Abstract
Solid-state NMR spectroscopy gives a powerful avenue for investigating G protein-coupled receptors and other integral membrane proteins in a native-like environment. This article reviews the use of solid-state 2H NMR to study the retinal cofactor of rhodopsin in the dark state as well as the meta I and meta II photointermediates. Site-specific 2H NMR labels have been introduced into three regions (methyl groups) of retinal that are crucially important for the photochemical function of rhodopsin. Despite its phenomenal stability 2H NMR spectroscopy indicates retinal undergoes rapid fluctuations within the protein binding cavity. The spectral lineshapes reveal the methyl groups spin rapidly about their three-fold (C3) axes with an order parameter for the off-axial motion of SC3 ≈ 0.9. For the dark state, the 2H NMR structure of 11-cis-retinal manifests torsional twisting of both the polyene chain and the β-ionone ring due to steric interactions of the ligand and the protein. Retinal is accommodated within the rhodopsin binding pocket with a negative pretwist about the C11=C12 double bond. Conformational distortion explains its rapid photochemistry and reveals the trajectory of the 11-cis to trans isomerization. In addition, 2H NMR has been applied to study the retinylidene dynamics in the dark and light-activated states. Upon isomerization there are drastic changes in the mobility of all three methyl groups. The relaxation data support an activation mechanism whereby the β-ionone ring of retinal stays in nearly the same environment, without a large displacement of the ligand. Interactions of the β-ionone ring and the retinylidene Schiff base with the protein transmit the force of the retinal isomerization. Solid-state 2H NMR thus provides information about the flow of energy that triggers changes in hydrogen-bonding networks and helix movements in the activation mechanism of the photoreceptor.
INTRODUCTION
Currently, receptors, transporters and ion channels are the focus of intense efforts to establish the structures and actions of membrane proteins (1–14). Aside from their fundamental significance these classes of proteins constitute particularly important therapeutic targets (15,16). Knowledge of the mechanism of receptor activation is a prime objective in the field of biological signal transduction. Yet structural knowledge is not always forthcoming for membrane proteins, as they are often refractory to crystallization. Another important aspect is that protein dynamics along the reaction coordinate may be implicated in their mechanisms of action. In this context solid-state NMR spectroscopy can reveal structural and dynamical knowledge for biological systems that are largely inaccessible to other biophysical or biochemical methods. Solid-state NMR is becoming recognized as complementary to X-ray crystallography in that local conformation changes in proteins can be probed in a native-like environment. For membrane proteins information is obtained about the structure and dynamics of bound cofactors, as well as the polypeptide backbone and amino acid side chains (16–28). An added feature is that the membrane lipid bilayer can also be investigated (29,30). It is known that lipids significantly affect key biological activities in the case of the visual receptor rhodopsin and other proteins (31–34).
This review describes studies of rhodopsin as a prototype for the cellular G protein-coupled receptors (GPCRs) implicated in many biological responses in humans (35). Application of solid-state 2H NMR spectroscopy provides information about interactions involved in receptor activation that is unobtainable from crystallography or other biophysical methods. Following light absorption, the molecular rearrangements within retinal govern the transition from the inactive dark state to the preactive meta I state occurring within a few milliseconds. Effectively this is the time it takes retinal to relax and convey its photon-encoded information to the rhodopsin binding pocket, after which the fully activated meta II state is formed. Although the mechanism of rhodopsin activation remains unknown, interactions that can affect the conformation of retinal include the protonated Schiff base (PSB) with its carboxylate counterion, the C9-methyl group of the polyene chain and the β-ionone ring within its hydrophobic binding pocket. Interestingly, the retinylidene methyl groups may play a crucial role in rhodopsin function, e.g. by directing conformational changes upon transition into the active state.
Rhodopsin activation has been explored at an atomistic level by conducting 2H NMR studies of retinal in which the methyl groups are site-specifically labeled with deuterium. Each of the three sites investigated correspond to regions of the ligand known to be crucial for receptor function (36). The specific hypothesis tested is that dihedral twisting and fluctuations of the retinal cofactor of rhodopsin are tightly coupled to light activation of the receptor. This work has addressed the following questions. (1) How can solid-state NMR give information pertinent to the dynamical structure of cofactors bound to integral membrane proteins? (2) Are there site-specific variations in local dynamics of the retinal cofactor of rhodopsin that explain its activation mechanism? (3) What are the interactions of retinal with rhodopsin that change with light activation of the photoreceptor? To address such questions, planar-supported fully hydrated membranes were investigated versus the tilt angle to the magnetic field. The oriented membranes involve substantial alignment disorder, and thus a proper treatment of the mosaic spread is vital to determining accurate bond angle restraints. Simulation of the solid-state 2H NMR lineshapes gives both the methyl group orientations and the alignment disorder (mosaic spread) of the membrane stack. In this way the retinal structure within the protein binding pocket is established in the different intermediates. Most interestingly, the 2H NMR structure of 11-cis retinal in the dark state shows torsional twisting involving both the polyene chain and the β-ionone ring. For rhodopsin the retinal cofactor is locked within the binding pocket with a negative pretwist about the C11=C12 double bond. This explains both the dark-state stability as well as its rapid photochemistry, and indicates the trajectory of the light-induced 11-cis to trans isomerization. Further, solid-state 2H NMR relaxation studies show striking influences of the protein on the site-specific methyl dynamics. Little change is evident for the activated meta II state versus the meta I state, indicating the major structural relaxation of the retinal cofactor has occurred already at the meta I intermediate in the activation mechanism. These results suggest the β-ionone ring and the Schiff base end of the ligand transmit the force of retinal isomerization to yield movement of the helices that accompany receptor activation (37,38).
SOLID-STATE NMR SPECTROSCOPY OF MEMBRANE PROTEINS
How can solid-state NMR provide information of biological relevance to the activation mechanism of rhodopsin and other membrane receptors? Here angular and positional constraints are pertinent to biomolecular structure and dynamics (39). Knowledge of connectivities of the functional groups (40,41) and interatomic distances (42,43) provide important structural restraints for noncrystalline biomolecular systems. Applications include membrane proteins and peptides (16,21,23,25–27,44–49), biopolymer fibers (50,51) and amyloid fibrils (52–54). In solid-state NMR, information about biomolecular dynamics is obtained from order parameters (55) and relaxation rates (56) as model-free experimental observables. It is distinguished from solution NMR by the absence of unrestricted tumbling of the molecules, meaning that investigations into molecular solids, liquid crystals, membranes, and protein aggregates are all possible (16,41). One should recognize that solid-state NMR spectroscopy does not imply that actual solid samples are studied. Rather, the anisotropies of the magnetic or electrical interactions are not completely averaged away by the fluctuations, as in solid-like or liquid-crystalline samples with considerable internal mobility; or conversely, liquid-like samples that can have a local structure. It follows that NMR provides a rich source of information about biomolecular structure and dynamics that can be used in concert with other complementary approaches (such as X-ray crystallography) (41,57).
To continue further, solid-state NMR spectra manifest the dipolar, chemical shift, or quadrupolar interactions of the atomic nuclei of the molecules. The dipolar interaction represents the direct or through-space coupling between the magnetic moments of nuclear spins that are geometrically in close proximity. The chemical shift is due to shielding of the magnetic moment of a nucleus from the applied magnetic field, and arises from the orbital motions of the electrons within a molecule. Finally, the quadrupolar interaction originates from coupling of the quadrupole moment of a nonspherically symmetric nucleus with the local electric field gradient of its surroundings. An example is 2H NMR where the electric field gradient of the chemical bond interacts with the quadrupole moment of a 2H nucleus, as in the present application to rhodopsin. The dipolar, chemical shift, or quadrupolar interactions are generally formulated in terms of nuclear spin interaction tensors that describe either the interatomic distances, or the orientations of the functional groups of the molecules relative to an appropriate axis system. Distances between nuclear spins are estimated from dipolar couplings in either random or aligned samples. However, typically studies of unaligned samples (as in magic-angle spinning [MAS]) do not yield direct knowledge of the tensor orientations with respect to the membrane. Rather, for dipolar interactions the tensor principal values correspond to distance restraints, or in the case of rigid molecules the distances themselves. Alternatively one can obtain angular restraints by studying macroscopically aligned samples in analogy with X-ray crystallography.
Solid-state NMR of aligned biomolecules thus enables one to investigate the orientations of the coupling tensors (dipolar, chemical shift, quadrupolar) with respect to the alignment axis. For biomembranes the constituents do not undergo overall isotropic reorientation, in contrast to proteins in solution or detergent micelles. As a result, solid-state NMR spectra of aligned membranes can provide the bond orientations directly, e.g. the C–2H bonds of proteins, cofactors, or the membrane lipid bilayer itself. By investigating different 2H-labeled positions, the relative tensor orientations can be determined, which allows one to determine the torsional (dihedral) angles about the various bonds of interest. When mobility is present the residual quadrupolar couplings (RQCs) are measured without the introduction of a motional model, which are related to the segmental order parameters and to the average molecular structure. Finally, the 2H nuclear spin relaxation detects internal dynamics or mobility in the case of either the membrane proteins or membrane lipids (58). The nuclear spin relaxation rates, e.g. the relaxation of Zeeman order (R1Z) or quadrupolar order (R1Q) (29,30) are obtained as model-free observables. The dynamical parameters involve the spectral densities of motion, which depend on correlation times, and can be expressed in terms of a pre-exponential factor (for mobility within a potential well), together with the corresponding barrier height (activation energy Ea). In this way, information is obtained about both the structural properties and dynamics of the membrane constituents using a combination of 2H NMR spectral measurements and relaxation methods.
