Summary
RNA-directed DNA methylation (RdDM) is an important epigenetic mechanism for silencing transgenes and endogenous repetitive sequences such as transposons. The RD29A promoter-driven LUCIFERASE transgene and its corresponding endogenous RD29A gene are hypermethylated and silenced in the Arabidopsis DNA demethylase mutant ros1. By screening for second-site suppressors of ros1, we identified the RDM12 locus. The rdm12 mutation releases the silencing of the RD29A-LUC transgene and the endogenous RD29A gene by reducing the promoter DNA methylation. The rdm12 mutation also reduces DNA methylation at endogenous RdDM target loci including transposons and other repetitive sequences. In addition, the rdm12 mutation affects the levels of siRNAs from some of the RdDM target loci. RDM12 encodes a protein with XS and coiled-coil domains and is similar to SGS3, which is a partner protein of RDR6 and can bind to double-stranded RNAs with a 5' overhang and is required for several posttranscriptional gene silencing pathways. Our results show that RDM12 is a component of the RdDM pathway and suggest that RdDM may involve double stranded RNAs with a 5' overhang and the partnering between RDM12 and RDR2.
Keywords: RDM12, SGS3, epigenetics, DNA methylation, siRNA
Introduction
Transcriptional as well as posttranscriptional gene silencing is frequently caused by transgenes and virus infection (Baulcombe, 2004; Vaucheret et al., 2001). Transcriptional gene silencing (TGS) also occurs at endogenous loci, particularly transposons, retrotransposons and other repetitive sequences (Chan et al., 2004; Matzke and Birchler, 2005; Huettel et al., 2006). In the TGS pathway, the transgenes and endogenous sequences are usually associated with cytosine methylation and repressive histone modifications, epigenetic changes that maintain the silencing and ensure genome stability (Henderson and Jacobsen, 2007; Matzke et al., 2009). RNA-directed DNA methylation (RdDM) is a mechanism for establishing TGS. The RdDM pathway in plants involves three DNA-dependent RNA polymerases, Pol II (Zheng et al., 2009), Pol IV and Pol V (Wierzbicki et al., 2008). Pol IV is hypothesized to be responsible for transcribing methylated sequences to generate abberant RNA transcripts, which are converted to double-stranded RNAs by the RNA-dependent RNA polymerase RDR2 (Xie et al., 2004; Herr et al., 2005; Pontes et al., 2006). The double-stranded RNA is cleaved into 24-nt siRNAs by the Dicer-like protein DCL3 (Xie et al., 2004). Pol II and Pol V appear to be involved in the synthesis of scaffold RNA transcripts that help recruit the RdDM effector complex (Zheng et al., 2009; Wierzbicki et al., 2008). The RdDM effector complex contains the ARGONAUTE protein AGO4 that binds to the 24 nt siRNAs, the WG/GW repeats-containing protein KTF1 that binds the scaffold transcripts and AGO4, and the de novo DNA methyltransferase DRM2 (Zilberman et al., 2003; Wierzbicki et al., 2008; He et al., 2009b; Cao and Jacobsen, 2002). The effector is guided to specific genomic targets by the base pairing between the siRNA guide and the nascent scaffold transcripts, and leads to de novo DNA methylation (Cao and Jacobsen, 2002; Li et al., 2006; Pontes et al., 2006; He et al., 2009b). The chromatin remodeling protein DRD1 and the SMC-related protein DMS3 facilitate the process and function at downstream steps in the RdDM pathway (Kanno et al., 2004; Kanno et al., 2008; Ausin et al., 2009).
In several post-transcriptional gene silencing (PTGS) pathways, the double-stranded RNA is synthesized by the RNA-dependent RNA polymerase RDR6 (Mourrain et al., 2000; Vaucheret et al., 2001). The stabilization of the double-stranded RNA requires the RNA binding protein SGS3 (Mourrain et al., 2000; Fukunaga and Doudna, 2009). DCL4 is responsible for the dicing of the double-stranded RNA into 21 nt siRNAs, which are loaded onto the ARGONAUTE protein AGO1 to target complementary RNAs for cleavage (Yoshikawa et al., 2005; Morel et al., 2002; Vazquez et al., 2004). The PTGS and TGS pathways share the methyltransferase protein HEN1, which introduces a 2'-O-methyl group to the 3′-terminal nucleotide of small RNAs for increased stability (Boutet et al., 2003; Yu et al., 2005). In addition, the TGS and PTGS pathways both require ARGONAUTE proteins, Dicer-like proteins and RNA-dependent RNA polymerases, suggesting an extensive similarity between the two gene silencing pathways (Mourrain et al., 2000; Morel et al., 2002; Zilberman et al., 2003; Xie et al., 2004; Yoshikawa et al., 2005).
