Abstract
RNAs in cells are associated with RNA-binding proteins (RBPs) to form ribonucleoprotein (RNP) complexes. The RBPs influence the structure and interactions of the RNAs and play critical roles in their biogenesis, stability, function, transport and cellular localization. Eukaryotic cells encode a large number of RBPs (thousands in vertebrates), each of which has unique RNA-binding activity and protein-protein interaction characteristics. The remarkable diversity of RBPs, which appears to have increased during evolution in parallel to the increase in the number of introns, allows eukaryotic cells to utilize them in an enormous array of combinations giving rise to a unique RNP for each RNA. In this short review, we focus on the RBPs that interact with pre-mRNAs and mRNAs and discuss their roles in the regulation of post-transcriptional gene expression.
Keywords: RNA, ribonucleoproteins, RNPs, RNA-binding proteins, RNA processing, gene expression 1 Equal contributions
1. Introduction
In prokaryotes, transcription and translation are physically coupled. In eukaryotes, these two processes occur in separate compartments, the nucleus and the cytoplasm, respectively. This allows eukaryotes to carry out extensive post-transcriptional processing of pre-mRNA that produces a more diverse assortment of mRNAs from its genome and provides an additional layer of gene regulation. The pre-mRNA processing reactions, including splicing, editing and polyadenylation, commence as soon as pre-mRNAs emerge from their sites of transcription and are mediated by RBPs and trans-acting RNAs, themselves present as RNPs (e.g. snRNPs). Although all RBPs bind RNA, they do so with different RNA-sequence specificities and affinities. This activity is mediated by a relatively small number of RNA-binding scaffolds whose properties are further modulated by auxiliary domains. The auxiliary domains can also mediate the interactions of the RBP with other proteins and, in many cases, are subject to regulation by post-translational modification. As a result, cells are able to generate numerous RNPs whose composition and arrangement of components is unique to each mRNA and the RNPs are further remodeled during the course of the maturation of the mRNA into its functional form. While our focus here is on the RBPs that are associated with pre-mRNAs and mRNAs, we note that many RBPs are associated with other classes of RNAs (for a recent review see [1]), and all of these are important for cell physiology (Figure 1). Many of the features of RBPs that we discuss, however, are general and also apply to RBPs that are part of many different types of RNPs. In the following, we discuss select examples that illustrate general principles of the biochemistry and cell biology of RBPs to highlight their central role in gene expression.
Figure 1.
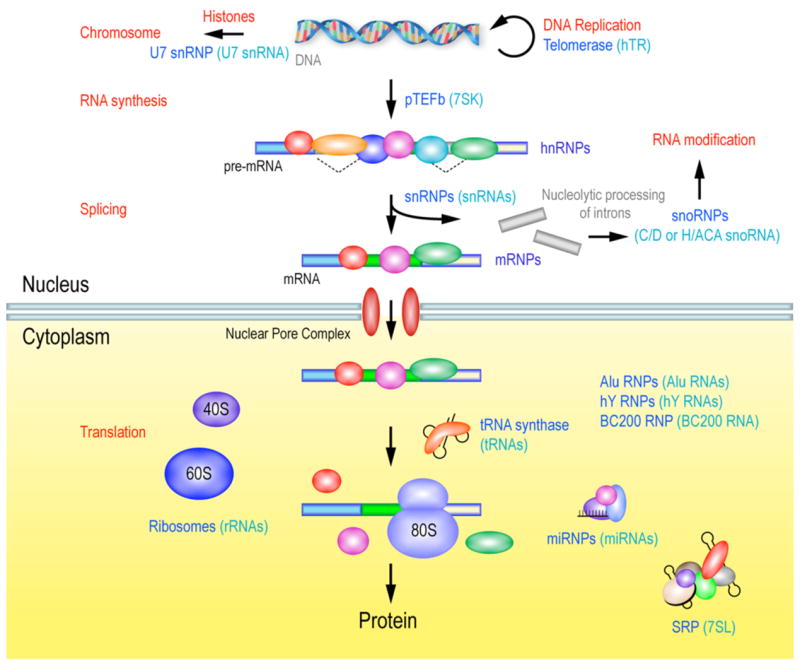
RNA-binding proteins (RBPs) function in multiple cellular processes. Genetic information stored in chromosomal DNA is translated into proteins through mRNAs. This allows for post-transcriptional control of gene expression conferring a crucial role to the mRNA-binding proteins in this regulation. In addition to the RBPs associated with mRNA, many different classes of RBPs interact with various small non-coding RNAs to form ribonucleoprotein (RNP) complexes that are actively involved in many different aspects of cell metabolism, such as DNA replication, expression of histone genes, regulation of transcription and translational control.
2. RNA-binding proteins are numerous and diverse
The discovery of the heterogeneous nuclear ribonucleoproteins (hnRNP) and other pre-mRNA/mRNA-binding proteins led to the identification of the first amino acid motifs and functional domains that confer binding to RNA [2]. RBPs contain one or, more often, multiple RNA-binding domains. Some well-characterized RNA-binding domains include the following: RNA-binding domain (RBD, also known as RNP domain and RNA recognition motif, RRM); K-homology (KH) domain (type I and type II); RGG (Arg-Gly-Gly) box; Sm domain; DEAD/DEAH box; zinc finger (ZnF, mostly C-x8-X-x5-X-x3-H); double stranded RNA-binding domain (dsRBD); cold-shock domain; Pumilio/FBF (PUF or Pum-HD) domain; and the Piwi/Argonaute/Zwille (PAZ) domain (Figure 2) (for review see [3,4]). Using these motifs, bioinformatic analyses revealed that eukaryotic genomes encode a large number of RBPs. In yeast, 5–8% of genes encode proteins predicted to function as RBPs, and in C. elegans and D. melanogaster, approximately 2% of the genome is annotated to encode RBPs [5–7]. However, it is likely that the number of RBPs is much higher, since there are probably other RNA-binding domains that remain to be uncovered. Why do eukaryotes need so many – hundreds and perhaps thousands of – RBPs? One possible explanation is that as eukaryotes evolved highly specific post-transcriptional processes to fine-tune gene expression, a concomitant expansion of the number of RBPs needed to function in these processes has occurred [8]. For example, in both vertebrates and plants, the emergence of alternative splicing during evolution drove the need for a corresponding increase in the number of RBPs [8].
Figure 2.
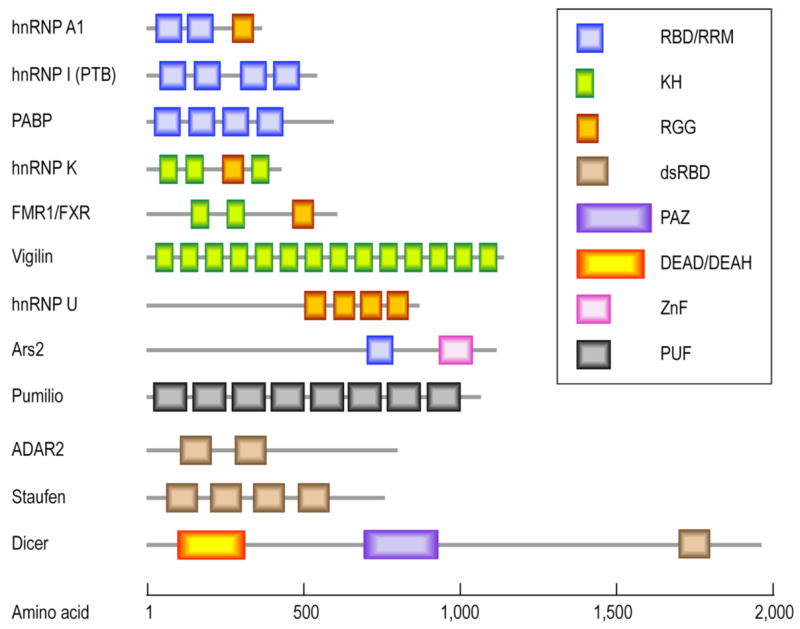
RNA-binding domains of RBPs. Often, several RNA-binding domains are found within one RBP. Different RNA-binding domains include the RNA-binding domain (RBD), K-homology (KH) domain, RGG (Arg-Gly-Gly) box, double stranded RNA-binding domain (dsRBD), Piwi/Argonaute/Zwille (PAZ) domain, RNA helicase DEAD/DEAH box, RNA-binding zinc finger (ZnF) and Puf RNA-binding repeats (PUF). All are presented as colored boxes.
