Summary
Objective
To assess the impact of seasonal variation on the distribution of the eba-175 allelic forms in the area where malaria transmission is markedly seasonal.
Methods
Blood samples were collected from 291 and 239 children under five years of age during the low and the high malaria transmission season, respectively, in four villages named Dawelgué, Kounda, Tanghin and Watenga of Saponé Health District, then screened for eba 175 F- and C- alleles by nested PCR analysis.
Results
F- alleles were more prevalent than C-alleles in the low [0.66 versus 0.34 (p<0.0001)] and high transmission season [0.67. versus 0.33(p<0.0001)]. No significant seasonal variation was observed in the distribution of the two alleles. However, according to Sewall Wright rules, the population pairwise FST values, between Dawelgué and Tanghin during the low transmission season (FST_ value= 0.10415, p-value= 0.0090 and during the high season (FST_value= 0.08244, p-value≪0.00001), between Tanghin and Watenga during the low season (FST value=0.07414, p-value=0.009) indicated a moderate but statistically significant genetic differentiation.
Conclusion
Although there was a moderate but significant genetic differentiation between some study villages at different times of the year, this study result in the seasonal stability of eba-175 allele’s distribution in the study area.
Keywords: EBA-175 allelic forms, Burkina Faso
Introduction
Malaria constitutes a major public health concern throughout sub-Saharan Africa. Worldwide deaths due to malaria are currently estimated at approximately 881,000 per year, 90% of which occur in sub-Saharan Africa and antimalarial drug and insecticide resistance continues to be on the rise (WHO 2006, 2008). During the last two decades there have been considerable efforts to develop vaccines against malaria (Rogier et al. 2006). The complexity of the life cycle and the high polymorphism level displayed by the parasite has hindered progress in the development of a malaria vaccine in spite of a worldwide effort.
Invasion of the human erythrocyte by Plasmodium merozoites represents a complex and crucial stage in malaria parasite life cycle, and significantly impacts survival and host pathogenesis (Miller & Greenwood 2002). This step involves several specific interactions between receptors on the red blood cells (RBCs) and parasite ligands. In the most virulent human malaria parasite, Plasmodium falciparum, only two invasion pathways have been well characterized, one involving Glycophorin A(GPA) and the 175 kDa Erythrocyte Binding Antigen (EBA-175) and a second pathway using glycophorin C (GPC) and a 140 kDa (EBA-140) also named BAEBL, a paralogue of EBA-175 (Camus 1985; Lobo et al. 2003; Maier et al. 2003; Sim et al. 1994). Other P. falciparum merozoites ligands involved in the erythrocytes invasion, like EBA-181 (also called JE-SEBL), PfNBP1, and PfNBP2b, have also been characterized (Duraisingh et al. 2003; Gilberger et al. 2003). However, little is know concerning the identity of the corresponding RBC receptors. The members of the erythrocyte binding-like (EBL) superfamily are highly diverse, providing the merozoite with high affinity binding ligands for a range of receptors on the surface of the erythrocyte (Adams et al. 2001). This genetic diversity likely explains why invasion of the erythrocyte by merozoite ligands appears to be strain-dependent(Hadley et al. 1987; Okoyeh et al. 1999).
The erythrocyte binding antigen-175 (eba-175) gene, located on chromosome seven, is one of the major genes in the Erythrocyte Binding Like (EBL) gene family that encodes for proteins which play a crucial role during erythrocyte invasion. The eba-175 gene is comprised of four exons and seven regions termed I–VII including three cysteine-rich regions (F1, F2 and C)(Sim et al. 1990; Adams et al. 1992; Toure et al. 2006). Binding regions F1 and F2 located at the N-terminus of the molecule, exhibit low polymorphism. Conversely, region III, which is centrally located, is characterized by two dimorphic segments termed FCR3 and CAMP (Kain et al. 1993). This dimorphism results from different sized insertions located at slightly different positions in the region III (Ware et al. 1993). A single parasite clone may include one or the other segment but never both. This dimorphic region has been implicated in the invasion process (Kain et al. 1993), and previous studies have analyzed the influence of this dimorphism on clinical disease and outcomes, as well as the distribution of the F and C genotypes in Africa (Cramer et al. 2004; Toure et al. 2006). As malaria epidemiology is known to differ between high and low transmission seasons (Molineaux L & Gramiccia G, 1980; Luxemburger et al. 1996; Theander 1998), and based on some previous studies carried out in endemic area, demonstrating seasonal changes in malaria parasite population (Roper et al.1998; Kobbe et al. 2006), we hypothized that the distribution of the two allelic forms (CAMP and FCR3) of eba-175 gene could be influenced by the season.
The objective of the present study was to assess the distribution of the F- and C-alleles in a malaria vaccine trial site of Burkina Faso where malaria transmission is endemic and markedly seasonal.
