Abstract
Pathogenic microorganisms encode proteins that antagonize specific aspects of innate or adaptive immunity. Just as the study of the HIV-1 accessory protein Vif led to the identification of cellular cytidine deaminases as host defense proteins, the study of HIV-1 Vpu recently led to the discovery of the interferon-induced transmembrane protein BST-2 (CD317; tetherin) as a novel component of the innate defense against enveloped viruses. BST-2 is an unusually structured protein that restricts the release of fully formed progeny virions from infected cells, presumably by a direct retention mechanism that is independent of any viral protein target. Its spectrum of activity includes at least four virus families: retroviruses, filoviruses, arenaviruses, and herpesviruses. Viral antagonists of BST-2 include HIV-1 Vpu, HIV-2 and SIV Env, SIV Nef, the Ebola envelope glycoprotein, and the K5 protein of KSHV. The mechanisms of antagonism are diverse and currently include viral cooption of cellular endosomal trafficking and protein degradation pathways, including those mediated by ubiquitination. Orthologs of human BST-2 are present in mammals. Primate BST-2 proteins are differentially sensitive to antagonism by lentiviral Vpu and Nef proteins, suggesting that BST-2 has subjected lentiviruses to evolutionary pressure and presents barriers to cross-species transmission. BST-2 functions not only as an effector of the interferon-induced antiviral response but also as a negative feedback regulator of interferon production by plasmacytoid dendritic cells. Future work will focus on the role and regulation of BST-2 during the innate response to viral infection, on the mechanisms of restriction and of antagonism by viral gene products, and on the role of BST-2 in primate lentiviral evolution. The augmentation of BST-2 activity and the inhibition of virally encoded antagonists, in particular Vpu, represent new approaches to the prevention and treatment of HIV-1 infection.
Discovery of the Antiviral Activity of BST-2
The antiviral activity of BST-2 was discovered through the investigation of the HIV-1 gene vpu. This viral gene encodes a small transmembrane protein that was known to enhance the release of HIV-1 virions from infected cells, but how it did so was unclear.1,2 Several features of the vpu phenotype led to BST-2. In the absence of vpu, newly assembled virions remain attached to the cell surface and/or accumulate in endosomal vesicles, rather than releasing efficiently into the extracellular space.3 This phenotype is cell-type dependent and reminiscent of the retention of virions induced by treatment of cells with type I interferon.4–6 Vpu enhances not only the release of HIV-1 but also the release of distantly related retroviruses, suggesting that it acts on a cellular rather than a viral factor.7 This cellular factor is an inhibitor of virion release that Vpu counteracts: when cells that support the vpu phenotype are fused with those that do not, the requirement for vpu is dominant.8 Virions retained at the cell surface in the absence of vpu can be released by proteolysis, indicating that the restriction of virion release is protein mediated.9 Finally, type I interferons induce a cellular environment that supports the vpu phenotype.10
These findings set the stage for the discovery of an interferon-stimulated gene product that blocked virion release and was counteracted by Vpu. When the question of this protein's identity came into full focus, the answer, though largely unnoticed, was already suggested in the literature: a proteomic analysis of changes induced in the content of the plasma membrane by the K5 protein of KSHV (a viral ubiquitin ligase that provides immune evasion by degrading class I MHC) had uncovered three novel cellular targets, one of which was bone marrow stromal antigen-2 (BST-2).11 Vpu, which was known to degrade CD4 by recruitment of a cellular ubiquitin ligase complex to membranes,12 was also shown to degrade BST-2.11 At that time, BST-2 had no known function. It was described as expressed on B cells, activated T cells, and plasmacytoid dendritic cells, and to be inducible on other cells by type I interferons.13,14 BST-2 was also predicted to have an unusual topology: it is a type II transmembrane protein (N-terminus in the cytoplasm) with a C-terminal glycosylphosphatidylinositol (GPI) anchor.15 This topology immediately suggested a model in which BST-2 spanned the cell and virion membranes to directly restrict release. Based on these data, we tested the hypothesis that BST-2 was the interferon-induced restriction factor counteracted by Vpu.16 At the same time, comparative gene expression analysis suggested that BST-2 was this elusive factor.17
These two independent reports, published in early 2008, showed that BST-2 restricted the release of HIV-1 virions and that Vpu counteracted this restriction.16,17 One designated BST-2 as a “tetherin” based on its ability to retain or tether nascent virions to the cell surface.17 BST-2 was both necessary and sufficient for cellular support of the vpu phenotype. Since BST-2 is induced by type I interferons, a novel effector protein of the innate immune response to HIV-1 and ultimately other enveloped viruses had been revealed, and HIV-1 Vpu was characterized as the prototype viral antagonist of this host response.
Mechanism of restriction of virion release by BST-2
Genetic, structural, and functional features of BST-2
The expression of BST-2 is induced by type I interferons.13 The promotor of the bst-2 gene contains response sequences suggesting induction by the inflammatory cytokine interleukin (IL)-6, but this has not yet been shown.18 A single copy of the gene is found on chromosome 19; the human genome database so far indicates only one nonsynonymous single nucleotide polymorphism in the region encoding the protein's ectodomain.
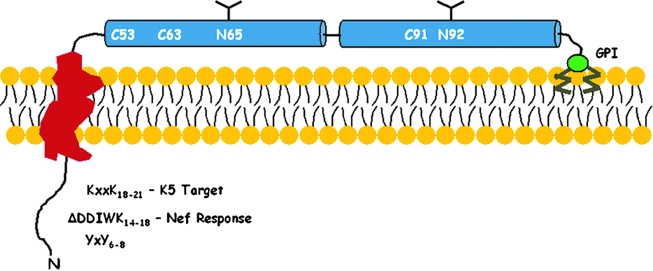
BST-2 is a 30- to 36-kDa, heterogeneously glycosylated, dimeric, type II integral membrane protein with an unusual topology.15,18 The protein contains an N-terminal cytoplasmic domain, a single membrane-spanning α-helix, an extracellular (ecto-) domain containing two predicted α-helices separated by a short loop, and a C-terminal GPI anchor (Fig. 1). The extracellular region contains residues conserved throughout mammalian orthologs: two asparagines that are glycosylation sites and three cysteines that are available for disulfide bonding. Indeed, BST-2 exists in cells as a disulfide-bonded homodimer.18 Additional residues are conserved within the ectodomain, which is predicted to form a dimeric coiled-coil structure. The cytoplasmic domain also contains conserved features: a YxY motif involved in clathrin-mediated endocytosis is present in mammalian orthologs,19,20 whereas a membrane proximal KxxK motif is present in primate orthologs. Lysines of the KxxK sequence are the targets of ubiquitination by the K5 protein of KSHV.21 Nonhuman primate BST-2 also contains the sequence DDIWK, which renders the molecule susceptible to the Nef protein of simian lentiviruses.22,23
FIG. 1.
Domain structure of BST-2. BST-2 is a type II (N-terminus in the cytoplasm) single-pass transmembrane protein whose C-terminus is attached to the lipid bilayer via a glycosylphosphatidylinositol (GPI) anchor. The cytoplasmic domain contains several conserved features including a YxY motif that binds the plasma membrane clathrin adaptor AP-2, a DDIWK sequence found in nonhuman primates that is required for response to Nef, and lysines (K18 and K21) that are targets for ubiquitination by the K5 protein of KSHV. The extracellular domain contains two α-helical regions (blue) that are predicted to form dimeric coiled-coils. Cysteines that contribute to disulfide-linked homodimerization and N-linked glycosylation sites are indicated.
BST-2 is found both at the plasma membrane as well as within the endosomal system, including the trans-Golgi network.15,19,20,24 Rodent BST-2 is endocytosed via the clathrin adaptor AP-2 and retrieved to the TGN via AP-1.19 At the plasma membrane, the protein localizes within cholesterol-enriched lipid rafts,15 presumably due to its C-terminal GPI modification. This optimally positions BST-2 to interfere directly with virion release, since several lipid-enveloped viruses, including HIV-1 and Ebola, bud selectively from raft domains.25–27
Mutagenesis of BST-2 indicates that the GPI anchor is required for the restriction of virion release.17 In contrast, the N-linked glycosylation sites are dispensable for activity.28,29 Interestingly, conflicting data exist regarding the role of cysteine-mediated dimerization; this feature appears dispensable for the restriction of Lassa virus but required for the restriction of HIV-1.29,30 No other conserved residues in the ectodomain are yet known to be essential for restrictive activity.
Spectrum of antiviral activity
The breadth of restrictive activity of BST-2 is currently emerging. Diverse viruses including Ebola, Marburg, Lassa, KSHV, as well as all retroviruses tested appear sensitive to BST-2.21,31–33 These viruses have few common features: they are all lipid enveloped, and some (HIV-1, Ebola) are known to bud from cholesterol-enriched domains of the plasma membrane,25–27 where BST-2 is itself enriched.15 Because BST-2 restricts the release of diverse viruses, a specific viral protein target seems unlikely. Instead, the binding partner for BST-2 may be the virion lipid envelope, less likely a ubiquitous cellular protein or sugar moiety, or BST-2 itself, as discussed below.