DEUTERIUM NMR IS USED TO STUDY RHODOPSIN IN A NATIVE-LIKE MEMBRANE ENVIRONMENT
For rhodopsin (20,59,60) organic synthesis (36,61–63) has been applied extensively to 2H and 13C labeling of retinal for solid-state NMR applications. Using this strategy one can apply 2H NMR to integral membrane proteins, e.g. channels, transporters or receptors. Retinal was site-specifically labeled by deuteration of the methyl groups followed by regeneration of the apoprotein. 2H NMR studies of aligned membrane samples were conducted under conditions where rotational and translational diffusion of the protein were absent on the NMR time scale (20,64). Hence, the focus is mainly on the equilibrium structures as investigated by solid-state 2H NMR lineshapes. For 2H NMR experiments with rhodopsin, the different methyl groups of the cofactor are specifically deuterated (Fig. 1). This isotopic labeling is an isomorphic replacement that does not change the chemistry of the photoisomerization. The 2H NMR method allows one to study the orientation of a single labeled group in an oriented sample. It can be used to study local structural changes and is complementary to X-ray diffraction (10,11,65,66).
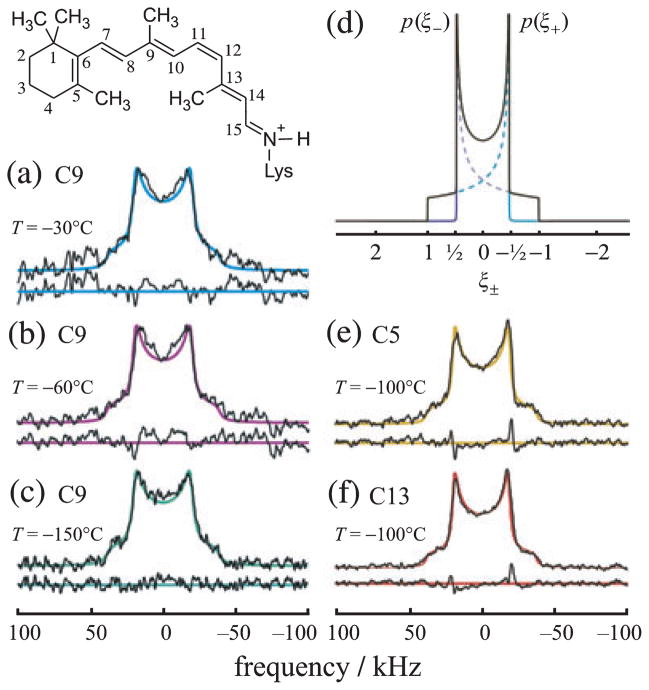
Figure 1.
Powder-type 2H NMR spectra for rhodopsin with 2Hlabeled retinal in the dark state indicate rotating methyl groups with large order parameters. (a–c) Experimental 2H NMR spectra for 11-Z-[9-C2H3]-retinylidene rhodopsin, i.e. having 11-cis-retinal deuterated at the C9-methyl group, in gel-phase POPC membranes (1:50 molar ratio). (d) Theoretical 2H NMR spectrum for randomly oriented C–C2H3 groups undergoing rapid three-fold rotation on the 2H NMR time scale (<(3χQ/8)−1 ≈ 10 μs). (e, f) Representative 2H NMR spectra for dark-state 11-Z-[5-C2H3]-retinylidene rhodopsin and 11-Z-[13-C2H3]-retinylidene rhodopsin, i.e. with 11-cis-retinal deuterated at the C5-methyl (yellow) or C13-methyl group (red), respectively, in POPC membranes (1:50). Theoretical 2H NMR spectra for C–C2H3 groups undergoing axial rotation (continuous color lines) are superimposed on the experimental spectra, with residuals below. Adapted with permission from Struts et al. (64).
Figure 1a–c shows illustrative 2H NMR spectra of 11-Z-[9-C2H3]-retinal bound to rhodopsin in random (nonaligned) 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) membrane dispersions. So-called powder-type samples have been investigated over a temperature range from −30 to −150°C, well below the lipid order–disorder transition temperature (Tm = −5°C for POPC). The lowest temperature studied, T = −150°C, is close to the cryogenic temperatures employed in X-ray crystallography of rhodopsin (3,6,10,11,14). Below Tm the 2H NMR spectra correspond to the lipid gel state, where rotational and translational diffusion of both the protein (67,68) and lipid components are diminished on the 2H NMR time scale (<10 μs for rotating methyls). The 2H NMR spectra were simulated using the well-known theoretical expression for a Pake doublet due to a spherical distribution of EFG principal axes (69). Beneath the powder pattern the residuals between the fit and the data are plotted. In Fig. 1d the spectral probability density p(ξ±) for the two spin I = 1 spectral branches is graphed against the reduced frequencies ξ±. For comparison, additional powder-type 2H NMR spectra of 11-Z-[5-C2H3]-retinylidene rhodopsin and 11-Z-[13-C2H3]-retinylidene rhodopsin in POPC membranes (1:50) are shown in Fig. 1e,f, respectively.
An immediate conclusion is that the quadrupolar coupling (electric field gradient; EFG tensor) is motionally averaged versus the static EFG coupling tensor for an aliphatic C–2H bond. The averaging is due to rapid three-fold rotation of the methyl groups on the 2H NMR time scale (<10 μs) at all temperatures studied, down to at least −150°C. For if there was a large off-axial motion of the methyl groups, then the RQC would be significantly less, as seen for liquid-crystalline lipid bilayers (69,70). As the observed coupling is near the theoretical limit for methyl three-fold rotation, it can be deduced there is little additional rotation of the protein on the 2H NMR time scale in the gel-state lipid bilayer. This finding agrees with spin-label EPR studies of rhodopsin rotational diffusion (67,68). Furthermore, comparison of the RQC to the theoretical value for a rotating C–C2H3 group indicates the order parameter for the methyl three-fold axis is SC3 = 38/41.75 ≈ 0.9 for each for the three methyl groups (64), corresponding to off-axial fluctuations of about 15°. Despite the spectacular dark-state stability of retinal, it nonetheless has appreciable mobility with the rhodopsin binding pocket, with rapidly rotating methyls and restricted off-axial fluctuations.
On the other hand, knowledge of the methyl orientation within the protein or membrane frame is largely unavailable from studies of nonaligned membrane dispersions. The same is true for dipolar recoupling experiments under MAS, which provide distances but not angles with respect to the membrane. To obtain such orientational restraints on the ligand conformation it is necessary to investigate aligned samples. By measuring a tilt series, one can determine the tensor orientation with respect to the laboratory frame in analogy with X-ray crystallography of oriented single crystals. Moreover by combining results from aligned samples (20,64) with distance restraints using MAS 13C NMR (71,72) one can eliminate multiple solutions and obtain a more complete picture of the retinal conformation, as well as the interaction with amino acid residues lining the rhodopsin binding pocket. In this regard, the approach corresponds to solution NMR spectroscopy whereby the nuclear Overhauser enhancements give the distance restraints, and angular restraints are provided by residual dipolar couplings (73,74).
The preparation of highly aligned membranes containing proteins is the counterpart of obtaining 3D crystals in X-ray crystallography (3,11,12,75–77). Rhodopsin was aligned on planar substrates by isopotential spin-dry centrifugation of the membranes onto glass slides (20,78). For 2H NMR the quadrupolar frequencies of the two I = 1 spectral branches depend on the orientation of the C–2H bond (the principal axis of the electric field gradient tensor) to the main magnetic field B0. The strategy is to decompose the overall angle of the C–C2H3 bond to the magnetic field in terms of various angles that characterize the biomembrane and receptor geometry. (Only static distributions are considered and motional rates appear only through the intrinsic linebroadening in the simulations.) By simulating the solid-state NMR spectra one can determine the retinal bond angles with respect to the protein, as well as the degree of alignment disorder (mosaic spread) of the oriented membrane stack.
SITE-DIRECTED DEUTERIUM NMR GIVES ORIENTATIONAL RESTRAINTS FOR RETINAL CONFORMATION
For the case of aligned biomolecules, the general aim is to determine the coupling tensor orientation within the alignment frame, e.g. the quadrupolar or dipolar coupling. One can then use the angular restraints for structure determination. However, hydrated membranes containing proteins or peptides can involve substantial alignment disorder, i.e. the individual membrane normals are not all completely aligned. Rather, they are distributed about the normal to the planar glass supports, so that the lineshape consists of inhomogeneously broadened lines due to a superposition of contributions from the various membrane domains. This distribution is known as the mosaic spread of the sample (79). Thus, an important aspect with regard to biomembrane applications is the correct treatment of the distribution of local membrane normals relative to the average alignment axis (79). For rhodopsin, the retinal structure within the binding cavity can be represented by three planes collinear with the methyl bonds of interest, which comprise the β-ionone ring and the polyene chain to either side of the C12–C13 bond. The relative orientations of the quadrupolar coupling tensors involve pairs of the methyl groups, and allow one to calculate effective torsional angles between the different planes of unsaturation of the retinal chromophore (20).
Angular restraints are obtained from 2H NMR spectroscopy of aligned membrane samples
The theory of the 2H NMR lineshapes has been previously worked out for such a uniaxial, immobile distribution (79). The lineshape in principle allows us to include any arbitrary distribution (18,79). The frequency separation of the single-quantum transitions (quadrupolar splitting) is given by , where θ̃ is the angle between the C–C2H3 bond axis and the magnetic field B0, which may vary due to rotation about the membrane normal for nonzero tilt. Here is the splitting of the two lines for θ = 90° in the case of a nonoriented powder-type sample. The residual quadrupolar coupling constant 〈χQ〉 is averaged by the methyl group motion, and depends on the static coupling constant χQ (=170 kHz), the tetrahedral geometry of the methyl group, and the order parameter for the off-axial motion (64), where is the effective quadrupolar coupling constant for the rotating methyl group. The value of for a rotating methyl group in the fast motional limit that applies to our samples (18,64,80). To obtain the C–C2H3 bond orientation for cases where it is necessary to simulate a tilt series of angular-dependent 2H NMR spectra, as noted above (18–20,64,79). Either an analytical solution in mathematical closed form or a numerical Monte Carlo procedure is used to simulate the lineshape with four free parameters (the bond orientation θB, membrane tilt angle to the magnetic field, mosaic spread and residual quadrupolar coupling 〈χQ〉), together with the intrinsic line broadening. Further description of the mathematical treatment can be found elsewhere (18,19,79).
