Abstract
Genetically encoded calcium indicators (GECIs) can be used to image activity in defined neuronal populations. However, current GECIs produce inferior signals compared to synthetic indicators and recording electrodes, precluding detection of low firing rates. We developed a single-wavelength GECI based on GCaMP2 (GCaMP3), with increased baseline fluorescence (3x), dynamic range (3x), and higher affinity for calcium (1.3x). GCaMP3 fluorescence changes triggered by single action potentials were detected in pyramidal cell dendrites, with signal-to-noise ratio and photostability significantly better than GCaMP2, D3cpVenus, and TN-XXL. In Caenorhabditis elegans chemosensory neurons and the Drosophila melanogaster antennal lobe, sensory stimulation-evoked fluorescence responses were significantly enhanced with the new indicator (4–6x). In somatosensory and motor cortical neurons in the intact mouse, GCaMP3 detected calcium transients with amplitudes linearly dependent on action potential number. Long-term imaging in the motor cortex of behaving mice revealed large fluorescence changes in imaged neurons over months.
INTRODUCTION
Calcium is a universal second messenger regulating essential cellular signaling events in a broad range of tissues and organisms. In neurons, action potentials (APs) trigger large and rapid changes in cytoplasmic free calcium. Similarly, activation of synaptic glutamate receptors during synaptic transmission produces [Ca2+] transients in dendritic spines. Calcium imaging using synthetic calcium indicators has been used to measure neuronal spiking and synaptic input across populations of neurons in vitro 1 and in vivo 2,3. However, synthetic indicators are difficult to target to specific cell types or sub-cellular locations (but see 4). The loading procedures are invasive and damaging to neural tissue, precluding repeated, chronic in vivo measurements.
Genetically encoded calcium indicators (GECIs) (also called fluorescent calcium indicator proteins; FCIPs) provide an alternative to synthetic indicators. GECIs can be easily targeted to specific cell types or sub-cellular compartments (for review see 5). They are compatible with long-term, repeated in vivo measurements 6. GECIs consist of a calcium-binding domain such as calmodulin or troponin C, fused to either one or two fluorescent proteins (FPs) (for review see 7,8). In single-FP GECIs, the fluorescence intensity of a circularly permuted FP (cpFP) is modulated by calcium binding-dependent changes in the chromophore environment 9,10. In two-FP GECIs, calcium binding modulates fluorescence resonance energy transfer (FRET) between FPs 11–13.
GECIs have been iteratively improved, and are becoming useful for quantitative imaging of neural activity in vivo. The calmodulin-based FRET indicator D3cpVenus (D3cpV) 13 has recently been reported to detect single APs in pyramidal neurons in organotypic mouse brain slices and in vivo 14. The troponin C-based indicator TN-XXL has been used for chronic in vivo activity imaging in the mouse brain 6. Among single-FP based GECIs, the GCaMP family has found the broadest use across multiple model organisms 15–17. However, the properties of all available GECIs are still inferior to synthetic indicators in terms of signal-to-noise ratio (SNR), response linearity, photostability, and properly tuned calcium affinity. The GCaMP indicators further suffer from poor protein stability. Improvements in each of these parameters would facilitate imaging of neural activity.
Recently, the Ca2+-bound and -free structures of GCaMP2 have been solved 18,19, forming the basis for rational improvement of indicator properties. Using a combination of protein structure-guided mutagenesis and semi-rational library screening (for review of GECI design, see 20), we developed improved GCaMP variants. The best mutant, GCaMP3, is brighter, possesses greater protein stability, and has a larger dynamic range and higher affinity for calcium compared to GCaMP2. GCaMP3 is more photostable than the FRET indicators D3cpV and TN-XXL and displays significantly greater sensitivity and faster kinetics, especially at higher levels of activity. GCaMP3 showed improved sensitivity in mammalian cell culture, pyramidal neurons in brain slices, and worms, flies, and mice in vivo.
RESULTS
Structure-guided engineering of GCaMP3
In HEK293 cells the fluorescence of GCaMP2 is one hundred fold lower than EGFP (Supplementary Fig. 1a). Addition of a proteasome inhibitor (10 μM lactacystin) increased the baseline fluorescence of HEK293 cells expressing GCaMP2 (Supplementary Fig. 1b). We reasoned that an N-terminal arginine, found immediately after the initiator methionine of GCaMP2, might destabilize the protein 21. Indeed, HEK293 cells transfected with a mutant lacking the arginine, named GCaMP2.1, showed 40% higher baseline fluorescence than those transfected with GCaMP2 (Supplementary Fig. 1b).
We then created small libraries of GCaMP2.1 variants via site-directed mutagenesis at many sites, both near the EGFP chromophore and at “superfolder GFP” positions 22 (Supplementary Fig. 2). Although screening of GCaMP mutants in bacterial lysate achieved high throughput, we found that the baseline fluorescence and dynamic range correlated only weakly with more intact preparations (Supplementary Fig. 3). Therefore, we designed a medium-throughput mammalian cell-based assay in HEK293 cells. Calcium transients were induced by activating endogenous muscarinic receptors with acetylcholine (Supplementary Fig. 3d). Acetylcholine titrations of GCaMP-transfected HEK293 cells revealed two point mutants with increased dynamic range and baseline fluorescence (T116V; GFP T203V and M66K; GFP M153K). One single (T116V) mutant and a double mutant (T116V and M66K) were named GCaMP2.2, GCaMP2.3, respectively (Fig. 1a, Supplementary Fig. 4 and Supplementary Table 1).
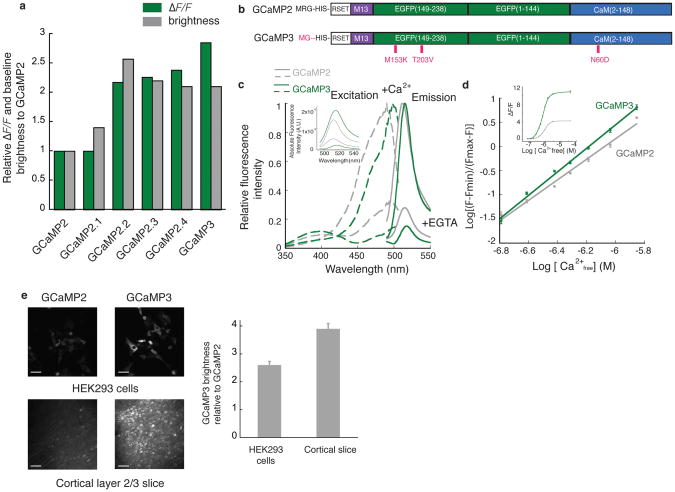
Figure 1. In vitro characterization of GCaMP3.
(a) Screening resulted in several mutants with improved baseline brightness and signal change in HEK293 cells. (b) Schematic representation of GCaMP2 and GCaMP3. Mutated residues are highlighted in red. (c) Fluorescence spectra of GCaMP3 and GCaMP2 (1 μM protein) with 1 mM Ca2+ or 10 mM EGTA in MOPS buffer (30 mM MOPS, 100 mM KCl, pH 7.5) (average of three independent measurements). The fluorescence intensity of each indicator was normalized to the peak of the calcium-saturated spectrum. The inset shows the un-normalized fluorescence emission spectra (485 nm excitation). (d) Ca2+ titration curve (1 μM protein) in MOPS buffer. Inset shows the dynamic range of the two indicators. (e) The improved baseline fluorescence of GCaMP3 compared to GCaMP2. Both indicators were either transfected into HEK293 cells or virally delivered to layer 2/3 cortical neurons. Images were taken either 48 hours post-transfection or 12 days post-viral injection, then analyzed with Volocity 5.0 (Improvision). 50 μm scale bar. Error bars indicate standard deviation of the mean.
To increase GCaMP’s affinity for calcium to allow better detection of the small and rapid calcium increases associated with individual APs, we analyzed mutations in the EF-hands and the interface between the M13 peptide and calmodulin (CaM) (Supplementary Fig. 2). The amino acid substitution N363D (CaM N60D) to both GCaMP2.2 and GCaMP2.3 increased the fluorescence change for small calcium transients, with little effect on baseline fluorescence (Fig. 1a and Supplementary Fig. 4). GCaMP2.2-N363D and GCaMP2.3-N363D were named GCaMP2.4 and GCaMP3, respectively (Fig. 1a,b and Supplementary Table 1). GCaMP3 showed the largest signal change in the acetylcholine assay (Fig. 1a,b and Supplementary Fig. 4) and was further characterized.
