Abstract
CCR5 antagonists are a new class of antiretroviral drugs that block viral entry by disrupting interactions between the viral envelope (Env) glycoprotein and coreceptor. During the CCR100136 (EPIC) Phase IIb study of the CCR5 antagonist aplaviroc (APL) in treatment-naive individuals, a patient was identified who harbored virus strains that exhibited partial resistance to APL at the time of virologic failure. Retrospectively, it was found that APL resistance was present at baseline as well. To investigate the mechanism of APL resistance in this patient, we cloned HIV-1 env genes from plasma obtained at baseline and after virologic failure. Approximately 85% of cloned Envs were functional, and all exhibited partial resistance to APL. All Envs were R5-tropic, were partially resistant to other CCR5 antagonists including maraviroc on cells with high CCR5 expression, but remained sensitive to the fusion inhibitor enfuvirtide. Competition studies with natural CCR5 ligands revealed that the mechanism of drug resistance entailed the use of the drug-bound conformation of CCR5 by the Env proteins obtained from this individual. The degree of drug resistance varied between Env clones, and also varied depending on the cell line used or the donor from whom the primary T cells were obtained. Thus, both virus and host factors contribute to CCR5 antagonist resistance. This study shows that R5 HIV-1 strains resistant to CCR5 inhibitors can arise in patients, confirming a mechanism of resistance previously characterized in vitro. In addition, some patients can harbor CCR5 antagonist-resistant viruses prior to treatment, which may have implications for the clinical use of this new class of antiretrovirals.
Introduction
A new group of antiretroviral drugs, collectively known as entry inhibitors, target discrete steps in the entry of HIV into target cells including CD4 attachment, binding to the CCR5 or CXCR4 coreceptors, and gp41-mediated membrane fusion.1,2 CCR5 is a promising pharmacological target as most infections in new hosts are caused by HIV-1 strains that utilize CCR5 (R5-tropic),3–7 R5 strains usually predominate for years after initial infection,7–9 and the absence of CCR5 in patients homozygous for the Δ32-CCR5 polymorphism results in substantial protection against HIV-1 infection.10–12 Several CCR5 antagonists have been shown to reduce virus load in HIV-infected patients including vicriviroc, aplaviroc (APL), and maraviroc.13–16 The binding site for most, if not all, CCR5 antagonists appears to be a lipophilic pocket near the interface of the extracellular loops (ECLs) and transmembrane helices of CCR5.17,18 CCR5 antagonists are believed to exert their antiviral effects by altering the conformation of CCR5 rather than by directly competing with the HIV envelope glycoprotein (Env) for binding to the coreceptor, making them allosteric inhibitors.19
With the recent approval of maraviroc for the treatment of HIV infection, it will be important to determine viral and host factors that influence the efficacy of these agents. For instance, current protocols recommend pretreatment screening for the presence of CXCR4-utilizing HIV variants, as these strains are insensitive to CCR5 inhibitors. Although there is a paucity of clinical data regarding resistance to CCR5 antagonists, the outgrowth of preexisting CXCR4-utilizing HIV has been described in patients receiving CCR5 inhibitors, including maraviroc, vicriviroc, or APL monotherapy.13,15,20 In vitro, viruses passaged in the presence of increasing concentrations of CCR5 antagonists rarely switch to utilize CXCR4; instead, HIV can acquire mutations in Env that enable it to utilize the drug-bound conformation of CCR5.21–24 This mechanism of HIV resistance to CCR5 antagonists has not yet been demonstrated for patient-derived viruses. It is possible that different selection criteria in vitro, including the gradual increase in the concentration of antagonist, the absence of humoral immune pressures, and homogeneous cell populations, enables viruses to evolve resistance in a manner that is not favored in vivo.
APL is a small molecule antagonist of CCR5 with potent anti-HIV activity in vivo, but clinical development was halted in 2005 due to signs of idiosyncratic hepatotoxicity in ∼1% of patients.25 In a recently published report of the EPIC/CCR100136 Phase IIb APL study, a patient was identified who harbored viruses that despite using CCR5 to infect cells were incompletely suppressed by APL in vitro. To examine the mechanism of in vivo resistance to APL, we cloned and analyzed Envs from this patient. All Envs were R5-tropic, and all exhibited a lack of complete suppression even by high concentrations of APL on multiple cell types, including Envs isolated prior to APL treatment. Thus, resistance to APL was preexisting in this patient. The degree to which APL inhibited infection varied depending on the cell line used, and for primary cells donor variability was also observed. These Envs demonstrated cross-resistance to other CCR5 antagonists, including maraviroc, but remained sensitive to the fusion inhibitor T20. Our results indicate that HIV-1 can acquire resistance to CCR5 antagonists in vivo by utilizing the drug-bound conformation of CCR5. The frequency with which this occurs either prior to treatment or during the course of therapy is not known.
Materials and Methods
Study population
A total of 191 treatment-naive patients were enrolled in the Phase IIb CCR100136 (EPIC) study, receiving LPV/r 400/100 mg twice daily (bid) in combination with either 200 mg APL bid, 400 mg APL bid, 800 mg APL once daily (qd), or 150 mg/300 mg Combivir bid. Virologic failure was defined as incomplete virologic response (less than a 1 log10 decrease in plasma HIV-1 RNA by week 4 from the baseline value) or virologic rebound to ≥400 viral RNA copies/ml on two consecutive measurements at least 2–4 weeks apart after previously being suppressed to <400 copies/ml on or after week 4, or the subject has two consecutive viral load determinations at least 2–4 weeks apart that are >0.5 log10 copies/ml plasma HIV-1 RNA from the nadir value on study where the nadir value is the lowest HIV-1 value ≥400 copies/ml on or after week 4. Plasma samples were collected for analysis at screening, day 1, week 2, week 4, and every 4 weeks thereafter. Previous analysis of viral phenotype done by Monogram Biosciences detected reduced susceptibility to APL manifesting as incomplete suppression/plateau in patient 5 (P5) using the PhenoSense HIV Entry Assay. Envelopes from two additional patients from the same study, P7 and P9, were completely or nearly completely sensitive to complete inhibition by APL in the PhenoSense assay. Informed consent was obtained from all patients or their parent/guardian and human experimentation guidelines in accordance with GlaxoSmithKline policies and standard operating procedures were followed. Additional details of this study population have been published.26
Cloning of patient envs
Cloning of envs from patient's plasma from baseline and week 12 time points was performed using 10 separate polymerase chain reaction (PCR) reactions using a high-fidelity polymerase with 3′-to-5′ proofreading exonuclease activity as previously described.27 Vectors were grown in XL-2 Escherichia coli at 30°C to minimize bacterially induced mutagenesis and recombination of env.
Cell–cell fusion assay
Cells for fusion and viral infection assays were cultured as previously described.28 For the fusion assay,29 “target” QT6 cells were transiently cotransfected with CD4, CXCR4, or CCR5, and a luciferase reporter plasmid under the control of a T7 promoter (pGEM2 T7-luc, Promega). “Effector” QT6 cells, transfected with env expression plasmids, were infected with a recombinant vaccinia virus expressing T7 polymerase (vTF1.1).30 Fusion of target and effector cells results in T7 promoter-driven luciferase expression.