Direct restriction models
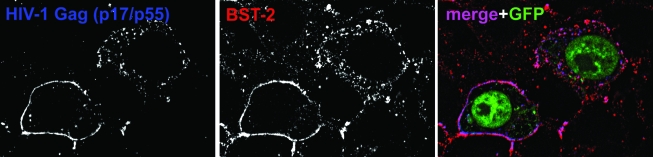
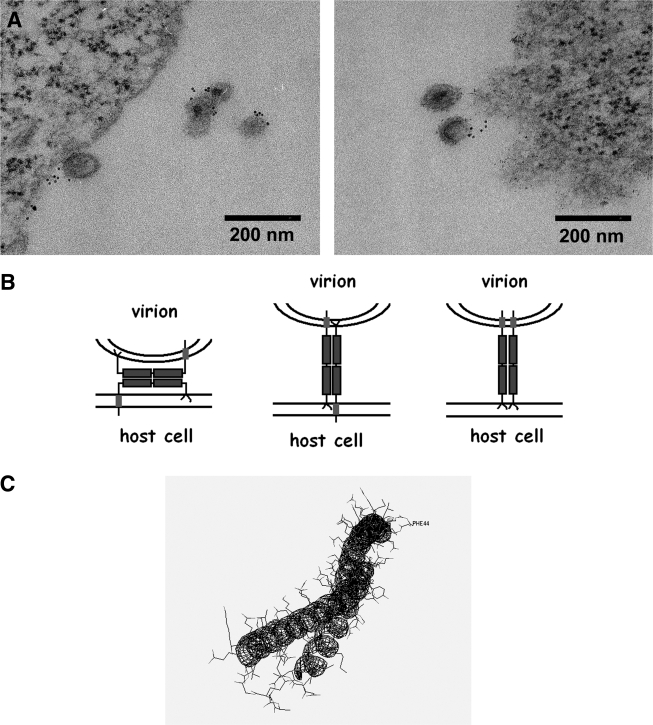
The designation of BST-2 as a tetherin evokes a model in which the protein directly retains virions on the cell surface. This model is supported by immunofluorescence data in which BST-2 and the HIV-1 structural protein Gag colocalize along the plasma membrane (Fig. 2), often in a punctate pattern.33,34 These puncta presumably represent nascent, tethered virions, although they may also represent endocytic pits containing Gag and BST-2 destined for internalization. Direct retention of virions by BST-2 at the plasma membrane has recently been supported unambiguously by immunoelectron microscopy.35,36 These data indicate that BST-2 is associated with virions, found between virions and the plasma membrane, and intercalated within clusters of virions (Fig. 3A).
FIG. 2.
BST-2 and HIV-1 Gag colocalize along the cell surface and in endosomes. Cells (HeLa), which express BST-2 constitutively, were transfected to express an HIV-1 genome lacking the vpu gene, as well as GFP (green in the color image) as a marker of the transfected cells, then stained for the structural HIV-1 protein Gag (p17/p55; left panel; blue in the color image) and BST-2 (middle panel; red in the color image). In the color (merge) image, the overlap of Gag and BST-2 appears purple.
FIG. 3.
Evidence for direct retention of nascent virions on the cell surface by BST-2 and topological models of restriction. (A) Electron microscopic evidence of direct retention and virion incorporation. HeLa cells expressing an HIV-1 genome lacking vpu were stained on their surface for BST-2 using an antibody to the protein's ectodomain, followed by a secondary labeling system using electron-dense cadmium selenide/zinc sulfide nanocrystals, which appear as small black dots in these transmission electron microscopic images. BST-2 is associated with virions and located between virions and the cell surface, supporting a direct tethering mechanism. (B) Schematic representations of direct tethering models. The left schematic depicts “ectodomain self-interaction,” whereas the middle and right depict “membrane spanning” mechanisms in which BST-2 homodimers are arranged in antiparallel or parallel orientation. (C) Predicted structure of the coiled-coil ectodomain. The structure of the monomeric ectodomain was predicted using the SAMT06 server77 and rendered using DeepView.
The general structural and membrane-binding features of BST-2 are both necessary and sufficient for restriction.36 Indeed, two lines of evidence indicate that restrictive activity is unlikely to require cellular or viral cofactors in addition to BST-2. First, human BST-2 can restrict virion release in a wide range of host cells, including cells of avian species whose genomes do not encode BST-2 orthologs.37 Second, an “artificial tetherin” containing the key structural features of BST-2 but composed of segments of different proteins (the cytoplasmic and transmembrane domains of the transferrin receptor, an ectodomain containing the dimeric coiled-coil from dystrophia myotonica protein kinase, and the GPI anchor sequence from urokinase plasminogen activator receptor) is fully competent as a restriction factor, though it shares no significant sequence homology with BST-2.36
Various topological models could account for direct restriction (Fig. 3B). One is a “membrane-spanning” model, in which BST-2 embeds one end in the cell membrane and the other in that of the virion. Homodimers could be arranged in either a parallel or antiparallel orientation. Notably, analysis of the residual BST-2 found in restricted virions after release from the cell surface by proteolysis suggests a parallel-dimeric orientation.36 An alternative is an “ectodomain self-interaction” model. In this model, the entirety of each BST-2 molecule is within either the plasma membrane or the virion membrane, and an interaction between the ectodomains of cell-associated and virion-associated BST-2 restricts the release of nascent virions.
Currently, we favor the ectodomain self-interaction model. Consistent with this model, BST-2 is fully incorporated into virions.35,36 Infectious particles spontaneously released from cells can be captured using specific antibody to the BST-2 ectodomain, and such virions contain the full spectrum of glycosylated BST-2 species.35 The ectodomain self-interaction model is consistent with a role for cysteine-linked dimerization in restriction,29 although the reducing agent DTT does not release restricted virions from the cell surface.31,35 Enzymatic cleavage of GPI anchors at the cell surface with phosphatidylinositol-specific phospholipase C does not release restricted virions35; it should do so if a membrane-spanning model with parallel orientation of subunits were correct. An ectodomain self-interaction model could involve either dimer–dimer interactions or dimerization between cell- and virion-associated monomers. Interestingly, in silico structure prediction suggests that the ectodomain of BST-2 forms a coiled-coil hairpin (Fig. 3C). If this hairpin were “straightened” and the same interactions formed between two molecules, then an antiparallel coiled-coil ectodomain dimer would result (Fig. 3B, left-most model).
Though attractive, a coiled-coil ectodomain self-interaction model is far from proven. No direct evidence indicates that the ectodomain has a propensity for self-interaction independent of disulfide-linked dimerization30; specific residues in the ectodomain have yet to be shown as important for restriction (although a dimeric coiled-coil domain seems required for activity36); and the structure of the ectodomain has not been solved. Notably, this model is not excluded by the activity of “artificial tetherin,” because the ectodomain of this construct contains, like native BST-2, a dimeric coiled-coil region,36 which could potentially mediate self-interaction.
Antagonism of BST-2 by viral proteins
The absence of a viral protein target for BST-2 mandates that viruses encode a specific antagonist of this host antiviral molecule. This relationship between the virus and its host is similar to that exemplified by the APOBEC family proteins, which target viral nucleic acids, and their antagonists, the lentiviral Vif proteins.38 It is distinct from that of the Trim5-α proteins, which target the lentiviral capsid protein, and whose restrictive action can be thwarted simply by mutation of the viral target without the acquisition of an antagonist gene.39 Vif proteins antagonize APOBEC3 proteins by inducing their degradation via the host ubiquitin–proteasome pathway.40,41 Viruses frequently use this approach to eliminate host proteins that restrict their replication, including proteins involved in innate and adaptive immunity.42 Not surprisingly, viruses have adopted this strategy to antagonize BST-2 (Table 1). Furthermore, because BST-2 seems likely to act directly at the cell surface, its removal from that location by most of the characterized viral antagonists is also not surprising (Table 1).
Table 1.
Viral Antagonists of BST-2: Mechanisms and Specificity
| Virally encoded antagonist | Downregulation of cell surface BST-2 | Region of BST-2 targeted | Role of ubiquitination | Species susceptibility |
|---|---|---|---|---|
| HIV-1 Vpu | Yes | Transmembrane | Yes | Human and chimpanzee |
| HIV-2 and SIV Env | Yes | Ectodomain? | Unlikely | Human and nonhuman primates |
| SIV Nef | Yes | Cytoplasmic | Unlikely | Nonhuman primates |
| KSHV K5 | Yes | Cytoplasmic | Yes | Human and ? |
| Ebola gp | Unknown | Ectodomain? | Unlikely | Human and mouse |
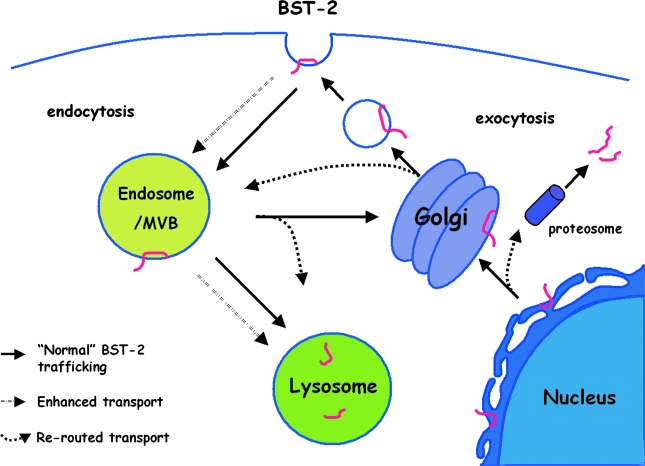
Six viral proteins have so far been reported to counteract BST-2: HIV-1 Vpu, HIV-2 and SIV Env, SIV Nef, KSHV K5, and Ebola glycoprotein (GP).16,17,21–23,31,43,44 All but Ebola GP are known to deplete BST-2 from the plasma membrane. Two of the antagonists, Vpu and K5, also decrease the total level of cellular BST-2 and utilize ubiquitin-mediated pathways.21,45,46 The effects of these viral proteins on the trafficking of BST-2 can be exerted at various steps (Fig. 4). These effects can be manifest and characterized in several ways: by mislocalization of BST-2 away from the plasma membrane; by a decrease in the total cellular level of BST-2; by the mode of interaction between BST-2, the viral antagonist, and the cellular trafficking machinery; and with respect to virion release, as an indicator of relevance to the antagonism of restriction.