We have previously shown that the RD29A promoter-driven LUCIFERASE transgene is expressed at high levels in wild-type genetic backgrounds, although 24 nt siRNAs are generated from the transgene promoter (Gong et al., 2002). However, in the ros1 mutant, the 24 nt siRNAs cause DNA hypermethylation at the RD29A promoter and consequent TGS of the transgene and endogenous gene (Gong et al., 2002). ROS1 encodes a bifunctional DNA glycosylase/lyase, which functions to demethylate DNA through a base excision repair pathway (Gong et al., 2002; Agius et al., 2006). The methylation status of the RD29A promoter is under dynamic regulation by RdDM and the ROS1-mediated DNA demethylation pathway (Agius et al., 2006; Zhu et al., 2007). By screening for second-site suppressors of ros1, we identified most of the previously known RdDM components such as NRPD1, NRPE1, NRPD2, AGO4, DRD1 and HEN1 (He et al., 2009a). Moreover, we found several additional RdDM components including AGO6, the histone 2B deubiquitination enzyme UBP26, NRPD/E4, a common subunit of Pol IV/Pol V, KTF1, a WG/GW motif containing protein, and RDM4, a transcription factor for both Pol II and Pol V (He et al., 2009a; He et al., 2009b; He et al., 2009c). The results suggest that screening for ros1 suppressor mutants is an excellent approach to identifying new components of the RdDM pathway. Here, we report another important RdDM component, RDM12. The rdm12 mutation releases the silencing of the RD29A-LUC transgene and endogenous RD29A gene in the ros1 mutant background. This effect is correlated with reductions in promoter DNA methylation. Moreover, the rdm12 mutation also substantially reduces DNA methylation and releases silencing at endogenous RdDM target loci including transposons and other repetitive DNA sequences. We show that RDM12 encodes a protein that is similar to SGS3, which is known to be a partner of RDR6 in PTGS and can bind double stranded RNAs with a 5' overhang (Fukunaga and Doudna, 2009). Our results suggest that RDM12 may be a partner protein of RDR2 and that the RdDM pathway may involve 5' overhang-containing double stranded RNAs.
Results
Isolation of rdm12 from ros1 suppressor screen
A construct containing the RD29A promoter-driven LUCIFERASE reporter gene (RD29A-LUC) and the CaMV 35S promoter-driven NPTII marker gene (CaMV 35S-NPTII) was introduced into wild-type (C24 ecotype) Arabidopsis plants (Ishitani et al., 1997). Both transgenes are expressed in the wild type, but are silenced in the ros1 mutant background (Gong et al., 2002). A T-DNA mutagenized library in the ros1 background was generated, and screened for second site suppressors of ros1 based on the luminescence phenotype (Kapoor et al., 2005; He et al., 2009a). Most RdDM components, including NRPD1, NRPE1, NRPD2, AGO4, DRD1 and HEN1, as well as RDM2/NRPD4/NRPE4, RDM3/KTF1 and RDM4 have been identified from the screen (He et al., 2009a; He et al., 2009b; He et al., 2009c).
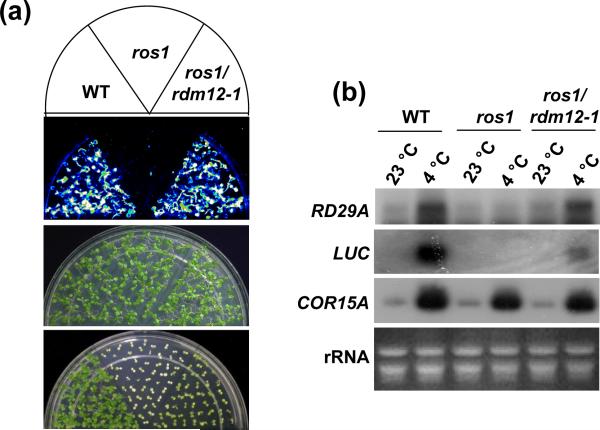
Further screening of the library led to the isolation of a novel suppressor mutant, ros1rdm12-1, which rescues the luminescence phenotype of ros1, but does not significantly affect the kanamycin sensitivity of ros1 (Figure 1a). Northern blot analysis shows that the expression of the RD29A-LUC transgene and endogenous RD29A gene in ros1rdm12-1 is substantially higher than that in ros1, although the LUC expression in ros1rdm12-1 is still much weaker than that in the wild type (Figure 1b). The results show that the rdm12 mutation partially releases the silencing of the RD29A-LUC transgene and corresponding endogenous RD29A gene, but does not release the silencing of the kanamycin resistance transgene NPTII. This phenotype is very similar to those of other suppressor mutants with lesions in RdDM components such as NRPD1, NRPE1, NRPD2 and DRD1 (He et al., 2009a; He et al., 2009b; He et al., 2009c).