Material and methods
Study area and Patients
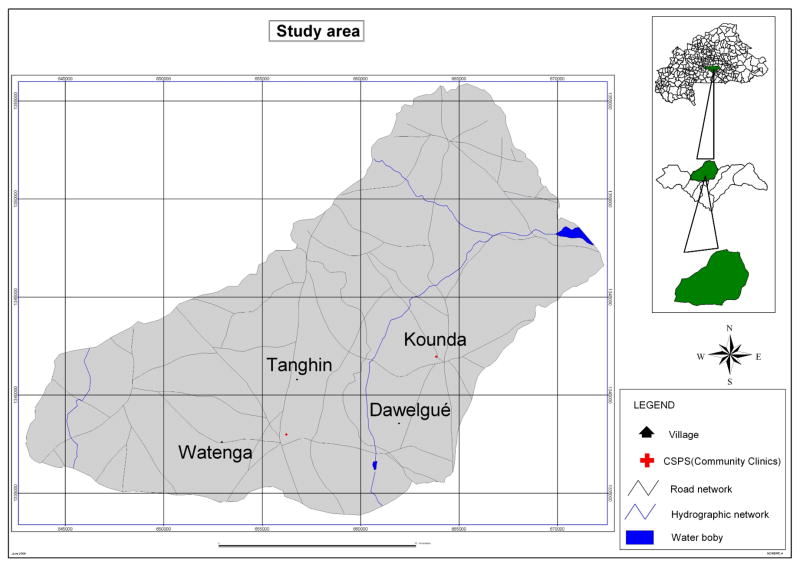
This study is part of a larger epidemiology study with one year longitudinal follow up. During this period children involved in the study received care of free on charge at the community health facilities involved in the study. All uncomplicated malaria cases were freely managed using artemisinin-based combination therapy (COARTEN®). The study was carried out in 2007 in Dawelgué, Kounda, Tanghin and Watenga, four villages of the Saponé Health District, located approximately 50 km Southwest of Ouagadougou, the capital city of Burkina Faso. The 4 study villages are located in the north-east part of the Saponé health district within an 8 km radius (Figure 1). The two farther villages (Kounda and Watenga) are about 11 km apart, while the two closest in distance (Dawelgué and Kounda) are 4 kilometres (Figure 1). The population is about 540, 1720, 1332 and 480 inhabitants in Dawelgué, Kounda, Tanghin and Watenga respectively. In all four villages the Mossi are the main ethnic group (> 95%). The estimated total population of children less than five years in the four study villages is about 465. Malaria transmission is perennial and seasonal in the district and peaks during the rainy season (from May to October). The entomological inoculation rate (EIR) was estimated at 200 infected/bites/person/year in the study area (Nebie et al. 2008). In 2007, the incidence of clinical malaria cases in children less than five years old living in Saponé Health District was estimated at 0.7 episode/child-year at risk (Sirima BS and others, unpublished data).
Figure 1.
Map of the study area indicating the four study villages
Ethical clearance
This study is part of a large epidemiology study whose protocol (DMID protocol 06-0020) was reviewed by the Office of Clinical Research Affairs (OCRA) within the Division of Microbiology and Infectious Disease (DMID at National Institute of Allergy and Infectious Diseases (NIAID) the United State of America. The Protocol was also approved by the Burkina Faso Health Research Ethics Review Committee. Written informed consent was obtained from the parents or guardians of all the children participating prior to the enrolment.
Sample collection
A finger prick blood sample was collected on filter paper (Whatman No 2) from 528 and 486 children aged 3 to 59 months living in the four study villages, during two cross-sectional surveys conducted in February and September 2007, constituting the middle of the low and the peak of malaria high transmission seasons, respectively. Before the genotyping, the blood filter paper samples were stored at room temperature with silica gel dessicant.
For the purpose of this study, 336 and 334 samples were randomized in low and high transmission season respectively, for DNA extraction and eba-175 genotyping. The samples were selectively randomized from each transmission season and each village to fulfil the statistical minimum size of 182 PCR positives samples required for each season for the data analysis. Thick and thin blood smears were also prepared for microscopic diagnosis and parasite count.
Asexual P. falciparum parasites density
Thick and thin blood smears were stained with Giemsa. The number of asexual P. falciparum parasites was estimated against 200 leukocytes and parasite density was calculated assuming a mean of 8,000 leukocytes per μl. A sample was declared negative after examining 200 thick film fields without observing any asexual parasites.
Parasite DNA extraction and genotyping
The DNA was extracted using QIAGEN commercial kits (QIAamp DNA blood Mini kit®) and conserved at minus 20°C until amplification was performed. eba-175 genotyping was performed as described elsewhere by a nested polymerase chain reaction (PCR) (Toure et al. 2001). Primers sequences for the first (nested) amplification were: eba1 5′-CAAGAAGCAGTTCCTGAGGAA-3′ (forward) and eba2 5′-TCTCAACATTCATATTAACAATTC-3′ (reverse). For the second (nested) amplification the following primers were used: eba3 5′-GAGGAAAACACTGAAATAGCACAC- 3′ (forward) and eba4 5′-CAATTCCTCCAGACTGTTGAACAT-3′ (reverse).
