1016/j.ygeno.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes F, Colot V, Jansen RC. Epigenome dynamics: A quantitative genetics perspective. Nat. Rev. Genet. 2008;9:883–890. doi: 10.1038/nrg2467. [DOI] [PubMed] [Google Scholar]
- Johnston SA, Hanneman REJ. Manipulations of endosperm balance number overcome crossing barriers between diploid solanum species. Science. 1982;217:446–448. doi: 10.1126/science.217.4558.446. [DOI] [PubMed] [Google Scholar]
- Kondrashov FA, Koonin EV. A common framework for understanding the origin of genetic dominance and evolutionary fates of gene duplications. Trends Genet. 2004;20:287–90. doi: 10.1016/j.tig.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Korbel JO, Kim PM, Chen X, Urban AE, Weissman S, Snyder M, Gerstein MB. The current excitement about copy-number variation: how it relates to gene duplications and protein families. Curr. Opin. Structural Biol. 2008;18:366–374. doi: 10.1016/j.sbi.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry CR, Wittkopp PJ, Taubes CH, Ranz JM, Clark AG, Hartl DL. Compensatory cis-trans evolution and the dysregulation of gene expression in interspecific hybrids of Drosophila. Genetics. 2005;171:1813–1822. doi: 10.1534/genetics.105.047449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie CC, Chasalow SD, LeDeaux JR, McCarroll R, Bush D, Hauge B, Lai C, Clark D, Rocheford TR, Dudley JW. The genetic architecture of response to long-term artificial selection for oil concentration in the maize kernel. Genetics. 2004;168:2141–2155. doi: 10.1534/genetics.104.029686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie CC, True JR, Liu J, Mercer JM. An introgression analysis of quantitative trait loci that contribute to a morphological difference between Drosophila simulans and D. mauritiana. Genetics. 1997;145:339–348. doi: 10.1093/genetics/145.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos B, Araripe LO, Fontanilla P, Hartl DL. Dominance and the evolutionary accumulation of cis- and trans-effects on gene expression. Proc. Natl. Acad. Sci. USA. 2008;105:1813–1822. doi: 10.1073/pnas.0805160105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine MT, Holloway AK, Arshad U, Begun DJ. Pervasive and largely lineage-specific adaptive protein evolution in the dosage compensation complex of Drosophila melanogaster. Genetics. 2007;177:1959–1962. doi: 10.1534/genetics.107.079459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L'Hote D, Serres C, Veitia RA, Montagutelli X, Oulmouden A, Vaiman D. Gene expression regulation in the context of mouse interspecific mosaic genomes. Genome Biol. 2008;9:R133. doi: 10.1186/gb-2008-9-8-r133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Rogale-Plazonic K, Chen J, Li WH, Fernandez A. Protein under-wrapping causes dosage sensitivity and decreases gene duplicability. PLoS Genetics. 2008;4:e11. doi: 10.1371/journal.pgen.0040011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley DL, Sandler L, Baker BS, Carpenter AT, Denell RE, Hall JC, Jacobs PA, Miklos GL, Davis BK, Gethman RC, et al. Segmental aneuploidy and the genetic gross structure of the Drosophila genome. Genetics. 1972;71:157–184. doi: 10.1093/genetics/71.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Van Eck J, Cong B, Tanksley SD. A new class of regulatory genes underlying the cause of pear-shaped tomato fruit. Proc. Natl. Acad. Sci. USA. 2002;99:13302–13306. doi: 10.1073/pnas.162485999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Mercer JM, Stam LF, Gibson GC, Zeng ZB, Laurie CC. Genetic analysis of a morphological shape difference in the male genitalia of Drosophila simulans and D. mauritiana. Genetics. 1996;142:1129–1145. doi: 10.1093/genetics/142.4.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Maere S, DeBodt S, Raes J, Casneuf T, Van Montagu M, Kuiper M, Y. Van de Peer Y. Modeling gene and genome duplications in eukaryotes. Proc. Natl. Acad. Sci. USA. 2005;102:5454–5459. doi: 10.1073/pnas.0501102102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley M, Molony CM, Weber TM, Devlin JL, Ewens KG, Spielman RS, Cheung VG. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller HJ. Isolating mechanisms, evolution and temperature. Biol Symp. 1942;6:71–125. [Google Scholar]
- Orr HA, Turelli M. The evolution of postzygotic isolation: accumulating Dobzhansky-Muller incompatibilities. Evolution. 2001;55:1085–1094. doi: 10.1111/j.0014-3820.2001.tb00628.x. [DOI] [PubMed] [Google Scholar]
- Papp B, Pal C, Hurst LD. Dosage sensitivity and the evolution of gene families in yeast. Nature. 2003;424:194–197. doi: 10.1038/nature01771. [DOI] [PubMed] [Google Scholar]
- Petretto E, Mangion J, Dickens NJ, Cook SSA, Kumaran MK, Lu M, Fischer J, Maatz H, Kren V, Pravenec M, et al. Heritability and tissue specificity of expression quantitative trait loci. PLoS Genetics. 2006;2:e172. doi: 10.1371/journal.pgen.0020172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinow L, Nguyen-Huynh AT, Birchler JA. A trans-acting regulatory gene that inversely affects the expression of the white, brown and scarlet loci in Drosophila melanogaster. Genetics. 1991;129:463–480. doi: 10.1093/genetics/129.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranz JM, Castillo-Davis CI, Meiklejohn CD, Hartl DL. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science. 2003;300:1742–1745. doi: 10.1126/science.1085881. [DOI] [PubMed] [Google Scholar]