The fluorescence spectra of purified GCaMP3 are similar to those of GCaMP2, with a slight red-shifting of the excitation maximum (Fig. 1c). GCaMP3 protein assayed in 3-(N-morpolino) propanesulfonic acid (MOPS) buffer had a dynamic range (Fmax/Fmin) of ~12, 3-fold larger than GCaMP2 (Fig. 1d-inset). This results from a 2-fold decrease of calcium-free fluorescence and a 1.5-fold increase of calcium-saturated fluorescence (Fig. 1c-inset). The affinity of GCaMP3 for Ca2+ was ~1.3-fold higher than GCaMP2 (660 ±19 nM versus 840 ± 25 nM, (p = 0.0017, paired t-test)) (Fig. 1d).
In HEK293 cells GCaMP3 showed ~2.6-fold higher baseline fluorescence than GCaMP2 (Fig. 1e-top). When expressed via viral gene transduction in cortical layer 2/3 neurons, baseline fluorescence was ~3.9-fold higher than GCaMP2 (Fig. 1e-bottom). Given the lower fluorescence of purified GCaMP3 in the apo state, the increase in baseline fluorescence is likely caused by increased protein expression and stability at 37°C.
Characterization of GCaMP3 in brain slice
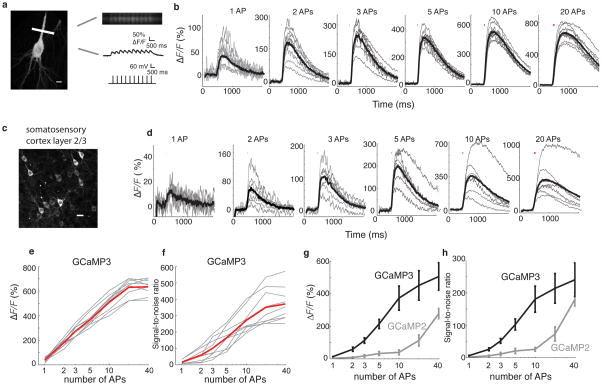
We measured the AP-triggered fluorescence responses of GCaMP3 in pyramidal neurons in cultured brain slices (Fig. 2a,b) 23,24 and acute neocortical brain slices at room temperature (Fig. 2c,d). In cultured slice, GCaMP3 was delivered by biolistic transfection. Increases in GCaMP3 fluorescence intensity (ΔF/F=46± 4.2%, n = 9 cells) at the base of the apical dendrite were detected reliably in response to single APs in all cells (100% single-trial detection; Supplementary Fig. 5a). The average ΔF/F of GCaMP3 (n = 9 cells) was 185± 13%, 250± 27%, 320 ± 35%, 480 ± 50%, 600 ± 100%, and 620 ± 130% for 2, 3, 5, 10, 20 and 40 AP, respectively (Fig. 2b,e). The signal-to-noise ratio (SNR; see Methods) of GCaMP3 was 16.3 ± 10.9, 167.1 ± 65.1 and 371.4 ± 102.8 for 1 AP, 5 AP and 40 AP, respectively (Fig. 2f). The fluorescence increase and single AP detection efficiency are significantly improved over GCaMP2 (1 AP ΔF/F = 17±10%; 38% single-trial detection) 24. The kinetics of GCaMP3 in cultured hippocampal slice are similar to those of GCaMP2 (GCaMP3: rise T1/2 = 83 ± 2 ms; decay T1/2= 610 ± 32 ms; GCaMP2: rise T1/2=95 ± 15 ms; decay T1/2= 480 ± 130 ms24; all measurements for 10 AP stimulus). The improved properties of GCaMP3 allow imaging spontaneous population activities in cultured hippocampal slice, as opposed to GCaMP2 (Supplementary Movies 1, 2 and Supplementary Fig. 6).
Figure 2. Action potential-evoked response of GCaMP3 in hippocampal pyramidal and layer 2/3 cortical neurons.
(a) Line-scan location at the base of the apical dendrite and evoked action potentials in the soma. 10 μm scale bar. Raw line-scan images showing fluorescence baseline and single action potential-evoked responses. (b) Average-trial responses of GCaMP3 for individual hippocampal pyramidal cells in organotypic slices (n = 9 cells, thin gray lines) and mean across all cells (thick black line) for each stimulus. Note different y-axis scales for each panel. (c) Expression of GCaMP3 in layer 2/3 cortical neurons (S1) via in utero electroporation. 20 μm scale bar. (d) Average-trial responses of GCaMP3 for individual layer 2/3 cortical cells (n = 9 cells, thin gray lines) in response to trains of action potentials given at 83 Hz, and the mean across cells (thick black line). Note different y-axis scales for each panel. (e, f) Amplitudes and SNR of GCaMP3 responses for individual hippocampal pyramidal cells (thin gray lines) in response to trains of action potentials given at 83 Hz, and the mean across cells (thick red line). (g) The average response of GCaMP3 is greater than GCaMP2. (h) The SNR of GCaMP3 is also greater than GCaMP2. Error bars indicate standard deviation of the mean.
We next tested the performance of GCaMP3 in layer 2/3 (L2/3) somatosensory cortical pyramidal neurons following long-term expression driven by the CAG promoter via in utero electroporation (Fig. 2c and Supplementary Fig. 5b). The average ΔF/F of GCaMP3 at the base of the apical dendrite was 14±2.7% (n = 9 cells) for single action potentials, and 505 ± 220% for 40 APs (Supplementary Fig. 5b, Fig. 2d,g and Supplementary Table 2). Compared to GCaMP2 (n = 8 cells), the ΔF/F and SNR of GCaMP3 were 2–5 fold larger (Fig. 2g,h and Supplementary Table 2). Individual action potentials in single trials could be resolved at rates up to 6 Hz (Supplementary Fig. 7). The threshold for 100% spike detection in acute brain slices was 2 APs, with a 1 AP detection rate of ~90% (Supplementary Figs. 5b and 8a), slightly inferior to the performance in cultured brain slices. The source of this discrepancy is unknown.
Comparison of GCaMP3 and FRET-based GECIs
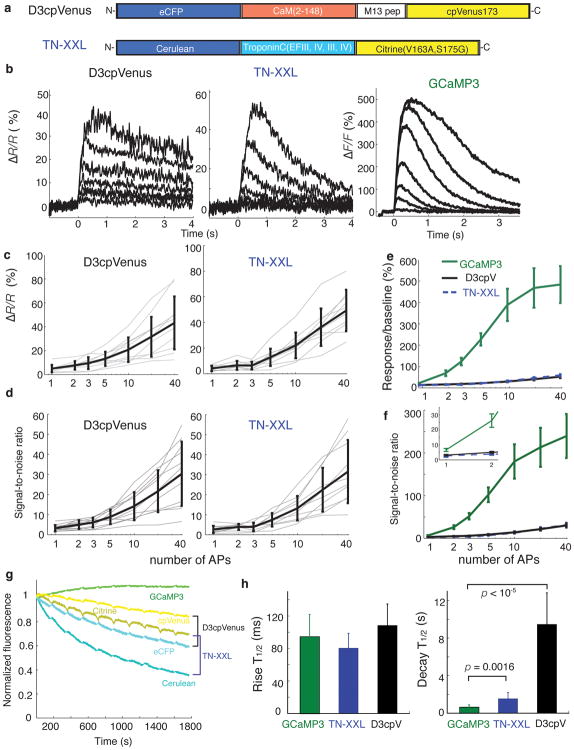
We compared the performance of D3cpV and TN-XXL (Fig. 3a) to GCaMP3 under identical experimental conditions. At baseline calcium levels, the FRET indicators (based on intact fluorescent proteins) were brighter than GCaMP3 (data not shown). However, the smaller fluorescence changes produced by the FRET indicators (Supplementary Fig. 9, Fig. 3b,c and Supplementary Table 2) resulted in lower SNR compared to GCaMP3 (Fig. 3d–f and Table 2). Furthermore, GCaMP3 was more photostable than the FRET indicators. Following 10 cycles of 150 seconds of frame-scan illumination of the soma and proximal dendrite (10 mW at the sample), interspersed by 30 seconds of darkness, GCaMP3 fluorescence remained unchanged (109% of starting fluorescence), whereas TN-XXL (36% CFP; 70% YFP) and D3cpV (59% CFP; 84% YFP) showed reduced fluorescence (Fig. 3g). The mean fluorescence rise times were similar: 95± 27 ms, 80 ± 18 ms, and 108± 26 ms for GCaMP3, TN-XXL and D3cpV (Fig. 3h-left). The fluorescence decay time of GCaMP3 (650 ± 230 ms, n = 7 cells), was significantly shorter than for the FRET indicators (TN-XXL, 1550 ± 640 ms, n = 10 cells (p = 0.0016, paired t-test); D3cpV, 9500 ± 3400 ms, n = 10 cells (p = 1.7e-05, paired t-test)) (Fig. 3h- right).
Figure 3. Comparison of GECI responses in pyramidal cell principal dendrite in acute cortical slice to back-propagating action potentials.