Figure 1. The rdm12 mutation releases the silencing of RD29A-LUC transgene in ros1.
(a) The two-week-old plants on MS plate were imaged after cold treatment at 4 °C for 24 h. The plants were sprayed with luciferin for luminescence imaging. The plants were grown on MS medium supplemental with 50 μg μl−1 kanamycin for two weeks and photographed. (b) The expression of LUC, RD29A and COR15A was detected by Northern blotting. The ethidium bromide-stained rRNA was used as a loading control.
The rdm12 mutation reduces DNA methylation at RdDM target loci
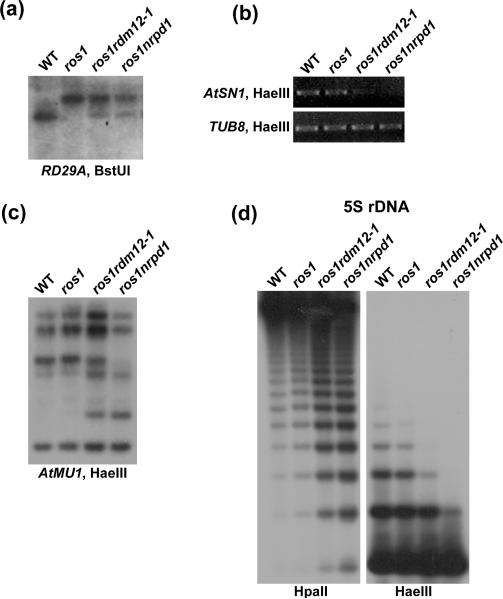
To investigate whether the reactivation of the RD29A-LUC transgene and endogenous RD29A gene in ros1rdm12-1 is related to changes in RD29A promoter DNA methylation, we examined the DNA methylation status of the RD29A promoter by Southern blotting. The result shows that the RD29A promoter DNA methylation level is partially reduced in ros1rdm12-1 compared to that in ros1 (Figure 2a). This partial reduction in DNA methylation in ros1rdm12-1 is similar to what was observed in ros1nrpd1 (He et al., 2009a). The result suggests that the rdm12 mutation suppresses the TGS of the RD29A-LUC transgene and endogenous RD29A gene by blocking DNA hypermethylation.
Figure 2. The rdm12 mutation reduces DNA methylation at RdDM target loci.
(a) Genomic DNA from the indicated genotypes was digested with the DNA methylation sensitive enzyme BstUI, followed by Southern blotting with the RD29A coding sequence as a probe. (b) The genomic DNA was digested with the DNA methylation sensitive enzyme HaeIII, and AtSN1 was amplified. TUB8 was amplified as an internal control. (c) The HaeIII-digested genomic DNA was used for Southern hybridization using the AtMU1 probe. (d) The genomic DNA was digested with HpaII and HaeIII, and followed by Southern hybridization using the 5S rDNA probe.
The DNA methylation of endogenous RdDM targets was also examined in the ros1rdm12-1 mutant. The genomic DNA from the indicated genotypes was digested by the DNA methylation sensitive enzyme HaeIII, followed by amplification of the AtSN1 transposon. The result shows that AtSN1 methylation was reduced in ros1rdm12-1 as well as in ros1nrpd1, although the effect of rdm12 is not as strong as that of nrpd1 (Figure 2b). The DNA methylation status of the transposon AtMU1 was tested by Southern blotting. The result shows that AtMU1 genomic DNA is partially digested by HaeIII in ros1rdm12-1 and is completely digested in ros1nrpd1, whereas there is little digestion in the wild type or ros1 (Figure 2c). The result suggests that rdm12-1 reduces DNA methylation at AtMU1.
The effect of rdm12 on 5S rDNA methylation was also investigated by Southern blotting. The result suggests that in ros1rdm12-1 and ros1nrpd1, the 5S rDNA methylation is reduced at non-symmetric cytosine sites as detected by HaeIII digestion, as well as at symmetric cytosine sites as detected by HpaII digestion (Figure 2d). However, no differences in DNA methylation were found among wild type, ros1, ros1rdm12-1 and ros1nrpd1 at the highly repetitive 180-bp centromeric repeat, which is not an RdDM target (Figure S1).