FIG. 4.
Cellular pathways potentially involved in the antagonism of BST-2 via removal from the cell surface by viral proteins. Upon synthesis at the endoplasmic reticulum (ER), BST-2 is normally delivered to the plasma membrane (PM), internalized from the PM via AP-2-dependent endocytosis, and presumably recycled from endosomes to the PM. The viral proteins that decrease the expression of BST-2 at the cell surface (HIV-I Vpu, HIV-2 and SIV Env, KSHV K5, and SIV Nef ) and that decrease the total cellular expression of BST-2 (Vpu and K5) may act at various steps. Certain mechanisms would block newly synthesized BST-2 from reaching the PM. For example, during exocytosis, BST-2 could be rerouted from the ER to the proteosome for degradation (possibly for Vpu) or rerouted from the Golgi to endosomes and lysosomes. BST-2 could also be sequestered after synthesis in the trans-Golgi network (possibly for Vpu and HIV-2 and SIV Env). Alternative mechanisms would directly remove BST-2 from the plasma membrane. For example, the rate of endocytosis of BST-2 could be enhanced (likely for SIV Nef and possibly for HIV-2 and SIV Env). Alternatively or in addition, BST-2 could be rerouted after endocytosis to late endosomes/multivesicular bodies (MVB) with further trafficking to the lysosome (likely for Vpu and K5). The latter pathway would be at the expense of recycling to the cell surface. Mechanisms involving the direct removal of BST-2 from the PM are appealing mechanistically. They are potentially the most rapid, because, unlike mechanisms in which newly synthesized BST-2 is prevented from reaching the PM, they do not rely on the endogenous rate of turnover at the cell surface to clear BST-2 from its site of action as a tethering factor. See text for references.
HIV-1 Vpu
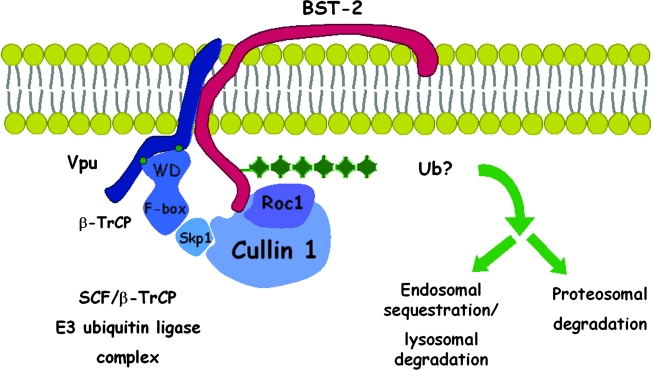
Vpu reduces the expression of BST-2 at the cell surface, and this reduction is at least conceptually sufficient to explain the antagonism of restriction.11,16 Vpu is a small transmembrane protein that resides predominantly within the endosomal system, in large part within the trans-Golgi network (TGN).1,47,48 Current data suggest that surface downregulation involves an interaction between Vpu and BST-2 and that this interaction is mediated by the transmembrane domains of each protein.49 Via this interaction, Vpu could, in principle, directly sequester BST-2 within the endosomal system, or it could recruit cellular proteins to facilitate the net removal of BST-2 from the cell surface. In particular, Vpu interacts via the sequence DpSGxxpS (pS indicates phosphoserine) in its cytoplasmic domain with β-TrCP, a substrate adaptor for an SCF (Skp-cullin-F-box) E3 ubiquitin ligase complex.12 This suggests that Vpu could recruit the E3 ligase complex to ubiquitinate BST-2, leading to one or more fates: increased endocytosis, targeting to late endosomes and lysosomes, or targeting to proteasomes (Fig. 5).
FIG. 5.
Vpu recruits a multisubunit ubiquitin ligase complex to BST-2. Vpu interacts with BST-2 via its transmembrane domain, while recruiting an SCF-E3 multisubunit ubiquitin ligase complex via a conserved DSGxxS motif in its cytoplasmic domain (green dots). The DSGxxS sequence binds β-TrCP, the substrate adaptor for the complex, via its WD domain. The presumed consequence of this interaction is the ubiquitination of BST-2, which may lead to endocytosis, endosomal sequestration, lysosomal degradation, and/or proteasomal degradation. Any of these mechanisms would remove BST-2 from the cell surface, it site of activity as a tethering factor, and so counteract restriction virion release. See text for references.
Three studies have documented a key role for β-TrCP as a cofactor for the downregulation of BST-2 and the relief of restricted release by Vpu.34,50,51 Each study used a combination of approaches to establish this role: characterization of Vpu mutants incapable of binding β-TrCP, expression of dominant negative β-TrCP mutants, and suppression of endogenous β-TrCP by RNA interference. These studies indicate that β-TrCP is required for optimal downregulation of BST-2 at the level of both surface and total cellular expression, and they further indicate that β-TrCP is required for optimal relief of restriction by Vpu. The data also suggest that a component of Vpu activity is independent of β-TrCP. As noted, binding alone may be sufficient for Vpu to sequester BST-2 within the endosomal system, away from its site of action at the cell surface. Vpu may induce such sequestration in the TGN, where it appears to colocalize extensively with BST-2.24
Presumably, Vpu recruits β-TrCP to modify BST-2 with ubiquitin, although this has yet to be shown. This modification could lead directly to endocytosis, to degradation in lysosomes, or to degradation by cytoplasmic proteasomes. Several studies have reported that the expression of Vpu decreases the steady-state levels of BST-2, consistent with a mechanism of degradation.45,46,51,52 However, the magnitude of this effect and its proximate mechanism varies with the cells tested and the experimental format. In HEK 293T cells that overexpress exogenous BST-2, Vpu induces a dramatic decrease in the steady-state levels of BST-2 that is due to decreased protein stability.45,46,52,53 In contrast, a more modest decrease in total cellular BST-2 is detected in HeLa cells, which express endogenous BST-2 constitutively.34 In HeLa cells, the decrease in the total level of BST-2 induced by Vpu is less than the decrease in surface expression, implying that the latter may result from sequestration of BST-2 in an endosomal compartment in addition to degradation.34
Several lines of evidence support the possibility that Vpu downregulates BST-2 at least in part via proteosomal degradation. First, the proteosome inhibitors MG132, ALLN, and clasto-lactacystin β-lactone rescue BST-2 levels in the presence of Vpu.45,46,51 Second, the ubiquitin mutant K48R, which blocks the formation of a specific form of polyubiquitin that target proteins to proteosomal degradation, partially restores the level of BST-2 in the presence of Vpu.51 Third, the endoplasmic reticulum-associated degradation (ERAD) ATPase p97 is required for optimal degradation of BST-2 by Vpu.51 A caveat to all of these studies is the overexpression of BST-2 in HEK 293T cells by transient transfection; this can potentially bias the apparent mechanism to ERAD-based pathways, as Vpu needs only to act on newly synthesized BST-2.
On the other hand, at least two lines of evidence support a mechanism of ubiquitin-dependent endosomal trafficking and subsequent lysosomal degradation. First, the plasma membrane-associated clathrin adaptor protein complex AP-2 and the GTPase dynamin-2 are each required for optimal downregulation of BST-2 from the surface of HeLa cells; this suggests that the influence of Vpu is exerted at least partly via direct removal of BST-2 from the plasma membrane.34,54 Second, drugs that inhibit acidification of endosomes, namely bafilomycin A and concanamycin A, inhibit the downregulation of BST-2 by Vpu in HeLa cells.34,50 In contrast, the proteasomal inhibitor MG-132 is minimally able to rescue the surface level of BST-2 in HeLa cells expressing Vpu, unless used for prolonged periods that are predicted to cause depletion of the cellular pool of ubiquitin.34
Vpu does not seem to affect the rate of endocytosis of BST-2.34 This suggests that Vpu affects the trafficking of BST-2 at a postendocytic step; this step could involve the targeting of BST-2 to late endosomes and lysosomes via the ESCRT pathway, whose components recognize ubiquitin as a sorting signal. Alternatively, Vpu could alter the mechanism of endocytosis, such that BST-2 is diverted from a physiological route associated with recycling to the plasma membrane to a route associated with degradation. In potential support of this model, the YxY endocytic motif in the cytoplasmic domain of BST-2 is dispensable for Vpu-mediated downregulation, suggesting that endocytosis of BST-2 in the presence of Vpu occurs by a mechanism distinct from that of constitutive endocytosis.54 Presumably, Vpu-mediated ubiquitination of BST-2 would again provide the signal for altered trafficking, the end result of which is removal from the plasma membrane.