Virus infection assays
Patient envs digested with KpnI and XbaI were subcloned into a pCI expression construct containing hepatitis B virus PRE to enable high-level, rev-independent Env expression. Pseudotyped viruses produced from 293T cells (30 μg of pCI-PRE-env vector and 10 μg of pNL-luc-Env–) and 5 or 25 ng p24 equivalent were utilized to infect cell lines or primary cells, respectively, amounts empirically determined to be in the linear range of the infection assay. Three days postinfection cells were lysed and luciferase activity was analyzed on a luminometer.
CD4+ T cells isolated from leukophereses (RosetteSep CD4+ T-cell kit; Stemcell Technologies) were stimulated at 4 × 106 cells/ml with 1 μl/ml anti-CD3 (eBioscience), 1 μl/ml anti-CD28 (Becton Dickinson), and 20 U/ml of interleukin-2 (IL-2, Sigma) for 3 days. For inhibition experiments with CCR5 antagonists or enfuvirtide, 1.25 × 105 CD4+ T cells were preincubated with drug for 30 min, infected with 25 ng virus by spinoculation (450 × g, 2 h), and incubated at 37°C for 72 h before lysis and analysis. In chemokine inhibition experiments, CD4+ T cells were preincubated with drug, incubated with a mix of 50 nM MIP-1α, MIP-1β, and LD78-βCCL3L1 (PeproTech) for 1 h, then infected, washed twice to remove unbound virus, and resuspended in media containing APL and/or chemokines at the indicated concentrations prior to incubation.
Molecular evolutionary analyses
Sequences have been submitted to GenBank under accession numbers FJ998049–FJ998131. Homologous DNA sequence alignments and phylogenetic trees of envelope clones were generated using previously reported methods31 with slightly modified bootstrap parameters [1000 bootstrap replicates for neighbor joining (NJ) and 200 replicates for maximum likelihood (ML)]. To evaluate nucleotide diversity across time points in the envelope clones, nucleotide diversity calculations were performed using DnaSP4.10.32 Nucleotide diversity (Pi) is defined as the average number of nucleotide differences per site between two sequences drawn at random from the population of sequences.33
Flow cytometry
Determination of APL occupancy of CCR5 was performed by staining 1.0 × 105 U87/CD4/CCR5 cells that had been preincubated with or without APL with the anti-CCR5 monoclonal antibody 45531 (R&D systems) or mouse isotype control, followed by the addition of phycoerythrin (PE)-conjugated goat antimouse antibody (Invitrogen). Between 30,000 and 50,000 events were collected on a FACScalibur cytometer (Becton Dickinson) and data analysis was performed using FlowJo software (TreeStar).
Statistical analysis
Comparisons between the effects of chemokines on viral infection in the presence or absence of APL were made using ANOVA models with adjustment for drug and viral replicates. Analyses were performed using the Prism software package (GraphPad Software) and validated by the University of Pennsylvania Biostatistics Core. In figures, error bars represent the standard error of the mean, with at least four replicates per experiment.
Results
Isolation of functional env clones from plasma before and after treatment with APL
In the EPIC study of optimized dosing of APL, treatment-naive subjects were randomized into groups receiving APL 200 mg bid, 400 mg bid, 800 mg qd, or 150/300 mg lamivudine/zidovudine bid in the presence of lopinavir/ritonavir. One patient (P5) in the 800 mg qd arm had a viral load of 45,100 copies/ml at baseline that was reduced to 3200 copies/ml after 4 weeks of therapy, and then rebounded to 38,100 copies/ml at week 12. This patient was found to have R5 virus strains that were incompletely suppressed by APL both at baseline and after virologic failure.26 To investigate the mechanism of APL resistance in P5, we obtained HIV env clones directly from plasma from baseline and week 12 time points by isolating RNA, synthesizing cDNA, and cloning full-length env genes from 10 independent PCR reactions. Input cDNA concentrations were minimized by titration to reduce chances of recombination during PCR. Env functionality, determined using a cell–cell fusion and virus infection assays as described below, was 85% (29/34 of env clones), with at least 10 unique, functional clones from each sample time point.
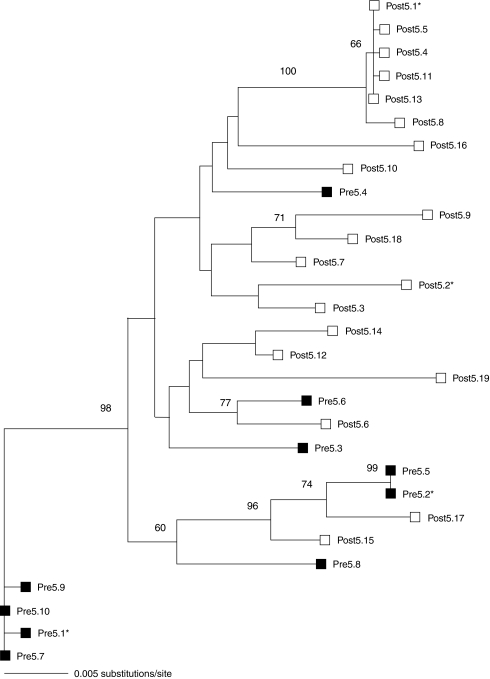
The V1–V5 regions contain the majority of sequence diversity in the env gene, are associated with determining coreceptor usage, and have been implicated in resistance to the CCR5 antagonists maraviroc and AD101.22,24 Phylogenetic analysis of V1–V5 from the cloned envs revealed a slight increase in genotypic diversity during the 12-week period of APL therapy from pretreatment Pi values of 0.0146 to posttreatment values of 0.0179. Phylogenetic trees revealed intermixing of pre- and posttreatment env clones (Fig. 1). No evidence of selective pressure on the env locus as a result of APL treatment was observed, as would be expected if resistance emerged during the course of treatment. In addition, all V3 loop sequences had genotypic signatures associated with CCR5 use. Basic residues associated with CXCR4 use at positions 11 and 24/25 of the V3 loop were not observed in any of the env clones. The V3 loop of Envs from P5 were 34 amino acids in length, with a deletion at residue 24, and did not contain amino acids previously identified with resistance to CCR5 antagonists.22–24 The Envs fell into clade B and are closely related to JRFL. All Envs pseudotyped well in our assays, with RLU values >4,000 × background levels. JRFL-pseudotyped viruses gave signals approximately 2-fold higher than P5 Envs from pretreatment time points, which in turn were 6- to 10-fold higher than the Envs from the virologic failure time point. However, the viral load at week 12 was similar to the pretreatment value in P5, suggesting that the Envs retained much of their initial fitness in vivo.
FIG. 1.