The effect of rdm4 on siRNA accumulation and transposon silencing
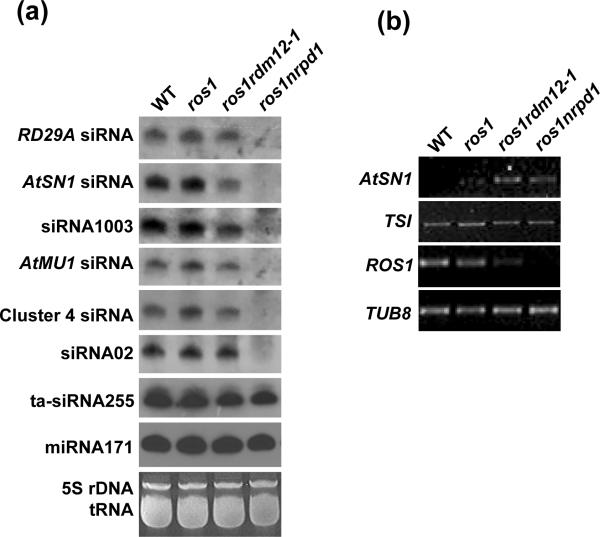
The siRNA accumulation was assessed by small RNA Northern blotting. The results show that the 24-nt siRNAs from the RD29A promoter are not substantially affected by the rdm12-1 mutation (Figure 3a). For endogenous siRNAs, AtSN1 siRNA and siRNA1003 (from 5S rDNA) are partially reduced in ros1rdm12-1 compared to those in the wild type and ros1, whereas the other tested siRNAs including AtMU1 siRNA, Cluster4 siRNA and siRNA02 appear not affected by the rdm12-1 mutation (Figure 3a). However, all these siRNAs are abolished in ros1nrpd1. The ta-siRNA255 and microRNA171 accumulate to a similar levels in wild type, ros1, ros1rdm12-1 and ros1nrpd1 (Figure 3a). The results show that rdm12-1 only affects some 24-nt siRNAs and does not affect miRNAs or ta-siRNAs, and suggest that RDM12 may function at a downstream step in the RdDM pathway.
Figure 3. The effect of rdm12 on the accumulation of siRNAs and RNA transcripts.
(a) The small RNA from floral tissiues was used for the Northern blot assay. The ethidium bromide-stained 5S rRNA and tRNA is shown as a loading control. (b) The total RNA from indicated genotypes was used for semi-quantitative RT-PCR to assess the transcript levels of AtSN1 and TSI. TUB8 was used as an internal control.
The effect of rdm12 on transposon expression was investigated by semi-quantitative RT-PCR. The results show that the AtSN1 transcript is increased in ros1rdm12-1 and ros1nrpd1, compared to that in the wild type and ros1 (Figure 3b). However, the TSI transcript is not affected by rdm12-1 or nrpd1. As reported previously (Huettel et al. 2006; Mathieu et al. 2007; He et al., 2009c), mutations in RdDM components reduce the transcript level of ROS1. Here, we found that similar to nrpd1, the rdm12-1 mutation also decreases the ROS1 transcript level, although the effect of rdm12-1 is not as strong as that of nrpd1 (Figure 3b).
RDM12 encodes an SGS3-like protein
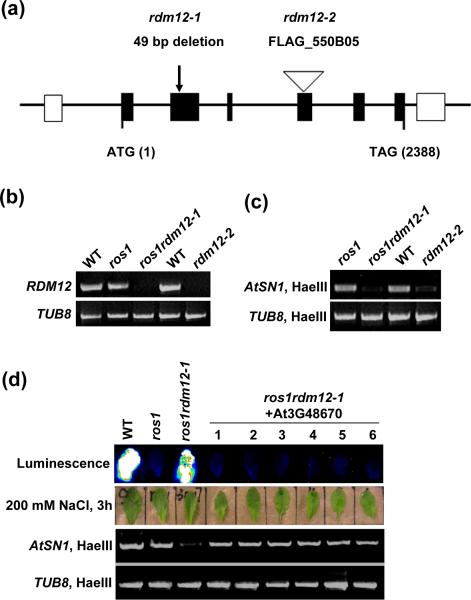
Since the ros1rdm12-1 mutant was isolated from a T-DNA insertion library, we tried and succeeded in finding an Arabidopsis genomic sequence flanking the T-DNA by TAIL-PCR. However, the T-DNA insertion was found not to co-segregate with the luminescence phenotype of ros1rdm12-1 (data not shown). Thus, we attempted to identify the RDM12 gene by map-based cloning. We generated a mapping population by crossing ros1rdm12-1 in the C24 ecotype to ros1-4 in the Col-0 ecotype. In the segregating selfed F2 population, ros1rdm12 mutants were selected based on the luminescence phenotype. We mapped the rdm12-1 mutation to a short region on Chromsome 3, to within the BAC clone T8P19 (Figure S2). Candidate genes in this region were sequenced and a 49-bp deletion was found in the second exon of At3G48670 in ros1rdm12-1 (Figure 4a). The At3G48670 transcript is abolished in ros1rdm12-1 (Figure 4b).
Figure 4. Identification of the RDM12 gene and mutant complementation assay.