- Rodriquez MA, Vermaak D, Bayes JJ, Malik HS. Species-specific positive selection of the male specific lethal complex that participates in dosage compensation in Drosophila. Proc. Natl. Acad. Sci. USA. 2007;104:15412–15417. doi: 10.1073/pnas.0707445104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabl JF, Birchler JA. Dosage dependent modifiers of white alleles in Drosophila melanogaster. Genetical Research. 1993;62:15–22. doi: 10.1017/s0016672300031517. [DOI] [PubMed] [Google Scholar]
- Satina S, Blakeslee AF, Avery AG. Balanced and unbalanced haploids in Datura. J. Hered. 1937;28:192–202. [Google Scholar]
- Sauer F, Jackle H. Concentration-dependent transcriptional activation or repression by Kruppel from a single binding site. Nature. 1991;353:563–566. doi: 10.1038/353563a0. [DOI] [PubMed] [Google Scholar]
- Schadt EE, Monks SA, Drake TA, Lusis AJ, Che N, Colinayo V, Ruff TG, Milligan SB, Lamb JR, Cavet G, et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- Schulz C, Tautz D. Autonomous concentration-dependent activation and repression of Kruppel by hunchback in the Drosophila embryo. Development. 1994;120:3043–3049. doi: 10.1242/dev.120.10.3043. [DOI] [PubMed] [Google Scholar]
- Seidman JG, Seidman C. Transcription factor haploinsufficiency: when half a loaf is not enough. J Clin Invest. 2002;109:451–455. doi: 10.1172/JCI15043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simillion C, Vandepoele K, Montagu MC, Zabeau M, Van de Peer Y. The hidden duplication past of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2002;99:13627–13632. doi: 10.1073/pnas.212522399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G, Struhl K, Mcdonald PM. The gradient morphogen bicoid is a concentration-dependent transcriptional activator. Cell. 1989;57:1259–1273. doi: 10.1016/0092-8674(89)90062-7. [DOI] [PubMed] [Google Scholar]
- Tanksley SD. Mapping polygenes. Annu. Rev. Genetics. 1993;27:205–233. doi: 10.1146/annurev.ge.27.120193.001225. [DOI] [PubMed] [Google Scholar]
- Tirosh I, Reikhav S, Levy AA, Barkai N. A yeast hybrid provides insight into the evolution of gene expression regulation. Science. 2009;324:659–662. doi: 10.1126/science.1169766. [DOI] [PubMed] [Google Scholar]
- True JR, Weir BS, Laurie CC. A genome-wide survey of hybrid incompatibility factors by the introgression of marked segments of Drosophila mauritiana chromosomes into Drosophila simulans. Genetics. 1996;142:819–837. doi: 10.1093/genetics/142.3.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veitia RA. Exploring the etiology of haploinsufficiency. Bioessays. 2002;24:175–184. doi: 10.1002/bies.10023. [DOI] [PubMed] [Google Scholar]
- Veitia RA. Nonlinear effects in macromolecular assembly and dosage sensitivity. J. Theor. Biol. 2003;220:19–25. doi: 10.1006/jtbi.2003.3105. [DOI] [PubMed] [Google Scholar]
- Veitia RA. Gene dosage balance in cellular pathways: implications for dominance and gene duplicability. Genetics. 2004;168:569–574. doi: 10.1534/genetics.104.029785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veitia RA, Bottani S, Birchler JA. Cellular reactions to gene dosage imbalance: genomic, transcriptomic and proteomic effects. Trends in Genetics. 2008;24:390–397. doi: 10.1016/j.tig.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Walia H, Josefsson C, Dilkes B, Kirkbride R, Harada J, Comai L. Dosage-dependent deregulation of an AGAMOUS-LIKE gene cluster contributes to interspecific incompatibility. Current Biology. 2009;19:1128–1132. doi: 10.1016/j.cub.2009.05.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Sung H-M, Wang T-Y, Huang C-J, Yang P, Chang T, Wang Y-C, Tseng D-L, Wu J-P, Lee T-C, et al. Expression evolution in yeast genes of single-input modules is mainly due to changes in trans-acting factors. Genome Research. 2007;17:1161–1169. doi: 10.1101/gr.6328907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrior R, Levine M. Dose-dependent regulation of pair-rule stripes by gap proteins and the initiation of segment polarity. Development. 1990;110:759–767. doi: 10.1242/dev.110.3.759. [DOI] [PubMed] [Google Scholar]
- Wayne ML, Pan Y-J, Nuzhdin SV, McIntyre LM. Additivity and trans-acting effects on gene expression in male Drosophila simulans. Genetics. 2004;168:1413–1420. doi: 10.1534/genetics.104.030973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West MAL, Kim K, Kliebenstein DJ, van Leeuwen H, Michelmore RW, Doerge RW, St. Clair DA. Global eQTL mapping reveals the complex genetic architecture of transcript-level variation in Arabidopsis. Genetics. 2007;175:1441–1450. doi: 10.1534/genetics.106.064972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams GC. Adaptation and natural selection. Princeton University Press; Princeton, NJ, USA: 1966. [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG. Evolutionary changes in cis and trans gene regulation. Nature. 2004;430:85–88. doi: 10.1038/nature02698. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG. Regulatory changes underlying expression differences within and between Drosophila species. Nature Genetics. 2008;40:346–350. doi: 10.1038/ng.77. [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- Yvert G, Brem RB, Whittle J, Akey JM, Foss E, Smith EN, Mackelprang R, Kruglyak L. Trans-acting regulatory variation in Saccharomyces cerevisiae and the role of transcripton factors. Nat. Genetics. 2003;35:57–64. doi: 10.1038/ng1222. [DOI] [PubMed] [Google Scholar]
- Yook KJ, Proulx SR, Jorgensen EM. Rules of nonallelic noncomplementation at the synapse in Caenorhabditis elegans. Genetics. 2001;158:209–220. doi: 10.1093/genetics/158.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]