Application of solid-state 2H NMR to G protein-coupled receptor rhodopsin
The visual pigment rhodopsin is an example of a GPCR, for which the ligand is 11-cis-retinal. The activation of rhodopsin in the visual process and the X-ray crystal structure (3,5,6,10,11,14) have been reviewed (81–85). Briefly, rhodopsin has seven transmembrane helices with a ligand-binding pocket in its center. An additional helix that runs approximately parallel to the membrane surface is also found, to which posttranslational lipid modifications are attached. Here the retinal chromophore functions as a light-activated switch, and generates a signaling state in a time-ordered sequence that is thermodynamically irreversible. For rhodopsin, the site-directed 2H NMR approach was used for structural analysis of retinal within its binding cavity in the dark-adapted (11-cis) and preactivated meta I (all-trans) states. The meta I and meta II states of rhodopsin correspond to the low-affinity and high-affinity forms of ligand-activated GPCRs. Three sites of interaction within the binding cavity are implicated, viz. the PSB and its associated counterion, the polyene chain with its C9-methyl group and the β-ionone ring which is in a hydrophobic pocket. Our hypothesis is that specific methyl binding sites are necessary for rhodopsin pigment formation and photoreceptor activation (36). The interaction sites are probed by 2H NMR of the C5-, C9- and C13-methyl groups. Solid-state 2H NMR of the planar-supported bilayers provides the orientation of retinal as well as its conformation within the protein binding pocket. Differences in the retinal conformation and orientation as revealed by solid-state 2H NMR spectroscopy are significant for explaining the mechanism of action of rhodopsin. In particular, we are able to capture individually subtle changes along the reaction coordinate, and most important their dynamical fingerprints.
Solid-state 2H NMR of rhodopsin in oriented membranes allows structural analysis for retinal cofactor
In this work, the opsin apoprotein was regenerated with 11-Z-[5-C2H3]-, 11-Z-[9-C2H3]- and 11-Z-[13-C2H3]-retinals, i.e. 11-cis-retinal deuterated at the C5-, C9- or C13-methyl groups. The selectively deuterated retinals were prepared by total organic synthesis (86). Thereafter, rhodopsin was recombined with POPC (1:50 protein/lipid molar ratio) followed by alignment of the membrane bilayers on planar glass substrates by isopotential ultracentrifugation (20,87). From a tilt series of the oriented sample spectra in the magnetic field and an analysis of the 2H NMR line shapes (necessary to distinguish complementary angle solutions), the angles between the individual C–C2H3 bonds and the membrane normal could be determined, even in the presence of a substantial degree of orientational disorder. The angular-dependent 2H NMR spectra were measured below the order–disorder transition temperature (Tm) of the membrane lipid bilayers. Below Tm rhodopsin is in a state of substantially reduced mobility, due to the presence of gel state lipids, as shown by freeze-fracture electron microscopy studies (88) and spin-label EPR spectroscopy (67,68). The 2H NMR studies show that the retinylidene methyl groups are all rotating at temperatures down to at least −150°C (20,80). In Fig. 2 we show a tilt series of angular-dependent 2H NMR spectra for rhodopsin/POPC membranes in the dark state having retinal deuterated at the C5-, C9- or C13-methyl groups at T = −150°C. As can be seen, characteristic lineshape changes are evident as a function of the tilt angle. The lineshapes differ for the various deuterated methyl positions due to the different C–C2H3 bond orientations with respect to the membrane plane.
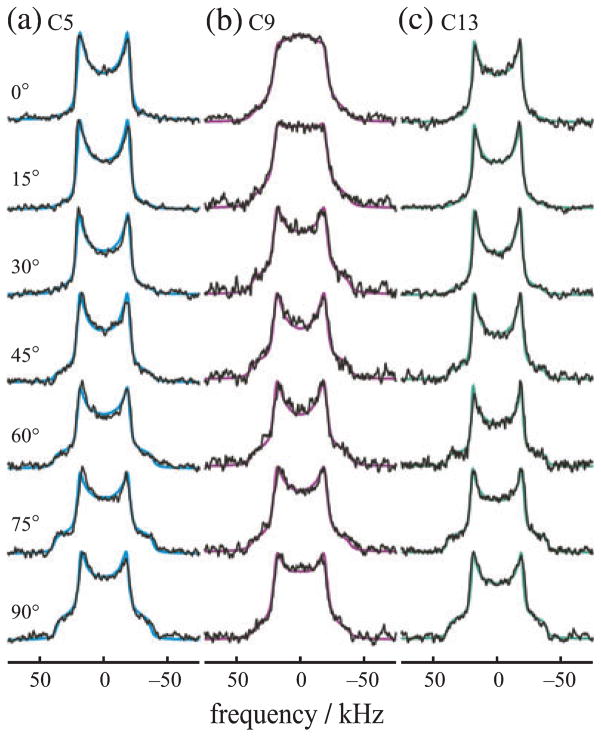
Figure 2.
Orientation-dependent 2H NMR spectra for aligned rhodopsin POPC (1:50) recombinant membranes provide angular restraints for retinylidene ligand in the dark state. (a–c) 2H NMR spectra for 11-Z-[5-C2H3]-retinylidene rhodopsin (blue), 11-Z-[9-C2H3]-retinylidene rhodopsin (magenta) and 11-Z-[13-C2H3]-retinylidene rhodopsin (green) at pH 7 and T = −150°C. Theoretical lineshapes for an immobile uniaxial distribution (solid lines) are superimposed on the experimental 2H NMR spectra. Note that characteristic lineshape changes are observed as a function of the tilt angle, which manifest the different methyl bond orientations with respect to the membrane frame. Reproduced with permission from Struts et al. (64).
Some qualitative deductions can be made by simply considering the angular dependence of the 2H NMR spectra. From the appearance of the θ = 90° spectral shoulders one can conclude that the bond orientation θB must be greater than ≈ 30–45° in each case (79,89). However due to the alignment disorder, the lineshapes must be numerically calculated to obtain accurate information. Theoretical simulations in Fig. 2 assume a static uniaxial distribution (79) of the rhodopsin molecules with rotating methyl groups, and are superimposed on the experimental 2H NMR data. Similar results are obtained at temperatures of T = −60 and −30°C (86). Simultaneous fitting of the 2H NMR lineshapes gives the orientations of the various methyl C–C2H3 axes relative to the bilayer normal, together with the mosaic spread of the aligned samples. For a given sample orientation the spectra were all fit with the same values of the adjustable fitting parameters. In accord with results obtained for powder-type samples (Fig. 1), off-axial order parameters were obtained for the rotating methyl groups of SC3 ≈ 0.9. Excellent agreement between the experimental and simulated lineshapes was found in all cases as evinced by the flat residuals (data not shown).
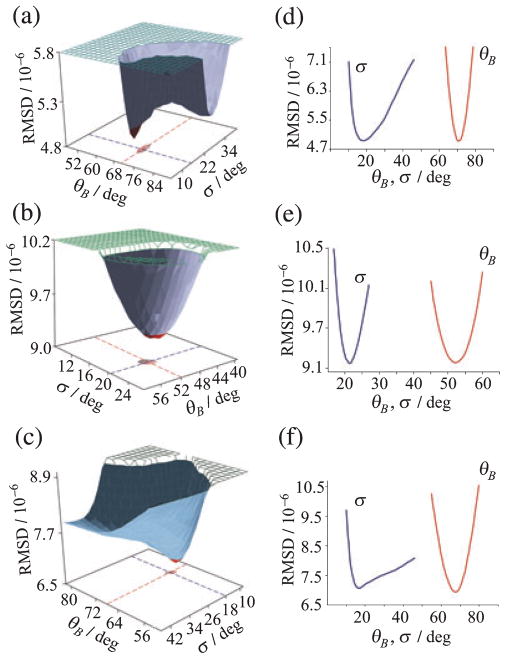
The outcome of the global fitting of 2H NMR lineshapes is shown in Fig. 3 (left) for the C5-, C9- or C13-methyl groups of retinal bound to rhodopsin in POPC membranes in the dark state. Here the root mean square deviation (RMSD) of the calculated 2H NMR spectra from the experimental results for the full tilt series is plotted versus the methyl bond orientation θB and the mosaic spread σ, and is seen to differ appreciably for the 2H-labeled sites. Figure 3 (right) shows cross-sections through the RMSD surfaces at the global minimum for the C5-, C9- and C13-methyl groups, respectively. Accurate bond orientations θB of the 2H-labeled methyl groups are obtained, despite the large mosaic spread (σ= 18–21°). The mosaic spread obtained from the 2H NMR lineshape simulations is greater than for aligned purple membranes containing bacteriorhodopsin (18). This may be due to the fact that rhodopsin (Mr = 40 kDa) is a larger molecule than bR (Mr = 26 kDa), with appreciable extramembranous domains (90). In this way, we determined values of the C–C2H3 bond orientations of 70 ± 3°, 52 ± 3° and 68 ± 2° for the C5-, C9- and C13-methyl groups of retinal in the dark state of rhodopsin (20).