It is certain that many RBPs remain yet to be characterized. Several methods have been developed to identify and characterize the RBPs and the RNAs with which they interact. The hnRNP and mRNP complexes were initially isolated by ultraviolet (UV) cross-linking of RNA-protein complexes in vivo [9–15]. This is a reliable and effective method to detect RNA-protein interactions, as it circumvents the adventitious association of proteins with RNAs that could occur after cell lysis [16]. Recently, this method has been adapted, using tagged proteins and including an immunoprecipitation step following cross-linking (cross-linking and immunoprecipitation or CLIP) [17]. Procedures to detect and delineate RNA-protein interactions in vitro include systematic evolution of ligands by exponential enrichment (SELEX) and electrophoretic mobility shift assay (EMSA) [18]. A yeast-three hybrid system has been devised as a screening method to identify RBPs and their target RNAs [19–21]. Several approaches have been utilized to identify RNA targets. For example, the RIP assay, which combines reversible cross-linking with formaldehyde followed by immunoprecipitation and RT-PCR, has been used to identify hepatitis delta antigen (HDAg) interactions with HDV RNAs and U1 snRNP protein-RNA interactions [22]. An affinity tag may also be introduced to facilitate the isolation of an RBP of interest, followed by analysis of associated RNAs using microarrays, an approach that has been successfully used to identify RNAs that associate with PUF proteins in S. cerevisiae [23]. Bioinformatics approaches can also be used to identify RNA targets if a consensus and non-degenerate RNA-binding sequence is known. In addition, traditional genetic approaches and reverse genetics can be employed to identify both RBPs and their target RNAs. For example, RNAi screening in cultured D. melanogaster cells using a candidate gene approach has been successfully used to examine which RBPs are involved in alternative splicing [24]. Taken together, a considerable array of technologies is now available to discover and further study the many RBPs that bioinformatics predicts to be present.
At the structural level, RBPs often exhibit a high degree of modularity, as most contain one or more RNA-binding and auxiliary domains (for review see [4]). This modularity creates both RNA-binding and functional diversity within the RBPs. The most extensively studied RNA-binding domain, the RBD, is often found as multiple repeats within a single protein, exemplified by the polypyrimidine tract-binding protein (PTB/hnRNP I), poly(A) binding protein (PABP), U2AF65 and U1A [4]. Although a single RBD, which typically can bind 2 – 6 nucleotides, is sufficient for binding RNA, having multiple copies of this domain enables the recognition of larger, more complex RNA targets, enhancing the specificity and affinity of binding [25]. A similar principle is found in PUF proteins. These typically contain eight consecutive Puf RNA-binding repeats, each of which consists of approximately 40 amino acids that form three α-helices [26–28]. The crystal structure of human Pumilio bound to RNA revealed that each of the eight repeats recognizes a single nucleotide in its target RNA, to bind a total of eight consecutive nucleotides [27]. This specific and high affinity interaction, in combination with its modular design, enables a unique and remarkably predictable PUF-RNA interaction that can be exploited to engineer proteins that bind sequences other than wild-type [27,29,30].
A further diversity of RBPs is achieved by combining RNA-binding domains with auxiliary functional domains. ADAR2 and PKR are two RBPs that have similar RNA-binding domains, the dsRBD, but differ in their auxiliary domains and their associated functions. ADAR2 combines its dsRBD with a deaminase domain that converts adenosine to inosine in its target RNAs, while PKR incorporates a kinase domain [31,32]. As PKR binds double-stranded RNA, it is converted to an active state where subsequent autophosphorylation triggers many downstream events [33]. The dsRBD of PKR is thus able to autoregulate its kinase domain due to the modularity of its structure.
Alternative splicing is yet another mechanism by which cells can expand its repertoire of RBPs. For example, alternative splicing of the polypyrimidine tract binding protein (PTB/hnRNP I) mRNA generates a splice variant that lacks the first two RBDs, and the corresponding PTB isoform may affect the stability of the CD154 mRNA [34]. Another example of alternatively spliced RBPs is the poly(C) binding protein family, which includes hnRNPs K/J and the αCPs (αCP−1 to −4) (for review see [35]). HnRNP K appears to have at least four alternative splice variants [36]. αCP-2 and αCP-4, two KH domain RNA-binding proteins, are both alternatively spliced [35]. However, for these examples, isoform-specific functions remain to be determined.
Post-translational modification of RBPs generates additional layers of complexity, as it can modify the RNA-binding, function and localization of the RNP. Three types of modifications have been described for RBPs: phosphorylation, arginine methylation and small ubiquitin-like modification (SUMO). Phosphorylation of αCP-1 and αCP-2 decreases their poly(rC)-binding activity [37]. Growth factors, oxidative stress and other stimuli can alter the phosphorylation status of hnRNP K [38–40]. Methylation of RGG repeats is found in several RBPs, including the hnRNPs (for review see [41]). In S. cerevisiae, two RBPs involved in mRNA processing and export, Hrp1 and Yra1 (Aly/REF in metazoans), have been shown to be methylated by the major type I arginine methyltransferase, Hmt1 [42,43]. It is possible that this methylation plays a role in the formation of Hrp1- and Yra1-containing RNPs. SUMO modification of hnRNP C and hnRNP M results in conformational and/or compositional changes in these RNPs at the nuclear pore and could therefore play a role in the regulation of nucleocytoplasmic transport [44].
Cell and developmental specific expression also serves to alter the stoichiometry of a cell’s RBPs. Changes in the relative amounts of hnRNP A/B proteins have been suggested to regulate alternative splicing, for example that of the 4.1R transcript during mouse erythropoiesis [45]. Specifically, the hnRNP A/B proteins interact with a conserved splicing silencing element (CE16) in exon 16 (E16) of the 4.1R transcript, leading to increased exclusion of E16. In turn, down-regulation of hnRNP A/B proteins during erythropoiesis correlates with E16 inclusion. This illustrates the importance of RBPs as modulators of a process, in this case alternative splicing, in the broader context of cellular differentiation.
3. Diverse functions of RNA-binding proteins
RBPs function in every aspect of RNA biology, from transcription, pre-mRNA splicing and polyadenylation to RNA modification, transport, localization, translation and turnover. The RBPs not only influence each of these processes, but also provide a link between them [46–49]. Proper functioning of these intricate networks is essential for the coordination of complex post-transcriptional events, and their perturbation can lead to disease.
3.1. Alternative splicing
At least 74% of human genes express multiple mRNAs through alternative splicing [50]. RBPs also function in the regulation of this process. For example, the neuronal specific Nova proteins, each containing three KH domains, control the alternative splicing of a subset of pre-messenger RNAs (e.g. gephyrins 1–2, JNK2, flamingo 1, neogenin) by recognizing intronic YCAY elements (Y indicates a pyrimidine, U or C). The majority of Nova target mRNAs encode proteins that function in the synapse thus linking Nova proteins to the regulation of factors involved in maintaining neuronal plasticity. Loss of Nova proteins, as a result of autoimmune paraneoplastic neurologic disorder (PND), manifests itself in neurologic symptoms of excess motor movements (Paraneoplastic Opsoclonus Myoclonus Ataxia, POMA) [51,52]. The TAR DNA binding protein (TDP43), which interacts with (UG)6–12 motifs in single-stranded RNA through its two RBDs [53], is involved in the regulation of splicing of the cystic fibrosis CFTR (cystic fibrosis transmembrane conductance regulator) mRNA, which encodes a Cl− channel [54]. TDP43 binds an extended stretch of UG repeats in a (UG)U-rich polymorphic region upstream of the 3′ splice site in intron 8, which causes exon 9 skipping in the CFTR mRNA, consequently producing nonfunctional chloride channels in patients with cystic fibrosis [53,54]. In the case of CFTR, the repeats in the transcript affect the function of the encoded protein. However, there are a number of diseases associated with repeats where the aberrant RNA mediates the disease by a gain-of-function mechanism. This is the case for myotonic dystrophy (DM). DM type I (DM1) is caused by a CUG triplet-repeat expansion (from 50 to >1500 repeats) in the 3′UTR of the DMPK mRNA [ 55,56]. This mutant mRNA is retained in the nucleus through its interaction with two splicing regulators, muscleblind-like protein 1 (MBNL1) and CUG-binding protein 1 (CUG-BP1) [56,57], causing splicing defects. MBNL1 becomes sequestered on the mislocalized repeat-containing RNAs which results in nuclear depletion and loss of function [56]. CUG-BP1 steady state-levels, on the other hand, are increased in DM1 due to hyperphosphorylation of the protein [58]. The resulting change in the ratio of MBNL1 to CUG-BP1 is correlated with aberrant splicing of their target pre-mRNAs [59].