Phylogenetic tree of env genes from patient P5 constructed using V1–V5 sequences. Pre- and posttreatment clones are marked with closed and open symbols, respectively. Envelopes selected for detailed characterization are denoted by an asterisk (*). Maximum likelihood (ML) and neighbor joining trees were highly similar in topology and there were no conflicting nodes with high bootstrap support. Therefore, only the ML tree is shown with ML bootstrap values greater than the 60% reported.
Patient-derived Envs utilize CCR5 for entry
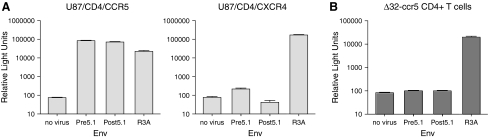
Resistance to CCR5 antagonists has been demonstrated to occur by two mechanisms. In vivo, patients have been described who failed CCR5 antagonist therapy due to the outgrowth of preexisting CXCR4-tropic HIV. In vitro, viruses passaged in the presence of increasing concentrations of CCR5 antagonists have evolved to utilize drug-bound CCR5 for entry, and coreceptor switching is rarely observed.34 The initial characterization of viruses from this patient by Kitrinos and colleagues26 indicated that some Envs used only CCR5 to mediate infection, whereas others could also use CXCR4 to a limited extent, with relative light units close to the threshold of the assay employed. In this study, we examined the tropism of the cloned envs by using pseudotyped viruses on cell lines expressing CD4 and either CCR5 or CXCR4. In our studies, we found that pseudotyped viruses from P5 were far more efficient at utilizing CCR5 for entry (Fig. 2A), consistent with our genotypic analysis. The use of CXCR4 was slightly above the background level of this assay, but the ability of Env to utilize CCR5 or CXCR4 is dependent in part on receptor expression levels and other as yet unidentified host factors.35–37 To determine coreceptor tropism under more physiological conditions, namely on primary cells, we infected CD4+ T cells from a Δ32-CCR5 homozygous donor. These cells were refractory to infection by all of the patient-derived Envs tested, while a control virus that can use CXCR4 gave robust signals (Fig. 2B). These data indicate that Envs from this patient exclusively utilize CCR5 for entry on primary cells.
FIG. 2.
(A) Infection of U87/CD4/CCR5 and U87/CD4/CXCR4 cells with pseudotyped viruses bearing representative pretreatment and posttreatment Envs cloned from P5. The dual-tropic R3A env was included as a positive control. (B) Infection of CD4+ T cells from a CCR5-Δ32 homozygous normal donor with viruses pseudotyped with envs from P5 and the R3A control. Pseudoviruses bearing envs from P5 are unable to use CXCR4 for entry on primary cells. These data are the result of three independent experiments.
Resistant Envs demonstrate incomplete inhibition by APL
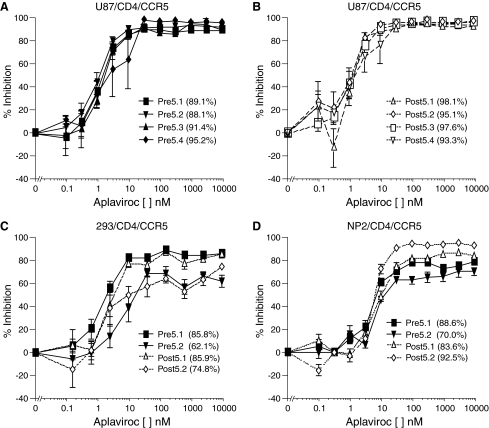
To determine APL susceptibility of the patient Envs, HIV pseudoviruses using a pNL-luc-Env– proviral reporter construct were produced. Pseudotype infection assays were performed on U87/CD4/CCR5 cells in the presence or absence of increasing concentrations of APL. Viruses pseudotyped with Envs cloned from this patient demonstrated incomplete suppression by 10 μM APL, with maximal percent inhibition (MPI) of between 88% and 98% (Fig. 3 and data not shown). These levels of infection in the presence of APL were significantly higher than for eight Envs cloned from two patients (P7 and P9) from the EPIC study that were completely inhibited by 10 μM APL in our assay, with an average MPI of >99%. The phenotype of incomplete suppression was observed for all P5 Envs from both the pretreatment and virologic failure plasma samples, and the maximal percent inhibition remained unchanged during APL treatment.
FIG. 3.
Infection of U87/CD4/CCR5 cells with pseudoviruses cloned with envs from P5 from (A) pretreatment and (B) posttreatment time points in the presence of APL. Infection of (C) 293/CD4/CCR5 and (D) NP2/CD4/CCR5 cell lines in the presence of APL. The maximal percent inhibitions (MPI) of each virus are listed in parentheses following the Env designation. Viruses pseudotyped with Envs from P5 demonstrate varying plateau levels to aplaviroc on different cell types. These data are the result of four independent experiments.
To determine if the concentrations of APL utilized in these assays were sufficient to completely occupy the available CCR5 receptors, binding studies using the monoclonal anti-CCR5 antibody 45531 were undertaken. This antibody binds to free CCR5 but does not bind to APL-bound CCR5.38,39 Consistent with previous studies,40 the addition of 10 nM APL to U87/CD4/CCR5, NP2/CD4/CCR5, 293/CD4/CCR5, or primary CD4+ T cells was sufficient to completely abrogate binding of 45531 to CCR5 as determined by flow cytometry (data not shown). This indicates that Envs from patient P5 can infect U87/CD4/CCR5 cells in the presence of APL concentrations up to 3 logs higher than necessary to completely saturate the CCR5 coreceptor.
Resistance to complete suppression by APL occurs in multiple cell types, but varies in magnitude
To determine whether cell type contributed to the presence or magnitude of the resistance plateau, 293/CD4/CCR5 and NP2/CD4/CCR5 cells were infected with pseudotyped viruses bearing either pretreatment or virologic failure Env proteins. Inhibition was diminished in both 293 and NP2 cells, with MPIs of 62–86% and 70–92% of no drug, respectively, in the presence of 10 μM APL (Fig. 3C and D). Control viruses were completely suppressed by APL in both cell lines (data not shown). The decreased sensitivity of Envs from P5 to inhibition by APL in NP2/CD4/CCR5 and 293/CD4/CCR5 indicates that cellular factors can regulate the magnitude of APL inhibition. One factor that has been identified as contributing to the magnitude of CCR5 antagonist resistance is the level of CCR5 expression.41,42 FACS analysis revealed that U87/CD4/CCR5 cells express much lower levels of CCR5 than either NP2/CD4/CCR5 or 293/CD4/CCR5 cells (CCR5 geometric mean fluorescence: 99 vs. 1048 and 1037, respectively). CD4 levels were similar between the three cell lines and did not correlate with the degree of resistance to APL. The differences in CCR5 expression levels likely account for the varied magnitude of APL resistance in these cell lines.