(a) Schematic representation of the FRET-based calcium indicators D3cpV and TN-XXL. (b) Mean of fluorescence responses for action potential trains across cells (n = 7 cells, 1 trial each cell). Traces from bottom to top represent the response to trains of 1, 2, 3, 5, 10, 20 and 40 APs. (c) Ratio change of D3cpV and TN-XXL for individual hippocampal pyramidal cells (thin gray lines) in response to trains of action potentials given at 83 Hz, and the mean across cells (thick black line). (d) SNR of D3cpV and TN-XXL. (e, f) Comparison of mean responses (ΔF/F or ΔR/R) and SNR of GCaMP3, D3cpV and TN-XXL. Zoom of lower stimuli is shown in inset. (g) Mean cellular fluorescence during periodic two-photon frame scans (n = 3–4 cells per GECI). (h) Rise and decay time comparison of all three indicators at 10 APs. Error bars indicate standard deviation of the mean.
In terms of absolute response and SNR, GCaMP3 performed better than both FRET indicators over the entire stimulus range, particularly from 2–20 APs (Fig. 3f and Table 2). GCaMP3 also showed greater photostability and faster kinetics (Fig. 3g,h). These factors translate into improved detection and measurement of physiologically relevant calcium signals (Supplementary Fig. 8a).
Imaging sensory-evoked Ca2+ transients in C. elegans
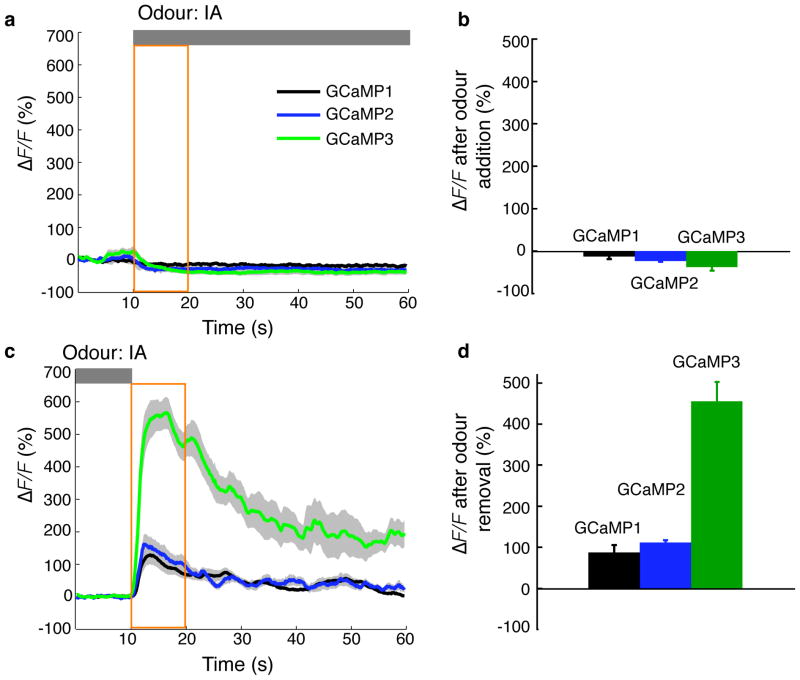
To compare the performance of GCaMP3 with previous GCaMPs in response to sensory stimulation-evoked activity in sensory neurons, we created stable C. elegans lines expressing GCaMP1, GCaMP2 and GCaMP3 in one of the two AWC neurons (AWCON). Expression of GCaMP1 and GCaMP2 in AWC neurons caused behavioral perturbations in some of the transgenic lines, reflected by decreased local search turning. In contrast, GCaMP3-expressing worms showed no detectable cytotoxicity or behavioral perturbation (Supplementary Fig. 10 and Supplementary Table 3). Individual worms were imaged following an odour addition-removal sequence 16. Presentation of isoamyl alcohol inhibited AWCON, causing a decrease in fluorescence for all three GCaMP indicators (Fig. 4a,b). The fluorescence change was larger for the two newer GCaMPs relative to GCaMP1 (−13 ± 6% for GCaMP1, −27 ± 8% for GCaMP2, −38 ± 8% for GCaMP3). Subsequent removal of the attractive odour resulted in an average of 455± 48% fluorescence increase in AWCON neurons expressing GCaMP3, a ~4–5-fold improvement over GCaMP2 (113± 25% ΔF/F) and GCaMP1 (88± 19% ΔF/F) (Fig. 4c,d). Variation in sensor expression levels due to the mosaic nature of transgenesis precluded quantitative comparison of indicator baseline brightness.
Figure 4. In vivo imaging of sensory-evoked Ca2+ transients with GCaMPs in C. elegans.
Odour-evoked responses of GCaMP1, GCaMP2 and GCaMP3 in C. elegans olfactory neurons. Transgenic worm lines expressing GCaMPs were imaged following an odour addition-removal sequence. (a,b) Upon odour presentation, GCaMP3 and GCaMP2 showed a similar decrease in fluorescence intensity. (c,d) Upon odour removal, GCaMP3 showed a 4- to 5-fold increase of fluorescence response compared to GCaMP2 and GCaMP1. Grey bars denote odour presence. Yellow intervals were analyzed in b and d. Grey shading of each trace and error bars indicate standard error of the mean (S.E.M., n = 12 animals for each genotype).
Imaging sensory-evoked Ca2+ transients in Drosophila
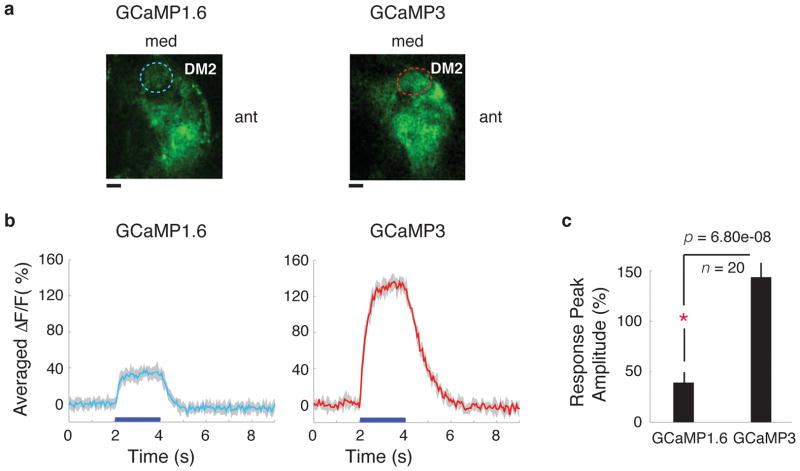
We then expressed GCaMP1.6 and GCaMP3 in a broad subset of Drosophila olfactory projection neurons (PNs) in the antennal lobe (AL) and compared their responses to odour application (GCaMP2 does not express well in Drosophila 25). Single copies of GCaMP were sufficient to produce visible fluorescence (data not shown) in glomeruli of the antennal lobe (AL), but we used two copies to allow imaging at low laser intensities. We imaged neural activity in an identified glomerulus, DM2 (Fig. 5a) in response to the presentation of two odours, vinegar and isoamyl acetate. We found a ~4-fold increased fluorescence change in DM2 for GCaMP3 compared to GCaMP1.6, as measured by frame-scans (Fig. 5b,c) in response to vinegar (average ΔF/F of GCaMP3 is 143.7± 16.7%; average ΔF/F of GCaMP1.6 is 39.3 ± 10.9%). Similar results were obtained with glomerulus DM2 when the fly was stimulated with isoamyl acetate odour (data not shown). These data show that GCaMP3 is a major improvement over existing GCaMPs for measuring sensory-evoked Ca2+ transients in invertebrates.
Figure 5. In vivo imaging of sensory-evoked Ca2+ transients with GCaMPs in Drosophila.
Odour-evoked responses of GCaMP1.6 and GCaMP3 in projection neurons of the Drosophila antennal lobe (AL). (a) Expression of GCaMP1.6 and GCaMP3 in DM2 glomeruli of the AL. DM2 ROI, circled with dashed line, was used for frame-scans. 10 μm scale bar. (b) Comparison of DM2 frame-scan responses of GCaMP1.6 and GCaMP3 to presentations of vinegar. 5-trial average response of a single animal each, expressing GCaMP1.6 (left panel) or GCaMP3 (right panel). (c) Peak response of GCaMP1.6 (4ALs from 3 animals) and GCaMP3 (4ALs from 4 animals) across all trials and animals. The response of GCaMP3 was increased ~4-fold compared to GCaMP1.6. Comparisons shown here are significant (p = 6.80e-08, Mann-Whitney). Error bars indicate standard deviation of the mean.
Imaging evoked and spontaneous Ca2+ transients in the mouse cortex in vivo
To test GCaMP3 in vertebrates we delivered it to layer 2/3 somatosensory or motor cortical neurons via infection with adeno-associated virus (AAV2/1; synapsin-1 promoter). Twelve days after infection, we observed robust expression of GCaMP3 in layer 2/3 pyramidal neurons (Fig. 1e). We used two-photon microscopy to image labeled cell bodies while simultaneously recording action potentials in whole-cell or cell-attached configurations (Fig. 6a).