3.2. RNA modification
RNA editing is the most prevalent type of RNA modification, involving the conversion of adenosine (A) to inosine (I). This post-transcriptional modification changes an RNA’s nucleotide content through the deamination of A to I, in a reaction catalyzed by the ADAR proteins [31]. This processing results in an RNA sequence that is different from that encoded by the genome and extends the diversity of the gene products. While the majority of RNA editing occurs in non-coding regions, a few genes have been identified that are subject to editing in their coding sequences [60]. The pre-mRNA substrate required by an ADAR enzyme is often an imperfect duplex RNA formed by base-pairing between the exon that contains the adenosine to be edited and an intronic non-coding element [61]. A classic example of A-I editing is the glutamate receptor GluR-B mRNA, where a glutamine at the editing site is converted to an arginine. This modification changes the conductance properties of the altered channel [61]. Most of the A-I modifications described to date are limited to transcripts in the nervous system encoding ion channels, G-protein coupled receptors and the glutamate and serotonin receptors [62]. Mutations in the Drosophila ADAR gene result in neuronal dysfunction, whereas a homozygous Adar null mutation in mice results in embryonic lethality [63–65]. In humans, a heterozygous functional-null mutation in the ADAR1 gene is less severe and leads to a skin disease, human pigmentary genodermatosis [66].
3.3. Polyadenylation
Polyadenylation of an mRNA has a strong effect on its nuclear transport, translation efficiency and stability, and all of these, as well as the process of polyadenylation, depend on specific RBPs. All eukaryotic mRNAs, with the exception of replication-dependent histone mRNAs, are processed to receive 3′ poly(A) tails of ~ 200 nucleotides. Polyadenylation is a tightly coupled two-step process in which the transcript is first cleaved between the highly conserved AAUAAA sequence upstream and a degenerate U/GU rich sequence downstream of the cleavage site, after which the poly(A) polymerase adds the poly(A) tail to the cleavage product [67]. One of the necessary protein complexes in the polyadenylation process is CPSF, which consists of at least 4 polypeptides and binds the canonical AAUAAA site, of which CPSF-160 and CPSF-30 appear to be the key RNA-binding subunits [68]. CPSF, together with the nuclear poly(A) binding protein (PABPN1), stimulates the activity of the poly(A) polymerase, which is essentially inactive on its own [69]. For PABN1 to interact with the poly(A) tail it needs both the RBD and the arginine-rich C-terminal domain [70]. Short GCG expansions in the coding region of PABPN1 mRNA have been found to cause oculopharyngeal muscular dystrophy (OPMD) [71]. These triplet-repeats give rise to an expanded polyalanine tract in the protein that likely causes mutated PABPN1 oligomers to accumulate as filament inclusions in the nuclei of skeletal muscle fibers, thus eliciting nuclear toxicity [71]. PABPN1 is post-translationally modified by arginine methylation, and it was recently shown that unmethylated PABPN1 oligomerizes more readily than methylated PABPN1 [72,73]. This suggests that the methylation state of the protein also influences the extent of nuclear aggregation in OPMD.
3.4. mRNA export
Normally, once pre-mRNA processing is complete, the translation-ready mRNA is exported from the nucleus to the cytoplasm. The cell therefore requires a mechanism to ensure that only fully processed mRNPs are exported. That is, transcription, splicing and 3′end processing of the mRNAs must be completed before export can occur. mRNA export is an excellent example of the dynamic network of rearrangements in which RBPs participate. It is a three step process involving the generation of a cargo-carrier complex in the nucleus, followed by translocation of the complex through the nuclear pore complex, and finally, release of the cargo in the cytoplasm with subsequent recycling of the carrier. The TAP/NXF1:p15 heterodimer is a key player in mRNA export. TAP (known as Mex67 in S. cerevisiae) was first shown to bind to the constitutive transport element (CTE), an element required for export of retroviral transcripts, and it was later demonstrated that TAP also has a role in mRNA export [74,75]. Overexpression of TAP in Xenopus oocytes increases the export of transcripts that are otherwise inefficiently exported suggesting a direct role for TAP in mRNA export. As both TAP and p15 show low affinity for RNA, they require adaptor proteins to mediate the interaction [76,77]. The Aly/REF protein which directly interacts with TAP, recruits TAP to mRNA, although the precise mechanistic details of mRNA export remain unclear [78,79].
3.5. mRNA localization
mRNA localization is critical for gene expression by allowing spatially regulated protein production. Localization of transcripts to a specific region of the cell during development has been particularly well studied in S. cerevisiae and D. melanogaster. For example, during cell division in S. cerevisiae, ASH1 mRNA is actively localized to the bud of the daughter cell by its association with myosin (Myo4) and actin [80]. This interaction depends on two other proteins, She2 and She3 [81]. She2 binds as a dimer to localization elements located partly in the coding region and in the 3′UTR of the ASH1 mRNA [82]. Binding to RNA increases the affinity of She2 for the C-terminus of She3, which then binds Myo4 through its N-terminus [81]. The resultant localized expression of the Ash1 protein is necessary for the suppression of mating type switching in the daughter cell by repressing the transcription of the HO endonuclease gene [80]. Another example that highlights how nuclear-acquired factors impact cytoplasmic mRNA metabolism is the localization of β-actin to the lamella region in several asymmetric cell types by the zipcode-binding protein (ZBP1) [83,84]. ZBP1 contains four KH domains and one RBD. It binds to β-actin mRNA at the site of transcription through a 54 nt localization element in the 3′ UTR of β-actin, termed the zipcode, and moves with the mRNA into the cytoplasm. This interaction is essential for proper β-actin mRNA localization in the cytoplasm [83,84].
3.6. Translation
Translational regulation provides a rapid mechanism to control gene expression, and numerous regulatory proteins target the initiation step, often in a way that couples translation to mRNA localization. ZBP1, in addition to its role in the localization of β-actin mRNA, is involved in the translational repression of β-actin mRNA by blocking translation initiation [85]. It is thought that phosphorylation of ZBP1 by the Src tyrosine kinase leads to decreased binding affinity to β-actin mRNA, and ultimately derepression of translation [85]. The dual role of ZBP1 makes it a valid candidate in linking transport and translational repression of β-actin mRNA.
Many species depend on distinct regulatory systems to keep mRNAs translationally silent during different stages of development. In the C. elegans germ line, for example, the KH domain protein GLD-1 represses the translation of pal-1 mRNA by binding to a germline repression element (GRE) in its 3′UTR [86]. The PAL-1 protein initiates a transcription regulatory network in the later blastomere lineages, and therefore needs to be translationally repressed in oocytes and early embryos [87].
3.7. mRNA turnover
Translation is tightly coupled to mRNA turnover and regulated mRNA stability. The ELAV/Hu proteins are involved in the stability and translation of early response gene and AU-rich transcripts predominantly in neurons [88]. HuB, HuC and HuD are neuron-specific ELAV proteins, whereas HuR is ubiquitously expressed [89]. Each contains three RBDs, the first two of which confer binding to AU-rich elements (AREs) [90]. These proteins stabilize many of their AU-rich target mRNAs (e.g. c-fos, GM-CSF, EGF) [88]. HuR appears to be stabilizing its target transcripts by protecting the messages from degradation in the cytoplasm [91]. In addition, HuR colocalizes with polysomes, suggesting that it binds to ARE-containing mRNAs undergoing translation [92]. Patients with paraneoplastic neurological disorder (PND) develop autoantibodies against HuC and HuD in tumors outside of the central nervous system [51,52]. These antibodies, as well as inflammatory cells, are able to cross the blood-brain barrier resulting in PND-associated encephalomyelitis and neuronopathy [88].