Figure 3.
Global fitting of 2H NMR spectra for 11-cis-retinal in the dark state of rhodopsin gives methyl bond orientations and mosaic spread of aligned membranes. (a–c) RMSD of calculated versus experimental 2H NMR spectra for retinal deuterated at C5-, C9- or C13-methyl groups, respectively and (d–f) cross-sections through hypersurfaces. Distinct minima are found in the bond orientation θB and mosaic spread σ of aligned membranes. Reproduced with permission from Struts et al. (64).
Analysis of 2H NMR data reveals torsional twisting of retinal chromophore in dark-adapted state
For rhodopsin, interpretation of the 2H NMR lineshapes was focused on determining the conformation of the retinal cofactor. What is the 3D structure of the retinal chromophore in rhodopsin as viewed by 2H NMR? Is distortion of the retinal linked to its photochemistry? Analysis of the retinal conformation employed a simple model with three planes of unsaturation (20,91), encompassing the β-ionone ring and the polyene chain to either side of the C12–C13 bond (20,36,79,91,92). Figure 4 shows that the relative orientations of the C5-, C9- and C13-methyl groups specify the C6–C7 and C12–C13 dihedral angles. In fact four (4) orientations are found for each plane giving 4 × 4 = 16 combinations for each torsion angle connecting the adjacent planes. The problem of multiple solutions due to symmetry of NMR interactions has been discussed previously (20,24,64,86). Comparison of the 2H NMR data with the results of other methods (linear and circular dichroism; solid-state MAS 13C NMR) enables calculation of the unique retinal structure within the rhodopsin binding cavity. Knowing the relative orientations of coupling tensors (bonds) one can then obtain the torsion angles that define the retinal conformation within the rhodopsin binding cavity (20).
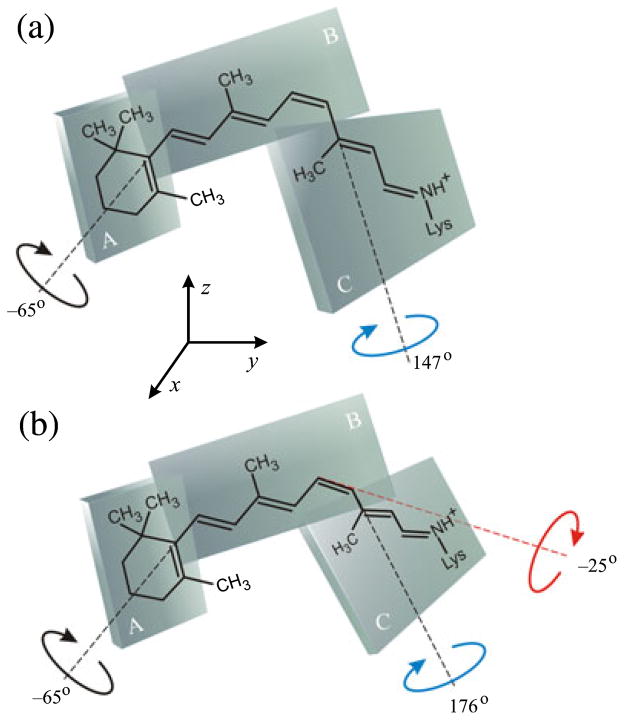
Figure 4.
Solid-state 2H NMR spectroscopy yields conformation and orientation of 11-cis-retinal ligand in the dark state of rhodopsin. Retinal is described by three planes of unsaturation (designated A, B, C). (a) Simple three-plane model with torsional twisting only about C6–C7 and C12–C13 bonds. (b) Extended three-plane model with additional pretwisting about the C11=C12 double bond. Reproduced with permission from Struts et al. (64).
Angular and distance restraints were combined in the structural analysis, as in solution NMR spectroscopy (74). The angular restraints were from 2H NMR studies (20,64) and linear dichroism (93) and distance restraints were from rotational-resonance 13C NMR studies (72). Circular dichroism data are also available for the dark state of rhodopsin, which yield information about the enantiomeric selectivity of the retinal pocket of the protein (92,94). The most likely physical solution is shown in Fig. 4, corresponding to a positively twisted, 12-s-trans conformation. Here the C13- and C9-methyl groups point oppositely from the membrane plane, and the retinylidene β-ionone ring has a negatively twisted 6-s-cis conformation. In Fig. 4a, a simple three-plane model is assumed which gives dihedral angles of χ9,13 = +147 ± 4° and χ5,9 = −65 ± 6°. Analysis of the conformation of retinal in rhodopsin from the 2H NMR data indicates that the chromophore is highly nonplanar, in contrast to bacteriorhodopsin (18). The 11-cis-retinylidene structure derived from 2H NMR can be further refined by inserting it into the binding pocket of the X-ray structure of rhodopsin in the dark state (2.2 Å) (10). On account of steric clashes a simple model is not readily accommodated within the rhodopsin binding cavity. Figure 4b shows that an extended three-plane model can then be introduced to allow for pretwisting of the C11=C12 cis-double bond in the direction of isomerization (64). The positive helical twist about the C12–C13 bond is in agreement with CD and bioorganic studies of locked retinoids (92) and with our previous 2H NMR results (20). Pretwisting about the 11-cis double bond is compatible with X-ray (10) and quantum-mechanical calculations (95) and can direct the chromophore movements upon photoexcitation, in this way affecting the ultrafast reaction dynamics (64,96).
The structure of retinal obtained in this work is depicted inserted into the binding pocket of the rhodopsin X-ray structure (10) in Fig. 5. Specific methyl sites are occupied which contributes to the dihedral twisting of the retinal chromophore in the rhodopsin dark state. Torsional twisting of the polyene arises from localization of the β-ionone ring within its hydrophobic pocket at one end of the chromophore, together with the salt bridge of the retinylidene Schiff base at the other end. In addition, the C9-methyl is positioned between Thr118 and Tyr268 and also contributes to the torsional twist by preventing rotation of the polyene about the molecular long axis. The absolute sense of the helical twists of the 11-cis, 12-s-trans and 6-s-cis conformations manifest the chiral selectivity of the binding site. It is proposed that this is related to the trajectory of the 11-cis to trans photoisomerization. The hydrogen bonding network and salt bridges lock the binding pocket and explain the phenomenal dark stability. Interactions of the C13-methyl group of retinal with Trp265 help to stabilize the dark state conformation (97). The configuration of the –C=NH+– bond of the PSB is anti and its hydrogen points oppositely from the C13 methyl, i.e. toward the extracellular side in the direction of the counterion Glu113.
Figure 5.
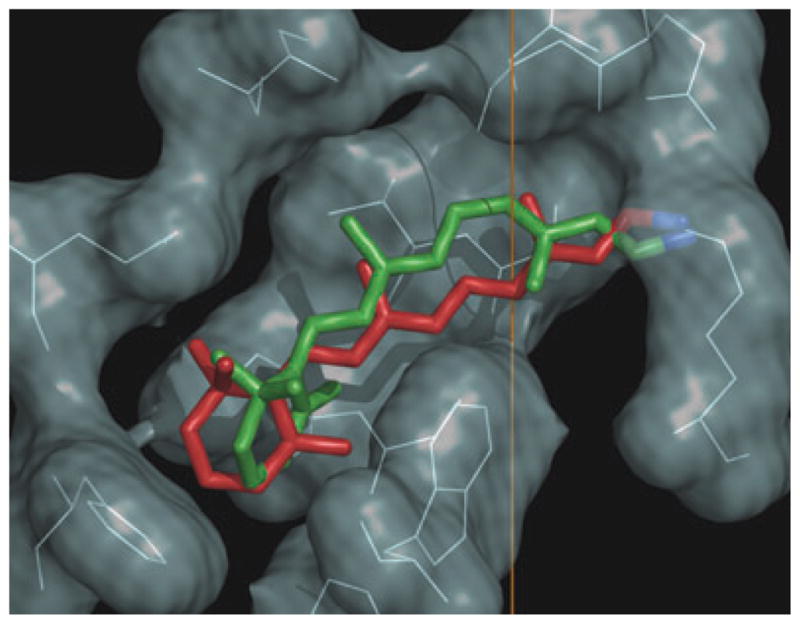
Solid-state NMR provides the structure of retinal ligand of the canonical GPCR rhodopsin. Vertical direction corresponds to the membrane normal, where the extracellular side is at top and the cytoplasmic side is below. The 2H NMR structure of 11-cis-retinal in the dark state (green) is compared to retinal structure in the preactivated metarhodopsin I state (red). Figure prepared with PyMOL [http://pymol.sourceforge.net/].
SOLID-STATE NMR ILLUMINATES CHANGES IN RETINAL CONFORMATION UPON LIGHT ACTIVATION
The next question addressed is how 2H NMR can be applied to understand the molecular mechanism of rhodopsin activation. What are the changes in the conformation and orientation of the retinal chromophore of rhodopsin that are coupled to light activation of the receptor? At present X-ray structures are available for bleached rhodopsin in the bathorhodopsin and lumirhodopsin states (77) as well as the opsin apoprotein (98). In addition, an electron density map has been derived for the preactivated meta I state from electron crystallography at relatively low resolution (5.5 Å in the membrane plane and 11.7 Å perpendicular to the plane) (99). No crystallographic data are currently available for the activated meta II state. The next step was to measure the tilt series of 2H NMR spectra for aligned membranes containing rhodopsin in the all-trans meta I state. The 2H NMR lineshape analysis for meta I gave the orientation θB of the various methyl bonds. Deuterium NMR structures were determined for meta I using a three-plane model as described (24). In addition to the C5-, C9- and C13-methyl bond orientations, the electronic transition dipole moment from linear dichroism studies was introduced as a further orientational restraint (100). Several NMR structures (86) were eliminated based on electron crystallography data which suggest that in the meta I state the β-ionone ring occupies its ground state position (64,86,99). The calculated meta I structures were further restrained by inserting them into the dark-state rhodopsin structure (10). We assumed that the properties of the retinal binding cavity were nearly the same in meta I as in the dark state, which is in accord with electron crystallographic data (99). The structure with χ5,9 = −32° and χ9,13 = 173° fits best within the binding cleft of the dark-state X-ray structure, having its only close contact with the side chain of Trp265(86). Although the polyene chain is relaxed in meta I, the β-ionone ring retains its torsional strain as in the dark state.