(a) Diagram of the RDM12 genomic sequence. Shown are the positions of exons (solid boxes) and introns, the deletion site in rdm12-1, and the T-DNA insertion site in rdm12-2. (b) Detection of the RDM12 transcript by RT-PCR in the wild type (C24), ros1, ros1rdm12-1, and rdm12-2 and its wild type control (Ws). (c) Genomic DNA from ros1, ros1rdm12-1, wild-type (Ws), and rdm12-2 was digested with the methylation-sensitive enzyme HaeIII and used for amplification of AtSN1. (d) Complementation assay in ros1rdm12-1. The leaves from wild type, ros1, ros1rdm12-1 and six independent RDM12 transgenic T1 lines in the ros1rdm12-1 background were used for luminescence imaging after treatment with 200 mM NaCl for 3 h, and for assaying AtSN1 methylation.
To confirm that At3G48670 is the RDM12 gene, a T-DNA insertion allele of At3G48670 (FLAG_550B05, named as rdm12-2) (Figure 4a) was obtained from the ABRC stock center. The At3G48670 transcript is also abolished in the rdm12-2 allele (Figure 4b). This allele shows a reduced DNA methylation at AtSN1, similar to the effect of rdm12-1 (Figure 4c). Moreover, the wild-type At3G48670 genomic sequence was cloned and introduced into ros1rdm12-1 for a complementation assay (Figure 4d). The result shows that in all six randomly selected T1 transformants the silencing of RD29A-LUC and AtSN1 hypermethylation were restored (Figure 4d). Taken together, the results show that At3G48670 is the RDM12 gene.
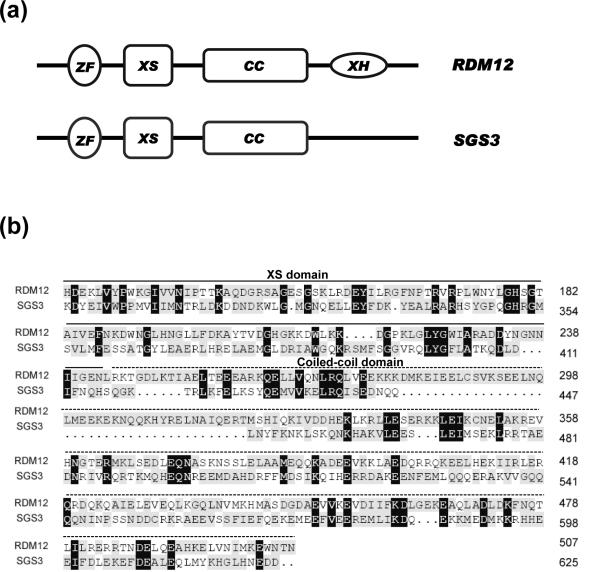
RDM12 is predicted to encode a protein of 648 amino acids. The protein contains a zinc finger domain, an XS domain and a coiled-coil domain, which are conserved in SGS3, a protein required for the post-transcriptional gene silencing (PTGS) pathway (Figure 5a, 5b). SGS3 is capable of binding double stranded RNAs with a 5' overhang, and the XS domain appears to be responsible for this RNA binding activity (Fukunaga and Doudna, 2009). RDM12 is highly similar to two other predicted proteins that are encoded by At3G12550 and At4G01780, respectively (Figure S3). In addition to the XS and coiled-coil domains, RDM12 and the two paralogs contain the XH domain that is not present in SGS3. The function of the XH domain is currently not known.
Figure 5. Characterization of the RDM12 protein.
(a) Diagram of the RDM12 and SGS3 proteins. The XS domain and a coiled-coil domain are conserved in RDM12 and SGS3. (b) Sequence alignment of RDM12 and SGS3 in the XS domain and coiled-coil domain.
Discussion
In our RD29A-LUC transgene system, 24 nt siRNAs are generated from the RD29A promoter, but cannot cause the hypermethylation and silencing of the promoter due to the DNA demethylation activity of ROS1 (Gong et al., 2002; Agius et al., 2006). Only in DNA demethylation mutants like ros1, the 24 nt siRNAs can lead to DNA hypermethylation of the RD29A promoter and consequently the RD29A-LUC transgene and the corresponding endogenous RD29A gene are silenced (Gong et al., 2002; Agius et al., 2006). The DNA methylation of a number of endogenous loci is also dynamically regulated by the opposing activities of RdDM and active DNA demethylation (Zhu et al., 2007; Penterman et al., 2007). This dynamic regulation of the RD29A-LUC transgene provides an excellent genetic system for identifying both active DNA demethylation pathway components as well as components of the RdDM pathway (Gong et al., 2002; Zheng et al., 2008; He et al., 2009a). In this study, we discovered RDM12 by employing the RD29A-LUC system in ros1. The phenotypes of the rdm12 mutants are similar to those of mutants defective in the known components of the RdDM pathway. Our results suggest that RDM12 is an important new component of the RdDM pathway. The DNA methylation and TGS phenotypes of the rdm12 mutants are relatively weak compared to those of many other RdDM pathway mutants. This is probably due to a partial redundancy between RDM12 and closely related proteins. There are two other proteins (At3G12550 and At4G01780) that are highly similar to RDM12, and these paralogs may be partially redundant with RDM12.