3.8. Multi-functional proteins
Many RBPs, for example the abundant hnRNP and serine/arginine-rich (SR) proteins, bind to multiple sites on numerous RNAs to function in diverse processes. The hnRNP A1 protein can bind to exonic splicing silencer sequences and regulate alternative splicing by antagonizing the SR splicing factors [45]. Additionally, hnRNP A1 has been shown to stimulate telomerase activity by associating with telomere ends [93]. Recently, hnRNP A1 was found to bind to human pri-mir18a, the precursor of miR-18a, and to facilitate its Drosha-mediated processing [94]. This is the first time an RBP has been implicated in miRNA maturation.
4. Assembly of RNPs
RNA-protein, and hence the sequence or structure of the RNA target, and protein-protein interactions are critical factors in determining the formation of an RNP. However, often more than one RBP has the capacity to bind to a specific sequence on the target RNA. The complement of RBPs present at a particular locale where the RNA is transcribed or changes in the post-translational modifications of these proteins would affect the resulting RNP complex, modulating its downstream functional activity. The recruitment of additional proteins to the RNP can result in the regulated formation of a highly dynamic complex. Here, we discuss two well-characterized examples of RNP assembly, the exon-junction complex (EJC) and the CPE-binding protein (CPEB) RNP.
The EJC is a large (~335 kDa in vitro) RNP that preferentially binds mRNAs produced by splicing [95–97]. It binds these newly spliced mRNAs approximately 20–24 nucleotides upstream of exon-exon junctions [96,97]. Proteins known to comprise the core EJC include eIF4AIII, Y14, magoh and MLN51/Barentsz [98–100]. Other proteins that associate with the core complex include RNPS1, SRm160, Aly/REF, PYM and Upf3 [95,97,100–107]. Of the core components, the best characterized interaction occurs between Y14 and magoh, two proteins that are found in spliceosomes following the first step of splicing [108,109]. While Y14, which contains an RBD, was initially a candidate for binding mRNA directly, the crystal stucture of human and Drosophila Y14:magoh revealed quite unexpectedly that the RBD is masked through its interaction with magoh and, thus, appears unable to directly contact mRNA [95,110–112]. Rather, it is more likely that eIF4AIII, a spliceosome-associated ATP-dependent DEAD-box RNA helicase, acts as the central factor in the initiation of EJC formation because it binds specifically to mRNA during the late stages of splicing and also binds Y14:magoh, perhaps serving to recruit these proteins to the exon-exon junction [98–100,107,113]. The EJC also enhances the association with the mRNP of the mRNA nuclear export factor TAP/NXF1:p15, and this promotes its transport of the complex through the nuclear pore to the cytoplasm [78,105,106,114,115]. Interestingly, while most EJC proteins dissociate from the mRNA either during or immediately following export, Y14 and magoh remain bound to the mRNA until it is translated, suggesting that they may play an additional role in this process [116]. Consistent with this idea, tethering of Y14, magoh or RNPS1 to mRNA can enhance the translation efficiency [117]. Recently, PYM has been shown to bind the cytoplasmic Y14:magoh complex, in addition to the 48S preinitiation complex and the small (40S) ribosomal subunit [118]. In addition, knockdown of PYM results in a decrease in translation of spliced mRNAs [118]. This data suggests that PYM may deliver spliced mRNAs containing the EJC to the translational apparatus in the cytoplasm to enhance protein production. The EJC can also serve as a marker to indicate mRNAs that have premature stop codons located upstream of the EJC (for review see [119]). Upf3, a protein that functions in nonsense-mediated decay (NMD), is also a component of the EJC [105,120]. It appears that information encoded by mRNAs, such as the presence or absence of premature termination codons, may be marked in the nucleus for subsequent communication to the translation or NMD apparatus in the cytoplasm. The dynamic nature of the association of proteins with mRNA in the EJC RNP over a history of processes – from splicing to export, translation and mRNA degradation – implies that this highly ordered RNP assembly is important for timely and coordinated gene expression.
The CPE-binding protein (CPEB) RNP is a large, dynamic complex that functions in cytoplasmic polyadenylation and translational regulation (for review see [121]). CPEB itself contains two RNA-binding domains, an RBD and a zinc finger, and is highly conserved in both vertebrates and invertebrates [122,123]. In the cytoplasm, CPEB first binds to the cytoplasmic polyadenylation element (CPE; UUUUUAU consensus sequence), located within the 3′ UTR of some mRNAs, and then initiates the assembly of an RNP complex that contains the following proteins: CPSF; PARN, a deadenylating enzyme that contains two RBDs; Gld2, a poly(A) polymerase; and symplekin [124–128]. When the CPE-containing mRNA is translati onally repressed, PARN deadenylation is more active than Gld2 polyadenylation, resulting in shortening of the poly(A) tail [129]. However, in the case of oocyte maturation, phosphorylation of CPEB Ser174 by Aurora A kinase results in the dissociation of PARN from the RNP complex, allowing Gld2 polyadenylation of the CPE-containing mRNA [128,129]. The maskin protein appears to provide a direct link to translation regulation and the CPEB RNP. Maskin interacts with both CPEB and the cap-binding factor eIF4E [130,131]. When the poly(A) tail is short, maskin binds eIF4E, therefore occluding the binding of eIF4G. As a result, the 40S ribosomal subunit cannot be recruited to the mRNA and translation is repressed. However, when the poly(A) tail is elongated, the poly(A)-binding protein (PABP) binds the poly(A) tail and interacts directly with eIF4G to abrogate maskin’s interaction with eIF4E, allowing the mRNA to be translated [130,132]. Subtle changes in the protein composition and mRNA polyadenylation status of this cytoplasmic RNP complex can determine the fate of the mRNA to which it is bound. These changes ultimately dictate whether the poly(A) tail in the CPE-containing mRNA is deadenylated and therefore translationally repressed, or is elongated and consequently subject to translation initiation. The repertoire of RBPs that binds a particular RNA is often highly influential in determining which RNPs form and, ultimately, the functional roles they play.
5. Perspectives
Since the definitive identification of the hnRNP proteins and the discovery of the first consensus motifs in RBPs more than two decades ago, the list of RBPs and the multitude of functions in which they participate has expanded enormously. In recent years, biochemical and genetic experiments as well as bioinformatic analysis of several sequenced genomes revealed a vast array of RBPs about which little is known. It is very likely that the inventory of RBPs is much larger, as it is doubtful that all of the RNA-binding motifs have already been discovered. From what has been learned so far, it is clear that RBPs are critical components of the gene expression pathway in eukaryotes. Their capacity to regulate every aspect of the biogenesis and function of RNAs is remarkable. It is also clear, however, that a great deal of information is lacking about the structure of RBPs, their mode of interaction with RNAs and the specific arrangements of these proteins in the complex RNP assemblies that they form on pre-mRNAs and mRNAs. Given the impressive progress that has already been made, the enormous number of RBPs that remain to be characterized and the rich arsenal of tools available to study them, the promise of what the study of RBPs still has in store for understanding biology and many diseases is tremendously exciting.
Figure 3.