Structural relaxation of the retinal chromophore in metarhodopsin I state
A central question is how the conformational distortion following retinal isomerization is propagated through the protein allosteric network, affecting the cytoplasmic loops and yielding activation of the receptor (90). In Fig. 5 the 2H NMR structure proposed for all-trans retinal in the meta I state (red) is compared to the NMR structure of 11-cis retinal in the dark-adapted state of rhodopsin (green). Isomerization of retinal from 11-cis to all-trans is in the negative direction due to the pretwist about the C11=C12 double bond in the dark state, together with the steric hindrance within the binding pocket (96). The Schiff base hydrogen switches from pointing toward the extracellular side in the dark state to the cytoplasmic side, which may have implications for a (possibly complex) counterion switch in meta I (101–103). In the case of rhodopsin, the C13-methyl also changes its orientation to the membrane upon photon absorption, rotating toward the extracellular side and becoming parallel to the C9-methyl. This may be essential for directing the retinal movement during activation (21,97). Steric hindrance between trans-retinal and Trp265 can be involved in triggering the transition into the active meta II state. In this way, 2H NMR spectroscopy provides a basis for investigating the changes in retinal conformation and orientation giving rise to the meta II signaling state in visual excitation.
Combining 2H NMR data with molecular dynamics simulations further illuminates rhodopsin function
A further aspect entails comparison of the experimental 2H NMR results with molecular dynamics (MD) simulations (90,103–107). Ultra-large-scale MD simulations have simulated the chromophore environment in both the dark state (106) as well as the preactivated meta I state (103). For the dark state, a total of 23 independent, 100-ns all-atom MD simulations were carried out for rhodopsin embedded in a lipid bilayer and were analyzed in the microcanonical (N,V,E) ensemble (106). We found that the polyene chain of retinal is rigidly locked into a single, twisted conformation, consistent with its function as an inverse agonist in the dark state. Yet the β-ionone ring is mobile within its binding pocket, as the cavity is sufficiently large to enable structural heterogeneity. It was found that the β-ionone ring can occupy two distinct conformations in the dark state (106). The β-ionone ring can rotate relative to the polyene chain, thereby populating both positively and negatively twisted 6-s-cis enantiomers. The influences of 11-cis to trans isomerization of the retinal were investigated by long-time scale simulations of rhodopsin having trajectories of 1000–2000 ns (103). Analysis of the MD and NMR data in the meta I state gave insight into the counterion switch mechanism that stabilizes the PSB. Retinal PSB deprotonation is thermodynamically the most important event that characterizes the subsequent activated meta II state. Comparison of the simulated 2H NMR spectra with experimental data supports a complex counterion mechanism in which both Glu113 and Glu181 stabilize the retinal PSB in the meta I state prior to forming the activated meta II state of rhodopsin (103).
FUNCTIONAL DYNAMICS OF RETINAL IN META I AND META II STATES OF RHODOPSIN ARE ELUCIDATED BY DEUTERIUM NMR RELAXATION
Besides obtaining knowledge of the retinal conformation, solid-state 2H NMR spectroscopy can be applied to provide information about fluctuations and structural dynamics of the retinal cofactor in the dark state and the light-activated meta I and meta II states (56). The unique features of the NMR approach are not replicated by other biophysical techniques and include the following: First, NMR provides atomic site-resolved information about dynamics that cannot be obtained with other experimental methods. For instance, X-ray crystallography gives information about protein mobility only indirectly, e.g. through Debye-Waller factors that do not distinguish static from dynamic disorder. Only MD simulations can provide such comprehensive information; yet they are theoretical and in effect capture a posteriori the results of experimental measurements. Second, rhodopsin is studied in a membrane environment and information about the activated meta II state is accessible through solid-state NMR spectroscopy (56,97).
Structural relaxation analysis in solid-state NMR spectroscopy
Protein dynamics are of broad interest in biophysics including NMR spectroscopy and computational biology. Taken together, such approaches are powerful in complementing the structural information from equilibrium studies. In the case of solution NMR, relaxation methods have yielded information about the dynamics of proteins during catalysis and signaling (108). Analogously, solid-state NMR relaxation methods have been applied to establish how the mobility of the retinal ligand is related to the structure of the chromophore and the binding pocket in the different rhodopsin photointermediates, and to shed light on the functional dynamics. In this way one can build up snapshots of the retinal photoactivation process. The motional averaged 2H NMR spectral lineshapes reveal directly that the retinylidene methyl groups undergo rapid spinning about their C3 axes with order parameters of SC3 ≈ 0.9. A summary of the results of the combined 2H NMR spectral lineshape and relaxation analysis is provided in Fig. 6. For rhodopsin in the dark state, the spin-lattice relaxation (T1Z) times were significantly less and corresponding activation energies Ea were greater for the C5- and C13-methyls than for the C9-methyl group. Nonbonded interactions yield a lower rate of methyl spinning for the C5- and C13-methyl groups. The longer T1Z value for the C9 methyl group indicates a shorter rotational correlation time due to a smaller pre-exponential factor (for motion within a potential well) and a smaller activation energy (for motion over the potential barrier). Analysis of the relaxation data shows that the dynamics of the C9- and C13-methyl groups are determined mostly by intra-retinal interactions, while the mobility of the C5-methyl may be affected by interaction of the β-ionone ring with Glu122 and potentially also Trp265.
Figure 6.
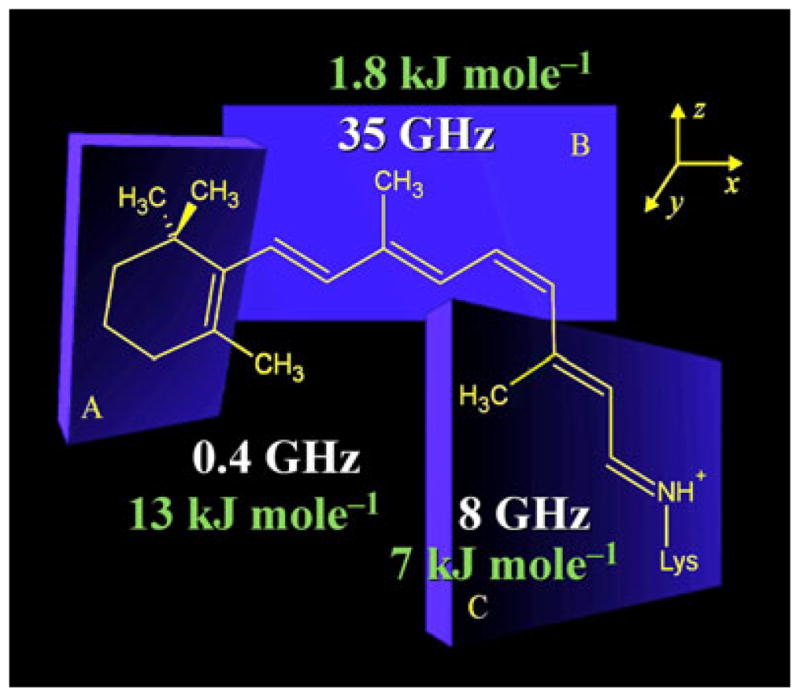
Structure and dynamics of retinal within the binding cavity of rhodopsin in the dark state at −60°C. Analysis of 2H NMR data reveals torsional twisting of retinal that accompanies nonbonded interactions of the C5- and C13-methyls with the polyene B-plane (for retinal numbering, see Fig. 1). Average structure of the retinylidene ligand corresponds to a distorted 6-s-cis,11-cis,12-s-trans,15-anti conformation. Rapid spinning of the methyl groups about their three-fold axes occurs with correlation times in the range ≈ 1–20 ps and off-axial order parameters of SC3 ≈ 0.9. The rates of three-fold rotation (in GHz) of the C5-, C9- and C13-methyl groups and the corresponding activation energies (in kJ mol−1) are indicated in the figure. Methyl rotation may be coupled to fluctuations of the C6–C7 and C12–C13 dihedral angles which connect the different planes of unsaturation (A, B and C). Correlation times and activation barriers for fluctuations of the C5-methyl of the β-ionone ring are considerably greater than for the polyene C9- or C13-methyl groups. The C9-methyl group may be implicated in the activation process through a nearly frictionless environment manifested by the low activation barrier. We propose that site-specific differences in mobility of the retinal ligand underlie movement of retinal in the activation mechanism of rhodopsin.