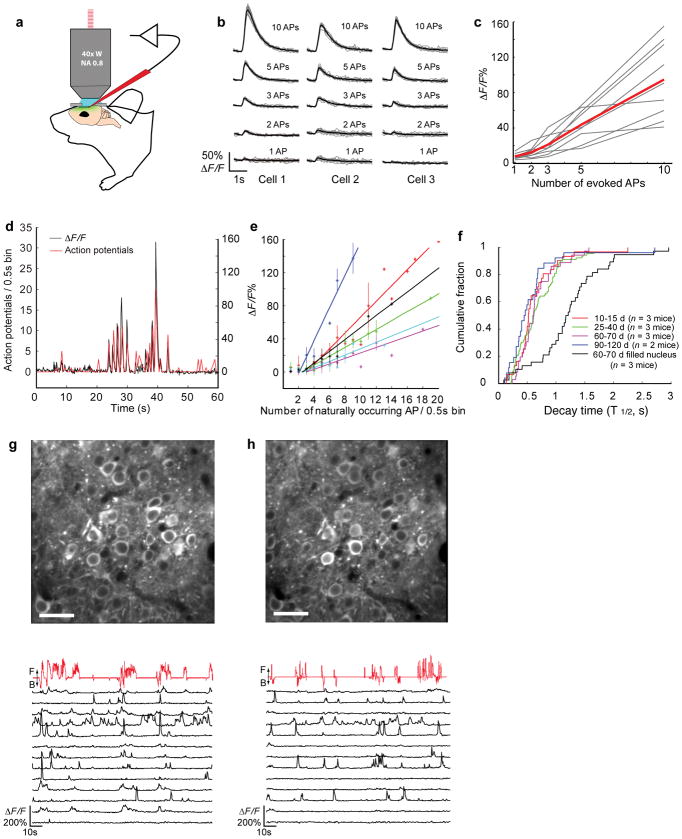
Figure 6. In vivo Ca2+ imaging of evoked and spontaneous activity with GCaMP3 in awake, behaving mice.
(a) Schematic illustrating simultaneous two-photon imaging and electrophysiology in virally infected L2/3 neurons in vivo. (b) Examples of single-trial responses (gray line) and average across 10 trials (black line) from three neurons to evoked APs at 50 Hz under anesthesia. (c) GCaMP3 showed linear ΔF/F in response to evoked APs (n = 9 cells, thin gray lines; average of 10 trials per neuron, thick red line). (d) Example trace of simultaneous recording of fluorescence changes (black) and spiking activity (red, number of APs per 0.5 sec bin) during head-fixed behavior. APs were recorded in loose seal cell-attached mode. (e) Fluorescence change in response to action potentials, binned over 0.5 s intervals. 6 cells from 3 animals were indicated with different colors. Error bars indicate standard deviation of the mean. (f) Cumulative distribution of the decay times (T½, single-exponential fit from last fluorescence maximum). Decay times of neurons with nuclear exclusion are similar at 10 to 120 days (colored lines, p = 0.22, Kolmogorov–Smirnov test). Nuclear-filled neurons have significantly longer decay times (black line, p = 5.78e-10, Kolmogorov–Smirnov test). (g) GCaMP3 expression in L2/3 neurons of the primary motor cortex at 72 days post injection (top, 30 μm scale bar) and ΔF/F traces of individual cells (bottom, black lines). Relative treadmill movement indicated by red line (F: forward, B: backward). (h) The same field of view and fluorescent traces as (g) at 120 days post injection.
We first tested the fluorescence changes of GCaMP3 evoked by action potentials that were triggered by brief current pulses in anesthetized mice. The average fluorescence response of GCaMP3 was nearly linearly related to the number of action potentials in trains of 1, 2, 3, 5 or 10 at 50 Hz (Fig. 6b,c). A single AP caused a fluorescence increase of 7.9± 2.8% (n = 9 cells) (Fig. 6c). For bursts of 2, 3, 5 and 10 AP, the corresponding responses were 12.5± 6.4%, 21.2± 6.4%, 43.7± 18.0% and 94.7± 42.5%, respectively (n = 9 cells) (Fig. 6c). The detection rate was 70% for single pulses, 90% for trains of 3 AP, and 100% for longer trains (Supplementary Fig. 8b). Consistent with rapid calcium extrusion at physiological temperature (37°C) 26, GCaMP3 showed faster kinetics in vivo (decay T1/2 at 10 pulses: 384± 76 ms) compared to slice preparations (p = 0.0015, paired t-test).
We next imaged the fluorescence changes of GCaMP3 in response to sensory-evoked and spontaneous calcium transients in the primary motor cortex of awake mice running on a treadmill while recording action potentials in a loose seal cell-attached configuration (Fig. 6d). The fluorescence change of GCaMP3 was linearly correlated with the number of action potentials from 3 APs up to 20 APs per 0.5 s (n = 6 cells from 3 animals) (Fig. 6e). Single and double action potentials were not reliably detected, likely due to movement noise and elevated baseline calcium levels in the awake brain.
Chronic imaging with GCaMP3 in mice
We found that long-term expression of GECIs in cortical neurons introduced via in utero electroporation caused altered physiology in some cells. We observed bright nuclear fluorescence at P25–P28 in ~8.3% of GCaMP3-, 13% of D3cpV- and 5% of TN-XXL- labeled L2/3 pyramidal neurons (Supplementary Fig. 11a). Neurons with filled nuclei had attenuated GCaMP fluorescence responses (data not shown), and reduced calcium changes evoked by neural activity (Supplementary Fig. 11b). Anti-His6 immuno-staining detected predominantly cytosolic GCaMP3, suggesting that the nuclei are filled with N-terminally cleaved GCaMP3 (Supplementary Fig. 11c). These results suggest that both calcium homeostasis and GECI function are impaired in neurons with labeled nuclei.
When GCaMP3 was introduced post-natally via viral transduction (under the control of the synapsin-1 promoter), some neurons near the injection site were bright and nuclear-filled (Supplementary Fig. 11d). The spontaneous fluorescence transients of nuclear-filled neurons had long decay times, another signature of abnormal physiology (Fig. 6f). These perturbed cells were easily visually identified for exclusion. Basal fluorescence decreased with distance from the injection site and was nuclear-excluded over large areas of the brain, even after 120 days of expression (Fig. 6f and Supplementary Fig. 11d). We used a variety of physiological methods to test for altered properties of nuclear-excluded, GCaMP3-positive neurons. We recorded in brain slices from neurons expressing GCaMP3 for 2–3 weeks. GCaMP3-positive neurons had similar resting potential and excitability compared to GCaMP3-negative cells (Supplementary Fig. 12a,b). We used laser scanning photo-stimulation circuit mapping 27 to test for changes in synaptic properties of GCaMP3-positive neurons. We found that GCaMP3-positive and GCaMP3-negative neurons had indistinguishable total synaptic input (Supplementary Fig. 12c,d). Thus, selection of neurons displaying nuclear-excluded G-CaMP3 expression allows quantitative optical physiology on broad swathes of cortical surface.
As a demonstration of chronic in vivo calcium imaging we used adenovirus-delivered GCaMP3 to image the in vivo calcium activity of motor cortical neurons repeatedly through a cranial window (Fig. 6g). Numerous neurons displayed large-amplitude fluorescence transients during a 140 s imaging period while head-fixed mice ran on a treadmill 28 (Fig. 6g and Supplementary Movie 3). Population activity in the motor cortex was correlated with locomotor activity (Fig. 6g). The fluorescence decay rates of spontaneous calcium transients in imaged cells with nuclear exclusion were stable 120 days post-infection (Fig. 6f). Repeated imaging of the same neuronal population at 72 and 120 days post-infection showed remarkably constant GCaMP3 expression and signal change (Fig. 6h). These results demonstrate that GCaMP3 is suitable for long-term imaging of behaviorally correlated activity in large neuronal populations over extended periods of time.
DISCUSSION
The improved properties of GCaMP3 allow new types of neuroscience applications in multiple model organisms. In C. elegans, expression of GECIs has caused behavioral phenotypes 29, likely due to altered neural physiology caused by indicator over-expression (Supplementary Fig. 10 and Supplementary Table 3). Neurons expressing GCaMP3 showed high SNR without detectable cytotoxicity or behavioral perturbations (Supplementary Fig. 10). The much higher signal levels provided by GCaMP3 (Fig. 4) may allow experiments with lower expression levels, ameliorating problems associated with calcium buffering.
In Drosophila, the poor expression of GCaMP2 25 has left GCaMP1.3 and 1.6 as the state-of-the-art single-FP indicators, but their poor sensitivity and non-linear response has precluded detection of low firing rates 30. GCaMP3 showed greatly improved expression and signal levels in vivo (Fig. 5) with no apparent cytotoxicity or behavioral phenotype (data not shown), and could permit imaging experiments not previously possible.