Overall, extensive progress has been made in understanding the mechanism of the downregulation of BST-2 by Vpu and the antagonism of restriction, but several questions remain open. What is the basis of the interaction between Vpu and BST-2? Can Vpu trigger both proteosomal and lysosomal degradation of BST-2? Is β-TrCP-dependent ubiquitination of BST-2 required for these pathways? Is the predominant mechanism of surface downregulation in CD4-positive T lymphocytes related to degradation of BST-2 or to specific removal from the cell surface? Does Vpu decrease the surface levels of BST-2 quickly enough to provide timely antagonism of BST-2 during the viral replication cycle?
HIV-2 and SIV Env
HIV-2 Env was long known to provide a Vpu-like activity, and like Vpu, it downregulates BST-2 from the cell surface.43,55,56 An intact GYxxθ motif in the cytoplasmic domain of gp41, which binds to AP-2, is required for this downregulation, whereas the ectodomain of gp41 Env provides the recognition of BST-2.43 Unlike Vpu, HIV-2 Env does not affect the intracellular level of BST-2 but rather redistributes it from the plasma membrane to a juxtanuclear compartment that overlaps with the TGN. HIV-2 Env causes BST-2 to accumulate in the TGN, even when the AP-recognizing tyrosines in the cytoplasmic domain of BST-2 (Fig. 1) are replaced with alanines.43 This is consistent with a model in which Env interacts with BST-2 and provides the AP-2 recognition signal for internalization from the cell surface. Further consistent with this model, HIV-2 Env coimmunoprecipitates with BST-243; the proteins likely form a tripartite complex with AP-2.
Similarly, SIV Env, at least from Tantalus monkeys, also antagonizes restriction by removing BST-2 from the cell surface via endosomal sequestration.44 A single amino acid substitution in the BST ectodomain, A100D, renders BST-2 unresponsive to SIV-Env, supporting a model of interaction between the ectodomains of the proteins.44
In summary, to remove BST-2 from the cell surface, Vpu and HIV-2 Env (and likely SIV Env) use two distinct effector mechanisms. Vpu links BST-2 to the ubiquitination machinery via its DpSGxxpS motif, whereas Env links BST-2 to clathrin adaptor protein complexes via its GYxxθ motif. Downregulation of BST-2 by Env provides lentiviruses lacking Vpu, such as HIV-2 and most SIVs, with the ability to overcome BST-2-mediated restriction of virion release.
SIV Nef
The Nef proteins of SIV strains, most of which lack Vpu, are yet a third class of lentiviral BST-2 antagonists.22,23 SIV Nef counteracts rhesus and sooty mangabey, but not human, BST-2. HIV-1 and HIV-2 Nef can also counteract rhesus and sooty mangabey BST-2, though less efficiently than SIV Nef.22 The mechanism of this counteraction is unclear, although like HIV-1 Vpu and HIV-2 Env, SIV Nef downregulates BST-2 from the cell surface.22 Within Nef, the myristoylation and putative cholesterol recognition sites are required for this downregulation. Some determinants in Nef for the counteraction of BST-2 overlap those required for the downregulation of CD4,23 suggesting that the modulation of BST-2 by SIV Nef may, like HIV-1 Vpu and HIV-2 Env, involve the endocytic adaptor AP-2. The specificity of lentiviral Nef proteins for the BST-2 proteins of nonhuman primates maps to a five-amino acid motif in the cytoplasmic domain of BST-2, G/DDIWK.22,23 This motif is absent in human BST-2, rendering human BST-2 nonresponsive to Nef.
KSHV K5
The Kaposi's sarcoma-associated herpesvirus (KSHV; HHV8) K5 protein is a membrane-associated ubiquitin ligase. K5 downregulates BST-2 from the surface of HeLa cells, along with other host defense molecules such as class I MHC.11,21 This viral protein directly ubiquitinates BST-2.21 This modification results in a reduction of both intracellular and surface levels of BST-2. Like Vpu, the exact mode of degradation is unclear, but the preponderance of evidence suggests an endolysosomal process.21 Only glycosylated forms of BST-2 are affected by K5, indicating a post- endoplasmic reticulum (ER) mechanism. Furthermore, inhibition of the ESCRT-mediated late endosomal pathway by expression of a dominant-negative mutant of the VPS4 ATPase restores the surface expression of BST-2 in the presence of K5. On the other hand, the proteosomal inhibitor MG132 also blocks the degradation of BST-2 by K5. Two lysines in the cytosolic tail of BST-2, K18 and K21 (Fig. 1), are required for K5-dependent ubiquitination.21
Unlike lentiviruses, herpesvirus egress includes the envelopment of capsids at the ER and TGN followed by transport to the cell surface in secretory vesicles.57 For BST-2 to restrict viral release, it would either need to capture virions as they are released from secretory vesicles or it would need to capture virions just after envelopment along the biosynthetic/secretory pathway. In the later case, K5 would need to act at the level of the ER or TGN. Notably, BST-2 is able to restrict the release of KSHV, though to a lesser degree than it restricts the release of lentiviruses.21
Ebola glycoprotein
Ebola GP counteracts BST-2 as efficiently as HIV-1 Vpu, when measured by the ability to enhance the release of virus-like particles (VLPs) from cells.31 In addition, Ebola GP may have evolved to counteract a wide variety of mammalian hosts, since its expression restores the release of Ebola VLPs in the presence of murine as well as human BST-2. (HIV-1 Vpu is minimally if at all active against murine BST-2.31,46,58) The mechanism by which Ebola GP counteracts BST-2 is unknown, although the full-length protein and proper subcellular localization are required. The intracellular level of BST-2 is unaffected by Ebola GP, but whether surface levels are affected is unclear. An interaction between BST-2 and Ebola GP has been documented by coimmunoprecipitation,31 and this interaction is likely required for the relief of restriction. One intriguing possibility is that Ebola GP physically disrupts the interaction between BST-2 and virions, for example, by disrupting the putative self-interaction of BST-2 ectodomains, but this is speculative.
In summary, a broad spectrum of viruses is restricted by BST-2, and consequently various viral proteins have evolved to counteract this host restriction factor. So far, six viral proteins have been described to antagonize BST-2 by at least two different mechanisms: ubiquitin-dependent degradation and/or trafficking (HIV-1 Vpu and KSHV K5)21,34,46 and AP-2-dependent mislocalization (HIV-2 Env and probably SIV Nef ).23,43 How Ebola GP counteracts BST-2 remains to be elucidated, but it could involve yet a third mechanism based on direct interaction with the BST-2 ectodomain and interference with the restriction mechanism.
BST-2 as a restriction factor during spreading infections in vitro
The assessment of BST-2 as a restriction factor during spreading infections of HIV-1 in vitro is complicated by the relative contribution of cell-free versus cell-associated virions to the infection of new target cells, as well as by the potential contribution of other Vpu activities such as the downregulation of CD4 to the replication rate. Historically, the replication rate of vpu-negative HIV-1 using T cell lines in vitro has appeared only mildly decreased.3,59 Such attenuation is more apparent in primary cultures of interferon-treated lymphocytes, consistent with a model of BST-2 as an effector of the antiviral state induced by interferons.10 On the other hand, the accumulation of virions on the cell surface by BST-2 has the potential to enhance the fusogenicity of infected cells to uninfected target cells, and so paradoxically to enhance the viral replication rate via facilitation of cell-to-cell spread.60 Recent data suggest that HIV-1 virions preferentially enter cells via endocytosis rather than by fusion at the plasma membrane61; if so, then BST-2 may actually inhibit cell–cell spread, but this remains to be shown. Although the degradation of BST-2 by Vpu had been reported as dispensable for a vpu phenotype in certain T cell lines,52 use of an inducible BST-2 expression system in SupT1 T cells, which lack endogenous BST-2 but express high levels of CD4, revealed a growth rate phenotype for vpu whose magnitude was directly related to the level of BST-2 expression and that correlated with the downregulation of BST-2.49
The ability of BST-2 to restrict spreading infection may depend on the extent to which virions retained at the cell surface are endocytosed and subsequently degraded. This pathway appears to account for the accumulation of virions in endosomal vesicles in the absence of Vpu9; such vesicles are largely clathrin coated and contain late endosomal proteins such as CD63 and ALIX.48 Recent data indicate that BST-2 binds directly to the late endosomal ubiquitin-ligase Rabring7.62 This interaction directs BST-2 and virions bound to it to late endosomes. Consequently, Rabring7 facilitates the net removal of nascent virions from the cell surface and their ultimate degradation in the presence of BST-2.62 This pathway contributes directly to the virion-release phenotype,62 but it may play an even greater role during spreading infection by antagonizing the cell-to-cell transfer of virions retained at the cell surface by BST-2.