We also performed infection assays on primary human CD4+ T cells. As seen with cell lines, viral pseudotypes with P5 Envs were able to infect activated primary T cells even in the presence of very high APL concentrations. Infection assays of CD4+ T cells from 10 donors in the presence of APL gave a mean IC50 of 2.93 nM (range 0.4–7.2 nM) and 1.96 nM (range 0.2–4.1 nM) for Envs pre5.2 and post5.1, respectively (Table 1). Mean plateau levels for these Envs in the presence of drug were 14% (range 4–47%). Thus, the magnitude of the plateau varied by approximately a log depending on the donor used, suggesting that host cell factors influence the efficiency with which virus can infect cells in the presence of saturating concentrations of APL. By comparison, infection with control viruses was completely suppressed by 1 μM APL (mean 0.62%, range 0.2–0.8%).
Table 1.
Infection of Primary CD4+ T Cells from Normal Donors
| |
Env pre5.2 |
Env post5.1 |
Env post7.1 |
|||
|---|---|---|---|---|---|---|
| Normal donor | IC50a | Plateaub | IC50 | Plateau | IC50 | Plateau |
| 1 | 0.4 | 7.5 ± 4.4% | 0.2 | 11.9 ± 4.6% | ND | ND |
| 2 | 4.5 | 4.3 ± 1.3% | 1.0 | 10.2 ± 3.3% | 7.2 | 0.4 ± 0.1% |
| 3 | 1.6 | 25.2 ± 3.1% | 2.7 | 47.4 ± 10.5% | ND | ND |
| 4 | 1.1 | 9.9 ± 2.2% | 0.9 | 4.9 ± 2.5% | ND | ND |
| 5 | 2.8 | 6.8 ± 2.1% | 2.4 | 4.3 ± 1.7% | ND | ND |
| 6 | 2.7 | 38.6 ± 8.6% | 4.1 | 22.1 ± 4.4% | 8.3 | 0.8 ± 0.1% |
| 7 | 2.6 | 6.4 ± 3.2% | 1.1 | 10.8 ± 7.4% | 17.6 | 0.7 ± 0.5% |
| 8 | 2.6 | 18.8 ± 2.4% | 2.0 | 8.6 ± 4.6% | 3.1 | 0.8 ± 0.5% |
| 9 | 7.2 | 15.4 ± 5.8% | ND | ND | 36.1 | 0.8 ± 0.3% |
| 10 | 3.8 | 6.8 ± 1.0% | 3.2 | 7.0 ± 2.5% | 9.1 | 0.2 ± .01% |
IC50 values are in nM concentration.
Plateau values are infection at 1 μM aplaviroc as a percentage of infection in the absence of drug ± the standard error of the mean. ND, not determined.
Envs resistant to APL are cross-resistant to other CCR5 antagonists, but not enfuvirtide
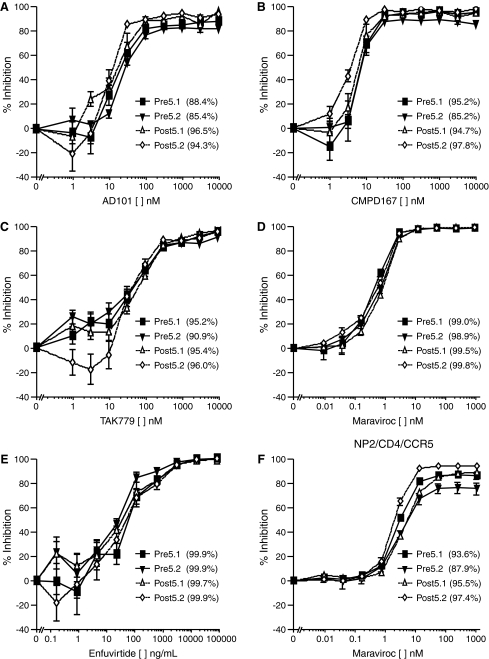
Pseudoviruses bearing Env proteins derived from P5 were also tested for sensitivity to the CCR5 antagonists maraviroc, AD101, CMPD-167, and TAK-779 and to the fusion inhibitor enfuvirtide. The pseudoviruses were incompletely suppressed by AD101, CMPD-167, and TAK-779 (Fig. 4A–C), with residual infectivity similar to that seen with APL. In contrast, infection was inhibited by approximately 98% by maraviroc (Fig. 4D). However, as described below, we subsequently found that on other cell types inhibition by maraviroc was incomplete (Fig. 4F). As observed with APL, the IC50 values for AD101, CMPD-167, maraviroc, and TAK-779 for env clones from baseline and virologic failure were similar. All of the env clones from P5 were completely inhibited by enfuvirtide (Fig. 4E), and no changes from baseline to virologic failure were observed. These data indicate that the incomplete suppression phenotype was not unique to APL, but was restricted to CCR5 antagonists.
FIG. 4.
Infection of U87/CD4/CCR5 cells in the presence of the CCR5 antagonists (A) AD101, (B) CMPD167, (C) TAK779, (D) maraviroc, and (E) the fusion inhibitor enfuvirtide. Viruses pseudotyped with envs from P5 are resistant to complete suppression by AD101, CMPD167, and TAK779 but are sensitive to maraviroc and enfuvirtide. (F) Infection of NP2/CD4/CCR5 cells in the presence of maraviroc. Percent inhibition values at 10 μM AD101, CMPD167, TAK779, 1 μM maraviroc, and 62.5 μg/ml of enfuvirtide are listed in parentheses following the Env designation. These data are the result of three independent experiments.
Since the magnitude of infection in the presence of APL was found to vary depending on the cell lines used, we tested the ability of maraviroc to suppress infection on the NP2/CD4/CCR5 cell line that was readily infected by viruses in the presence of APL. In contrast to the infection of U87 cells, NP2 cells (Fig. 4F) and 293 cells (data not shown) were readily infected by pseudoviruses bearing envs from P5 in the presence of saturating levels of maraviroc. These findings correlate with the higher expression of CCR5 on NP2 and 293 cells, suggesting that the Envs from P5 are more sensitive to inhibition by maraviroc and so need higher levels of CCR5 to compensate.
Envs resistant to complete suppression by APL utilize drug-bound CCR5
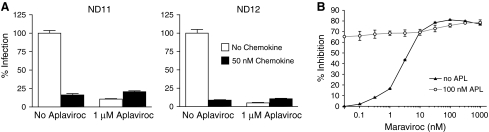
The ability of Envs from patient P5 to infect cells in the presence of saturating levels of APL suggested that these viruses could use the drug-bound conformation of CCR5 for entry, albeit less efficiently than the drug-free receptor. An alternative explanation is that infection in the presence of APL occurs via a small population of CCR5 molecules that remains drug free despite saturating levels of the inhibitor. We took two approaches in an attempt to distinguish between these possibilities. The chemokines CCL3, CCL4, and CCL3L1 inhibit infection of HIV by blocking interactions between gp120 and CCR5 and by inducing the internalization of CCR5 from the cell surface, reducing the amount available for viral entry.43 Since APL blocks CCR5 binding of CCL3 and CCL4 and presumably of CCL3L1, we hypothesized that if Envs from P5 could utilize drug-bound receptor, pretreatment with APL would reduce the inhibitory effect of chemokines on infection. In contrast, if the virus is utilizing a form of the receptor that is not bound by APL but could support binding of chemokines, then infection should be sensitive to chemokine inhibition both in the presence or absence of APL. We treated primary CD4+ T cells from two normal donors with a combination of 50 nM CCL3, CCL4, and CCL3L1 and found that this reduced pre5.2 and post5.1 virus infection by 89.8% and 95% (p < 0.001 for both comparisons) in the absence of APL (Fig. 5). However, pretreatment with 1 μM APL completely abrogated the inhibitory effect of these chemokines (p = 0.141 and p = 0.154), findings consistent with the hypothesis that Envs from P5 are utilizing drug-bound CCR5 for entry.