RDM12 and the two paralogs are similar to SGS3 in that all contain the conserved XS and coiled-coil domains. SGS3 is an important component of PTGS pathways, and is required for the accumulation of viral siRNAs, trans-acting siRNAs and nat-siRNAs (Mourrain et al., 2000; Yoshikawa et al., 2004; Borsani et al., 2005). It contains a zinc finger domain, an XS domain and a coiled-coil domain (Bateman, 2002). Of these, the XS domain is probably involved in binding double stranded RNAs with a 5' overhang (Fukunaga and Doudna, 2009) and the coiled-coil domain is likely involved in protein dimerization (Elmayan et al., 2009). SGS3 interacts with and co-localizes with RDR6 in the cytoplasm (Kumakura et al., 2009). Like SGS3, RDR6 is also a critical PTGS component and is required for the production of viral siRNAs, trans-acting siRNAs and nat-siRNAs (Mourrain et al., 2000; Yoshikawa et al., 2004; Borsani et al., 2005). The binding of SGS3 to double stranded RNAs might lead to the stabilization of RDR6-produced double-stranded RNAs and facilitate subsequent PTGS steps.
Unlike SGS3, RDM12 is not required for ta-siRNAs. However, like SGS3, RDM12 and its paralogs may also be able to bind double-stranded RNAs with a 5' overhang. The XH domain in RDM12 may confer additional activities to this protein. The double stranded RNAs are presumably produced by RDR2. In addition, the base-pairing between guide siRNAs and complementary nascent scaffold RNA transcripts produced by Pol II or Pol V could also generate double stranded RNAs with a 5' overhang. The binding of RDM12 may help stabilize this base-pairing interaction. As a component of the nuclear RdDM pathway, RDM12 is presumably a nuclear protein and may interact with the nucleus-localized RDR2. Just as SGS3 is a partner protein of RDR6 in PTGS, we suggest that RDM12 may be a partner protein of RDR2 in TGS. During the preparation of this manuscript, Ausin et al (2009) reported the IDN2 gene that is required for de novo DNA methylation. IDN2 is identical to RDM12. The identification of RDM12/IDN2 as a component of RdDM required for de novo DNA methylation from two completely independent genetic screens further underlies the importance of RDM12/IDN2 in the RdDM pathway. Future studies will reveal whether and how RDM12/IDN2 may partner with RDR2 to function in RdDM.
Materials and methods
Plant growth, mutant screening and cloning
The wild-type C24 and ros1 mutant plants carry the homozygous stress-inducible RD29A-LUC transgene (He et al., 2009a). A T-DNA mutagenized library in the ros1 mutant background were generated (Kapoor et al., 2005). Plants were grown in a controlled room at 23°C with 16 h of light and 8 h of darkness. The library screening was as described previously (He et al., 2009a). The identified ros1rdm12-1 mutant in the C24 ecotype was crossed to the ros1-4 mutant in the Col-0 ecotype (Salk_045303) to generate a F2 mapping population. About 800 F2 progenies with a high luminescence phenotype were selected for mapping. For complementation assay, The RDM12 genomic sequence were amplified, and cloned into the binary vector pCAMBIA1303. The RDM12 construct was introduced to ros1rdm12-1 using the Agrobacterium tumefaciens strain GV3101.
RNA analysis
Arabidopsis seedlings were grown on MS plates at 23°C for two weeks, and harvested after cold treatment (4°C, 1 d) or no treatment. Total RNA was extracted from the plants of each genotype using Trizol (Invitrogen). Twenty microgram of RNA for each sample was separated on 1.2% denaturing agarose gels, and transferred onto Hybond-N+ membranes (Amersham) for Northern hybridization. Small RNA was extracted from floral tissues. Small RNA Northern blotting was as described previously (He et al., 2009a). The DNA oligos used for DNA probe preparation are listed in Supplemental Table S1.
Five microgram of total RNA from the indicated genotypes was reverse transcribed to synthesize the first-strand cDNA with Superscript III System (Invitrogen). The cDNA templates were used for semi-quantitative RT-PCR or real-time PCR. For real-time PCR, the amplified DNA was labeled by Sybon (BioRad). The TUB8 was used as an internal control. The primer sequences are listed in Supplemental Table S1.
DNA methylation assays
For DNA methylation assays, the genomic DNA from each genotype was digested with the DNA methylation sensitive enzyme BstU1, HpaII or HaeIII. The digested DNA was used for Chop-PCR and Southern hybridization. For Southern hybridization, 2 ug of digested DNA was separated on 1.2% argorose gels at 40 V overnight, and transferred to Hybond-N+ membrance. The primer sequences used for chop-PCR and probe preparation are listed in Supplemental Table S1.