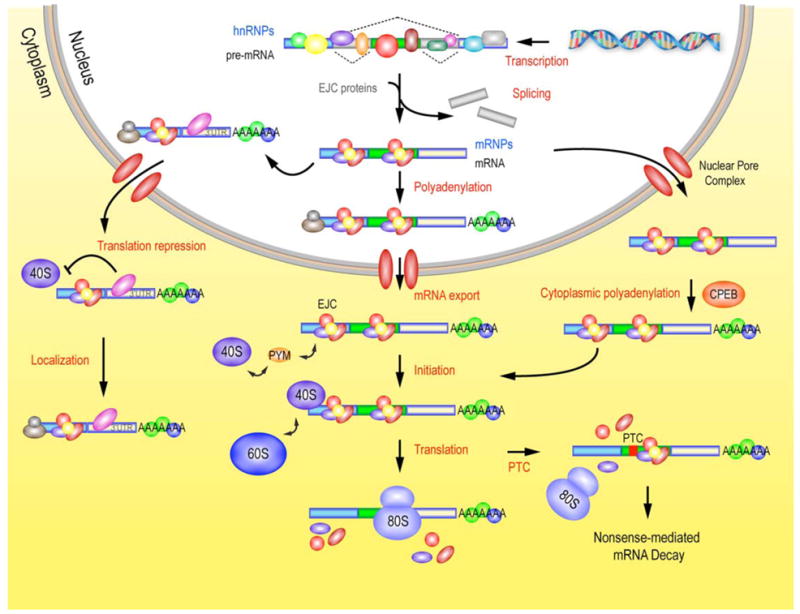
The function of RBPs in the regulation of post-transcriptional gene expression. Once pre-mRNAs are transcribed by RNA polymerase II in the nucleus, they undergo many different processing steps that can determine the fate of the transcript. During splicing, molecular imprinting of the exon-junction complex (EJC) occurs specifically on spliced mRNAs, and this event affects the fate of the mRNPs in the following steps, such as recruitment of ribosomal subunits for translation initiation, or surveillance of mRNA for nonsense-mediated mRNA decay (NMD). RBPs bound, for example, to the 3′UTR of an mRNA can repress the initiation of translation and direct the subcellular localization of the mRNAs. Some mRNAs, upon transport to the cytoplasm, are further modified by the cytoplasmic polyadenylation RNP, CPEB.
Acknowledgments
We thank the members of our laboratory, especially Dan Battle, Mumtaz Kasim, Chi-kong Lau, Lili Wan and Ihab Younis for helpful discussions and comments on this manuscript, and Sharon Kontra for secretarial assistance. G.D. is an investigator of the Howard Hughes Medical Institute.
Abbreviations
- RBP
RNA-binding protein
- RNP
ribonucleoprotein
- hnRNP
heterogeneous ribonucleoprotein
- mRNP
messenger ribonucleoprotein
- EJC
exon-junction complex
- UTR
untranslated region
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol. 2007;8:209–20. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 2.Burd CG, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–21. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 3.Chen Y, Varani G. Protein families and RNA recognition. FEBS J. 2005;272:2088–97. doi: 10.1111/j.1742-4658.2005.04650.x. [DOI] [PubMed] [Google Scholar]
- 4.Lunde BM, Moore C, Varani G. RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol. 2007;8:479–90. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keene JD. Ribonucleoprotein infrastructure regulating the flow of genetic information between the genome and the proteome. Proc Natl Acad Sci USA. 2001;98:7018–24. doi: 10.1073/pnas.111145598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lasko P. The drosophila melanogaster genome: translation factors and RNA binding proteins. J Cell Biol. 2000;150:F51–6. doi: 10.1083/jcb.150.2.f51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee MH, Schedl T. RNA-binding proteins. WormBook. 2006:1–13. doi: 10.1895/wormbook.1.79.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anantharaman V, Koonin EV, Aravind L. Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res. 2002;30:1427–64. doi: 10.1093/nar/30.7.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dreyfuss G, Adam SA, Choi YD. Physical change in cytoplasmic messenger ribonucleoproteins in cells treated with inhibitors of mRNA transcription. Mol Cell Biol. 1984;4:415–23. doi: 10.1128/mcb.4.3.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dreyfuss G, Choi YD, Adam SA. Characterization of heterogeneous nuclear RNA-protein complexes in vivo with monoclonal antibodies. Mol Cell Biol. 1984;4:1104–14. doi: 10.1128/mcb.4.6.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mayrand S, Pederson T. Nuclear ribonucleoprotein particles probed in living cells. Proc Natl Acad Sci USA. 1981;78:2208–12. doi: 10.1073/pnas.78.4.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mayrand S, Setyono B, Greenberg JR, Pederson T. Structure of nuclear ribonucleoprotein: identification of proteins in contact with poly(A)+ heterogeneous nuclear RNA in living HeLa cells. J Cell Biol. 1981;90:380–4. doi: 10.1083/jcb.90.2.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pinol-Roma S, Adam SA, Choi YD, Dreyfuss G. Ultraviolet-induced cross-linking of RNA to proteins in vivo. Methods Enzymol. 1989;180:410–8. doi: 10.1016/0076-6879(89)80114-4. [DOI] [PubMed] [Google Scholar]
- 14.van Eekelen CA, Riemen T, van Venrooij WJ. Specificity in the interaction of hnRNA and mRNA with proteins as revealed by in vivo cross linking. FEBS Lett. 1981;130:223–6. doi: 10.1016/0014-5793(81)81125-8. [DOI] [PubMed] [Google Scholar]
- 15.Wagenmakers AJ, Reinders RJ, van Venrooij WJ. Cross-linking of mRNA to proteins by irradiation of intact cells with ultraviolet light. Eur J Biochem. 1980;112:323–30. doi: 10.1111/j.1432-1033.1980.tb07207.x. [DOI] [PubMed] [Google Scholar]
- 16.Mili S, Steitz JA. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA. 2004;10:1692–4. doi: 10.1261/rna.7151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ule J, Jensen K, Mele A, Darnell RB. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods. 2005;37:376–86. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 18.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–10. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 19.Seay D, Hook B, Evans K, Wickens M. A three-hybrid screen identifies mRNAs controlled by a regulatory protein. RNA. 2006;12:1594–600. doi: 10.1261/rna.145306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.SenGupta DJ, Zhang B, Kraemer B, Pochart P, Fields S, Wickens M. A three-hybrid system to detect RNA-protein interactions in vivo. Proc Natl Acad Sci USA. 1996;93:8496–501. doi: 10.1073/pnas.93.16.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang B, Gallegos M, Puoti A, Durkin E, Fields S, Kimble J, Wickens MP. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature. 1997;390:477–84. doi: 10.1038/37297. [DOI] [PubMed] [Google Scholar]
- 22.Niranjanakumari S, Lasda E, Brazas R, Garcia-Blanco MA. Reversible cross-linking combined with immunoprecipitation to study RNA-protein interactions in vivo. Methods. 2002;26:182–90. doi: 10.1016/S1046-2023(02)00021-X. [DOI] [PubMed] [Google Scholar]
- 23.Gerber AP, Herschlag D, Brown PO. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004;2:E79. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park JW, Parisky K, Celotto AM, Reenan RA, Graveley BR. Identification of alternative splicing regulators by RNA interference in Drosophila. Proc Natl Acad Sci USA. 2004;101:15974–9. doi: 10.1073/pnas.0407004101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–31. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 26.Edwards TA, Pyle SE, Wharton RP, Aggarwal AK. Structure of Pumilio reveals similarity between RNA and peptide binding motifs. Cell. 2001;105:281–9. doi: 10.1016/s0092-8674(01)00318-x. [DOI] [PubMed] [Google Scholar]
- 27.Wang X, McLachlan J, Zamore PD, Hall TM. Modular recognition of RNA by a human pumilio-homology domain. Cell. 2002;110:501–12. doi: 10.1016/s0092-8674(02)00873-5. [DOI] [PubMed] [Google Scholar]
- 28.Wang X, Zamore PD, Hall TM. Crystal structure of a Pumilio homology domain. Mol Cell. 2001;7:855–65. doi: 10.1016/s1097-2765(01)00229-5. [DOI] [PubMed] [Google Scholar]
- 29.Cheong CG, Hall TM. Engineering RNA sequence specificity of Pumilio repeats. Proc Natl Acad Sci USA. 2006;103:13635–9. doi: 10.1073/pnas.0606294103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Opperman L, Hook B, DeFino M, Bernstein DS, Wickens M. A single spacer nucleotide determines the specificities of two mRNA regulatory proteins. Nat Struct Mol Biol. 2005;12:945–51. doi: 10.1038/nsmb1010. [DOI] [PubMed] [Google Scholar]
- 31.Valente L, Nishikura K. ADAR gene family and A-to-I RNA editing: diverse roles in posttranscriptional gene regulation. Prog Nucleic Acid Res Mol Biol. 2005;79:299–338. doi: 10.1016/S0079-6603(04)79006-6. [DOI] [PubMed] [Google Scholar]