The first and second amplification were performed under the following conditions: 2 μL of DNA template for the first and 1 μL PCR-product for the second reaction; 1 μM of each Primer, 1 U Taq DNA polymerase (SIGMA-Aldrich CHEME GMBH), 2 μL of 10x PCR-Buffer (SIGMA-Aldrich CHEME GMBH) as supplied by the manufacturer and 200 μM dNTP (SIGMA-Aldrich CHEME GMBH) were used for the both reaction for a 20 μL final volume. The amplification reaction was performed on a PTC 100 Thermocycler (MJ Research, Inc) using the following reaction conditions: 29 cycles (first reaction) and 24 cycles (second reaction) of denaturation at 94°C for 1 minute, annealing at 56°C for 1 minute and extension at 72°C for 2 minutes followed by a final extension period of 3 minutes at 72°C.
The second amplification products were separated on 1.5% ethidium bromide stained agarose gel and were visualized on UV-translumination.
Quality control and data interpretation
P. falciparum CAMP (MRA-149) and FCR3 (MRA-102) lines were provided by MR4, ATCC® Manassas Virginia and were used as positives controls during the amplification reactions. To confirm the results, 20% of the sample extracts were re-amplified using the nested primers (eba3 and eba4) only, following whole genome amplification (Qiagen).
The CAMP allele was identified as a single fragment of ~714 bp in length while the FCR3 allele was a single fragment of ~795 bp relative to CAMP and FCR3 controls. Mixed infections were defined as the simultaneous presence of the F and C fragment in the same sample (Figure 2).
Figure 2.
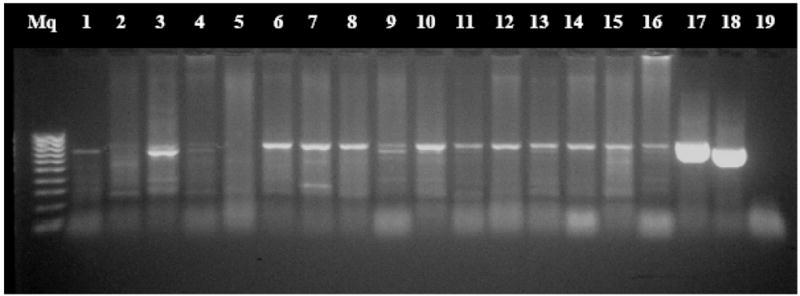
Nested polymerase chain reaction of eba-175 products
Lane Mq: DNA 100 bp ladder; Lanes 1, 6–8, 11–16: FCR-3 genotypes; Lanes 3, 4, 9: Mixed infections (FCR-3 plus CAMP genotypes); Lane 17: FCR-3 positive control; Lane 18: CAMP positive control; Lane 19: Nested PCR negative controls.
Statistical analysis
Allelic population data were analyzed using Epi Info v6.04a (http://www.cdc.gov/epiinfo/Epi6/EI6dnjp.htm) and Arlequin v3.1 (Excoffier et al. 2005). Due to the presence of mixed infections in the population (approximately 1/3 of all the infections), some samples displayed more than one eba-175 allele, while the rest showed a single allele (Figure 2). Comparisons were made using the Chi squared or Fisher’s exact test, and the Student’s t-test and ANOVA for normally distributed continuous data.
To analyse whether allele frequencies varied across the two transmission seasons regardless of geographical location (village), data were pooled from the four villages and allele frequencies compared between the high and low transmission seasons.
Secondly, to investigate whether geographical location (i.e. village) or transmission season within a particular village explained a significant proportion of the variation in the data, the data were organized into 8 populations and 4 groups. Each group represents one village while each population represents a further subdivision of the data into high and low transmission season samples, such that each group contains two populations. For this purpose the Analysis of Molecular Variance (AMOVA) was used implemented in Arlequin v3.1 (Excoffier et al. 1992, 2005). Va estimates the amount of variation in the data explained by transmission season, Vb, estimates the amount of variation due to different transmission season within individual villages, and finally Vc calculate the amount of variation found within each village/transmission season combination in the study population. 10,000 permutations were run for the AMOVA analysis and populations were considered to be significantly differentiated at P < 0.05 (Excoffier et al. 2005).
To explore genetic differentiation based on genetic distance method between the fur study villages, population pairwise FST values (computed genetic distance, ranges from 0–1) were assessed and the results interpreted according to guidelines suggested by Sewall Wright (Hartl & Clark 1989). Briefly: “The range of FST values 0 to 0.05 may be considered as indication little genetic differentiation; the range of 0.05 to 0.15 indicates moderate genetic differentiation; The range of 0.15 to 0.25 indicates great genetic differentiation; Values of FST above 0.25 indicate very great genetic differentiation.”