Targeting the HIV-1 BST-2 antagonist protein Vpu for antiviral drug discovery
Role of the Vpu transmembrane domain (TMD)
HIV-1 Vpu is a 16-kDa, 81-amino acid, homooligomeric type I transmembrane phosphoprotein.1,2 Historically, the cytoplasmic domain of Vpu was associated with the ER-associated degradation of newly synthesized CD4, whereas the TMD was associated with the enhancement of virion release.59 As described above, the cytoplasmic and TMDs each contributes to the current model in which Vpu antagonizes the restriction of virion release by Vpu. The cytoplasmic domain contains a DSGxxS β-TrCP-binding motif required for the recruitment of a multisubunit E3 ubiquitin ligase complex.12 This interaction is important not only for the degradation of CD4 but also for the downregulation of BST-2 from the cell surface,16,34,50 presumably through ubiquitin-mediated mechanisms as noted above. The α-helical Vpu TMD can form homooligomeric cation-selective channels that are likely pentameric.63,64 Whether this ion channel activity is causally related to the enhancement of virion release is unknown, but the sequence of the TMD is important for both ion channel and virion release activities.59,63
The correct sequence of the Vpu TMD is required for the downregulation of BST-2 as well as for the enhancement of virion release.16 Vpu and BST-2 colocalize in endosomal vesicles.16,17 These data suggest that the actual function of the Vpu TMD with respect to virion release is to interact within the lipid bilayer with the TMD of BST-2. Subsequently, the BST-2 proteins of nonhuman primates (with the exception of chimpanzees) were found to be resistant to antagonism by HIV-1 Vpu, and this resistance mapped to the BST-2 TMD.22,45,49,58 These reciprocal data supported the idea of a TMD–TMD interaction between Vpu and BST-2.
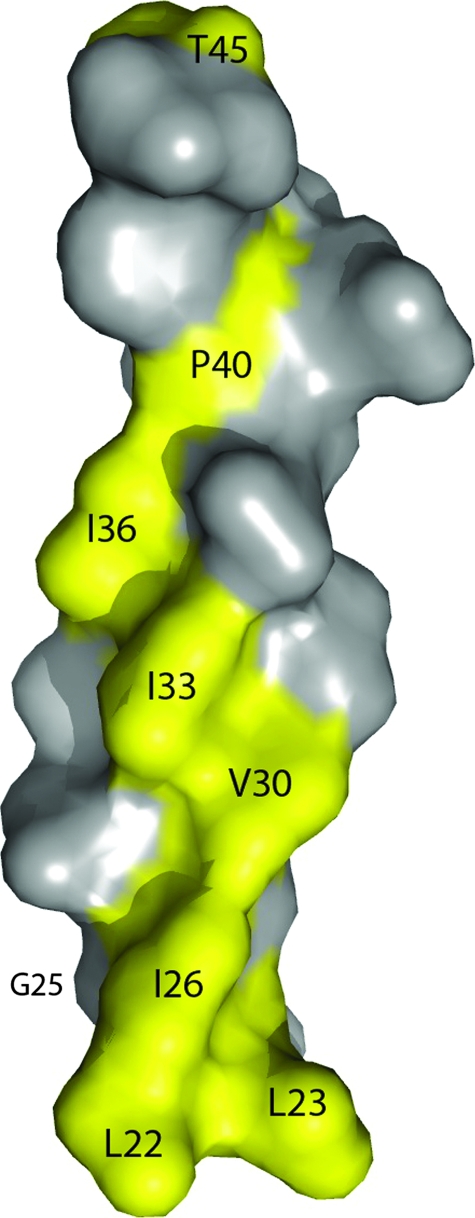
Several groups demonstrated that the TMD of BST-2 is sufficient to determine sensitivity or resistance to antagonism by Vpu.22,45,49,58 In each case, BST-2 proteins from nonhuman primates (rhesus macaque or African green monkey) were able to restrict virion release but were not counteracted by Vpu. Chimeric BST-2 proteins containing the TMD region of human BST-2 were Vpu responsive, whereas proteins containing the TMD region of rhesus or African green monkey BST-2 were Vpu resistant. No single mutation within the human TMD conferred complete resistance to Vpu.45,49,58 Instead, high-level resistance required several mutations that rendered the human protein more “monkey-like,” including deletion of two residues near the N-terminus of the TMD, as well as V30G, I33V, P40L, and T45I substitutions (Fig. 6). Two studies found evidence of positive selection within the TMD region of primate BST-2,45,58 further supporting the hypothesis that viral proteins such as Vpu have targeted this region. Strikingly, residues under positive selection generally reside on one side of the transmembrane helix of BST-2 (Fig. 6), and many of them contributed to Vpu resistance when mutated. These results suggested an interaction with Vpu along one side of the BST-2 transmembrane helix.
FIG. 6.
Residues of the BST-2 TMD potentially involved in an interaction with Vpu. Both genetic studies mapping residues under positive selection and functional studies have indicated that residues in the transmembrane domain (TMD) of human BST-2 are required for responsiveness to Vpu. Changes in these residues found in the BST-2 orthologs of nonhuman primates relative to the human protein account for the inactivity of HIV-1 Vpu against these proteins (see text for details). Remarkably, these residues are predominantly on one face of the TMD α-helix (highlighted in yellow), suggesting an interaction along that face with the TMD of Vpu. Image drawn using PyMOL.
Several groups have shown that Vpu and BST-2 interact, at least at the level of coimmunoprecipitation from human cells.49–51 In one study, the interaction was mapped at least in part to the TMD of BST-2.49 Interestingly, the interaction between Vpu and BST-2 can be stabilized by perturbations that block the activity of the Vpu–β-TrCP E3 ligase complex, such as using a Vpu mutated in its β-TrCP-binding sequence or expressing a dominant negative mutant of β-TrCP.50,51 These data substantiate a model in which ubiquitination targets BST-2 for degradation or sequestration, separating it from Vpu.
A molecular model for the interaction of BST-2 and Vpu remains to be determined. Potentially, such a model could be derived from nuclear magnetic resonance (NMR) studies conducted in a lipid environment, in which the structure of the α-helical Vpu TMD is already known.65
Approaches to the discovery of Vpu inhibitors using BST-2
The studies reviewed above suggest an interaction in which the TMD of Vpu forms heterooligomers with the TMD of BST-2. Such heterooligomerization is presumably a prerequisite for the counteraction of restriction. This relationship may be a viable target for antiviral drug design, in which synthetic transmembrane peptide decoys preferentially bind the α-helical TMD of Vpu.66 The sequestration of Vpu with peptide decoys would decrease the formation of Vpu/BST-2 heterooligomers and potentially restore restriction mediated by BST-2. This approach has been exemplified using the protein TASK-1 (TWIK related acid-sensitive K+ channel-1).67 Overexpression of the TASK-1 TMD, which binds the TMD of Vpu, inhibits Vpu-mediated enhancement of virion release. Computational analysis can be used to aid in the design of novel peptide decoys. For example, the CHAMPS (computed helical antimembrane protein) algorithm was used to predict a peptide sequence that might block the formation of Vpu/BST-2 TMD oligomers.66
In addition to enabling rational drug design, insights into the Vpu/BST-2 relationship provide new opportunities to develop high-throughput screening assays for inhibitors of HIV-1 Vpu. One possible readout is the destabilization of the interaction between BST-2 and Vpu, which could potentially be measured using a fluorescence resonance energy transfer (FRET) assay. Another approach might be the use of a flow cytometric assay to measure the Vpu-mediated downregulation of BST-2 from the cell surface. Such assays typically show a 10-fold or greater decrease in surface BST-2 levels within 18–24 h of Vpu expression.16 As noted above, the ability of Vpu to downregulate BST-2 is a close correlate of its ability to antagonize BST-2 virologically. Consequently, this assay could be used as a primary high-throughput screen, with measurement of Vpu-mediated enhancement of virion release as a secondary screen.
Role of BST-2 in the evolution of primate lentiviruses
Evidence for positive selection at specific residues of BST-2
Human and nonhuman primate BST-2 sequences are sufficiently homologous to be analyzed for evidence of positive selection. Two such analyses have revealed that certain positions in each domain of BST-2 are under positive selection; specific codons in the cytoplasmic, transmembrane, and ectodomains have high ratios of nonsynonymous to synonymous mutations.45,58 As noted above, experimental mutation at some of these sites renders human BST-2 resistant to counteraction by Vpu. These data suggest that the TMD region of BST-2 has evolved to escape viral antagonists such as Vpu. The presence of sites within the cytoplasmic and ectodomains of BST-2 that are under positive selection may similarly reflect escape from viral antagonists such as SIV Nef and KSHV K5, which target the cytoplasmic domain,21–23 and the glycoproteins of HIV-2, SIV, and Ebola virus, which presumably target the BST-2 ectodomain.31,43 Indeed, one such residue in the ectodomain of BST-2, A100, is required for antagonism of restriction by SIV Env.44
Species-specific activities of virally encoded BST-2 antagonists
Primate lentiviral antagonists are, to various extents, specific for the cognate BST-2 ortholog expressed in their hosts. For example, HIV-1 Vpu antagonizes human and chimpanzee BST-2, but not BST-2 from rhesus macaques, pigtailed macaques, or African green monkeys; this specificity maps to the transmembrane domain of BST-2.23,45,49,58 Similarly, Nefs from SIVs antagonize to various extents the BST-2 from their nonhuman primate hosts, but they do not antagonize human BST-2; this specificity maps to a short sequence in the cytoplasmic domain of BST-2 (DDIWK) that is missing in the human protein.22,23 Interestingly, SIVagm Nef appears to only minimally antagonize BST-2 from African green monkeys, potentially consistent with the lack of an inflammatory state in these infected animals and the hypothesis that the antiviral activity of BST-2 in vivo requires induction by the interferon response.53 Notably, Ebola GP antagonizes both human and murine BST-2.31 We have observed that HIV-2 Env antagonizes both human and rhesus BST-2 (unpublished data). Similarly, SIVtan Env is broadly active against human and nonhuman primate BST-2.44
The specificity of virally encoded antagonists for cognate host BST-2 presents potential barriers to cross-species transmission. Such species specificity, like that of lentiviral Vif for APOBEC3 proteins, may also complicate the design of HIV-1 variants able to replicate in nonhuman primates.68 Interestingly, one species of owl monkey (Aotus lemurinus griseimembra) encodes a BST-2 ortholog that is defective in restriction due to a single missense mutation near the C-terminus of the protein.69 This mutation causes a defect in processing consistent with a failure of the molecule to leave the ER. Whether this species is characterized by enhanced susceptibility to retroviruses or other BST-2-sensitive viruses is unknown.