- 32.Garcia MA, Meurs EF, Esteban M. The dsRNA protein kinase PKR: virus and cell control. Biochimie. 2007;89:799–811. doi: 10.1016/j.biochi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 33.Clemens MJ. PKR--a protein kinase regulated by double-stranded RNA. Int J Biochem Cell Biol. 1997;29:945–9. doi: 10.1016/s1357-2725(96)00169-0. [DOI] [PubMed] [Google Scholar]
- 34.Hamilton BJ, Genin A, Cron RQ, Rigby WF. Delineation of a novel pathway that regulates CD154 (CD40 ligand) expression. Mol Cell Biol. 2003;23:510–25. doi: 10.1128/MCB.23.2.510-525.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Makeyev AV, Liebhaber SA. The poly(C)-binding proteins: a multiplicity of functions and a search for mechanisms. RNA. 2002;8:265–78. doi: 10.1017/s1355838202024627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dejgaard K, Leffers H, Rasmussen HH, Madsen P, Kruse TA, Gesser B, Nielsen H, Celis JE. Identification, molecular cloning, expression and chromosome mapping of a family of transformation upregulated hnRNP-K proteins derived by alternative splicing. J Mol Biol. 1994;236:33–48. doi: 10.1006/jmbi.1994.1116. [DOI] [PubMed] [Google Scholar]
- 37.Leffers H, Dejgaard K, Celis JE. Characterisation of two major cellular poly(rC)-binding human proteins, each containing three K-homologous (KH) domains. Eur J Biochem. 1995;230:447–53. [PubMed] [Google Scholar]
- 38.Ostrowski J, Sims JE, Sibley CH, Valentine MA, Dower SK, Meier KE, Bomsztyk K. A serine/threonine kinase activity is closely associated with a 65-kDa phosphoprotein specifically recognized by the kappa B enhancer element. J Biol Chem. 1991;266:12722–33. [PubMed] [Google Scholar]
- 39.Schullery DS, Ostrowski J, Denisenko ON, Stempka L, Shnyreva M, Suzuki H, Gschwendt M, Bomsztyk K. Regulated interaction of protein kinase Cdelta with the heterogeneous nuclear ribonucleoprotein K protein. J Biol Chem. 1999;274:15101–9. doi: 10.1074/jbc.274.21.15101. [DOI] [PubMed] [Google Scholar]
- 40.Van Seuningen I, Ostrowski J, Bustelo XR, Sleath PR, Bomsztyk K. The K protein domain that recruits the interleukin 1-responsive K protein kinase lies adjacent to a cluster of c-Src and Vav SH3-binding sites. Implications that K protein acts as a docking platform. J Biol Chem. 1995;270:26976–85. doi: 10.1074/jbc.270.45.26976. [DOI] [PubMed] [Google Scholar]
- 41.Godin KS, Varani G. How arginine-rich domains coordinate mRNA maturation events. RNA Biol. 2007;4:69–75. doi: 10.4161/rna.4.2.4869. [DOI] [PubMed] [Google Scholar]
- 42.Shen EC, Henry MF, Weiss VH, Valentini SR, Silver PA, Lee MS. Arginine methylation facilitates the nuclear export of hnRNP proteins. Genes Dev. 1998;12:679–91. doi: 10.1101/gad.12.5.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu MC, Bachand F, McBride AE, Komili S, Casolari JM, Silver PA. Arginine methyltransferase affects interactions and recruitment of mRNA processing and export factors. Genes Dev. 2004;18:2024–35. doi: 10.1101/gad.1223204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vassileva MT, Matunis MJ. SUMO modification of heterogeneous nuclear ribonucleoproteins. Mol Cell Biol. 2004;24:3623–32. doi: 10.1128/MCB.24.9.3623-3632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hou VC, et al. Decrease in hnRNP A/B expression during erythropoiesis mediates a pre-mRNA splicing switch. EMBO J. 2002;21:6195–204. doi: 10.1093/emboj/cdf625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hilleren P, McCarthy T, Rosbash M, Parker R, Jensen TH. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature. 2001;413:538–42. doi: 10.1038/35097110. [DOI] [PubMed] [Google Scholar]
- 47.Kyburz A, Friedlein A, Langen H, Keller W. Direct interactions between subunits of CPSF and the U2 snRNP contribute to the coupling of pre-mRNA 3′ end processing and splicing. Mol Cell. 2006;23:195–205. doi: 10.1016/j.molcel.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 48.Millevoi S, Loulergue C, Dettwiler S, Karaa SZ, Keller W, Antoniou M, Vagner S. An interaction between U2AF 65 and CF I(m) links the splicing and 3′ end processing machineries. EMBO J. 2006;25:4854–64. doi: 10.1038/sj.emboj.7601331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rigo F, Martinson HG. Functional coupling of last-intron splicing and 3′-end processing to transcription in vitro: the poly(A) signal couples to splicing before committing to cleavage. Mol Cell Biol. 2008;28:849–62. doi: 10.1128/MCB.01410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson JM, et al. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–4. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 51.Buckanovich RJ, Posner JB, Darnell RB. Nova, the paraneoplastic Ri antigen, is homologous to an RNA-binding protein and is specifically expressed in the developing motor system. Neuron. 1993;11:657–72. doi: 10.1016/0896-6273(93)90077-5. [DOI] [PubMed] [Google Scholar]
- 52.Szabo A, Dalmau J, Manley G, Rosenfeld M, Wong E, Henson J, Posner JB, Furneaux HM. HuD, a paraneoplastic encephalomyelitis antigen, contains RNA-binding domains and is homologous to Elav and Sex-lethal. Cell. 1991;67:325–33. doi: 10.1016/0092-8674(91)90184-z. [DOI] [PubMed] [Google Scholar]
- 53.Ayala YM, Pantano S, D’Ambrogio A, Buratti E, Brindisi A, Marchetti C, Romano M, Baralle FE. Human, Drosophila, and C.elegans TDP43: nucleic acid binding properties and splicing regulatory function. J Mol Biol. 2005;348:575–88. doi: 10.1016/j.jmb.2005.02.038. [DOI] [PubMed] [Google Scholar]
- 54.Anderson MP, Gregory RJ, Thompson S, Souza DW, Paul S, Mulligan RC, Smith AE, Welsh MJ. Demonstration that CFTR is a chloride channel by alteration of its anion selectivity. Science. 1991;253:202–5. doi: 10.1126/science.1712984. [DOI] [PubMed] [Google Scholar]
- 55.Davis BM, McCurrach ME, Taneja KL, Singer RH, Housman DE. Expansion of a CUG trinucleotide repeat in the 3′ untranslated region of myotonic dystrophy protein kinase transcripts results in nuclear retention of transcripts. Proc Natl Acad Sci USA. 1997;94:7388–93. doi: 10.1073/pnas.94.14.7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miller JW, Urbinati CR, Teng-Umnuay P, Stenberg MG, Byrne BJ, Thornton CA, Swanson MS. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–48. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Timchenko LT, et al. Identification of a (CUG)n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Res. 1996;24:4407–14. doi: 10.1093/nar/24.22.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kuyumcu-Martinez NM, Wang GS, Cooper TA. Increased steady-state levels of CUGBP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol Cell. 2007;28:68–78. doi: 10.1016/j.molcel.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ranum LP, Cooper TA. RNA-mediated neuromuscular disorders. Annu Rev Neurosci. 2006;29:259–77. doi: 10.1146/annurev.neuro.29.051605.113014. [DOI] [PubMed] [Google Scholar]
- 60.Nishikura K. Editor meets silencer: crosstalk between RNA editing and RNA interference. Nat Rev Mol Cell Biol. 2006;7:919–31. doi: 10.1038/nrm2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Higuchi M, Single FN, Kohler M, Sommer B, Sprengel R, Seeburg PH. RNA editing of AMPA receptor subunit GluR-B: a base-paired intron-exon structure determines position and efficiency. Cell. 1993;75:1361–70. doi: 10.1016/0092-8674(93)90622-w. [DOI] [PubMed] [Google Scholar]
- 62.Hoopengardner B, Bhalla T, Staber C, Reenan R. Nervous system targets of RNA editing identified by comparative genomics. Science. 2003;301:832–6. doi: 10.1126/science.1086763. [DOI] [PubMed] [Google Scholar]