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Health grants R01GM070795 to J.-K. Zhu.
References
- Agius F, Kapoor A, Zhu JK. Role of the Arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation. Proc. Natl. Acad. Sci. U S A. 2006;103:11796–11801. doi: 10.1073/pnas.0603563103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausin I, Mockler TC, Chory J, Jacobsen SE. IDN1 and IDN2 are required for de novo DNA methylation in Arabidopsis thaliana. Nat. Struct. Mol. Biol. 2009;16:1325–1327. doi: 10.1038/nsmb.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A. The SGS3 protein involved in PTGS finds a family. BMC Bioinformatics. 2002;3:21. doi: 10.1186/1471-2105-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell. 2005;123:1279–1291. doi: 10.1016/j.cell.2005.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutet S, Vazquez F, Liu J, Béclin C, Fagard M, Gratias A, Morel JB, Crété P, Chen X, Vaucheret H. Arabidopsis HEN1: a genetic link between endogenous miRNA controlling development and siRNA controlling transgene silencing and virus resistance. Curr. Biol. 2003;13:843–848. doi: 10.1016/s0960-9822(03)00293-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Jacobsen SE. Role of the Arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr. Biol. 2002;12:1138–1144. doi: 10.1016/s0960-9822(02)00925-9. [DOI] [PubMed] [Google Scholar]
- Chan SW, Zilberman D, Xie Z, Johansen LK, Carrington JC, Jacobsen SE. RNA silencing genes control de novo DNA methylation. Science. 2004;303:1336. doi: 10.1126/science.1095989. [DOI] [PubMed] [Google Scholar]
- Elmayan T, Adenot X, Gissot L, Lauressergues D, Gy I, Vaucheret H. A neomorphic sgs3 allele stabilizing miRNA cleavage products reveals that SGS3 acts as a homodimer. FEBS J. 2009;276:835–844. doi: 10.1111/j.1742-4658.2008.06828.x. [DOI] [PubMed] [Google Scholar]
- Fukunaga R, Doudna JA. dsRNA with 5' overhangs contributes to endogenous and antiviral RNA silencing pathways in plants. EMBO J. 2009;28:545–555. doi: 10.1038/emboj.2009.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Morales-Ruiz T, Ariza RR, Roldan-Arjona T, David L, Zhu JK. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell. 2002;111:803–814. doi: 10.1016/s0092-8674(02)01133-9. [DOI] [PubMed] [Google Scholar]
- He XJ, Hsu YF, Pontes O, Zhu J, Lu J, Bressan RA, Pikaard C, Wang CS, Zhu JK. NRPD4, a protein related to the RPB4 subunit of RNA polymerase II, is a component of RNA polymerase IV and V and is required for RNA-directed DNA methylation. Genes Dev. 2009b;23:31–30. doi: 10.1101/gad.1765209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XJ, Hsu YF, Zhu S, Liu HL, Pontes O, Zhu J, Cui X, Wang CS, Zhu JK. A conserved transcriptional regulator is required for RNA-directed DNA methylation and plant development. Genes Dev. 2009c Nov 10; doi: 10.1101/gad.1851809. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XJ, Hsu YF, Zhu S, Wierzbicki AT, Pontes O, Pikaard CS, Liu HL, Wang CS, Jin H, Zhu JK. An effector of RNA-directed DNA methylation in arabidopsis is an ARGONAUTE 4- and RNA-binding protein. Cell. 2009a;137:498–508. doi: 10.1016/j.cell.2009.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Jacobsen SE. Epigenetic inheritance in plants. Nature. 2007;447:418–424. doi: 10.1038/nature05917. [DOI] [PubMed] [Google Scholar]
- Herr AJ, Jensen MB, Dalmay T, Baulcombe DC. RNA polymerase IV directs silencing of endogenous DNA. Science. 2005;308:118–120. doi: 10.1126/science.1106910. [DOI] [PubMed] [Google Scholar]
- Huettel B, Kanno T, Daxinger L, Aufsatz W, Matzke AJ, Matzke M. Endogenous targets of RNA-directed DNA methylation and Pol IV in Arabidopsis. EMBO J. 2006;25:2828–2836. doi: 10.1038/sj.emboj.7601150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani M, Xiong L, Stevenson B, Zhu JK. Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis: interactions and convergence of abscisic acid-dependent and abscisic acid-independent pathways. Plant Cell. 1997;9:1935–1949. doi: 10.1105/tpc.9.11.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T, Bucher E, Daxinger L, Huettel B, Böhmdorfer G, Gregor W, Kreil DP, Matzke M, Matzke AJ. A structural-maintenance-of-chromosomes hinge domain-containing protein is required for RNA-directed DNA methylation. Nat Genet. 2008;40:670–675. doi: 10.1038/ng.119. [DOI] [PubMed] [Google Scholar]