The lineages of Vpu and Env as BST-2 antagonists
The vpu gene is present in HIV-1, SIVcpz, SIVgsn (found in greater spot-nosed monkeys), SIVmon (found in mona monkeys), SIVmus (found in mustached monkeys), and SIVden (found in a Dent's Mona monkey).70,71,72 SIVcpz itself appears to be a recombinant virus whose 3′ half, including vpu, derives from SIVgsn.73 If the primary function of Vpu were to relieve the restriction imposed by BST-2, then the vpu gene products from these viruses would be expected to antagonize their host's BST-2. This hypothesis has yet to be fully tested and may not be correct in all cases, because the nef or env genes in these SIVs can alternatively provide this function. So far, data addressing whether the Vpu proteins of nonhuman primate lentiviruses counteract BST-2 are limited to the reported antagonism of African green monkey BST-2 by SIVmus Vpu.53 Remarkably, BST-2 from chimpanzees, though responsive to HIV-1 Vpu,58 contains the cytoplasmic DDIWK sequence that confers responsiveness to the Nef proteins of the simian lentiviruses. Indeed, SIVcpz Vpu appears not to antagonize chimpanzee (or human) BST-2; instead, the antagonism of chimpanzee BST-2 is provided by SIVcpz Nef (P. Cannon, personal communication).
Why should the lentiviruses of nonhuman primates, including SIVcpz, have acquired a vpu gene when their nef genes may encode a BST-2 antagonist? One answer is that vpu provides activities in addition to antagonizing BST-2, such as the degradation of CD4 or other host molecules. The attenuated phenotype of vpu mutant pathogenic SHIVs in pigtailed macaques, whose BST-2 appears resistant to HIV-1 Vpu, supports this possibility.74 This alternative hypothesis suggests that BST-2 antagonist activity is relatively unique to the Vpu proteins encoded by HIV-1, and is specifically required because human BST-2, unlike that of all other examined nonhuman primates, lacks the sequence targeted by Nef in its cytoplasmic domain.
Similarly, why HIV-1 antagonizes human BST-2 via Vpu, when the HIV-2 or SIV envelope glycoprotein can accomplish the same task, is also unclear. A clue may be obtained from the observation that the lack of activity of at least one HIV-2 Env protein maps to the ectodomain of gp41.75 Potentially, selective forces on the envelope ectodomain, such as could be imposed by neutralizing antibodies, may have favored the acquisition of vpu as an alternative antagonist of BST-2.
Overall, although the lineage of vpu as a BST-2 antagonist remains an open question, it seems conceivable if not likely that the acquisition of this activity provided relief of restricted replication by human BST-2 to HIV-1 strains, while freeing the env and nef genes from a functional constraint. If so, then this event may have played a key role in the emergence of pathogenic HIV-1 strains in humans.
Additional roles of BST-2 in the regulation of the immune response
BST-2 is likely to play as yet undefined roles in the innate immune response beyond its function as a direct antiviral restriction factor. Indeed, the antiviral activity of “artificial tetherin” weighs in favor of this conclusion, as this molecule lacks conserved features of native BST-2, such as the glycosylation sites in the ectodomain, yet is highly active in restriction.36 What, then, can be the reason for evolutionary conservation of such sites in the native protein? The obvious answer is that they serve important host functions beyond direct restriction of virion release. What these functions are and whether they are sufficient to induce the evolution of viral antagonists remain to be fully evaluated.
BST-2 as a specific ligand for ILT7: negative feedback for interferon production by plasmacytoid dendritic cells (PDCs)
BST-2 clearly functions as an interferon-induced restriction factor and as such is an effector protein of the innate immune response. Remarkably, recent data revealed a distinct function for BST-2: it is also a negative feedback regulator of interferon production by PDCs.76 BST-2 binds the ILT7 receptor on PDCs, transducing a signal that inhibits the release of interferons.76 This role of BST-2 has important implications for the shut-off of the interferon response, and it provokes the intriguing idea that virion-associated BST-2 could play a role in such a negative feedback loop.
Is BST-2 an uptake receptor on antigen-presenting cells?
Another intriguing possibility is that BST-2 could play a role as an attachment receptor for virions, especially on antigen-presenting cells. This idea, though highly speculative, is supported by the ectodomain self-interaction model of restriction discussed above, because it incorporates the binding of virions to cells via BST-2/BST-2 interactions.77 It is also supported by the documented constitutive expression of BST-2 on B cells, PDCs, and macrophages.13,14
A remarkable possibility is that the infected cell “labels” virions with BST-2 for a dual purpose, not only to inhibit their release, but also to “flag” them for uptake into antigen-presenting cells, while simultaneously enabling them to shut off the interferon response. These combined functions would place BST-2 at the interface of the innate and adaptive immune response to enveloped viruses.
Conclusion
The discovery of the antiviral activity of BST-2 exemplifies how the study of a pathogen and its encoded proteins, in this case HIV-1 and Vpu, can uncover novel aspects of the host defense against infection. Though cloned over 10 years ago, BST-2 has been recognized as a protein targeted by viruses for just over 3 years, and the discovery of its novel ability to restrict the release of budding virions is nearing only its second anniversary. Yet, in this short time, the known spectrum of action of BST-2 has expanded to include not only HIV-1 but almost all retroviruses tested, as well as members of the filovirus (Ebola and Marburg), arenavirus (Lassa), and herpesvirus (KSHV) families. Each of these viruses (with the current exceptions of Marburg and Lassa) encodes specific antagonists of BST-2.
A frequent feature of this antagonism, described for HIV-1 Vpu, HIV-2 and SIV Env, SIV Nef, and KSHV K5, is the removal of BST-2 from the cell surface, the presumed site of the protein's action as a tetherin. The diversifying action of positive selection on specific residues in primate BST-2 and the specificity of primate lentiviral antagonist proteins for cognate host BST-2 suggest that the protein has contributed to the evolution of these viruses and has presented barriers to cross-species transmission. Remarkably, the current pandemic HIV-1 strains all encode the BST-2 antagonist Vpu, as does SIVcpz, from which these strains were derived. The alias “tetherin” aptly evokes the ability of BST-2 to directly retain newly formed virions at the cell surface, but it does not capture the full scope of the protein's function: in addition to restricting virion release, BST-2 mediates a specific feedback mechanism to turn off interferon production by plasmacytoid dendritic cells.
Many questions remain open regarding the complete role of BST-2 in the innate immune response to viruses, its mechanism of action as a restriction factor, and the mechanisms by which it is counteracted by viral antagonists such as Vpu. Answering these questions may enable the design of novel strategies for the prevention and treatment of infections due to enveloped viruses, including HIV-1.
Acknowledgments
We thank Thomas Deerinck, John Crum, and Mark Ellisman of the National Center for Microscopy and Imaging Research at UCSD (NIH Grant RP41004050) for collaboration on the electron microscopy and Nanette Wong for technical assistance. This work was supported by NIH Grant AI081668 to J.G. Mark Skasko was supported by AIDS Training Grant T32AI007384. Kathleen Fitzpatrick is in the Biomedical Sciences Graduate Program at UCSD. The authors dedicate this review to the memory of Richard S. Mitchell.
Disclosure Statement
No financial interests exist.