- 63.Hartner JC, Schmittwolf C, Kispert A, Muller AM, Higuchi M, Seeburg PH. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J Biol Chem. 2004;279:4894–902. doi: 10.1074/jbc.M311347200. [DOI] [PubMed] [Google Scholar]
- 64.Palladino MJ, Keegan LP, O’Connell MA, Reenan RA. A-to-I pre-mRNA editing in Drosophila is primarily involved in adult nervous system function and integrity. Cell. 2000;102:437–49. doi: 10.1016/s0092-8674(00)00049-0. [DOI] [PubMed] [Google Scholar]
- 65.Wang Q, Miyakoda M, Yang W, Khillan J, Stachura DL, Weiss MJ, Nishikura K. Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J Biol Chem. 2004;279:4952–61. doi: 10.1074/jbc.M310162200. [DOI] [PubMed] [Google Scholar]
- 66.Miyamura Y, Suzuki T, Kono M, Inagaki K, Ito S, Suzuki N, Tomita Y. Mutations of the RNA-specific adenosine deaminase gene (DSRAD) are involved in dyschromatosis symmetrica hereditaria. Am J Hum Genet. 2003;73:693–9. doi: 10.1086/378209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Minvielle-Sebastia L, Keller W. mRNA polyadenylation and its coupling to other RNA processing reactions and to transcription. Curr Opin Cell Biol. 1999;11:352–7. doi: 10.1016/S0955-0674(99)80049-0. [DOI] [PubMed] [Google Scholar]
- 68.Ryan K, Calvo O, Manley JL. Evidence that polyadenylation factor CPSF-73 is the mRNA 3′ processing endonuclease. RNA. 2004;10:565–73. doi: 10.1261/rna.5214404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bienroth S, Wahle E, Suter-Crazzolara C, Keller W. Purification of the cleavage and polyadenylation factor involved in the 3′-processing of messenger RNA precursors. J Biol Chem. 1991;266:19768–76. [PubMed] [Google Scholar]
- 70.Wahle E, Ruegsegger U. 3′-End processing of pre-mRNA in eukaryotes. FEMS Microbiol Rev. 1999;23:277–95. doi: 10.1111/j.1574-6976.1999.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 71.Brais B, et al. Short GCG expansions in the PABP2 gene cause oculopharyngeal muscular dystrophy. Nat Genet. 1998;18:164–7. doi: 10.1038/ng0298-164. [DOI] [PubMed] [Google Scholar]
- 72.Perreault A, Lemieux C, Bachand F. Regulation of the nuclear poly(A)-binding protein by arginine methylation in fission yeast. J Biol Chem. 2007;282:7552–62. doi: 10.1074/jbc.M610512200. [DOI] [PubMed] [Google Scholar]
- 73.Smith JJ, Rucknagel KP, Schierhorn A, Tang J, Nemeth A, Linder M, Herschman HR, Wahle E. Unusual sites of arginine methylation in Poly(A)-binding protein II and in vitro methylation by protein arginine methyltransferases PRMT1 and PRMT3. J Biol Chem. 1999;274:13229–34. doi: 10.1074/jbc.274.19.13229. [DOI] [PubMed] [Google Scholar]
- 74.Ernst RK, Bray M, Rekosh D, Hammarskjold ML. A structured retroviral RNA element that mediates nucleocytoplasmic export of intron-containing RNA. Mol Cell Biol. 1997;17:135–44. doi: 10.1128/mcb.17.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gruter P, et al. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol Cell. 1998;1:649–59. doi: 10.1016/s1097-2765(00)80065-9. [DOI] [PubMed] [Google Scholar]
- 76.Katahira J, Strasser K, Podtelejnikov A, Mann M, Jung JU, Hurt E. The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J. 1999;18:2593–609. doi: 10.1093/emboj/18.9.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Santos-Rosa H, Moreno H, Simos G, Segref A, Fahrenkrog B, Pante N, Hurt E. Nuclear mRNA export requires complex formation between Mex67p and Mtr2p at the nuclear pores. Mol Cell Biol. 1998;18:6826–38. doi: 10.1128/mcb.18.11.6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stutz F, Bachi A, Doerks T, Braun IC, Seraphin B, Wilm M, Bork P, Izaurralde E. REF, an evolutionary conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA. 2000;6:638–50. doi: 10.1017/s1355838200000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Taniguchi I, Ohno M. ATP-dependent recruitment of export factor Aly/REF onto intronless mRNAs by RNA helicase UAP56. Mol Cell Biol. 2008;28:601–8. doi: 10.1128/MCB.01341-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bobola N, Jansen RP, Shin TH, Nasmyth K. Asymmetric accumulation of Ash1p in postanaphase nuclei depends on a myosin and restricts yeast mating-type switching to mother cells. Cell. 1996;84:699–709. doi: 10.1016/s0092-8674(00)81048-x. [DOI] [PubMed] [Google Scholar]
- 81.Bohl F, Kruse C, Frank A, Ferring D, Jansen RP. She2p, a novel RNA-binding protein tethers ASH1 mRNA to the Myo4p myosin motor via She3p. EMBO J. 2000;19:5514–24. doi: 10.1093/emboj/19.20.5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gonsalvez GB, Urbinati CR, Long RM. RNA localization in yeast: moving towards a mechanism. Biol Cell. 2005;97:75–86. doi: 10.1042/BC20040066. [DOI] [PubMed] [Google Scholar]
- 83.Oleynikov Y, Singer RH. Real-time visualization of ZBP1 association with beta-actin mRNA during transcription and localization. Curr Biol. 2003;13:199–207. doi: 10.1016/s0960-9822(03)00044-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ross AF, Oleynikov Y, Kislauskis EH, Taneja KL, Singer RH. Characterization of a beta-actin mRNA zipcode-binding protein. Mol Cell Biol. 1997;17:2158–65. doi: 10.1128/mcb.17.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huttelmaier S, et al. Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature. 2005;438:512–5. doi: 10.1038/nature04115. [DOI] [PubMed] [Google Scholar]
- 86.Mootz D, Ho DM, Hunter CP. The STAR/Maxi-KH domain protein GLD-1 mediates a developmental switch in the translational control of C. elegans PAL-1. Development. 2004;131:3263–72. doi: 10.1242/dev.01196. [DOI] [PubMed] [Google Scholar]
- 87.Evans TC, Hunter CP. Translational control of maternal RNAs. WormBook. 2005:1–11. doi: 10.1895/wormbook.1.34.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Keene JD. Why is Hu where? Shuttling of early-response-gene messenger RNA subsets. Proc Natl Acad Sci U S A. 1999;96:5–7. doi: 10.1073/pnas.96.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Brennan CM, Steitz JA. HuR and mRNA stability. Cell Mol Life Sci. 2001;58:266–77. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Inoue M, Muto Y, Sakamoto H, Yokoyama S. NMR studies on functional structures of the AU-rich element-binding domains of Hu antigen C. Nucleic Acids Res. 2000;28:1743–50. doi: 10.1093/nar/28.8.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Peng SS, Chen CY, Xu N, Shyu AB. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 1998;17:3461–70. doi: 10.1093/emboj/17.12.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gallouzi IE, Brennan CM, Stenberg MG, Swanson MS, Eversole A, Maizels N, Steitz JA. HuR binding to cytoplasmic mRNA is perturbed by heat shock. Proc Natl Acad Sci U S A. 2000;97:3073–8. doi: 10.1073/pnas.97.7.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang QS, Manche L, Xu RM, Krainer AR. hnRNP A1 associates with telomere ends and stimulates telomerase activity. RNA. 2006;12:1116–28. doi: 10.1261/rna.58806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Guil S, Caceres JF. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat Struct Mol Biol. 2007;14:591–6. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- 95.Kataoka N, Yong J, Kim VN, Velazquez F, Perkinson RA, Wang F, Dreyfuss G. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol Cell. 2000;6:673–82. doi: 10.1016/s1097-2765(00)00065-4. [DOI] [PubMed] [Google Scholar]