Changes in retinal dynamics are detected upon photoactivation of rhodopsin
Upon isomerization, there are noticeable changes in the motion of all three methyl groups. Results for the meta I and meta II states are similar and overall dynamics of the C9- and C13-methyl groups indicate the absence of significant steric clashes of these groups with the surrounding amino acids. These results can be explained as follows. After photoisomerization of retinal, both the C9-methyl and the C13-methyl groups have (1,6) steric interactions with the contiguous vinyl hydrogens to either side of the substituent. Twisting about the C11=C12 double bond is reduced due to isomerization of the retinal cofactor (64) and both methyl groups are in similar environment. Interactions of the C9-methyl group with Tyr268 and Thr118 do not affect its mobility significantly as on average they are about 4 Å away; however, they maintain the retinal orientation. A steric clash between the ligand polyene chain and Trp265 (64) occurs due to straightening of highly distorted all-trans retinal following isomerization. This leads to displacement of the β-ionone ring toward helix H3 (77) and the disruption of the hydrogen bonding network around Glu122 which stabilizes the inactive state. The absence of significant steric clashes of the C9- and C13-methyl groups with the side-chain amino acids of the binding pocket in metarhodopsin II may indicate a smaller translation of the ligand toward helix H5 than indicated by 13C solid-state NMR studies (21). Perhaps more surprising, however, with light activation there is little change in the mobility of the β-ionone ring. The β-ionone ring occupies a similar environment up to and including the meta II state. In both the meta I and meta II states the β-ionone ring maintains the constraints as in the dark state. Its expulsion from its hydrophobic pocket upon receptor activation is therefore rendered unlikely. However, this does not rule out helical movements that keep the environment of the β-ionone ring essentially intact.
Summary
Solid-state deuterium NMR spectroscopy has been applied to study the G-protein-coupled receptor rhodopsin in the dark, meta I and meta II states. Information about the energy landscape and mobility of the retinal cofactor is obtained that cannot be obtained with X-ray crystallography or solution NMR. The findings have implications for the flow of energy in the activation mechanism of rhodopsin, and are pertinent to triggering changes in hydrogen-bonding networks and helix movements. For rhodopsin an intriguing aspect is torsional twisting of the ligand involving both the β-ionone ring and the polyene chain. Retinal distortion is implicated in the ultra-fast isomerization and high quantum yield. Combining the solid-state 2H NMR data with the X-ray crystal structure suggests that an extended three-plane model is applicable that involves torsional twisting about the activated C11=C12 bond. The C11=C12 double bond is pretwisted within the rhodopsin binding pocket in the direction of the isomerization. The solid-state 2H NMR structure of retinal explains its dark-state stability, and moreover indicates the direction of the rapid photoisomerization. In the preactivated meta I state, the major changes in retinal upon isomerization involve the Schiff base end of the ligand. The β-ionone ring and the polyene with its crucial C9-methyl group occupy similar binding sites in meta I as in the dark state. Straightening of all-trans retinal after isomerization yields a steric clash with Trp265 that displaces the β-ionone ring toward helix H3, and disrupts the hydrogen-bonding network around Glu122. This triggers the helical movements that yield the receptor activation. Finally, 2H NMR relaxation studies show remarkable differences in the mobility of retinal that occur along the reaction coordinate as the ligand switches from an inverse agonist to an agonist. Isomerization leads to dramatic changes in the motion of the retinylidene methyl groups. The relaxation data support an activation mechanism whereby the environment of retinal in the preactivated meta I state is similar to the active meta II state. Sequential relief of the ligand frustration (conformational distortion) as detected by solid-state NMR is linked to the activation mechanism of the membrane-bound visual receptor.
Acknowledgments
This work was supported in part by the U.S. National Institutes of Health (grants EY 12049 and EY18891 to M.F.B. and GM 36564 to K.N.). We are most grateful to our colleagues and laboratory members for insightful discussions and many contributions to this research.
Footnotes
This paper is part of the Proceedings of the 13th International Conference on Retinal Proteins, Barcelona, Spain, 15–19 June 2008.
References
- 1.Pebay-Peyroula E, Rummel G, Rosenbusch JP, Landau EM. X-ray structure of bacteriorhodopsin at 2.5 Angstroms from microcrystals grown in lipid cubic phases. Science. 1997;277:1676–1681. doi: 10.1126/science.277.5332.1676. [DOI] [PubMed] [Google Scholar]
- 2.Chang G, Spencer RH, Lee AT, Barclay MT, Rees DC. Structure of the MscL homolog from Mycobacterium tuberculosis: A gated mechanosensitive ion channel. Science. 1998;282:2220–2226. doi: 10.1126/science.282.5397.2220. [DOI] [PubMed] [Google Scholar]
- 3.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 4.Toyoshima C, Nakasako M, Nomura H, Ogawa H. Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 Å resolution. Nature. 2000;405:647–655. doi: 10.1038/35015017. [DOI] [PubMed] [Google Scholar]
- 5.Teller DC, Okada T, Behnke CA, Palczewski K, Stenkamp RE. Advances in determination of a high-resolution three-dimensional structure of rhodopsin, a model of G-protein-coupled receptors (GPCRs) Biochemistry. 2001;40:7761–7772. doi: 10.1021/bi0155091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okada T, Fujiyoshi Y, Silow M, Navarro J, Landau EM, Shichida Y. Functional role of internal water molecules in rhodopsin revealed by X-ray crystallography. Proc Natl Acad Sci USA. 2002;99:5982–5987. doi: 10.1073/pnas.082666399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Toyoshima C, Nomura H. Structural changes in the calcium pump accompanying the dissociation of calcium. Nature. 2002;418:605–611. doi: 10.1038/nature00944. [DOI] [PubMed] [Google Scholar]
- 8.Luecke H, Lanyi JK. Structural clues to themechanism of ionpumpingin bacteriorhodopsin. Adv Prot Chem. 2003;63:111–130. doi: 10.1016/s0065-3233(03)63005-6. [DOI] [PubMed] [Google Scholar]
- 9.Abramson J, Smirnova I, Kasho V, Verner G, Kaback HR, Iwata S. Structure and mechanism of the lactose permease of Escherichia coli. Science. 2003;301:610–615. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 10.Okada T, Sugihara M, Bondar AN, Elstner M, Entel P, Buss V. The retinal conformation and its environment in rhodopsin in light of a new 2.2 Å crystal structure. J Mol Biol. 2004;342:571–583. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 11.Li J, Edwards PC, Burghammer M, Villa C, Schertler GFX. Structure of bovine rhodopsin in a trigonal crystal form. J Mol Biol. 2004;343:1409–1438. doi: 10.1016/j.jmb.2004.08.090. [DOI] [PubMed] [Google Scholar]
- 12.Salom D, Lodowski DT, Stenkamp RE, Le Trong I, Golczak M, Jastrzebska B, Harris T, Ballesteros JA, Palczewski K. Crystal structure of a photoactivated deprotonated intermediate of rhodopsin. Proc Natl Acad Sci USA. 2006;103:16123–16128. doi: 10.1073/pnas.0608022103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rasmussen SGF, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VRP, Sanishvili R, Fischetti RF, Schertler GFX, Weis WI, Kobilka BK. Crystal structure of the human β2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 14.Standfuss J, Xie G, Edwards PC, Burghammer M, Oprian DD, Schertler GFX. Crystal structure of a thermally stable rhodopsin mutant. J Mol Biol. 2007;372:1179–1188. doi: 10.1016/j.jmb.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drews J. Drug discovery: A historical perspective. Science. 2000;287:1960–1964. doi: 10.1126/science.287.5460.1960. [DOI] [PubMed] [Google Scholar]
- 16.Watts A. Solid-stateNMRin drug design and discovery for membrane-embedded targets. Nat Rev Drug Discov. 2005;4:555–568. doi: 10.1038/nrd1773. [DOI] [PubMed] [Google Scholar]
- 17.Ulrich AS, Wallat I, Heyn MP, Watts A. Re-orientation of retinal in the M-photointermediate of bacteriorhodopsin. Nat Struct Biol. 1995;2:190–192. doi: 10.1038/nsb0395-190. [DOI] [PubMed] [Google Scholar]
- 18.Moltke S, Nevzorov AA, Sakai N, Wallat I, Job C, Nakanishi K, Heyn MP, Brown MF. Chromophore orientation in bacteriorhodopsin determined from the angular dependence of deuterium NMR spectra of oriented purple membranes. Biochemistry. 1998;37:11821–11835. doi: 10.1021/bi980676v. [DOI] [PubMed] [Google Scholar]