Standard diversity indices (gene diversity) were also assessed to estimate genetic diversity, the probability that two randomly chosen haplotypes are different in each population (Nei 1987).
Results
Seventy-one of 336 samples (21.1%) in low and 105 of 334 samples (31.4%) in high transmission season analyzed in this study were negative slides. With the PCR reactions 45 of 336 samples (13.4%) and 95 of 334 samples (28.4%) were negatives in low and high transmission season respectively.
Finally, the eba-175 genotypes were obtained for malaria parasites from a total of 530 samples from four villages, 291 and 239 during low and high transmission respectively that were used for the subsequent analysis. The proportion of male and female participants in total population was 1.2:1 (Male/Female). The mean participant age was 3.1 years (95% confidence interval [CI], 3.1–3.3). The geometric mean of parasite density, although slightly higher in CAMP infections, was not statistically different between CAMP, FCR3 and mixed infections (p-value= 0.69) (Table 1). Genetic diversity was moderately high for all 4 villages (Table 2). Unsurprisingly, the 2 villages with the largest sample sizes, Kounda and Tanghin, showed the greatest diversity, though diversity did not differ significantly among villages.
Table 1.
Proportion of eba-175 genotypes and Geometric Means of parasites densities (P. falciparum trophozoites/μl)
| Types of infections | Low transmission season | High transmission season | Total population | Geometric Mean of Parasite Density/Confidence intervals |
|---|---|---|---|---|
| (N= 291) | (N= 239) | N= 530 | ||
| n (%) | n (%) | N (%) | ||
| CAMP infections | 35 (12) | 40 (16.7) | 75 (14.2) | 2404 [1480–4024] |
| FCR3 infections | 158 (54.3) | 142 (59.4) | 300 (56.6) | 2346 [1808–2921] |
| Mixed infections | 98 (33.7)* | 57 (23.8)* | 155 (29.2) | 2043 [1480–2697] |
| All infection | 291 (100) | 239 (100) | 530 (100) | 2257 [1998–2697] |
| p-values | < 0.0001** | < 0.0001*** | < 0.0001$ | 0.69£ |
The prevalence of the mixes infections (FCR3 plus CAMP) cases decrease significantly (p= 0.01) from the low season to the high season;
p- value [comparison of the proportion of FCR3, CAMP and Mixed (FCR3+CAMP) infections during the low transmission season];
p- value (comparison of the proportion of FCR3, CAMP and Mixed (FCR3+CAMP) infections during the high transmission season);
p- value [comparison of the proportion of FCR3, CAMP and Mixed (FCR3+CAMP) infections in the total (low and high season) population];
p- value [comparison of the Geometric Mean of Parasite Density between FCR3, CAMP and mixed (FCR3+CAMP) infections in the total population]
Table 2.
Allele frequencies and molecular diversity indexes in the four study villages during the high and low transmission seasons.
| Population allele frequencies | χ2 | |||||
| Low season | Alleles | Dawelgué (n= 61) | Kounda (n= 130) | Tanghin (n= 166) | Watenga (n= 32) | P |
| FCR3 | 0.70 | 0.66 | 0.62 | 0.75 | 0.42* | |
| CAMP- | 0.30 | 0.34 | 0.38 | 0.25 | 0.42** | |
| P | ≪ 0.0001 | ≪ 0.0001 | 0.00001 | ≪ 0.0001 | ||
| High season | Alleles | Dawelgué (n= 68) | Kounda (n= 112) | Tanghin (n= 98) | Watenga (n= 18) | P |
| FCR3- | 0.78 | 0.65 | 0.61 | 0.62 | 0.13$ | |
| CAMP- | 0.22 | 0.35 | 0.39 | 0.38 | 0.13£ | |
| P | ≪ 0.0001 | 0.0002 | 0.002 | 0.008 | ||
| Molecular diversity Indexes | ||||||
| Number of gene copies | 103 | 192 | 193 | 42 | p-value | |
| Gene diversity | 0.4744 | 0.5834 | 0.6165 | 0.4843 | 0.07§ | |
| Standard Deviation of Gene diversity | 0.0440 | 0.0250 | 0.0163 | 0.0765 | ||
n represents the total number of alleles in each village
p- value comparing the frequency of F alleles between the four study villages during the low transmission season;
p- value comparing the frequency of C alleles between the four study villages during the low transmission season;
p- value compare the frequency of F alleles between the four study villages during the high transmission season;
p- value compare the frequency of C alleles between the four study villages during the high transmission season;
p- value comparing the genetic diversity between study villages.