In behaving mice, GCaMP3 allows the simultaneous recording of the activity of dozens of identified neurons over months (Fig. 6g,h). Thus, GCaMP3 should allow tracking learning-induced changes in circuit-level activity over multiple recording sessions. Pan-neuronal expression of GCaMP3 might allow wide-field imaging of whole-cortex activity and signal propagation akin to super-resolution functional MRI 31.
Long-term, high-level expression of GECIs in the mouse brain can result in nonfunctional indicators32 and abnormal physiology (Supplementary Fig. 11). Though expression of GCaMP3 in the postnatal brain, mediated by viral gene transfer, limited such adverse effects, the proportion of nuclear-filled neurons still increased with proximity to the injection site, and slowly increased with time after injection. Therefore, optimizing the timing and magnitude of expression to balance signal levels and cytotoxicity needs further study.
Ratiometric indicators have potential advantages over single-FP GECIs, including higher baseline brightness and insensitivity to motion artifacts via wavelength ratioing. However, GCaMP3 is more photostable, likely because of reduced bleaching in the low fluorescence state (Fig. 3g). Ratiometric measurements could be achieved with GCaMP3 by co-expression or fusion of a reference fluorescent protein. The smaller size of GCaMP3 potentially facilitates a wider array of targeted protein fusions for calcium imaging in subcellular compartments. The faster kinetics of GCaMP3 may allow more faithful detection of the number and timing of action potentials in spike trains.
GCaMP3 may not be the most suitable GECI for all applications. Single action potentials can also be detected using D3cpV 14; indeed, the slow kinetics of D3cpV might be suitable for situations characterized by very low and sparse spike rates. Troponin-based indicators, such as TN-XXL, may cause less perturbation of endogenous calcium signaling 6. Prudence dictates testing each of these indicators in the context of specific applications.
Although GCaMP3 is a major improvement over GCaMP2, additional avenues for protein engineering remain. Mutagenesis to ablate interactions of the CaM and M13pep domains of GCaMP, such as that done to the D2/D3/D4 indicators 13, may improve performance at high expression levels. A single-FP indicator based on troponin C 33 might combine the benefits of a non-endogenous calcium binding protein with the high signal-to-noise and fast kinetics of GCaMP. Quantitative modeling studies 20 have suggested that increasing the fluorescence on-rate would further improve detection of the brief calcium transients associated with single APs. Expression cassettes that maintain steady, moderate levels of GCaMP3 expression for months would facilitate signal calibration and further reduce toxicity concerns.
METHODS
Construct and virus production
The original GCaMP2 expression construct was obtained from M. Kotlikoff 34, TN-XXL from O. Griesbeck 6, and D3cpV from A. Palmer 13. GCaMPs were sub-cloned into pRSETa for expression and purification in E. coli. GCaMPs were sub-cloned into pCMV for HEK293 cell assays and cultured brain slice experiments. GCaMP variants, TN-XXL and D3cpV were sub-cloned into the pCAGGS vector with a CAG promoter (CMV-enhancer, β-actin promoter, and regulatory element from the woodchuck hepatitis virus35 (WPRE)) for in utero electroporation 36. pCAG-mCherry 37 was co-transfected with GCaMPs for cultured hippocampal slices and in utero electroporation for better control of expression level. To make transgenic worms and flies, GCaMPs were sub-cloned into pSM under control of the str-2 promoter (from C.I. Bargmann) and pMUH (a gift from Barret Pfeiffer, Janelia Farm Research Campus), respectively. pMUH-GCaMPs were incorporated into an attP40 integrase site on the second Drosophila chromosome 38 (Genetic Services, Inc.). For in vivo calcium imaging in mice, GCaMP2 and GCaMP3 were expressed using an adeno-associated virus 2/1 (AAV2/1) driving the sensor under control of the pan-neuronal human synapsin-1 promoter 39. GCaMP2 and GCaMP3 were sub-cloned into the rAAV-hSYN expression vector, and live virus was produced (University of Pennsylvania Vector Core Services). All constructs were verified by sequencing.
Bacterial protein expression, purification, and testing
GCaMPs in pRSETa were transformed into chemically competent BL21(DE3)-pLysS, and purified via the N-terminal His tag. Protein concentration was determined by intrinsic tryptophan fluorescence. Calcium clamping was performed at pH 7.2 with 10 mM blends of K2H2EGTA and Ca2EGTA from the Calcium Calibration Kit #1 (Invitrogen). Free [Ca2+] levels were calculated using MAXCHELATOR (maxchelator.stanford.edu). Fluorescence spectra were recorded on a Safire2 fluorescence plate reader (Tecan). The dynamic range here is calculated as Fmax/Fmin. Fmax is the fluorescence intensity at saturating [Ca 2+] and Fmin is the fluorescence intensity at zero [Ca 2+].
HEK293 cell-based screen
GCaMPs in pCMV were transfected into HEK293 cells using Lipofectamine 2000 (Invitrogen), and imaging experiments were performed 48h post transfection. 293 cells transfected with GCaMPs in 96-well plate were imaged on an FDSS plate reader (Hamamatsu). Acetylcholine (Ach) was automatically added 10s after read initiation. Brightness was quantified using Volocity 5.0 (Improvision).
Calcium imaging in worms
Calcium imaging of GCaMP-expressing worms was performed as described previously 16. A total of 12 animals for each genotype were studied in a custom-designed microfluidic device, and the fluorescence response to odour stimulation was averaged. For odour presentation, each animal was first starved in the imaging chip for 20 min. Odours were presented at t = 10 s in a 60s recording, and removed five minutes later, at t = 10 s in a second 60 s recording.
Mice brain slice preparation
Hippocampal slice cultures were prepared using standard methods 24,40. For biolistic gene transfer, 10 μg of DNA was used per full tube. Imaging experiments were performed 24 – 48 hours after biolistic transfection. For acute slice experiments, GECIs were introduced into E-16 mouse embryos by in utero electroporation 35, and acute slices were prepared at P14–17 as described before 24.
Fly stocks, preparation and odour delivery
Flies were reared on standard cornmeal agar medium. We used the Gal4/UAS system 41 to direct the expression of the calcium sensors to PNs. GH146-Gal4 flies were a gift from L. Luo (Stanford University, Stanford, CA). UAS-GCaMP1.6 flies were a gift from D. Reiff and A. Borst (MPI, Martinsried, Germany). All experimental animals were adult females, 3–5 days after eclosion. Adult flies were dissected using previously described methods 11. Flies were anaesthetized in a vial on ice just until movement stopped (<15 second) and then gently inserted into a hole in a piece of aluminum foil. Small drops of wax (55°C) were used to suspend the fly in the hole, with the edge of foil defining a horizontal plane around the head and thorax, from the first antennal segment anteriorly to the scutellum posteriorly. The dorsal side of the foil was bathed in saline, while the ventral side (including antennae and maxillary palps) remained dry and accessible to odours. A window was cut in the dorsal head cuticle between the eyes, extending from the ocelli to the first antennal segment. Fat and air sacs dorsal and anterior to the brain were removed, but the perineural sheath was left intact. The proboscis was affixed with a small drop of wax to a strand of human hair to limit brain movement. Spontaneous leg movements were typically observed in this preparation for the duration of the recording (1.5–3 hr). The saline composition used in all experiments was 42 (in mM): 103 NaCl,3 KCl, 5 N-tris(hydroxymethyl) methyl-2-aminoethane-sulfonicacid, 10 trehalose, 10 glucose, 2 sucrose, 26 NaHCO3, 1 NaH2PO4,1.5 CaCl2, and 4 MgCl2, adjusted to 275 mOsm, pH 7.3 when bubbled with 95% O2/5%CO2.
Odours (cis-3-hexen-1-ol (cis), and isoamyl acetate (ia)) were delivered using a custom-made odour-delivery system and a Teflon nozzle (entry diameter 1/8″) directed towards the antennae. Odours were delivered in a constant stream of air (1 l/min) at final concentrations of ca. 15%. Odour delivery times were measured using a mini-PID (Aurora Scientific Inc., Ontario, Canada). Odours were presented for either 3s or 5s. All comparisons of sensor performance were made using experiments with identical odour presentation times. The results reported are based on data obtained from 3 GCaMP1.6-expressing flies (4 ALs) and 4 GCaMP3-expressing flies (4 ALs).
Calcium imaging in fly
We imaged on a two-photon laser-scanning microscope (Prairie Technologies) using an Olympus 0.8 NA LUMPlFI40XW/IR2 objective. A mode-locked Ti:Sapphire Chameleon Ultra II laser (Coherent, Santa Clara, CA) tuned to 920 nm was used as excitation source. Fluorescence was collected using photomultiplier tubes (Hamamatsu, Hamamatsu City, Japan) after bandpass filtering using a 525/70 nm emission filter. Images were acquired using PrairieView software in frame-scan mode (4–16 Hz) for a single plane of one antennal lobe.