FIG. 5.
(A) Chemokine inhibition of HIV infection in primary CD4+ T cells from two normal donors pretreated with APL. Cells were pretreated with no or 1 μM APL, followed by stimulation with 50 nM CCL3, CCL4, and CCL3L1 prior to infection with viruses bearing Env pre5.2. Pretreatment with APL completely blocked the inhibitory effects of chemokines on HIV infection. These data are the result of two independent experiments for each donor. (B) Maraviroc inhibition of pseudoviruses bearing Env pre5.2 in the presence or absence of 100 nM aplaviroc on NP2/CD4/CCR5 cells. Higher concentrations of maraviroc were required to reach plateau levels and 50% of the maximal inhibitory potential in the presence of 100 nM APL than in its absence. These data suggest that pre5.2 Env can utilize both aplaviroc- and maraviroc-bound CCR5 but uses the former more efficiently. These results are from three independent experiments.
In a second approach, we took advantage of the fact that APL and maraviroc bind to similar regions of CCR5, but that Envs from patient P5 exhibit different plateau values on NP2/CD4/CCR5 cells in the presence of these drugs. Therefore, we examined the effect of a fixed, saturating dose of APL on the pre5.2 Env in a maraviroc sensitivity curve. In the absence of APL, maraviroc had an MPI of 81.0 ± 0.3% on pre5.2, which was stable over a range of 31–1000 nM of drug. The concentration of maraviroc at which 50% of the MPI was reached was approximately 3.0 nM. Compared to infection in the absence of drug, 100 nM APL inhibited 66.5 ± 0.6% of infection by Env pre5.2. With increasing concentrations of maraviroc, increasing inhibition was observed, reaching an MPI of 78.3 ± 1.4%. However, the MPI plateau was not reached until at least 310 nM maraviroc, and the concentration of maraviroc required to reach 50% of the MPI in the presence of 100 nM APL was approximately 30 nM. Both of these values are approximately 10-fold higher than in the absence of APL. Additionally, at maraviroc concentrations between 10 and 317 nM, the infectivity of Env pre5.2 in the presence of 100 nM APL was higher than in the absence of APL, indicating that the presence of 100 nM APL counteracted some of the inhibitory action of maraviroc at these doses. The most likely explanation for these data is that the pre5.2 Env can utilize both APL- and maraviroc-bound CCR5, but uses the former more efficiently.
Discussion
The clinical use of maraviroc and other CCR5 antagonists makes it important to identify the pathways by which HIV-1 can acquire resistance to these entry inhibitors, and the implications of drug resistance for viral tropism and sensitivity to other classes of drugs. While clinical development of APL was halted, its in vivo potency was confirmed in clinical studies. Thus far, one patient (P5) has been identified who responded to therapy but subsequently exhibited virologic failure with virus strains that were incompletely suppressed by APL in vitro: viral infectivity diminished as the drug concentration increased, but residual levels of viral infectivity were maintained even at saturating drug concentrations. In our assays, Envs from this patient were R5-tropic on U87, NP2, and 293 cell lines, with no or extremely poor use of CXCR4. These data are slightly different than the initial characterization of Envs from P5, in which the magnitude of APL resistance was greater in CCR5-expressing cells, and some clones were able to utilize CXCR4 for entry.26 One explanation for these differences is that the pseudoviruses were produced separately using different cloning vectors and may have different characteristics. Additionally, although both sets of assays were performed on the same cell line, namely U87/CD4/CCR5, the CD4 and CCR5 expression levels may have been different in cells cultured in two separate laboratories. Similarly, CXCR4 expression level differences may account for the isolation of X4 tropic clones in the initial characterization of these Envs. This possibility is supported by the observation that some of the Envs isolated in this study were able to utilize CXCR4 in the cell–cell fusion assay, where Env and coreceptor levels are expressed at very high levels, but used CXCR4 very poorly or not at all on cell lines that have much lower coreceptor expression levels. However, the most definitive experiment entailed the use of peripheral blood mononuclear cells (PBMCs) from a Δ32-CCR5 homozygous individual: these cells were completely refractory to infection by viruses bearing Env proteins from this patient, but were readily infected by control viruses that can use CXCR4, indicating that Envs from P5 are strictly R5-tropic on primary cells.
A virus that is resistant to a CCR5 antagonist despite being R5-tropic could theoretically utilize the drug-bound form of the receptor or interact with drug-free CCR5 molecules. Our results argue that the HIV-1 strains studied here most likely utilize the drug-bound conformation of CCR5, demonstrating that a mechanism for resistance to CCR5 antagonists that occurs in vitro24,44 can also occur in patients. As CCR5 antagonists induce conformational changes in the receptor,19 this type of resistance must arise from mutations that enable the viral Env protein to recognize CCR5 differently. How this occurs is not yet clear, though there is baseline variability in precisely how different HIV-1 isolates interact with CCR5, since mutations introduced into CCR5 can inhibit infection by some but not all viruses.45–47 Likewise, in vitro-derived CCR5 antagonist-resistant viruses have varied cross-resistance profiles, with some viruses demonstrating broad cross-resistance and others showing very narrow cross-resistance, again revealing variability in CCR5 interactions.23,24 We have characterized a virus with a partially deleted V3 loop that is resistant to all CCR5 antagonists tested, and that appears to interact primarily with the N-terminal domain of CCR5,28 a region of the receptor that may not be greatly impacted by this class of entry inhibitors that bind to the transmembrane domains immediately contiguous with the extracellular loops. The viruses derived from the patient studied here were broadly cross-resistant to other CCR5 antagonists, although they remained sensitive to enfuvirtide, which targets a separate stage of the entry process.
An interesting aspect of this study is that drug-resistant viruses were present prior to treatment with APL, which may be a reflection of the underlying plasticity of Env–CCR5 interactions. Sequence analyses indicated that in this patient there was no obvious genetic pressure on the env gene, as revealed by intermixing of pre- and posttreatment clones in the phylogenetic tree, a finding that is unexpected in cases in which drug-resistant Envs emerge and are selected for during treatment. One potential explanation for this finding is lack of adherence to the treatment regimen. During the EPIC study, patient compliance was monitored by measuring serum levels of APL and with pill counting. Pharmacokinetic studies on patient P5 showed that serum APL levels at week 4 of treatment were near the mean for patients in the corresponding dosing arm, as were trough levels.48 These data indicate that noncompliance is unlikely to be responsible for the absence of evidence of selective pressure on the env gene for P5. The pretreatment ability of HIV Envs isolated from P5 to utilize the drug-bound form of the receptor likely abrogated any selective pressure on env that the drug may have exerted.