eba-175 allele’s distribution in four study villages across two transmission seasons
In the entire study population, F and C alleles were present as single genotype infections in 56.6% (300/530) and 14.2 % (75/530) of the participants, respectively. The remaining enrolled children, 29.2% (155/530), showed mixed infections (F+C alleles) (Table 1). Moreover, the proportion of samples carrying mixed infections showed a statistically significance decrease (p= 0.01, χ2 test) between the low transmission season, [33.7% (98/291)] and the high transmission season [23.8% (57/239); Table 1]
Comparison of allele frequency between high and low transmission seasons
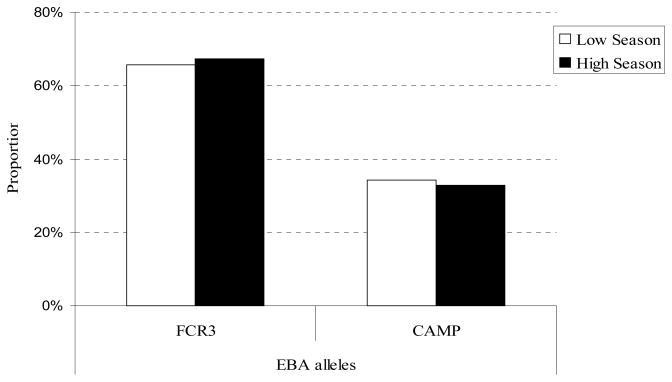
For this analysis data were pooled from the four villages in order to compare allele frequencies from the high and low transmission seasons. The pairwise population FST indicated no statistical differences in allele frequencies between the two seasons (FST = 0.00277; p-value = 0.12, Figure 3).
Figure 3.
Temporal distribution of eba-175 alleles
FST = 0.00277; p= 0.12
Village-seasonal distribution and genetic differentiation in the eba-175 alleles
Significant differences were observed in allele frequencies, with the F-allele being twice as common as the C-allele within each study village and across seasons (Table 2). No difference in F- and C- allele distribution was observed between villages or transmission seasons.
Analysis of the pairwise FSTs between the 8 populations according to Stewall guidelines showed little genetic differentiation (FST values between 0 and 0.05) between all study villages except between Dawelgué in high season and in Tanghin low season (Fst-value= 0.10415); Dawelgué and Tanghin in high season (Fst-value= 0.08244), and Tanghin and Watenga in low season (Fst-value=0.07414) where the genetic differentiation was moderate. There is no major genetic differentiation between study villages with a FST > 0.15 (Table 3).
Table 3. Population pairwise FST.
Distance method: Pairwise difference
| 1(D_L) | 2(D_H) | 3(K_L) | 4(K_H) | 5(T_L) | 6(T_H) | 7(W_L) | 8(W_H) | |
|---|---|---|---|---|---|---|---|---|
| 1(D_L) | 0.00000 | |||||||
| 2(D_H) | 0.00938 | 0.00000 | ||||||
| 3(K_L) | 0.00180 | 0.03762 | 0.00000 | |||||
| 4(K_H) | 0.00655 | 0.04515* | −0.00923 | 0.00000 | ||||
| 5(T_L) | 0.02328 | 0.10415* | 0.01314 | 0.01716 | 0.00000 | |||
| 6(T_H) | 0.02925 | 0.08244* | −0.00069 | −0.00643 | 0.01489 | 0.00000 | ||
| 7(W_L) | −0.00173 | −0.01686 | 0.00596 | 0.00733 | 0.07414* | 0.03371 | 0.00000 | |
| 8(W_H) | −0.04672 | −0.02920 | −0.02406 | −0.01657 | 0.01454 | 0.00859 | −0.03973 | 0.00000 |
Significance Level= 0.0500;
Significant test p≤0.05; p (D_H/K_H)= 0.018; p (D_H/T_L)=0.009; p (D_H/T_H)≪0.00001; p (T_L/W_L)=0.009
D_L (Dawelgué Low season); D_H (Dawelgué High season); K_L (Kounda Low season); K_H (Kounda High season); T_L (Tanghin Low season); T_H (Tanghin High season); W_L (Watenga Low season); W_H (Watenga High season)
Analysis of molecular variance (AMOVA) within populations and transmission seasons
The variance analysis indicated that the vast majority of the variance (98.2%) in the data is contained within individual populations, or village/season combinations (Table 4). In addition, when looking within transmission seasons, villages account for a small but significant proportion of the variance (2.22%; p-value= 0.04). In contrast, transmission season did not appear to contribute to the variance (Table 4).
Table 4.
Variance in eba-175 alleles frequencies
| Hierarchical level | Variance | % total variation | P-value | φ-statistic* |
|---|---|---|---|---|
| Among transmission season | −0.0015 | −0.41 | 0.601 | φCT** = −0.00408 |
| Among village/within transmission season | 0.00808 | 2.22 | 0.042 | φ SC$ = 0.02211 |
| Within populations | 0.36166 | 98.18 | 0.0108 | φ ST£ = 0.01812 |
φ-statistic (average F-statistic Indices over loci);
φCT, F-statistic indices by permuting populations among transmission seasons.