- 19.Moltke SM, Wallat I, Sakai N, Nakanishi K, Brown MF, Heyn MP. The angles between the C1-, C5-, and C9-methyl bonds of the retinylidene chromophore and the membrane normal increase in the M intermediate of bacteriorhodopsin: Direct determination with solid-state 2H NMR. Biochemistry. 1999;38:11762–11772. doi: 10.1021/bi990593u. [DOI] [PubMed] [Google Scholar]
- 20.Salgado GFJ, Struts AV, Tanaka K, Fujioka N, Nakanishi K, Brown MF. Deuterium NMR structure of retinal in the ground state of rhodopsin. Biochemistry. 2004;43:12819–12828. doi: 10.1021/bi0491191. [DOI] [PubMed] [Google Scholar]
- 21.Patel AB, Crocker E, Eilers M, Hirshfeld A, Sheves M, Smith SO. Coupling of retinal isomerization to the activation of rhodopsin. Proc Natl Acad Sci USA. 2004;101:10048–10053. doi: 10.1073/pnas.0402848101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hiller M, Krabben L, Vinothkumar KR, Castellani F, van Rossum BJ, Kühlbrandt W, Oschkinat H. Solid-state magic-angle spinning NMR of outer-membrane protein G from Escherichia coli. Chem Bio Chem. 2005;6:1679–1684. doi: 10.1002/cbic.200500132. [DOI] [PubMed] [Google Scholar]
- 23.Lange A, Giller K, Hornig S, Martin-Eauclaire MF, Pongs O, Becker S, Baldus M. Toxin-induced conformational changes in a potassium channel revealed by solid-state NMR. Nature. 2006;440:959–962. doi: 10.1038/nature04649. [DOI] [PubMed] [Google Scholar]
- 24.Salgado GFJ, Struts AV, Tanaka K, Krane S, Nakanishi K, Brown MF. Solid-state 2H NMR structure of retinal in metarhodopsin I. J Am Chem Soc. 2006;128:11067–11071. doi: 10.1021/ja058738+. [DOI] [PubMed] [Google Scholar]
- 25.Etzkorn M, Martell S, Andronesi OC, Seidel K, Engelhard M, Baldus M. Secondary structure, dynamics, and topology of a seven-helix receptor in native membranes, studied by solid-state NMR spectroscopy. Angew Chem Int Ed Engl. 2007;46:459–462. doi: 10.1002/anie.200602139. [DOI] [PubMed] [Google Scholar]
- 26.Varga K, Tian L, McDermott AE. Solid-state NMR study and assignments of the KcsA potassium ion channel of S. lividans. Biochim Biophys Acta. 2007;1774:1604–1613. doi: 10.1016/j.bbapap.2007.08.029. [DOI] [PubMed] [Google Scholar]
- 27.Williamson PTF, Verhoeven A, Miller KW, Meier BH, Watts A. The conformation of acetylcholine at its target site in the membrane-embedded nicotinic acetylcholine receptor. Proc Natl Acad Sci USA. 2007;104:18031–18036. doi: 10.1073/pnas.0704785104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hiller M, V, Higman A, Jehle S, van Rossum BJ, Kühlbrandt W, Oschkinat H. [2,3-13C]-labeling of aromatic residues—Getting a head start in the magic-angle-spinning NMR assignment of membrane proteins. J Am Chem Soc. 2008;130:408–409. doi: 10.1021/ja077589n. [DOI] [PubMed] [Google Scholar]
- 29.Brown MF, Lope-Piedrafita S, Martinez GV, Petrache HI. Solid-state deuterium NMR spectroscopy of membranes. In: Webb GA, editor. Modern Magnetic Resonance. Springer; Heidelberg: 2006. pp. 245–256. [Google Scholar]
- 30.Petrache HI, Brown MF. X-ray scattering and solid-state deuterium nuclear magnetic resonance probes of structural fluctuations in lipid membranes. In: Dopico AM, editor. Methods in Molecular Biology. Humana Press; Totowa, NJ: 2007. pp. 341–353. [DOI] [PubMed] [Google Scholar]
- 31.Brown MF. Modulation of rhodopsin function by properties of the membrane bilayer. Chem Phys Lipids. 1994;73:159–180. doi: 10.1016/0009-3084(94)90180-5. [DOI] [PubMed] [Google Scholar]
- 32.Mitchell DC, Niu SL, Litman BJ. Optimization of receptor-G protein coupling by bilayer lipid composition I—Kinetics of rhodopsin-transducin binding. J Biol Chem. 2001;276:42801–42806. doi: 10.1074/jbc.M105772200. [DOI] [PubMed] [Google Scholar]
- 33.Botelho AV, Gibson NJ, Wang Y, Thurmond RL, Brown MF. Conformational energetics of rhodopsin modulated by nonlamellar forming lipids. Biochemistry. 2002;41:6354–6368. doi: 10.1021/bi011995g. [DOI] [PubMed] [Google Scholar]
- 34.Botelho AV, Huber T, Sakmar TP, Brown MF. Curvature and hydrophobic forces drive oligomerization and modulate activity of rhodopsin in membranes. Biophys J. 2006;91:4464–4477. doi: 10.1529/biophysj.106.082776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fanelli F, De Benedetti PG. Computational modeling approaches to structure-function analysis of G protein-coupled receptors. Chem Rev. 2005;105:3297–3351. doi: 10.1021/cr000095n. [DOI] [PubMed] [Google Scholar]
- 36.Nakanishi K, Crouch R. Application of artificial pigments to structure determination and study of photoinduced transformations of retinal proteins. Isr J Chem. 1995;35:253–272. [Google Scholar]
- 37.Knierim B, Hofmann KP, Ernst OP, Hubbell WL. Sequence of late molecular events in the activation of rhodopsin. Proc Natl Acad Sci USA. 2007;104:20290–20295. doi: 10.1073/pnas.0710393104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Altenbach C, Kusnetzow AK, Ernst OP, Hofmann KP, Hubbell WL. High-resolution distance mapping in rhodopsin reveals the pattern of helix movement due to activation. Proc Natl Acad Sci USA. 2008;105:7439–7444. doi: 10.1073/pnas.0802515105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brown MF, Heyn MP, Job C, Kim S, Moltke S, Nakanishi K, Nevzorov AA, Struts AV, Salgado GFJ, Wallat I. Solid-state 2H NMR spectroscopy of retinal proteins in aligned membranes. Biochim Biophys Acta. 2007;1768:2979–3000. doi: 10.1016/j.bbamem.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Igumenova TI, McDermott AE, Zilm KW, Martin RW, Paulson EK, Wand AJ. Assignments of carbon NMR resonances for microcrystalline ubiquitin. J Am Chem Soc. 2004;126:6720–6727. doi: 10.1021/ja030547o. [DOI] [PubMed] [Google Scholar]
- 41.McDermott AE. Structural and dynamic studies of proteins by solid-state NMR spectroscopy: Rapid movement forward. Curr Opin Struct Biol. 2004;14:554–561. doi: 10.1016/j.sbi.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 42.Castellani F, van Rossum B, Diehl A, Schubert M, Rehbein K, Oschkinat H. Structure of a protein determined by solid-state magic-angle-spinning NMR spectroscopy. Nature. 2002;420:98–102. doi: 10.1038/nature01070. [DOI] [PubMed] [Google Scholar]
- 43.Franks WT, Wylie BJ, Schmidt HLF, Nieuwkoop AJ, Mayrhofer RM, Shah GJ, Graesser DT, Rienstra CM. Dipole tensor-based atomic-resolution structure determination of a nanocrystalline protein by solid-state NMR. Proc Natl Acad Sci USA. 2008;105:4621–4626. doi: 10.1073/pnas.0712393105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li C, Qin H, Gao FP, Cross TA. Solid-state NMR characterization of conformational plasticity within the transmembrane domain of the influenza A M2 proton channel. Biochim Biophys Acta. 2007;1768:3162–3170. doi: 10.1016/j.bbamem.2007.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dürr UHN, Yamamoto K, Im SC, Waskell L, Ramamoorthy A. Solid-state NMR reveals structural and dynamical properties of a membrane-anchored electron-carrier protein, cytochrome b5. J Am Chem Soc. 2007;129:6670–6671. doi: 10.1021/ja069028m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lange C, Müller SD, Walther TH, Bürck J, Ulrich AS. Structure analysis of the protein translocating channel TatA in membranes using a multi-construct approach. Biochim Biophys Acta. 2007;1768:2627–2634. doi: 10.1016/j.bbamem.2007.06.021. [DOI] [PubMed] [Google Scholar]
- 47.Komi N, Okawa K, Tateishi Y, Shirakawa M, Fujiwara T, Akutsu H. Structural analysis of pituitary adenylate cyclase-activating polypeptides bound to phospholipid membranes by magic angle spinning solid-state NMR. Biochim Biophys Acta. 2007;1768:3001–3011. doi: 10.1016/j.bbamem.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 48.Cady SD, Hong M. Amantadine-induced conformational and dynamical changes of the influenza M2 transmembrane proton channel. Proc Natl Acad Sci USA. 2008;105:1483–1488. doi: 10.1073/pnas.0711500105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mak-Jurkauskas ML, V, Bajaj S, Hornstein MK, Belenky M, Griffin RG, Herzfeld J. Energy transformations early in the bacteriorhodopsin photocycle revealed by DNP-enhanced solid-state NMR. Proc Natl Acad Sci USA. 2008;105:883–888. doi: 10.1073/pnas.0706156105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nevzorov AA, Moltke S, Brown MF. Structure of the A-form and B-form of DNA from deuterium NMR line shape simulation. J Am Chem Soc. 1998;120:4798–4805. [Google Scholar]
- 51.van Beek JD, Beaulieu L, Schäfer H, Demura M, Asakura T, Meier BH. Solid-state NMR determination of the secondary structure of Samia cynthia ricini silk. Nature. 2000;405:1077–1079. doi: 10.1038/35016625. [DOI] [PubMed] [Google Scholar]
- 52.Tycko R. Molecular structure of amyloid fibrils: Insights from solid-state NMR. Q Rev Biophys. 2006;39:1–55. doi: 10.1017/S0033583506004173. [DOI] [PubMed] [Google Scholar]