Electrophysiology and calcium imaging in brain slice
We made recordings from CA1 cells in hippocampal slice culture, and cortical layer 2/3 pyramidal cells (S1) in acute brain slices at room temperature (22–24 °C). Patch pipettes were pulled from borosilicate glass (standard wall with filament) and had 4–6 MΩ resistance when filled with internal solution (128 K-methylsulfate or K-gluconate, 10 HEPES, 10 Na-phosphocreatine, 4 MgCl2, 4 Na2ATP, 0.4 Na2GTP, 3 ascorbic acid (pH 7.25, 290 mOsm), in mM). Slice recording and simultaneous line scan imaging were performed as before 24. During recording, slices were bathed in ACSF (127 NaCl, 25 NaHCO3, 1.25 NaH2PO4, 25 glucose, 2.5 KCl, 2 CaCl2, 1 MgCl2, in mM) bubbled with carbogen. Cells were selected for data analysis if they had nuclear exclusion of GECI fluorescence, input resistances of at least 100 MΩ, and resting potentials ≤ −50 mV in cultured slice, or ≤ −65 mV in acute slice. For experiments with evoked-action potential stimuli, 10 μM (R)-CPP (Tocris) and 10 μM NBQX (Sigma) were added to the bath to block glutamate receptors. Action potentials were triggered at 83 Hz by current injection (1–4 nA, 2 ms) through the patch pipette.
Imaging was performed in line-scan mode (500 Hz) across the apical dendrite, 20–50 μm from the base (Fig. 2a). The Ti:Sapphire laser (Mai Tai, Spectro-Physics, CA) was tuned to 910 nm for GCaMPs imaging and 860 nm to excite FRET indicators. For GCaMPs co-expressed with mCherry, we separated fluorescence into green and red channels with a 565 nm dichroic, and BG22 (green channel) and HQ620/90 (red channel) emission filters. For the FRET-based GECIs, we separated fluorescence with a 505 nm dichroic, and HQ480/80 (cyan channel) and HQ535/50 (yellow channel) emission filters. The PMT dark current was subtracted from all traces. In slice culture recordings, mean baseline fluorescence (F0) was calculated from the filter raw trace (20 Hz) prior to the action potential stimuli, as in 24. Peak fluorescence was determined by averaging 30 ms of the raw fluorescence time series about the peak of the trace linearly filtered at 20 Hz. For acute slices, response baseline was defined as the mean of the 250 ms window immediately prior to stimulation. Peak response was calculated as the maximum value of the filtered trace (100 ms moving window) within 500 ms of stimulation cessation. This method gave ~3% ΔF/F for 0 AP traces. Noise was calculated on a per cell level, as the mean standard deviation of stimulation-free, one second, bleach-corrected trace segments. For display, example traces were filtered with a Savitzky-Golay filter (2nd order, 50 ms span). Action potential detection was quantified both by a double blind psychometric test and by algorithmic template matching. In the psychometric test, eight volunteers were shown a response template and asked if it was present in randomly ordered, sequentially presented traces. False positive rate was determined by the response to 0 AP traces. The algorithmic method computed the maximum cross-correlation between a template and the fluorescence trace lagged 200 ms about the stimulus onset. Detection success was defined as a cross-correlation value greater than 95% of baseline traces. The baseline trace set consisted of all recorded 0 AP traces plus those traces reversed and/or inverted. The template was the first 1.5 seconds of the mean 3 AP response (GCaMP3) or the mean 5AP response (D3cpV, TN-XXL). Rise T1/2 of hippocampal neurons was measured as the time between the onset of current injection and the half peak response. Decay T1/2 was measured as the time of half decay of a single exponential fit of the recovery from peak response to baseline. All analysis was performed with MATLAB (Mathworks, Natick, MA).
In vivo calcium imaging and electrophysiology in mice
rAAVs (AAV2/1; synapsin-1 promoter) were injected into the primary somatosensory cortex (S1) of 2–3 week old C57Bl/6Crl wild-type mice. Two weeks after injection, mice were anaesthetized with 2% isoflurane, and a 1.5mm circular craniotomy was performed over the injection site as previously described 43. Cells were recorded with a patch pipette containing (in mM): 10.0 KCl, 140 K-gluconate, 10.0 HEPES, 2.0 MgCl2, 2.0 CaCl2, 0.05 Alexa 594, pH 7.25, 290 mOsm. For recording and stimulation a MultiClamp 700B amplifier (Molecular Devices, Sunnyvale, California) was used. In whole cell mode, action potentials were evoked by 2–5 ms long current injections; in cell attached mode currents up to 100 nA were necessary. The Ti:Sapphire laser (Mai Tai, Spectro-Physics, CA) was tuned to 910 nm for GCaMP3 imaging. Fluorescence images were simultaneously acquired using a custom-built, two-photon laser-scanning microscope equipped with a 40X, 0.8 NA objective (Olympus, Tokyo, Japan). Frame scans were acquired at 15 Hz (256×32 pixels) for a period of 3 seconds.
For imaging awake, head-fixed running mice, virus injection and surgery were identical to the anesthetized condition, except that the injection and craniotomy were performed over the primary whisker and forelimb motor area (M1). In addition, local (Marcaine) and general (Buprenorphine, 0.1mg/kg IP and Ketoprofen, 5mg/kg SC) anesthetics were administered. After full recovery on a heating pad the animals were head restrained, but allowed to run freely on a linear treadmill. Action potentials were recorded using a loose-seal cell attached configuration with patch pipettes filled with buffer (in mM: 125 NaCl, 2.5 KCl, 25.0 glucose, 10.0 HEPES, 2.0 CaCl2, 2.0 MgSO4, 0.05 Alexa 594; pH 7.4, 285 mOsm), and signals were amplified using a MultiClamp 700B (Molecular Devices, Sunnyvale, California). To confirm the identity of recorded neurons, each recording was terminated by breaking into the cell and filling with red pipette solution. During the imaging sessions the animals were kept alert by sporadic acoustic stimuli (clapping) or by presenting a pole or mild air puffs to the whisker field. Images were acquired at frame rates of 4–8 Hz at a resolution of 256×512 pixels using a 16X, 0.8 NA water immersion objective (Nikon USA, Lewisville, TX). All images acquired while awake were corrected for movement artifacts using the ImageJ plug-in TurboReg (http://bigwww.epfl.ch/thevenaz/turboreg/). ΔF/F was calculated by subtracting the baseline fluorescence level (F0, 35th percentile of total fluorescence) from the actual fluorescence level and normalized to F0.
Chronic calcium imaging in behaving mice
For chronic imaging, surgery and craniotomy were carried out as described above, but the GCaMP3-AAV was injected into the cortex directly prior to sealing the imaging window with dental acrylic. Chronic imaging was performed on C57BL/6Crl wild-type (infected with AAV2/1-hsyn1-GCaMP3) and PV-CRE mice 44 (infected with CRE-dependent AAV2/1-hsyn1-GCaMP3) over periods from 10 to 120 days post infection. To keep the animals alert and active during imaging sessions, mice were water restricted and trained to lick for water rewards upon whisker deflection. Decay times (τ ½, time at half maximum) were calculated by fitting to a single exponential. All data analysis was performed with MATLAB (Mathworks, Natick, MA).
Imaging Data Analysis
For in vivo imaging in worms, fluorescence signals in the AWC cell body were analyzed using automated tracking software and MATLAB scripts as described 16.
In flies, fluorescence time series were then obtained by averaging across the spatial extent of the glomerulus in the frame. In all cases fluorescence changes were calculated relative to baseline fluorescence levels as determined by averaging over 2 seconds just before odour presentation.
For imaging data analysis in mice in vivo, the cell body without nucleus was used as ROI for fluorescent transient analysis. Recordings with spontaneous spikes were excluded. ΔF/F was the peak fluorescence increase within 500 ms of stimulus onset divided by the mean of the three frames preceding stimulus onset. Action potential detection was quantified using cross-correlation template-matching with the first six frames of the mean response to 3 APs as a template and the second half of 1 AP and 2 AP traces (1.5–2.83 s post stimulus, 100 total traces) as the baseline. In awake behaving mice, the ΔF/F of spontaneous fluorescence transients was calculated as the peak fluorescence increase divided by the mean of the 5th–10th percentile of fluorescent intensities. All data analysis was performed with MATLAB (MathWorks, Natick, MA).