φSC, F-statistic indices by permuting haplotypes among villages within transmission seasons; φ ST, F-statistic, indices by permuting haplotypes within villiage within transmission seasons.
Discussion
The Erythrocyte Binding Antigen 175, one of the promising candidates in the malaria vaccine pipeline, has been studied in different malaria endemic areas (Toure et al. 2001; Cramer et al. 2004; Toure et al. 2006). The central role of this antigen in erythrocyte invasion warrants a clearer understanding of genetic polymorphism in this gene (Camus & Hadley 1985; Klotz et al. 1992; Orlandi et al. 1992; Sim et al. 1994). The present study was designed to evaluate the seasonal variation of the frequencies of P. falciparum erythrocyte binding antigen-175 alleles in malaria positives children under age five, living in four endemic villages located in southwestern Burkina Faso.
The higher frequency of the F- compared with the C- allele that we observed in this study is consistent with findings from Ghana,(Cramer et al. 2004) and Gabon (Toure et al. 2006), suggesting that the F-allele may predominate throughout West and Central Africa, and that the distribution of alleles is relatively stable over 8 months, including a low and high transmission season. In contrast, in East Africa, where malaria endemicity is low, the C- allele has been shown to be the most common (Binks et al. 2001). By way of comparison, a study in Laos People’s Democratic Republic reported significant difference in the distribution of eba-175 alleles when the northern and the southern provinces were compared. There are several explanations as to why F- and C- allele frequencies differ among geographical regions. One explanation is random shifts in parasite allele frequencies due to genetic drift in genetically isolated populations (Cramer et al. 2004). In this case allele frequencies might be predicted to change over time. Another possibility is that differences in the host genetic background among study population may select for different eba-175 alleles in which case stable allele frequencies would be predicted (Dittrich et al. 2003).
To our knowledge, this study is one of the first to analyze seasonal variation of the eba-175 allelic dimorphism between low and high malaria transmission seasons in an endemic area. We found that fluctuations in transmission level had no effect on the distribution of eba-175 alleles except the decrease of mixed infection cases from low to high season which could be explained by the cases management facilities with care free on charge set up in the health community clinic during the one year passive follow up. Indeed the accessibility improved by the care free on charge increased the attendance rate at the community health clinics and consequently the reduction of the number of malaria episodes by regular and frequent management of cases. The artemisine-based combination therapy used at the community health clinics for uncomplicated malaria case treatments would clean the sensitive genotypes with a possible reduction of the number of mixed infection from children.
The lack of change in allele frequencies across transmission seasons supports the prediction that distribution of the two allelic forms of eba-175 gene is relatively stable over time. The separated comparison of allele distribution across villages further indicates stability throughout the geographic region sampled, despite moderate genetic differentiation among some villages.
The fact that the study villages were located close to each other (within an 8 kilometer radius) could explain the observed similarity in F-and C-alleles diversity and the absence of restricted gene flow demonstrated by the low or moderate genetic differentiation, such that we are essentially sampling a single panmictic parasite population. But, it is also possible that the genetic background (most of the study population are from the ethnic majority group, Mossi) may contribute to the homogeneity of the eba-175 alleles distribution, as it has been previously suggested by the findings in Laos People’s Democratic Republic study where a significant difference in eba-175 allele distribution was showed between the North and South province with different ethnic groups (Dittrich et al. 2003). The apparent stability of eba-175 allele frequencies in the four villages in this study suggests that it is a suitable area to test vaccine candidates targeting this antigen.
Broader regional sampling in sub-Saharan sites will address broader boundaries within Africa. A large scale geographic comparison study that includes different sub-Saharan African countries which takes into consideration both isolation by distance and differences in the genetic background of the host is recommended to further address the factors contributing to the distribution of eba-175 alleles.
Conclusion
This comparative analysis of the allelic dimorphism of the eba-175gene in P. falciparum isolates from low and high transmission seasons indicates that the dimorphism was not affected by seasonal fluctuations in transmission intensity. This analysis highlights the importance of assessing the distribution of eba-175 allelic forms in different sentinel sites within the same country as well as in neighbouring countries. These findings will inform strategies for the development of malaria vaccine trial sites.