- 96.Kim VN, Yong J, Kataoka N, Abel L, Diem MD, Dreyfuss G. The Y14 protein communicates to the cytoplasm the position of exon-exon junctions. EMBO J. 2001;20:2062–8. doi: 10.1093/emboj/20.8.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000;19:6860–9. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Andersen CB, et al. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313:1968–72. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- 99.Bono F, Ebert J, Lorentzen E, Conti E. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell. 2006;126:713–25. doi: 10.1016/j.cell.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 100.Shibuya T, Tange TO, Sonenberg N, Moore MJ. eIF4AIII binds spliced mRNA in the exon junction complex and is essential for nonsense-mediated decay. Nat Struct Mol Biol. 2004;11:346–51. doi: 10.1038/nsmb750. [DOI] [PubMed] [Google Scholar]
- 101.Chan CC, Dostie J, Diem MD, Feng W, Mann M, Rappsilber J, Dreyfuss G. eIF4A3 is a novel component of the exon junction complex. RNA. 2004;10:200–9. doi: 10.1261/rna.5230104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Degot S, et al. Association of the breast cancer protein MLN51 with the exon junction complex via its speckle localizer and RNA binding module. J Biol Chem. 2004;279:33702–15. doi: 10.1074/jbc.M402754200. [DOI] [PubMed] [Google Scholar]
- 103.Ferraiuolo MA, Lee CS, Ler LW, Hsu JL, Costa-Mattioli M, Luo MJ, Reed R, Sonenberg N. A nuclear translation-like factor eIF4AIII is recruited to the mRNA during splicing and functions in nonsense-mediated decay. Proc Natl Acad Sci U S A. 2004;101:4118–23. doi: 10.1073/pnas.0400933101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kataoka N, Diem MD, Kim VN, Yong J, Dreyfuss G. Magoh, a human homolog of Drosophila mago nashi protein, is a component of the splicing-dependent exon-exon junction complex. EMBO J. 2001;20:6424–33. doi: 10.1093/emboj/20.22.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kim VN, Kataoka N, Dreyfuss G. Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon-exon junction complex. Science. 2001;293:1832–6. doi: 10.1126/science.1062829. [DOI] [PubMed] [Google Scholar]
- 106.Le Hir H, Gatfield D, Braun IC, Forler D, Izaurralde E. The protein Mago provides a link between splicing and mRNA localization. EMBO Rep. 2001;2:1119–24. doi: 10.1093/embo-reports/kve245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Palacios IM, Gatfield D, St Johnston D, Izaurralde E. An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature. 2004;427:753–7. doi: 10.1038/nature02351. [DOI] [PubMed] [Google Scholar]
- 108.Jurica MS, Licklider LJ, Gygi SR, Grigorieff N, Moore MJ. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA. 2002;8:426–39. doi: 10.1017/s1355838202021088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Reichert VL, Le Hir H, Jurica MS, Moore MJ. 5′ exon interactions within the human spliceosome establish a framework for exon junction complex structure and assembly. Genes Dev. 2002;16:2778–91. doi: 10.1101/gad.1030602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fribourg S, Gatfield D, Izaurralde E, Conti E. A novel mode of RBD-protein recognition in the Y14-Mago complex. Nat Struct Biol. 2003;10:433–9. doi: 10.1038/nsb926. [DOI] [PubMed] [Google Scholar]
- 111.Lau CK, Diem MD, Dreyfuss G, Van Duyne GD. Structure of the Y14-Magoh core of the exon junction complex. Curr Biol. 2003;13:933–41. doi: 10.1016/s0960-9822(03)00328-2. [DOI] [PubMed] [Google Scholar]
- 112.Shi H, Xu RM. Crystal structure of the Drosophila Mago nashi-Y14 complex. Genes Dev. 2003;17:971–6. doi: 10.1101/gad.260403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ballut L, Marchadier B, Baguet A, Tomasetto C, Seraphin B, Le Hir H. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat Struct Mol Biol. 2005;12:861–9. doi: 10.1038/nsmb990. [DOI] [PubMed] [Google Scholar]
- 114.Rodrigues JP, Rode M, Gatfield D, Blencowe BJ, Carmo-Fonseca M, Izaurralde E. REF proteins mediate the export of spliced and unspliced mRNAs from the nucleus. Proc Natl Acad Sci U S A. 2001;98:1030–5. doi: 10.1073/pnas.031586198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhou Z, Luo MJ, Straesser K, Katahira J, Hurt E, Reed R. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature. 2000;407:401–5. doi: 10.1038/35030160. [DOI] [PubMed] [Google Scholar]
- 116.Dostie J, Dreyfuss G. Translation is required to remove Y14 from mRNAs in the cytoplasm. Curr Biol. 2002;12:1060–7. doi: 10.1016/s0960-9822(02)00902-8. [DOI] [PubMed] [Google Scholar]
- 117.Nott A, Le Hir H, Moore MJ. Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Genes Dev. 2004;18:210–22. doi: 10.1101/gad.1163204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Diem MD, Chan CC, Younis I, Dreyfuss G. PYM binds the cytoplasmic exon-junction complex and ribosomes to enhance translation of spliced mRNAs. Nat Struct Mol Biol. 2007;14:1173–9. doi: 10.1038/nsmb1321. [DOI] [PubMed] [Google Scholar]
- 119.Lejeune F, Maquat LE. Mechanistic links between nonsense-mediated mRNA decay and pre-mRNA splicing in mammalian cells. Curr Opin Cell Biol. 2005;17:309–15. doi: 10.1016/j.ceb.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 120.Gehring NH, Neu-Yilik G, Schell T, Hentze MW, Kulozik AE. Y14 and hUpf3b form an NMD-activating complex. Mol Cell. 2003;11:939–49. doi: 10.1016/s1097-2765(03)00142-4. [DOI] [PubMed] [Google Scholar]
- 121.Richter JD. CPEB: a life in translation. Trends Biochem Sci. 2007;32:279–85. doi: 10.1016/j.tibs.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 122.Hake LE, Mendez R, Richter JD. Specificity of RNA binding by CPEB: requirement for RNA recognition motifs and a novel zinc finger. Mol Cell Biol. 1998;18:685–93. doi: 10.1128/mcb.18.2.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hake LE, Richter JD. CPEB is a specificity factor that mediates cytoplasmic polyadenylation during Xenopus oocyte maturation. Cell. 1994;79:617–27. doi: 10.1016/0092-8674(94)90547-9. [DOI] [PubMed] [Google Scholar]
- 124.Barnard DC, Ryan K, Manley JL, Richter JD. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell. 2004;119:641–51. doi: 10.1016/j.cell.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 125.Mandel CR, Kaneko S, Zhang H, Gebauer D, Vethantham V, Manley JL, Tong L. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature. 2006;444:953–6. doi: 10.1038/nature05363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rouhana L, et al. Vertebrate GLD2 poly(A) polymerases in the germline and the brain. RNA. 2005;11:1117–30. doi: 10.1261/rna.2630205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rouhana L, Wickens M. Autoregulation of GLD-2 cytoplasmic poly(A) polymerase. RNA. 2007;13:188–99. doi: 10.1261/rna.333507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sarkissian M, Mendez R, Richter JD. Progesterone and insulin stimulation of CPEB-dependent polyadenylation is regulated by Aurora A and glycogen synthase kinase-3. Genes Dev. 2004;18:48–61. doi: 10.1101/gad.1136004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kim JH, Richter JD. Opposing polymerase-deadenylase activities regulate cytoplasmic polyadenylation. Mol Cell. 2006;24:173–83. doi: 10.1016/j.molcel.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 130.Cao Q, Richter JD. Dissolution of the maskin-eIF4E complex by cytoplasmic polyadenylation and poly(A)-binding protein controls cyclin B1 mRNA translation and oocyte maturation. EMBO J. 2002;21:3852–62. doi: 10.1093/emboj/cdf353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Stebbins-Boaz B, Cao Q, de Moor CH, Mendez R, Richter JD. Maskin is a CPEB-associated factor that transiently interacts with elF-4E. Mol Cell. 1999;4:1017–27. doi: 10.1016/s1097-2765(00)80230-0. [DOI] [PubMed] [Google Scholar]
- 132.Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–80. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]