Characterizing intrinsic and circuit properties of GCaMP3 expressing neurons
L2/3 progenitor cells were transfected via in utero electroporation in C57BL/6Crl E16 time pregnant mice with a plasmid expressing CRE recombinase under the CAGS promoter as previously described 36,45. At postnatal day P14, a CRE-dependent AAV virus expressing GCaMP3 under the human synapsin-1 promoter was injected into the neocortex. This combinatorial method allowed us to label a sparse subpopulation of L2/3 pyramidal neurons with GCaMP3. Cells were recorded at a depth of 50 to 95 μm. Immediately after breaking in, cells were depolarized by injection of graded current pulses. 14 to 21 days after the viral infection (P28 to P34), animals were anesthetized with an intraperitoneal injection of a ketamine/xylazine mixture (0.13 mg ketamine/0.01 mg xylazine/g body weight) and perfused through the heart with a small volume of ice cold ACSF containing (in mM): 130 NaCl, 25 NaHCO3, 25 D-glucose, 2.5 KCl, 1.0 MgCl2, 2.0 CaCl2, and 1.25 NaH2PO4, aerated with 95% O2/5% CO2. The brain was removed and placed in an ice-cold cutting solution containing (in mM): 110 choline chloride, 25 NaHCO3, 25 D-glucose, 11.6 sodium ascorbate, 7.0 MgCl2, 3.1 sodium pyruvate, 2.5 KCl, 1.25 NaH2PO4, and 0.5 CaCl2. 400 μm thick coronal slices of the barrel cortex were cut with a vibrating slicer (Microm, Walldorf, Germany) and incubated in oxygenated ACSF for 45 min at 37°C before the recordings. Pairs of L2/3 pyramidal neurons (within <100 μm; one GCaMP3+, the other GCaMP3−) were recorded sequentially. We compared the synaptic input impinging onto GCaMP3+ and GCaMP3− neurons by measuring the total excitatory input onto both recorded cells using laser-scanning photo-stimulation by glutamate uncaging. Briefly, stimulation was with an ultraviolet laser (DPSS Lasers) on a grid (16×16, spacing 75 μm). This area included the entire thickness of the cortical grey matter and adjacent barrel columns. MNI-glutamate was uncaged for 1 ms with 20 mW of laser power at the specimen plane. We verified that under our experimental conditions these stimulation parameters elicited action potentials only when the laser beam was close to the soma of the neurons. Only excitatory inputs were mapped as cells were held at −70 mV, close to the reversal for fast inhibition. Responses were analyzed within 100 ms after the UV stimulus. Direct and synaptic responses were separated according to their different onset time 3. Responses with an onset time below 7 ms were categorized as direct (i.e. purely postsynaptic) and later responses as synaptic. Synaptic input maps were calculated as the mean current in a response window from 7 to 75 ms.
Signal-to-noise (SNR) calculation
SNR was calculated as the ratio of ΔF/F or ΔR/R to standard deviation of the filtered trace (100ms moving window), 250 ms before the stimulus up to stimulus onset.
Statistical analysis
P-values were computed by a Mann-Whitney algorithm in MATLAB. All value ranges are given as mean ± standard deviation (SD), unless otherwise noted.
Supplementary Material
Supplementary Figure 1. Improving the protein stability of GCaMP2.
Supplementary Figure 2. Summary of positions where mutants were generated
Supplementary Figure 3. Screening of GCaMP2 mutants in a HEK293 cell based assay.
Supplementary Figure 4. Screening resulted in several mutants with improved baseline brightness and signal change in HEK 293 cells.
Supplementary Figure 5. Examples of single-trial responses of GCaMP3
Supplementary Figure 6. Imaging spontaneous activity of neurons in hippocampal slice using GCaMP2 and GCaMP3
Supplementary Figure 7. Examples of single-trial fluorescence responses of GCaMP3 to low frequency stimuli
Supplementary Figure 8. Examples of single-trial responses of D3cpV (top panel) or TN-XXL
Supplementary Figure 9. Action potential detection probability
Supplementary Figure 10. Expressing GECIs in C. elegans
Supplementary Figure 11. Long-term expression of GECIs can cause altered neuron morphology and sensor properties
Supplementary Figure 12. Intrinsic and circuit properties of neurons with or without expression of GCaMP3
Supplementary Table 1. Summary of the improved GCaMPs from HEK 293 cell-based screening
Supplementary Table 2. Comparing GCaMP3 with GCaMP2 and the FRET-based sensors
Supplementary Table 3. Expression of Genetically-encoded calcium indicators in AWC neurons of C. elegans
Supplementary Movie 1. Spontaneous neural activity visualized by GCaMP2 in cultured hippocampal brain slice (3x real-time)
Supplementary Movie 2. Spontaneous neural activity visualized by GCaMP3 in cultured hippocampal brain slice (3x real-time)
Supplementary Movie 3. Naturally occurring activity of populations of layer 2/3 neurons expressing GCaMP3 in the primary motor cortex (M1) of awake, behaving mouse during 140 sec sequence of two photon images (10X real time)
Acknowledgments
We would like to thank J. Simpson and S. Hampel for cloning GCaMPs into pMUH, and the Fly Core for making fly crosses. pMUH was a generous gift from B. Pfeiffer and G. Rubin. Thanks to J. Seelig who contributed to the Drosophila imaging experiments. Thanks to S. Viswanathan and S. Sternson for assistance in screening mutants in HEK293 cells, and J. Marvin for assistance in screening mutants in E. coli. Thanks to B. Zemelman for assistance in virus production. Thanks to H. Zhong, T. Sato and B. Borghuis for developing image analysis software. Thanks to D. K. O’Connor for in utero electroporation assistance. Thanks to H. White and S. Winfrey for cell culture, B. Shields and A. Hu for histology, and A. Arnold for imaging experiments. Thanks to the Molecular Biology Shared Resource for plasmid preparation and sequencing. Thanks to J. Marvin, J. Simpson and G. Tervo for critical reading of the manuscript. All affiliations are Howard Hughes Medical Institute, Janelia Farm Research Campus.
Footnotes
Author contributions
L.L.L., K.S., L.T., S.A.H., T.M., design of project; L.T., L.L.L., sensor design and screen in E. coli, HEK293 cells; L.T., T.M., S.A.H., L.P., GCaMP variant testing in brain slice; S.A.H., FRET sensor and GCaMP2 testing in brain slice; S.A.H., L.T., D.H., imaging data analysis of brain slice and in vivo; D.H., in vivo mouse brain imaging; S.C., C.B., worm imaging and data analysis; V.J., E.C., calcium imaging and data analysis in fly; S.A.M, S.A.H., data analysis for AP detection probability; J.A.,L.L.L, E.R.S., structure analysis; L.L.L. and L.T., led the project; L.L.L., K.S., L.T. and S.A.H., wrote the paper.
Competing interests statement
The authors declare no competing financial interests.