Acknowledgments
We express our gratitude to the population of the four villages for their kind cooperation and support of the study, the health staff of Saponé Health District as well as all the investigation staff of CNRFP. We also thank MR4 (ATCC®, Manassas, VA, USA) for providing us with malaria parasites and Timothy Stedman and Linda Amoah for providing training under the MR4 Workshop “Assessing the diversity of vaccine antigen and drug resistance markers in field isolates”, Ouagadougou Burkina Faso 2008; Dr Deirdre A. Joy, Program Officer, Parasite Genomics (Parasitology and International Programs Branch, Division of Microbiology and Infectious Diseases, NIAID\NIH\DHHS) for her assistance with the statistical analyses and critically reading the manuscript. This study was supported by the regular budget of National Institutes of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Department of Infectious Disease, MD (NIH/NIAID/DMID (DMID Protocol N°06-20, Contract HHSN266200400016C).
References
- Adams JH, Blair PL, Kaneko O, Peterson DS. An expanding ebl family of Plasmodium falciparum. Trends Parasitol. 2001;17:297–299. doi: 10.1016/s1471-4922(01)01948-1. [DOI] [PubMed] [Google Scholar]
- Adams JH, Sim BK, Dolan SA, Fang X, Kaslow DC, Miller LH. A family of erythrocyte binding proteins of malaria parasites. Proc Natl Acad Sci U S A. 1992;89:7085–7089. doi: 10.1073/pnas.89.15.7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binks RH, Baum J, Oduola AM, et al. Population genetic analysis of the Plasmodium falciparum erythrocyte binding antigen-175 (eba-175) gene. Mol Biochem Parasitol. 2001;114:63–70. doi: 10.1016/s0166-6851(01)00240-7. [DOI] [PubMed] [Google Scholar]
- Camus D, Hadley TJ. A Plasmodium falciparum antigen that binds to host erythrocytes and merozoites. Science. 1985;230:553–556. doi: 10.1126/science.3901257. [DOI] [PubMed] [Google Scholar]
- Cramer JP, Mockenhaupt FP, Mohl I, et al. Allelic dimorphism of the erythrocyte binding antigen-175 (eba-175) gene of Plasmodium falciparum and severe malaria: Significant association of the C-segment with fatal outcome in Ghanaian children. Malar J. 2004;3:11. doi: 10.1186/1475-2875-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittrich S, Schwobel B, Jordan S, et al. Distribution of the two forms of Plasmodium falciparum erythrocyte binding antigen-175 (eba-175) gene in Lao PDR. Malar J. 2003;2:23. doi: 10.1186/1475-2875-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duraisingh MT, Maier AG, Triglia T, Cowman AF. Erythrocyte-binding antigen 175 mediates invasion in Plasmodium falciparum utilizing sialic acid-dependent and -independent pathways. Proc Natl Acad Sci U S A. 2003;100:4796–4801. doi: 10.1073/pnas.0730883100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Estoup A, Cornuet JM. Bayesian Analysis of an Admixture Model With Mutations and Arbitrarily Linked Markers. Genetics. 2005;169:1727–1738. doi: 10.1534/genetics.104.036236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Smouse P, Quattro J. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilberger TW, Thompson JK, Triglia T, Good RT, Duraisingh MT, Cowman AF. A novel erythrocyte binding antigen-175 paralogue from Plasmodium falciparum defines a new trypsin-resistant receptor on human erythrocytes. J Biol Chem. 2003;278:14480–14486. doi: 10.1074/jbc.M211446200. [DOI] [PubMed] [Google Scholar]
- Hadley TJ, Klotz FW, Pasvol G, et al. Falciparum malaria parasites invade erythrocytes that lack glycophorin A and B (MkMk). Strain differences indicate receptor heterogeneity and two pathways for invasion. J Clin Invest. 1987;80:1190–1193. doi: 10.1172/JCI113178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl DL, Clark AG. Principles of Population Genetics. 2. Sinauer Associates; Sunderland, Massachusetts: 1989. pp. 118–119. [Google Scholar]
- Kain KC, Orlandi PA, Haynes JD, Sim KL, Lanar DE. Evidence for two-stage binding by the 175-kD erythrocyte binding antigen of Plasmodium falciparum. J Exp Med. 1993;178:1497–1505. doi: 10.1084/jem.178.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotz FW, Orlandi PA, Reuter G, et al. Binding of Plasmodium falciparum 175-kilodalton erythrocyte binding antigen and invasion of murine erythrocytes requires N-acetylneuraminic acid but not its O-acetylated form. Mol Biochem Parasitol. 1992;51:49–54. doi: 10.1016/0166-6851(92)90199-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobbe R, Neuhoff R, Marks F, et al. Seasonal variation and high multiplicity of first Plasmodium falciparum infections in children from a holoendemic area in Ghana, West Africa. Trop Med Int Health. 2006;11:613–619. doi: 10.1111/j.1365-3156.2006.01618.x. [DOI] [PubMed] [Google Scholar]