References
- 1.Strebel K. Klimkait T. Martin MA. A novel gene of HIV-1, vpu, and its 16-kilodalton product. Science. 1988;241:1221–1223. doi: 10.1126/science.3261888. [DOI] [PubMed] [Google Scholar]
- 2.Cohen EA. Terwilliger EF. Sodroski JG. Haseltine WA. Identification of a protein encoded by the vpu gene of HIV-1. Nature. 1988;334:532–534. doi: 10.1038/334532a0. [DOI] [PubMed] [Google Scholar]
- 3.Klimkait T. Strebel K. Hoggan MD. Martin MA. Orenstein JM. The human immunodeficiency virus type 1-specific protein vpu is required for efficient virus maturation and release. J Virol. 1990;64(2):621–629. doi: 10.1128/jvi.64.2.621-629.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sakai H. Tokunaga K. Kawamura M. Adachi A. Function of human immunodeficiency virus type 1 Vpu protein in various cell types. J Gen Virol. 1995;76(Pt 11):2717–2722. doi: 10.1099/0022-1317-76-11-2717. [DOI] [PubMed] [Google Scholar]
- 5.Yasuda Y. Miyake S. Kato S, et al. Interferon-alpha treatment leads to accumulation of virus particles on the surface of cells persistently infected with the human immunodeficiency virus type 1. J Acquir Immune Defic Syndr. 1990;3(11):1046–1051. [PubMed] [Google Scholar]
- 6.Dianzani F. Castilletti C. Gentile M, et al. Effects of IFN alpha on late stages of HIV-1 replication cycle. Biochimie. 1998;80(8–9):745–754. doi: 10.1016/s0300-9084(99)80028-5. [DOI] [PubMed] [Google Scholar]
- 7.Gottlinger HG. Dorfman T. Cohen EA. Haseltine WA. Vpu protein of human immunodeficiency virus type 1 enhances the release of capsids produced by gag gene constructs of widely divergent retroviruses. Proc Natl Acad Sci USA. 1993;90(15):7381–7385. doi: 10.1073/pnas.90.15.7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Varthakavi V. Smith RM. Bour SP. Strebel K. Spearman P. Viral protein U counteracts a human host cell restriction that inhibits HIV-1 particle production. Proc Natl Acad Sci USA. 2003;100(25):15154–15159. doi: 10.1073/pnas.2433165100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neil SJ. Eastman SW. Jouvenet N. Bieniasz PD. HIV-1 Vpu promotes release and prevents endocytosis of nascent retrovirus particles from the plasma membrane. PLoS Pathog. 2006;2(5):e39. doi: 10.1371/journal.ppat.0020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neil SJ. Sandrin V. Sundquist WI. Bieniasz PD. An interferon-alpha-induced tethering mechanism inhibits HIV-1 and Ebola virus particle release but is counteracted by the HIV-1 Vpu protein. Cell Host Microbe. 2007;2(3):193–203. doi: 10.1016/j.chom.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartee E. McCormack A. Fruh K. Quantitative membrane proteomics reveals new cellular targets of viral immune modulators. PLoS Pathog. 2006;2(10):e107. doi: 10.1371/journal.ppat.0020107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Margottin F. Bour SP. Durand H, et al. A novel human WD protein, h-beta TrCp, that interacts with HIV-1 Vpu connects CD4 to the ER degradation pathway through an F-box motif. Mol Cell. 1998;1(4):565–574. doi: 10.1016/s1097-2765(00)80056-8. [DOI] [PubMed] [Google Scholar]
- 13.Blasius AL. Giurisato E. Cella M, et al. Bone marrow stromal cell antigen 2 is a specific marker of type I IFN-producing cells in the naive mouse, but a promiscuous cell surface antigen following IFN stimulation. J Immunol. 2006;177(5):3260–3265. doi: 10.4049/jimmunol.177.5.3260. [DOI] [PubMed] [Google Scholar]
- 14.Vidal-Laliena M. Romero X. March S, et al. Characterization of antibodies submitted to the B cell section of the 8th Human Leukocyte Differentiation Antigens Workshop by flow cytometry and immunohistochemistry. Cell Immunol. 2005;236(1–2):6–16. doi: 10.1016/j.cellimm.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 15.Kupzig S. Korolchuk V. Rollason R, et al. Bst-2/HM1.24 is a raft-associated apical membrane protein with an unusual topology. Traffic. 2003;4(10):694–709. doi: 10.1034/j.1600-0854.2003.00129.x. [DOI] [PubMed] [Google Scholar]
- 16.Van Damme N. Goff D. Katsura C, et al. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe. 2008;3(4):245–252. doi: 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neil SJ. Zang T. Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451(7177):425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 18.Ohtomo T. Sugamata Y. Ozaki Y, et al. Molecular cloning and characterization of a surface antigen preferentially overexpressed on multiple myeloma cells. Biochem Biophys Res Commun. 1999;258(3):583–591. doi: 10.1006/bbrc.1999.0683. [DOI] [PubMed] [Google Scholar]
- 19.Rollason R. Korolchuk V. Hamilton C. Schu P. Banting G. Clathrin-mediated endocytosis of a lipid-raft-associated protein is mediated through a dual tyrosine motif. J Cell Sci. 2007;120(Pt 21):3850–3858. doi: 10.1242/jcs.003343. [DOI] [PubMed] [Google Scholar]
- 20.Masuyama N. Kuronita T. Tanaka R, et al. HM1.24 is internalized from lipid rafts by clathrin-mediated endocytosis through interaction with alpha-adaptin. J Biol Chem. 2009;284(23):15927–15941. doi: 10.1074/jbc.M109.005124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mansouri M. Viswanathan K. Douglas JL, et al. Molecular mechanism of BST2/tetherin downregulation by K5/MIR2 of Kaposi's sarcoma-associated herpesvirus. J Virol. 2009;83(19):9672–9681. doi: 10.1128/JVI.00597-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jia B. Serra-Moreno R. Neidermyer W, et al. Species-specific activity of SIV Nef and HIV-1 Vpu in overcoming restriction by tetherin/BST2. PLoS Pathog. 2009;5(5):e1000429. doi: 10.1371/journal.ppat.1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang F. Wilson SJ. Landford WC, et al. Nef proteins from simian immunodeficiency viruses are tetherin antagonists. Cell Host Microbe. 2009;6(1):54–67. doi: 10.1016/j.chom.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dube M. Roy BB. Guiot-Guillain P, et al. Suppression of tetherin-restricting activity upon human immunodeficiency virus type 1 particle release correlates with localization of Vpu in the trans-Golgi network. J Virol. 2009;83(9):4574–4590. doi: 10.1128/JVI.01800-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aloia RC. Tian H. Jensen FC. Lipid composition and fluidity of the human immunodeficiency virus envelope and host cell plasma membranes. Proc Natl Acad Sci USA. 1993;90(11):5181–5185. doi: 10.1073/pnas.90.11.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nguyen DH. Hildreth JEK. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J Virol. 2000;74:3264–3272. doi: 10.1128/jvi.74.7.3264-3272.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Panchal RG. Ruthel G. Kenny TA, et al. In vivo oligomerization and raft localization of Ebola virus protein VP40 during vesicular budding. Proc Natl Acad Sci USA. 2003;100(26):15936–15941. doi: 10.1073/pnas.2533915100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakuma T. Noda T. Urata S. Kawaoka Y. Yasuda J. Inhibition of Lassa and Marburg virus production by tetherin. J Virol. 2009;83(5):2382–2385. doi: 10.1128/JVI.01607-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andrew AJ. Miyagi E. Kao S. Strebel K. The formation of cysteine-linked dimers of BST-2/tetherin is important for inhibition of HIV-1 virus release but not for sensitivity to Vpu. Retrovirology. 2009;6(1):80. doi: 10.1186/1742-4690-6-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakuma T. Sakurai A. Yasuda J. Dimerization of tetherin is not essential for its antiviral activity against Lassa and Marburg viruses. PLoS One. 2009;4(9):e6934. doi: 10.1371/journal.pone.0006934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaletsky RL. Francica JR. grawal-Gamse C. Bates P. Tetherin-mediated restriction of filovirus budding is antagonized by the Ebola glycoprotein. Proc Natl Acad Sci U S A. 2009;106(8):2886–2891. doi: 10.1073/pnas.0811014106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakuma T. Noda T. Urata S. Kawaoka Y. Yasuda J. Inhibition of Lassa and Marburg virus production by tetherin. J Virol. 2008;83(5):2382–2385. doi: 10.1128/JVI.01607-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jouvenet N. Neil SJ. Zhadina M, et al. Broad-spectrum inhibition of retroviral and filoviral particle release by tetherin. J Virol. 2009;83(4):1837–1844. doi: 10.1128/JVI.02211-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitchell RS. Katsura C. Skasko MA, et al. Vpu antagonizes BST-2-mediated restriction of HIV-1 release via beta-TrCP and endo-lysosomal trafficking. PLoS Pathog. 2009;5(5):e1000450. doi: 10.1371/journal.ppat.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fitzpatrick K. Skasko M. Deerinck T, et al. Direct restriction of virus release, incorporation of the interferon-induced protein BST-2 into HIV-1 particles. PLoS Pathog. 2009 doi: 10.1371/journal.ppat.1000701. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perez-Caballero D. Zang T. Ebrahimi A, et al. Tetherin inhibits HIV-1 release by directly tethering virions to cells. Cell. 2009;139:499–511. doi: 10.1016/j.cell.2009.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sato K. Yamamoto SP. Misawa N, et al. Comparative study on the effect of human BST-2/tetherin on HIV-1 release in cells of various species. Retrovirology. 2009;6:53. doi: 10.1186/1742-4690-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malim MH. Emerman M. HIV-1 accessory proteins—ensuring viral survival in a hostile environment. Cell Host Microbe. 2008;3(6):388–398. doi: 10.1016/j.chom.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 39.Stremlau M. Owens CM. Perron MJ, et al. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427(6977):848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 40.Yu X. Yu Y. Liu B, et al. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science. 2003;302(5647):1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- 41.Mehle A. Strack B. Ancuta P, et al. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J Biol Chem. 2004;279(9):7792–7798. doi: 10.1074/jbc.M313093200. [DOI] [PubMed] [Google Scholar]