The efficiency with which Envs from P5 are able to utilize the drug-bound receptor is reflected in the magnitude of the plateau, which varied in different cell lines. PBMCs also exhibited host variability, with plateaus ranging from 4% to 47%. Such differences have been previously reported, with pseudoviruses producing modest plateau levels on U87 cells while replication competent virus bearing the same Envs are completely refractory to inhibition by R5 antagonists on PBMCs.44 The level of CCR5 expression on host cells has been demonstrated to modulate the magnitude of the plateau effect,42 and expression levels of CD4 or attachment factors may also affect drug-bound receptor use. Indeed, the expression levels of CCR5 in U87, NP2, and 293 cells correlated with the magnitude of APL resistance of Envs from P5 in these cell lines. Interestingly, certain Envs from P5 gave consistently higher plateau values across target cells, indicating that viral factors such as CCR5 affinity and/or activation energy required for triggering fusion might also be involved in determining the efficacy of entry. These host and viral factors that affect the efficiency of drug-bound R5 receptor usage are under investigation and may identify characteristics that affect the in vivo efficacy of R5 antagonist therapy.
A clinical concern regarding the use of CCR5 antagonists is that they can select for preexisting CXCR4-utilizing HIV isolates, which are associated with accelerated loss of CD4+ T cells and progression to AIDS when they emerge during the course of natural infection.7,8,49–51 However, in patients with an outgrowth of viruses using CXCR4 while on CCR5 antagonist monotherapy, nearly all patients reverted to R5-tropism following withdrawal of the CCR5 antagonist.13,15,20 P5 demonstrates that continued use of drug-bound CCR5 is an alternative viral resistance strategy to CCR5 antagonists in vivo. The resistance mechanism that HIV utilizes to evade CCR5 antagonists is likely to depend in part on the disease stage of the patient. Patients with more advanced disease have a greater likelihood of harboring X4-utilizing viruses52–54 and have R5-tropic viruses with reduced sensitivity to RANTES and the CCR5 antagonists TAK779 and AD101 compared with patients at earlier disease stages.55–58 As a consequence, these patients may fail R5 antagonist therapy via either outgrowth of minor preexisting X4-utilizing populations or the continued use of CCR5 in the presence of drug. In patients treated earlier during the course of infection, who have lower levels of circulating X4-tropic viral isolates, continued use of R5 may be a more common resistance pathway. Moreover, the viruses from P5 indicate that some patients can harbor viruses with preexisting resistance to CCR5 antagonists. At present, patients are screened for the presence of R5/X4 viruses prior to the initiation of therapy, with patients harboring only R5 viruses being candidates for CCR5 antagonists. This screening assay does not directly probe for resistance to this class of antiretroviral agents, so the observation of baseline resistance to CCR5 antagonists raises two important questions. First, what fraction of CCR5 antagonist-naive patients having only R5 viruses will harbor viruses resistant to R5 inhibitors? Second, can facile screens be developed to determine which patients are most likely to benefit from R5 inhibitor therapy? The findings here suggest that simply screening patients for the presence or absence of R5/X4 viruses may not be sufficient to determine if they are good candidates for CCR5 antagonists.
Acknowledgments
This work was supported by NIH Grants R01 AI040880 and NRSA F32 1F32AI077370-01. Portions of this work were presented at the following conference: Keystone Symposia on HIV Pathogenesis, March 27–April 1, 2008, Banff, Canada, Abstract #366.
Disclosure Statement
HA.-M., K.M.K., J.F.D., J.L.J., and C.C.L. are current or former employees of GlaxoSmithKline; it should be noted that the work described in this article was completed before their departure. All are consenting authors on the manuscript and there are no other potential conflicts of interest.
References
- 1.Moore JP. Doms RW. The entry of entry inhibitors: A fusion of science and medicine. Proc Natl Acad Sci USA. 2003;100(19):10598–10602. doi: 10.1073/pnas.1932511100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tilton JC. Doms RW. Entry Inhibitors in HIV Infection. 1st. Birkhäuser Verlag; >Basel: 2007. [Google Scholar]
- 3.Balotta C. Vigano A. Riva C, et al. HIV type 1 phenotype correlates with the stage of infection in vertically infected children. AIDS Res Hum Retroviruses. 1996;12(13):1247–1253. doi: 10.1089/aid.1996.12.1247. [DOI] [PubMed] [Google Scholar]
- 4.Casper CH. Clevestig P. Carlenor E, et al. Link between the X4 phenotype in human immunodeficiency virus type 1-infected mothers and their children, despite the early presence of R5 in the child. J Infect Dis. 2002;186(7):914–921. doi: 10.1086/342948. [DOI] [PubMed] [Google Scholar]
- 5.Long EM. Rainwater SM. Lavreys L. Mandaliya K. Overbaugh J. HIV type 1 variants transmitted to women in Kenya require the CCR5 coreceptor for entry, regardless of the genetic complexity of the infecting virus. AIDS Res Hum Retroviruses. 2002;18(8):567–576. doi: 10.1089/088922202753747914. [DOI] [PubMed] [Google Scholar]
- 6.Salvatori F. Scarlatti G. HIV type 1 chemokine receptor usage in mother-to-child transmission. AIDS Res Hum Retroviruses. 2001;17(10):925–935. doi: 10.1089/088922201750290041. [DOI] [PubMed] [Google Scholar]
- 7.Schuitemaker H. Koot M. Kootstra NA, et al. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: Progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J Virol. 1992;66(3):1354–1360. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Connor RI. Sheridan KE. Ceradini D. Choe S. Landau NR. Change in coreceptor use correlates with disease progression in HIV-1 infected individuals. J Exp Med. 1997;185(4):621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scarlatti G. Tresoldi E. Bjorndal A, et al. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nat Med. 1997;3(11):1259–1265. doi: 10.1038/nm1197-1259. [DOI] [PubMed] [Google Scholar]