- Kanno T, Mette MF, Kreil DP, Aufsatz W, Matzke M, Matzke AJ. Involvement of putative SNF2 chromatin remodeling protein DRD1 in RNA-directed DNA methylation. Curr. Biol. 2004;14:801–805. doi: 10.1016/j.cub.2004.04.037. [DOI] [PubMed] [Google Scholar]
- Kapoor A, Agarwal M, Andreucci A, Zheng X, Gong Z, Hasegawa PM, Bressan RA, Zhu JK. Mutations in a conserved replication protein suppress transcriptional gene silencing in a DNA-methylation-independent manner in Arabidopsis. Curr. Biol. 2005;15:1912–1918. doi: 10.1016/j.cub.2005.09.013. [DOI] [PubMed] [Google Scholar]
- Kumakura N, Takeda A, Fujioka Y, Motose H, Takano R, Watanabe Y. SGS3 and RDR6 interact and colocalize in cytoplasmic SGS3/RDR6-bodies. FEBS Lett. 2009;583:1261–1266. doi: 10.1016/j.febslet.2009.03.055. [DOI] [PubMed] [Google Scholar]
- Li CF, Pontes O, El-Shami M, Henderson IR, Bernatavichute YV, Chan SW, Lagrange T, Pikaard CS, Jacobsen SE. An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell. 2006;126:93–106. doi: 10.1016/j.cell.2006.05.032. [DOI] [PubMed] [Google Scholar]
- Mathieu O, Reinders J, Caikovski M, Smathajitt C, Paszkowski J. Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell. 2007;130:851–862. doi: 10.1016/j.cell.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Matzke MA, Birchler JA. RNAi-mediated pathways in the nucleus. Nat. Rev. Genet. 2005;6:24–35. doi: 10.1038/nrg1500. [DOI] [PubMed] [Google Scholar]
- Morel JB, Godon C, Mourrain P, Béclin C, Boutet S, Feuerbach F, Proux F, Vaucheret H. Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell. 2002;14:629–639. doi: 10.1105/tpc.010358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourrain P, Béclin C, Elmayan T, et al. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- Penterman J, Zilberman D, Huh JH, Ballinger T, Henikoff S, Fischer RL. DNA demethylation in the Arabidopsis genome. Proc. Natl. Acad. Sci. U S A. 2007;104:6752–6757. doi: 10.1073/pnas.0701861104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes O, Li CF, Nunes PC, Haag J, Ream T, Vitins A, Jacobsen SE, Pikaard CS. The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell. 2006;126:79–92. doi: 10.1016/j.cell.2006.05.031. [DOI] [PubMed] [Google Scholar]
- Pontier D, Yahubyan G, Vega D, Bulski A, Saez-Vasquez J, Hakimi MA, Lerbs-Mache S, Colot V, Lagrange T. Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev. 2005;19:2030–2040. doi: 10.1101/gad.348405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H, Béclin C, Fagard M. Post-transcriptional gene silencing in plants. J. Cell Sci. 2001;114:3083–3091. doi: 10.1242/jcs.114.17.3083. [DOI] [PubMed] [Google Scholar]
- Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert JL, Bartel DP, Crété P. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell. 2004;16:69–79. doi: 10.1016/j.molcel.2004.09.028. [DOI] [PubMed] [Google Scholar]
- Wierzbicki AT, Haag JR, Pikaard CS. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008;135:635–648. doi: 10.1016/j.cell.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Johansen L, Gustafson A, Kasschau K, Lellis A, Zilberman D, Jacobsen S, Carrington J. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M, Peragine A, Park MY, Poethig RS. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 2005;19:2164–2175. doi: 10.1101/gad.1352605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Wang Z, Li S, Yu B, Liu JY, Chen X. Intergenic transcription by RNA Polymerase II coordinates Pol IV and Pol V in siRNA-directed transcriptional gene silencing in Arabidopsis. Genes Dev. 2009 Nov 30; doi: 10.1101/gad.1868009. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Pontes O, Zhu J, Miki D, Zhang F, Li WX, Iida K, Kapoor A, Pikaard CS, Zhu JK. ROS3 is an RNA-binding protein required for DNA demethylation in Arabidopsis. Nature. 2008;455:1259–1262. doi: 10.1038/nature07305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Kapoor A, Sridhar VV, Agius F, Zhu JK. The DNA glycosylase/lyase ROS1 functions in pruning DNA methylation patterns in Arabidopsis. Curr. Biol. 2007;17:54–59. doi: 10.1016/j.cub.2006.10.059. [DOI] [PubMed] [Google Scholar]
- Zilberman D, Cao X, Jacobsen SE. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.