- 42.Barry M. Fruh K. Viral modulators of cullin RING ubiquitin ligases: Culling the host defense. Sci STKE. 2006;2006(335):e21. doi: 10.1126/stke.3352006pe21. [DOI] [PubMed] [Google Scholar]
- 43.Le Tortorec A. Neil SJ. Antagonism and intracellular sequestration of human tetherin by the HIV-2 envelope glycoprotein. J Virol. 2009;83:11966–11978. doi: 10.1128/JVI.01515-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gupta RK. Mlcochova P. Pelchen-Matthews A, et al. Simian immunodeficiency virus envelope glycoprotein counteracts tetherin/BST-2/CD317 by intracellular sequestration. Proc Natl Acad Sci USA. 2009 doi: 10.1073/pnas.0907075106. epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gupta RK. Hue S. Schaller T, et al. Mutation of a single residue renders human tetherin resistant to HIV-1 Vpu-mediated depletion. PLoS Pathog. 2009;5(5):e1000443. doi: 10.1371/journal.ppat.1000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goffinet C. Allespach I. Homann S, et al. HIV-1 antagonism of CD317 is species specific and involves Vpu-mediated proteasomal degradation of the restriction factor. Cell Host Microbe. 2009;5(3):285–297. doi: 10.1016/j.chom.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 47.Varthakavi V. Smith RM. Martin KL, et al. The pericentriolar recycling endosome plays a key role in Vpu-mediated enhancement of HIV-1 particle release. Traffic. 2006;7(3):298–307. doi: 10.1111/j.1600-0854.2005.00380.x. [DOI] [PubMed] [Google Scholar]
- 48.Van Damme N. Guatelli J. HIV-1 Vpu inhibits accumulation of the envelope glycoprotein within clathrin-coated, Gag-containing endosomes. Cell Microbiol. 2007;10:1040–1057. doi: 10.1111/j.1462-5822.2007.01101.x. [DOI] [PubMed] [Google Scholar]
- 49.Rong L. Zhang J. Lu J, et al. The transmembrane domain of BST-2 determines its sensitivity to down-modulation by human immunodeficiency virus type 1 Vpu. J Virol. 2009;83(15):7536–7546. doi: 10.1128/JVI.00620-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Douglas JL. Viswanathan K. McCarroll MN, et al. Vpu directs the degradation of the human immunodeficiency virus restriction factor BST-2/tetherin via a {beta}TrCP-dependent mechanism. J Virol. 2009;83(16):7931–7947. doi: 10.1128/JVI.00242-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mangeat B. Gers-Huber G. Lehmann M, et al. HIV-1 Vpu neutralizes the antiviral factor tetherin/BST-2 by binding it and directing its beta-TrCP2-dependent degradation. PLoS Pathog. 2009;5(9):e1000574. doi: 10.1371/journal.ppat.1000574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miyagi E. Andrew AJ. Kao S. Strebel K. Vpu enhances HIV-1 virus release in the absence of Bst-2 cell surface down-modulation and intracellular depletion. Proc Natl Acad Sci USA. 2009;106(8):2868–2873. doi: 10.1073/pnas.0813223106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lim E. Emerman M. Simian immunodeficiency virus from African green monkeys (SIVagm) does not antagonize endogenous levels of African green monkey tetherin/BST-2. J Virol. 2009;83(22):11673–11681. doi: 10.1128/JVI.00569-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iwabu Y. Fujita H. Kinomoto M, et al. HIV-1 accessory protein Vpu internalizes cell-surface BST-2/tetherin through transmembrane interactions leading to lysosomes. J Biol Chem. 2009 doi: 10.1074/jbc.M109.058305. epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bour S. Schubert U. Peden K. Strebel K. The envelope glycoprotein of human immunodeficiency virus type 2 enhances viral particle release: A Vpu-like factor? J Virol. 1996;70(2):820–829. doi: 10.1128/jvi.70.2.820-829.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abada P. Noble B. Cannon PM. Functional domains within the human immunodeficiency virus type 2 envelope protein required to enhance virus production. J Virol. 2005;79(6):3627–3638. doi: 10.1128/JVI.79.6.3627-3638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mettenleiter TC. Klupp BG. Granzow H. Herpesvirus assembly: An update. Virus Res. 2009;143(2):222–234. doi: 10.1016/j.virusres.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 58.McNatt MW. Zang T. Hatziioannou T, et al. Species-specific activity of HIV-1 Vpu and positive selection of tetherin transmembrane domain variants. PLoS Pathog. 2009;5(2):e1000300. doi: 10.1371/journal.ppat.1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schubert U. Bour S. Ferrer-Montiel AV, et al. The two biological activities of human immunodeficiency virus type 1 Vpu protein involve two separable structural domains. J Virol. 1996;70(2):809–819. doi: 10.1128/jvi.70.2.809-819.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gummuluru S. Kinsey CM. Emerman M. An in vitro rapid-turnover assay for human immunodeficiency virus type 1 replication selects for cell-to-cell spread of virus. J Virol. 2000;74(23):10882–10891. doi: 10.1128/jvi.74.23.10882-10891.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miyauchi K. Kim Y. Latinovic O. Morozov V. Melikyan GB. HIV enters cells via endocytosis, dynamin-dependent fusion with endosomes. Cell. 2009;137(3):433–444. doi: 10.1016/j.cell.2009.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miyakawa K. Ryo A. Murakami T, et al. BCA2/Rabring7 promotes tetherin-dependent HIV-1 restriction. PLoS Pathog. 2009 doi: 10.1371/journal.ppat.1000700. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schubert U. Ferrer-Montiel AV. Oblatt-Montal M, et al. Identification of an ion channel activity of the Vpu transmembrane domain and its involvement in the regulation of virus release from HIV-1-infected cells. FEBS Lett. 1996;398(1):12–18. doi: 10.1016/s0014-5793(96)01146-5. [DOI] [PubMed] [Google Scholar]
- 64.Lopez CF. Montal M. Blasie JK. Klein ML. Moore PB. Molecular dynamics investigation of membrane-bound bundles of the channel-forming transmembrane domain of viral protein U from the human immunodeficiency virus HIV-1. Biophys J. 2002;83(3):1259–1267. doi: 10.1016/S0006-3495(02)73898-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Park SH. De Angelis AA. Nevzorov AA. Wu CH. Opella SJ. Three-dimensional structure of the transmembrane domain of Vpu from HIV-1 in aligned phospholipid bicelles. Biophys J. 2006;91(8):3032–3042. doi: 10.1529/biophysj.106.087106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Montal M. Vpu matchmakers as a therapeutic strategy for HIV infection. PLoS Pathog. 2009;5(5):e1000246. doi: 10.1371/journal.ppat.1000246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hsu K. Seharaseyon J. Dong P. Bour S. Marban E. Mutual functional destruction of HIV-1 Vpu and host TASK-1 channel. Mol Cell. 2004;14(2):259–267. doi: 10.1016/s1097-2765(04)00183-2. [DOI] [PubMed] [Google Scholar]
- 68.Hatziioannou T. Ambrose Z. Chung NP, et al. A macaque model of HIV-1 infection. Proc Natl Acad Sci USA. 2009;106(11):4425–4429. doi: 10.1073/pnas.0812587106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wong SK. Connole M. Sullivan JS, et al. A New World primate deficient in tetherin-mediated restriction of human immunodeficiency virus type 1. J Virol. 2009;83(17):8771–8780. doi: 10.1128/JVI.00112-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barlow KL. Ajao AO. Clewley JP. Characterization of a novel simian immunodeficiency virus (SIVmonNG1) genome sequence from a mona monkey (Cercopithecus mona) J Virol. 2003;77(12):6879–6888. doi: 10.1128/JVI.77.12.6879-6888.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Courgnaud V. Salemi M. Pourrut X, et al. Characterization of a novel simian immunodeficiency virus with a vpu gene from greater spot-nosed monkeys (Cercopithecus nictitans) provides new insights into simian/human immunodeficiency virus phylogeny. J Virol. 2002;76(16):8298–8309. doi: 10.1128/JVI.76.16.8298-8309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dazza MC. Ekwalanga M. Nende M, et al. Characterization of a novel vpu-harboring simian immunodeficiency virus from a Dent's Mona monkey (Cercopithecus mona denti) J Virol. 2005;79(13):8560–8571. doi: 10.1128/JVI.79.13.8560-8571.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bailes E. Gao F. Bibollet-Ruche F, et al. Hybrid origin of SIV in chimpanzees. Science. 2003;300(5626):1713. doi: 10.1126/science.1080657. [DOI] [PubMed] [Google Scholar]
- 74.Stephens EB. McCormick C. Pacyniak E, et al. Deletion of the vpu sequences prior to the Env in a simian-human immunodeficiency virus results in enhanced Env precursor synthesis but is less pathogenic for pig-tailed macaques. Virology. 2002;293(2):252–261. doi: 10.1006/viro.2001.1244. [DOI] [PubMed] [Google Scholar]
- 75.Bour S. Akari H. Miyagi E. Strebel K. Naturally occurring amino acid substitutions in the HIV-2 ROD envelope glycoprotein regulate its ability to augment viral particle release. Virology. 2003;309(1):85–98. doi: 10.1016/s0042-6822(02)00128-9. [DOI] [PubMed] [Google Scholar]
- 76.Cao W. Bover L. Cho M, et al. Regulation of TLR7/9 responses in plasmacytoid dendritic cells by BST2 and ILT7 receptor interaction. J Exp Med. 2009;206(7):1603–1614. doi: 10.1084/jem.20090547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Karplus K. Karchin R. Draper J, et al. Combining local-structure, fold-recognition, and new fold methods for protein structure prediction. Proteins. 2003;53(Suppl 6):491–496. doi: 10.1002/prot.10540. [DOI] [PubMed] [Google Scholar]