- 10.Dean M. Carrington M. Winkler C, et al. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science. 1996;273(5283):1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 11.Liu R. Paxton WA. Choe S, et al. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86(3):367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 12.Samson M. Libert F. Doranz BJ, et al. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382(6593):722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 13.Gulick RM. Su Z. Flexner C, et al. Phase 2 study of the safety and efficacy of vicriviroc, a CCR5 inhibitor, in HIV-1-Infected, treatment-experienced patients: AIDS clinical trials group 5211. J Infect Dis. 2007;196(2):304–312. doi: 10.1086/518797. [DOI] [PubMed] [Google Scholar]
- 14.Fatkenheuer G. Pozniak AL. Johnson MA, et al. Efficacy of short-term monotherapy with maraviroc, a new CCR5 antagonist, in patients infected with HIV-1. Nat Med. 2005;11(11):1170–1172. doi: 10.1038/nm1319. [DOI] [PubMed] [Google Scholar]
- 15.Lalezari J. Thompson M. Kumar P, et al. Antiviral activity and safety of 873140, a novel CCR5 antagonist, during short-term monotherapy in HIV-infected adults. AIDS. 2005;19(14):1443–1448. doi: 10.1097/01.aids.0000183633.06580.8a. [DOI] [PubMed] [Google Scholar]
- 16.Strizki JM. Xu S. Wagner NE, et al. SCH-C (SCH 351125), an orally bioavailable, small molecule antagonist of the chemokine receptor CCR5, is a potent inhibitor of HIV-1 infection in vitro and in vivo. Proc Natl Acad Sci USA. 2001;98(22):12718–12723. doi: 10.1073/pnas.221375398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dragic T. Trkola A. Thompson DA, et al. A binding pocket for a small molecule inhibitor of HIV-1 entry within the transmembrane helices of CCR5. Proc Natl Acad Sci USA. 2000;97(10):5639–5644. doi: 10.1073/pnas.090576697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maeda K. Das D. Ogata-Aoki H, et al. Structural and molecular interactions of CCR5 inhibitors with CCR5. J Biol Chem. 2006;281(18):12688–12698. doi: 10.1074/jbc.M512688200. [DOI] [PubMed] [Google Scholar]
- 19.Tsamis F. Gavrilov S. Kajumo F, et al. Analysis of the mechanism by which the small-molecule CCR5 antagonists SCH-351125 and SCH-350581 inhibit human immunodeficiency virus type 1 entry. J Virol. 2003;77(9):5201–5208. doi: 10.1128/JVI.77.9.5201-5208.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Westby M. Lewis M. Whitcomb J, et al. Emergence of CXCR4-using human immunodeficiency virus type 1 (HIV-1) variants in a minority of HIV-1-infected patients following treatment with the CCR5 antagonist maraviroc is from a pretreatment CXCR4-using virus reservoir. J Virol. 2006;80(10):4909–4920. doi: 10.1128/JVI.80.10.4909-4920.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trkola A. Kuhmann SE. Strizki JM, et al. HIV-1 escape from a small molecule, CCR5-specific entry inhibitor does not involve CXCR4 use. Proc Natl Acad Sci USA. 2002;99(1):395–400. doi: 10.1073/pnas.012519099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuhmann SE. Pugach P. Kunstman KJ, et al. Genetic and phenotypic analyses of human immunodeficiency virus type 1 escape from a small-molecule CCR5 inhibitor. J Virol. 2004;78(6):2790–2807. doi: 10.1128/JVI.78.6.2790-2807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marozsan AJ. Kuhmann SE. Morgan T, et al. Generation and properties of a human immunodeficiency virus type 1 isolate resistant to the small molecule CCR5 inhibitor, SCH-417690 (SCH-D) Virology. 2005;338(1):182–199. doi: 10.1016/j.virol.2005.04.035. [DOI] [PubMed] [Google Scholar]
- 24.Westby M. Smith-Burchnell C. Mori J, et al. Reduced maximal inhibition in phenotypic susceptibility assays indicates that viral strains resistant to the CCR5 antagonist maraviroc utilize inhibitor-bound receptor for entry. J Virol. 2007;81(5):2359–2371. doi: 10.1128/JVI.02006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Currier J. Lazzarin A. Sloan L, et al. Antiviral activity and safety of aplaviroc with lamivudine/zidovudine in HIV-infected, therapy-naive patients: The ASCENT (CCR102881) study. Antivir Ther. 2008;13(2):297–306. [PubMed] [Google Scholar]
- 26.Kitrinos KM. Amrine-Madsen H. Irlbeck DM. Word JM. Demarest JF. Virologic failure in therapy-naive subjects on aplaviroc plus lopinavir-ritonavir: detection of aplaviroc resistance requires clonal analysis of envelope. Antimicrob Agents Chemother. 2009;53(3):1124–1131. doi: 10.1128/AAC.01057-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ray N. Harrison JE. Blackburn LA. Martin JN. Deeks SG. Doms RW. Clinical resistance to enfuvirtide does not affect susceptibility of human immunodeficiency virus type 1 to other classes of entry inhibitors. J Virol. 2007;81(7):3240–3250. doi: 10.1128/JVI.02413-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laakso MM. Lee FH. Haggarty B, et al. V3 loop truncations in HIV-1 envelope impart resistance to coreceptor inhibitors and enhanced sensitivity to neutralizing antibodies. PLoS Pathog. 2007;3(8):e117. doi: 10.1371/journal.ppat.0030117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rucker J. Doranz BJ. Edinger AL. Long D. Berson JF. Doms RW. Cell-cell fusion assay to study role of chemokine receptors in human immunodeficiency virus type 1 entry. Methods Enzymol. 1997;288:118–133. doi: 10.1016/s0076-6879(97)88011-1. [DOI] [PubMed] [Google Scholar]
- 30.Alexander WA. Moss B. Fuerst TR. Regulated expression of foreign genes in vaccinia virus under the control of bacteriophage T7 RNA polymerase and the Escherichia coli lac repressor. J Virol. 1992;66:2934–2942. doi: 10.1128/jvi.66.5.2934-2942.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Irlbeck DM. Amrine-Madsen H. Kitrinos KM. Labranche CC. Demarest JF. Chemokine (C-C motif ) receptor 5-using envelopes predominate in dual/mixed-tropic HIV from the plasma of drug-naive individuals. AIDS. 2008;22(12):1425–1431. doi: 10.1097/QAD.0b013e32830184ba. [DOI] [PubMed] [Google Scholar]
- 32.Rozas J. Sanchez-DelBarrio JC. Messeguer X. Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19(18):2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- 33.Nei M. Molecular Evolutionary Genetics. 1st. Columbia University Press; New York: 1987. [Google Scholar]
- 34.Moore JP. Kuritzkes DR. A piece de resistance: How HIV-1 escapes small molecule CCR5 inhibitors. Curr Opin HIV AIDS. 2009;4(2):118–124. doi: 10.1097/COH.0b013e3283223d46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heredia A. Gilliam B. DeVico A, et al. CCR5 density levels on primary CD4 T cells impact the replication and enfuvirtide susceptibility of R5 HIV-1. AIDS. 2007;21(10):1317–1322. doi: 10.1097/QAD.0b013e32815278ea. [DOI] [PubMed] [Google Scholar]