- Lobo CA, Rodriguez M, Reid M, Lustigman S. Glycophorin C is the receptor for the Plasmodium falciparum erythrocyte binding ligand PfEBP-2 (baebl) Blood. 2003;101:4628–4631. doi: 10.1182/blood-2002-10-3076. [DOI] [PubMed] [Google Scholar]
- Luxemburger C, White NJ, Webster HK, et al. The epidemiology of malaria in a Karen population on the western border of Thailand. Trans R Soc Trop Med Hyg. 1996;90:105–111. doi: 10.1016/s0035-9203(96)90102-9. [DOI] [PubMed] [Google Scholar]
- Maier AG, Duraisingh MT, Reeder JC, et al. Plasmodium falciparum erythrocyte invasion through glycophorin C and selection for Gerbich negativity in human populations. Nat Med. 2003;9:87–92. doi: 10.1038/nm807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller LH, Greenwood B. Malaria--a shadow over Africa. Science. 2002;298:121–122. doi: 10.1126/science.1078048. [DOI] [PubMed] [Google Scholar]
- Molineaux L, Gramiccia G. The Garki Project, Research on the Epidemiology and Control of Malaria in the Sudan Savanna of West Africa. WHO; Geneva: 1980. [Google Scholar]
- Nebie I, Diarra A, Ouedraogo A, et al. Humoral responses to Plasmodium falciparum blood-stage antigens and association with incidence of clinical malaria in children living in an area of seasonal malaria transmission in Burkina Faso, West Africa. Infect Immun. 2008;76:759–766. doi: 10.1128/IAI.01147-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Molecular Evolutionary Genetics. Columbia University Press; New York, NY, USA: 1987. [Google Scholar]
- Okoyeh JN, Pillai CR, Chitnis CE. Plasmodium falciparum field isolates commonly use erythrocyte invasion pathways that are independent of sialic acid residues of glycophorin A. Infect Immun. 1999;67:5784–5791. doi: 10.1128/iai.67.11.5784-5791.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlandi PA, Klotz FW, Haynes JD. A malaria invasion receptor, the 175-kilodalton erythrocyte binding antigen of Plasmodium falciparum recognizes the terminal Neu5Ac(alpha 2–3)Gal-sequences of glycophorin A. J Cell Biol. 1992;116:901–909. doi: 10.1083/jcb.116.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogier C, Orlandi-Pradines E, Fusai T, Pradines B, Briolant S, Almeras L. Malaria vaccines: prospects and reality. Med Mal Infect. 2006;36:414–422. doi: 10.1016/j.medmal.2006.05.016. [DOI] [PubMed] [Google Scholar]
- Roper C, Elhassan IM, Giha H, et al. Seasonal changes in the Plasmodium falciparum population in individuals and their relationship to clinical malaria: a longitudinal study in a Sudanese village. Parasitology. 1998;116:501–510. doi: 10.1017/s0031182098002650. [DOI] [PubMed] [Google Scholar]
- Sim BK, Chitnis CE, Wasniowska K, Hadley TJ, Miller LH. Receptor and ligand domains for invasion of erythrocytes by Plasmodium falciparum. Science. 1994;264:1941–1944. doi: 10.1126/science.8009226. [DOI] [PubMed] [Google Scholar]
- Sim BK, Orlandi PA, Haynes JD, et al. Primary structure of the 175K Plasmodium falciparum erythrocyte binding antigen and identification of a peptide which elicits antibodies that inhibit malaria merozoite invasion. J Cell Biol. 1990;111:1877–1884. doi: 10.1083/jcb.111.5.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theander T. Unstable malaria in Sudan. The influence of the dry season. Trans R Soc Trop Med Hyg. 1998;92:589–592. doi: 10.1016/s0035-9203(98)90775-1. [DOI] [PubMed] [Google Scholar]
- Toure FS, Bisseye C, Mavoungou E. Imbalanced distribution of Plasmodium falciparum EBA-175 genotypes related to clinical status in children from Bakoumba, Gabon. Clin Med Res. 2006;4:7–11. doi: 10.3121/cmr.4.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toure FS, Mavoungou E, Ndong JM, Tshipamba P, Deloron P. Erythrocyte binding antigen (EBA-175) of Plasmodium falciparum: improved genotype determination by nested polymerase chain reaction. Trop Med Int Health. 2001;6:767–769. doi: 10.1046/j.1365-3156.2001.00789.x. [DOI] [PubMed] [Google Scholar]
- Ware LA, Kain KC, Lee Sim BK, Haynes JD, Baird JK, Lanar DE. Two alleles of the 175-kilodalton Plasmodium falciparum erythrocyte binding antigen. Mol Biochem Parasitol. 1993;60:105–109. doi: 10.1016/0166-6851(93)90033-t. [DOI] [PubMed] [Google Scholar]
- WHO. WHO guideline for the treatment of malaria. WHO/HTM/MAL/2006; Geneva: 2006. [Google Scholar]
- WHO. World malaria report 2008. Vol. 1. WHO/HTM/GMP/2008; Geneva: 2008. [Google Scholar]