References
- 1.Yuste R, Peinado A, Katz LC. Neuronal domains in developing neocortex. Science. 1992;257:665–669. doi: 10.1126/science.1496379. [DOI] [PubMed] [Google Scholar]
- 2.Stosiek C, Garaschuk O, Holthoff K, Konnerth A. In vivo two-photon calcium imaging of neuronal networks. Proc Natl Acad Sci U S A. 2003;100:7319–7324. doi: 10.1073/pnas.1232232100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fetcho JR, Cox KJ, O’Malley DM. Monitoring activity in neuronal populations with single-cell resolution in a behaving vertebrate. Histochem J. 1998;30:153–167. doi: 10.1023/a:1003243302777. [DOI] [PubMed] [Google Scholar]
- 4.Tour O, et al. Calcium Green FlAsH as a genetically targeted small-molecule calcium indicator. Nat Chem Biol. 2007;3:423–431. doi: 10.1038/nchembio.2007.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palmer AE, Tsien RY. Measuring calcium signaling using genetically targetable fluorescent indicators. Nat Protoc. 2006;1:1057–1065. doi: 10.1038/nprot.2006.172. [DOI] [PubMed] [Google Scholar]
- 6.Mank M, et al. A genetically encoded calcium indicator for chronic in vivo two-photon imaging. Nat Methods. 2008 doi: 10.1038/nmeth.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCombs JE, Palmer AE. Measuring calcium dynamics in living cells with genetically encodable calcium indicators. Methods. 2008;46:152–159. doi: 10.1016/j.ymeth.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mank M, Griesbeck O. Genetically encoded calcium indicators. Chem Rev. 2008;108:1550–1564. doi: 10.1021/cr078213v. [DOI] [PubMed] [Google Scholar]
- 9.Baird GS, Zacharias DA, Tsien RY. Circular permutation and receptor insertion within green fluorescent proteins. Proc Natl Acad Sci U S A. 1999;96:11241–11246. doi: 10.1073/pnas.96.20.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagai T, Sawano A, Park ES, Miyawaki A. Circularly permuted green fluorescent proteins engineered to sense Ca2+ Proc Natl Acad Sci U S A. 2001;98:3197–3202. doi: 10.1073/pnas.051636098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyawaki A, et al. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 12.Heim N, Griesbeck O. Genetically encoded indicators of cellular calcium dynamics based on troponin C and green fluorescent protein. J Biol Chem. 2004;279:14280–14286. doi: 10.1074/jbc.M312751200. [DOI] [PubMed] [Google Scholar]
- 13.Palmer AE, et al. Ca2+ indicators based on computationally redesigned calmodulin-peptide pairs. Chem Biol. 2006;13:521–530. doi: 10.1016/j.chembiol.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 14.Wallace DJ, et al. Single-spike detection in vitro and in vivo with a genetic Ca(2+) sensor. Nat Methods. 2008 doi: 10.1038/nmeth.1242. [DOI] [PubMed] [Google Scholar]
- 15.He J, Ma L, Kim S, Nakai J, Yu CR. Encoding gender and individual information in the mouse vomeronasal organ. Science. 2008;320:535–538. doi: 10.1126/science.1154476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chalasani SH, et al. Dissecting a circuit for olfactory behaviour in Caenorhabditis elegans. Nature. 2007;450:63–70. doi: 10.1038/nature06292. [DOI] [PubMed] [Google Scholar]
- 17.Wang JW, Wong AM, Flores J, Vosshall LB, Axel R. Two-photon calcium imaging reveals an odor-evoked map of activity in the fly brain. Cell. 2003;112:271–282. doi: 10.1016/s0092-8674(03)00004-7. [DOI] [PubMed] [Google Scholar]
- 18.Akerboom J, et al. Crystal structures of the GCaMP calcium sensor reveal the mechanism of fluorescence signal change and aid rational design. J Biol Chem. 2008 doi: 10.1074/jbc.M807657200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Q, Shui B, Kotlikoff MI, Sondermann H. Structural Basis for Calcium Sensing by GCaMP2. Structure. 2008;16:1817–1827. doi: 10.1016/j.str.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hires SA, Tian L, Looger LL. Reporting neural activity with genetically encoded calcium indicators. Brain Cell Biol. 2008;36:69–86. doi: 10.1007/s11068-008-9029-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varshavsky A. The N-end rule at atomic resolution. Nat Struct Mol Biol. 2008;15:1238–1240. doi: 10.1038/nsmb1208-1238. [DOI] [PubMed] [Google Scholar]
- 22.Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 23.Pologruto TA, Yasuda R, Svoboda K. Monitoring neural activity and [Ca2+] with genetically encoded Ca2+ indicators. J Neurosci. 2004;24:9572–9579. doi: 10.1523/JNEUROSCI.2854-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mao T, O’Connor DH, Scheuss V, Nakai J, Svoboda K. Characterization and subcellular targeting of GCaMP-type genetically-encoded calcium indicators. PLoS ONE. 2008;3:e1796. doi: 10.1371/journal.pone.0001796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hendel T, et al. Fluorescence changes of genetic calcium indicators and OGB-1 correlated with neural activity and calcium in vivo and in vitro. J Neurosci. 2008;28:7399–7411. doi: 10.1523/JNEUROSCI.1038-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Markram H, Helm PJ, Sakmann B. Dendritic calcium transients evoked by single back-propagating action potentials in rat neocortical pyramidal neurons. J Physiol. 1995;485:1–20. doi: 10.1113/jphysiol.1995.sp020708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shepherd GM, Svoboda K. Laminar and columnar organization of ascending excitatory projections to layer 2/3 pyramidal neurons in rat barrel cortex. J Neurosci. 2005;25:5670–5679. doi: 10.1523/JNEUROSCI.1173-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dombeck DA, Khabbaz AN, Collman F, Adelman TL, Tank DW. Imaging large-scale neural activity with cellular resolution in awake, mobile mice. Neuron. 2007;56:43–57. doi: 10.1016/j.neuron.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferkey DM, et al. C. elegans G protein regulator RGS-3 controls sensitivity to sensory stimuli. Neuron. 2007;53:39–52. doi: 10.1016/j.neuron.2006.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jayaraman V, Laurent G. Evaluating a genetically encoded optical sensor of neural activity using electrophysiology in intact adult fruit flies. Front Neural Circuits. 2007;1:3. doi: 10.3389/neuro.04.003.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim SG, Ogawa S. Insights into new techniques for high resolution functional MRI. Curr Opin Neurobiol. 2002;12:607–615. doi: 10.1016/s0959-4388(02)00355-0. [DOI] [PubMed] [Google Scholar]
- 32.Hasan MT, et al. Functional fluorescent Ca2+ indicator proteins in transgenic mice under TET control. PLoS Biol. 2004;2:e163. doi: 10.1371/journal.pbio.0020163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garaschuk O, Griesbeck O, Konnerth A. Troponin C-based biosensors: a new family of genetically encoded indicators for in vivo calcium imaging in the nervous system. Cell Calcium. 2007;42:351–361. doi: 10.1016/j.ceca.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 34.Kotlikoff MI. Genetically encoded Ca2+ indicators: using genetics and molecular design to understand complex physiology. J Physiol. 2007;578:55–67. doi: 10.1113/jphysiol.2006.120212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gray NW, Weimer RM, Bureau I, Svoboda K. Rapid redistribution of synaptic PSD-95 in the neocortex in vivo. PLoS Biol. 2006;4:e370. doi: 10.1371/journal.pbio.0040370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saito T, Nakatsuji N. Efficient gene transfer into the embryonic mouse brain using in vivo electroporation. Dev Biol. 2001;240:237–246. doi: 10.1006/dbio.2001.0439. [DOI] [PubMed] [Google Scholar]
- 37.Shaner NC, et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 38.Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat Genet. 2008;40:476–483. doi: 10.1038/ng.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thiel G, Greengard P, Sudhof TC. Characterization of tissue-specific transcription by the human synapsin I gene promoter. Proc Natl Acad Sci U S A. 1991;88:3431–3435. doi: 10.1073/pnas.88.8.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stoppini L, Buchs PA, Muller D. A simple method for organotypic cultures of nervous tissue. J Neurosci Methods. 1991;37:173–182. doi: 10.1016/0165-0270(91)90128-m. [DOI] [PubMed] [Google Scholar]
- 41.Brand AH, Manoukian AS, Perrimon N. Ectopic expression in Drosophila. Methods Cell Biol. 1994;44:635–654. doi: 10.1016/s0091-679x(08)60936-x. [DOI] [PubMed] [Google Scholar]
- 42.Wilson RI, Laurent G. Role of GABAergic inhibition in shaping odor-evoked spatiotemporal patterns in the Drosophila antennal lobe. J Neurosci. 2005;25:9069–9079. doi: 10.1523/JNEUROSCI.2070-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huber D, et al. Sparse optical microstimulation in barrel cortex drives learned behaviour in freely moving mice. Nature. 2008;451:61–64. doi: 10.1038/nature06445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hippenmeyer S, et al. A developmental switch in the response of DRG neurons to ETS transcription factor signaling. PLoS Biol. 2005;3:e159. doi: 10.1371/journal.pbio.0030159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Petreanu L, Huber D, Sobczyk A, Svoboda K. Channelrhodopsin-2-assisted circuit mapping of long-range callosal projections. Nat Neurosci. 2007;10:663–668. doi: 10.1038/nn1891. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Improving the protein stability of GCaMP2.
Supplementary Figure 2. Summary of positions where mutants were generated
Supplementary Figure 3. Screening of GCaMP2 mutants in a HEK293 cell based assay.
Supplementary Figure 4. Screening resulted in several mutants with improved baseline brightness and signal change in HEK 293 cells.
Supplementary Figure 5. Examples of single-trial responses of GCaMP3
Supplementary Figure 6. Imaging spontaneous activity of neurons in hippocampal slice using GCaMP2 and GCaMP3
Supplementary Figure 7. Examples of single-trial fluorescence responses of GCaMP3 to low frequency stimuli
Supplementary Figure 8. Examples of single-trial responses of D3cpV (top panel) or TN-XXL
Supplementary Figure 9. Action potential detection probability
Supplementary Figure 10. Expressing GECIs in C. elegans
Supplementary Figure 11. Long-term expression of GECIs can cause altered neuron morphology and sensor properties
Supplementary Figure 12. Intrinsic and circuit properties of neurons with or without expression of GCaMP3
Supplementary Table 1. Summary of the improved GCaMPs from HEK 293 cell-based screening
Supplementary Table 2. Comparing GCaMP3 with GCaMP2 and the FRET-based sensors
Supplementary Table 3. Expression of Genetically-encoded calcium indicators in AWC neurons of C. elegans
Supplementary Movie 1. Spontaneous neural activity visualized by GCaMP2 in cultured hippocampal brain slice (3x real-time)
Supplementary Movie 2. Spontaneous neural activity visualized by GCaMP3 in cultured hippocampal brain slice (3x real-time)
Supplementary Movie 3. Naturally occurring activity of populations of layer 2/3 neurons expressing GCaMP3 in the primary motor cortex (M1) of awake, behaving mouse during 140 sec sequence of two photon images (10X real time)