- 36.Moonis M. Lee B. Bailer RT. Luo Q. Montaner LJ. CCR5, CXCR4 expression correlated with X4, R5 HIV-1 infection yet not sustained replication in Th1, Th2 cells. AIDS. 2001;15(15):1941–1949. doi: 10.1097/00002030-200110190-00005. [DOI] [PubMed] [Google Scholar]
- 37.Ketas TJ. Kuhmann SE. Palmer A, et al. Cell surface expression of CCR5 and other host factors influence the inhibition of HIV-1 infection of human lymphocytes by CCR5 ligands. Virology. 2007;364(2):281–290. doi: 10.1016/j.virol.2007.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maeda K. Nakata H. Koh Y, et al. Spirodiketopiperazine-based CCR5 inhibitor which preserves CC-chemokine/CCR5 interactions and exerts potent activity against R5 human immunodeficiency virus type 1 in vitro. J Virol. 2004;78(16):8654–8662. doi: 10.1128/JVI.78.16.8654-8662.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Demarest JF. Sparks SS. Schell K, et al. In vitro and clinical investigation of the relationship between CCR5 receptor occupancy and anti-HIV activity of Aplaviroc. J Clin Pharmacol. 2008;48(10):1179–1188. doi: 10.1177/0091270008322178. [DOI] [PubMed] [Google Scholar]
- 40.Demarest JF. Sparks SS. Schell K, et al. In vitro and clinical investigation of the relationship between CCR5 receptor occupancy and anti-HIV activity of aplaviroc. J Clin Pharmacol. 2008;48:1179–1188. doi: 10.1177/0091270008322178. [DOI] [PubMed] [Google Scholar]
- 41.Agrawal-Gamse C. Lee FH. Haggarty B, et al. Adaptive mutations in a V3-truncated HIV-1 envelope restore function by improving interactions with CD4. J Virol. 2009;83:11005–11015. doi: 10.1128/JVI.01238-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pugach P. Ray N. Klasse PJ, et al. Inefficient entry of vicriviroc-resistant HIV-1 via the inhibitor-CCR5 complex at low cell surface CCR5 densities. Virology. 2009;387:296–302. doi: 10.1016/j.virol.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alkhatib G. Locati M. Kennedy PE. Murphy PM. Berger EA. HIV-1 coreceptor activity of CCR5 and its inhibition by chemokines: Independence from G protein signaling and importance of coreceptor downmodulation. Virology. 1997;234(2):340–348. doi: 10.1006/viro.1997.8673. [DOI] [PubMed] [Google Scholar]
- 44.Pugach P. Marozsan AJ. Ketas TJ. Landes EL. Moore JP. Kuhmann SE. HIV-1 clones resistant to a small molecule CCR5 inhibitor use the inhibitor-bound form of CCR5 for entry. Virology. 2007;361(1):212–228. doi: 10.1016/j.virol.2006.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bieniasz PD. Fridell RA. Aramori I. Ferguson SS. Caron MG. Cullen BR. HIV-1-induced cell fusion is mediated by multiple regions within both the viral envelope and the CCR-5 co-receptor. EMBO J. 1997;16(10):2599–2609. doi: 10.1093/emboj/16.10.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rabut GE. Konner JA. Kajumo F. Moore JP. Dragic T. Alanine substitutions of polar and nonpolar residues in the amino-terminal domain of CCR5 differently impair entry of macrophage- and dualtropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72(4):3464–3468. doi: 10.1128/jvi.72.4.3464-3468.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Picard L. Simmons G. Power CA. Meyer A. Weiss RA. Clapham PR. Multiple extracellular domains of CCR-5 contribute to human immunodeficiency virus type 1 entry and fusion. J Virol. 1997;71(7):5003–5011. doi: 10.1128/jvi.71.7.5003-5011.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yeni P. Lamarca A. Berger D, et al. Antiviral activity and safety of aplaviroc, a CCR5 antagonist, in combination with lopinavir/ritonavir in HIV-infected, therapy-naive patients: Results of the EPIC study (CCR100136) HIV Med. 2009;10(2):116–124. doi: 10.1111/j.1468-1293.2008.00660.x. [DOI] [PubMed] [Google Scholar]
- 49.Richman DD. Bozzette SA. The impact of the syncytium-inducing phenotype of human immunodeficiency virus on disease progression. J Infect Dis. 1994;169(5):968–974. doi: 10.1093/infdis/169.5.968. [DOI] [PubMed] [Google Scholar]
- 50.Karlsson A. Parsmyr K. Sandstrom E. Fenyo EM. Albert J. MT-2 cell tropism as prognostic marker for disease progression in human immunodeficiency virus type 1 infection. J Clin Microbiol. 1994;32(2):364–370. doi: 10.1128/jcm.32.2.364-370.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maas JJ. Gange SJ. Schuitemaker H. Coutinho RA. van Leeuwen R. Margolick JB. Strong association between failure of T cell homeostasis and the syncytium-inducing phenotype among HIV-1-infected men in the Amsterdam Cohort Study. AIDS. 2000;14(9):1155–1161. doi: 10.1097/00002030-200006160-00012. [DOI] [PubMed] [Google Scholar]
- 52.Hunt PW. Harrigan PR. Huang W, et al. Prevalence of CXCR4 tropism among antiretroviral-treated HIV-1-infected patients with detectable viremia. J Infect Dis. 2006;194(7):926–930. doi: 10.1086/507312. [DOI] [PubMed] [Google Scholar]
- 53.Poveda E. Briz V. de Mendoza C, et al. Prevalence of X4 tropic HIV-1 variants in patients with differences in disease stage and exposure to antiretroviral therapy. J Med Virol. 2007;79(8):1040–1046. doi: 10.1002/jmv.20900. [DOI] [PubMed] [Google Scholar]
- 54.de Mendoza C. Rodriguez C. Garcia F, et al. Prevalence of X4 tropic viruses in patients recently infected with HIV-1 and lack of association with transmission of drug resistance. J Antimicrob Chemother. 2007;59(4):698–704. doi: 10.1093/jac/dkm012. [DOI] [PubMed] [Google Scholar]
- 55.Repits J. Oberg M. Esbjornsson J, et al. Selection of human immunodeficiency virus type 1 R5 variants with augmented replicative capacity and reduced sensitivity to entry inhibitors during severe immunodeficiency. J Gen Virol. 2005;86(Pt 10):2859–2869. doi: 10.1099/vir.0.81111-0. [DOI] [PubMed] [Google Scholar]
- 56.Karlsson I. Antonsson L. Shi Y, et al. Coevolution of RANTES sensitivity and mode of CCR5 receptor use by human immunodeficiency virus type 1 of the R5 phenotype. J Virol. 2004;78(21):11807–11815. doi: 10.1128/JVI.78.21.11807-11815.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koning FA. Kwa D. Boeser-Nunnink B, et al. Decreasing sensitivity to RANTES (regulated on activation, normally T cell-expressed and -secreted) neutralization of CC chemokine receptor 5-using, non-syncytium-inducing virus variants in the course of human immunodeficiency virus type 1 infection. J Infect Dis. 2003;188(6):864–872. doi: 10.1086/377105. [DOI] [PubMed] [Google Scholar]
- 58.Koning FA. Koevoets C. van der Vorst TJ. Schuitemaker H. Sensitivity of primary R5 HTV-1 to inhibition by RANTES correlates with sensitivity to small-molecule R5 inhibitors. Antivir Ther. 2005;10(2):231–237. [PubMed] [Google Scholar]