- 53.Brender JR, Dürr UHN, Heyl D, Budarapu MB, Ramamoorthy A. Membrane fragmentation by an amyloidogenic fragment of human islet amyloid polypeptide detected by solid-state NMR spectroscopy of membrane nanotubes. Biochim Biophys Acta. 2007;1768:2026–2029. doi: 10.1016/j.bbamem.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kloepper KD, Zhou DH, Li Y, Winter KA, George JM, Rienstra CM. Temperature-dependent sensitivity enhancement of solid-state NMR spectra of alpha-synuclein fibrils. J Biomol NMR. 2007;39:197–211. doi: 10.1007/s10858-007-9189-z. [DOI] [PubMed] [Google Scholar]
- 55.Lorieau JL, McDermott AE. Conformational flexibility of a microcrystalline globular protein: Order parameters by solid-state NMR spectroscopy. J Am Chem Soc. 2006;128:11505–11512. doi: 10.1021/ja062443u. [DOI] [PubMed] [Google Scholar]
- 56.Struts AV, Salgado GFJ, Martínez-Mayorga K, Job C, Tanaka K, Krane S, Nakanishi K, Brown MF. 2HNMRrelaxation and dynamics of retinal cofactor in dark, meta I, and meta II states of rhodopsin. Biophys J. 2007;92:150A–151A. [Google Scholar]
- 57.Baldus M. Molecular interactions investigated by multidimensional solid-state NMR. Curr Opin Struct Biol. 2006;16:618–623. doi: 10.1016/j.sbi.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 58.Brown MF. Theory of spin-lattice relaxation in lipid bilayers and biological membranes. 2H and 14N quadrupolar relaxation. J Chem Phys. 1982;77:1576–1599. [Google Scholar]
- 59.Smith SO, Palings I, Miley ME, Courtin J, de Groot H, Lugtenberg J, Mathies RA, Griffin RG. Solid-state NMR studies of the mechanism of the opsin shift in the visual pigment rhodopsin. Biochemistry. 1990;29:8158–8164. doi: 10.1021/bi00487a025. [DOI] [PubMed] [Google Scholar]
- 60.Gröbner G, I, Burnett J, Glaubitz C, Choi G, Mason AJ, Watts A. Observations of light-induced structural changes of retinal within rhodopsin. Nature. 2000;405:810–813. doi: 10.1038/35015604. [DOI] [PubMed] [Google Scholar]
- 61.Lugtenburg J. The synthesis of 13C-labeled retinals. Pure Appl Chem. 1985;57:753–762. [Google Scholar]
- 62.Wada A, Fujioka N, Tanaka Y, Ito M. A highly stereoselective synthesis of 11Z-retinal using tricarbonyliron complex. J Org Chem. 2000;65:2438–2443. doi: 10.1021/jo9916030. [DOI] [PubMed] [Google Scholar]
- 63.Creemers AFL, Lugtenburg J. The preparation of all-trans uniformly 13C-labeled retinal via a modular total organic synthetic strategy. Emerging central contribution of organic synthesis toward the structure and function study with atomic resolution in protein research. J Am Chem Soc. 2002;124:6324–6334. doi: 10.1021/ja012368h. [DOI] [PubMed] [Google Scholar]
- 64.Struts AV, Salgado GFJ, Tanaka K, Krane S, Nakanishi K, Brown MF. Structural analysis and dynamics of retinal chromophore in dark and meta I states of rhodopsin from 2HNMR of aligned membranes. J Mol Biol. 2007;372:50–66. doi: 10.1016/j.jmb.2007.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Luecke H, Schobert B, Richter HT, Cartailler JP, Lanyi JK. Structure of bacteriorhodopsin at 1.55 Å resolution. J Mol Biol. 1999;291:899–911. doi: 10.1006/jmbi.1999.3027. [DOI] [PubMed] [Google Scholar]
- 66.Lanyi JK. Bacteriorhodopsin. Annu Rev Physiol. 2004;66:665–688. doi: 10.1146/annurev.physiol.66.032102.150049. [DOI] [PubMed] [Google Scholar]
- 67.Kusumi A, Hyde JS. Spin-label saturation-transfer electron spin resonance detection of transient association of rhodopsin in reconstituted membranes. Biochemistry. 1982;21:5978–5983. doi: 10.1021/bi00266a039. [DOI] [PubMed] [Google Scholar]
- 68.Ryba NJP, Marsh D. Protein rotational diffusion and lipid/protein interactions in recombinants of bovine rhodopsin with saturated diacylphosphatidylcholines of different chain lengths studied by conventional and saturation-transfer electron spin resonance. Biochemistry. 1992;31:7511–7518. doi: 10.1021/bi00148a011. [DOI] [PubMed] [Google Scholar]
- 69.Brown MF. Membrane structure and dynamics studied with NMR spectroscopy. In: Merz KM Jr, Roux B, editors. Biological Membranes: A Molecular Perspective from Computation and Experiment. Birkhäuser; Basel: 1996. pp. 175–252. [Google Scholar]
- 70.Petrache HI, Dodd SW, Brown MF. Area per lipid and acyl length distributions in fluid phosphatidylcholines determined by 2H NMR spectroscopy. Biophys J. 2000;79:3172–3192. doi: 10.1016/S0006-3495(00)76551-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Verdegem PJE, Bovee-Geurts PHM, de Grip WJ, Lugtenburg J, de Groot HJM. Retinylidene ligand structure in bovine rhodopsin, metarhodopsin-I, and 10-methyl-rhodopsin from internuclear distance measurements using 13C-labeling and 1-D rotational resonance MAS NMR. Biochemistry. 1999;38:11316–11324. doi: 10.1021/bi983014e. [DOI] [PubMed] [Google Scholar]
- 72.Spooner PJR, Sharples JM, Verhoeven MA, Lugtenberg J, Glaubitz C, Watts A. Relative orientation between the β-ionone ring and the polyene chain for the chromophore of rhodopsin in native membranes. Biochemistry. 2002;41:7549–7555. doi: 10.1021/bi020007o. [DOI] [PubMed] [Google Scholar]
- 73.Bax A. Weak alignment offers new NMR opportunities to study protein structure and dynamics. Protein Sci. 2003;12:1–16. doi: 10.1110/ps.0233303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cavanaugh J, Fairbrother WJ, Palmer AG, III, Skelton NJ, Rance M. Protein NMR Spectroscopy: Principles and Practice. 2. Academic Press; New York: 2006. [Google Scholar]
- 75.Rees DC, Chang G, Spencer RH. Crystallographic analyses of ion channels: Lessons and challenges. J Biol Chem. 2000;275:713–716. doi: 10.1074/jbc.275.2.713. [DOI] [PubMed] [Google Scholar]
- 76.Fu D, Libson A, Miercke LJW, Weitzman C, Nollert P, Krucinski J, Stroud RM. Structure of a glycerolconducting channel and the basis for its selectivity. Science. 2000;290:481–486. doi: 10.1126/science.290.5491.481. [DOI] [PubMed] [Google Scholar]
- 77.Nakamichi H, Okada T. Local peptide movement in the photoreaction intermediate of rhodopsin. Proc Natl Acad Sci USA. 2006;103:12729–12734. doi: 10.1073/pnas.0601765103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gröbner G, Taylor A, Williamson PTF, Choi G, Glaubitz C, Watts JA, deGrip WJ, Watts A. Macroscopic orientation of natural and model membranes for structural studies. Anal Biochem. 1997;254:132–138. doi: 10.1006/abio.1997.2415. [DOI] [PubMed] [Google Scholar]
- 79.Nevzorov AA, Moltke S, Heyn MP, Brown MF. Solid-state NMR line shapes of uniaxially oriented immobile systems. J Am Chem Soc. 1999;121:7636–7643. [Google Scholar]
- 80.Copié V, McDermott AE, Beshah K, Williams JC, Spyker-Assink M, Gebhard RT, Lugtenberg J, Herzfeld J, Griffin RG. Deuterium solid-state NMR studies of methyl group dynamics in bacteriorhodopsin and retinal model compounds: Evidence for a 6-s-trans chromophore in the protein. Biochemistry. 1994;33:3280–3286. doi: 10.1021/bi00177a019. [DOI] [PubMed] [Google Scholar]
- 81.Okada T, Ernst OP, Palczewski K, Hofmann KP. Activation of rhodopsin: New insights from structural and biochemical studies. Trends Biochem Sci. 2001;26:318–324. doi: 10.1016/s0968-0004(01)01799-6. [DOI] [PubMed] [Google Scholar]
- 82.Sakmar TP, Menon ST, Marin EP, Awad ES. Rhodopsin: Insights from recent structural studies. Annu Rev Biophys Biomol Struct. 2002;31:443–484. doi: 10.1146/annurev.biophys.31.082901.134348. [DOI] [PubMed] [Google Scholar]
- 83.Hubbell WL, Altenbach C, Hubbell CM, Khorana HG. Rhodopsin structure, dynamics, and activation: A perspective from crystallography, site-directed spin labeling, sulfhydryl reactivity, and disulfide cross-linking. Adv Prot Chem. 2003;63:243–290. doi: 10.1016/s0065-3233(03)63010-x. [DOI] [PubMed] [Google Scholar]
- 84.Palczewski K. G protein-coupled receptor rhodopsin. Annu Rev Biochem. 2006;75:743–767. doi: 10.1146/annurev.biochem.75.103004.142743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ridge KD, Palczewski K. Visual rhodopsin sees the light: Structure and mechanism of G protein signaling. J Biol Chem. 2007;282:9297–9301. doi: 10.1074/jbc.R600032200. [DOI] [PubMed] [Google Scholar]
- 86.Tanaka K, Struts AV, Krane S, Fujioka N, Salgado GFJ, Martínez-Mayorga K, Brown MF, Nakanishi K. Synthesis of CD3-labeled 11-cis-retinals and applications to solid-state deuterium NMR spectroscopy of rhodopsin. Bull Chem Soc Jpn. 2007;80:2177–2184. [Google Scholar]
- 87.Gröbner G, Choi G, Burnett IJ, Glaubitz C, Verdegem PJE, Lugtenberg J, Watts A. Photoreceptor rhodopsin: Structural and conformational study of its chromophore 11-cis retinal in oriented membranes by deuterium solid state NMR. FEBS Lett. 1998;422:201–204. doi: 10.1016/s0014-5793(97)01591-3. [DOI] [PubMed] [Google Scholar]
- 88.Chen YS, Hubbell WL. Temperature- and light-dependent structural changes in rhodopsin-lipid membranes. Exp Eye Res. 1973;17:517–532. doi: 10.1016/0014-4835(73)90082-1. [DOI] [PubMed] [Google Scholar]
- 89.Ulrich AS, Watts A, Wallat I, Heyn MP. Distorted structure of the retinal chromophore in bacteriorhodopsin resolved by 2H-NMR. Biochemistry. 1994;33:5370–5375. doi: 10.1021/bi00184a003. [DOI] [PubMed] [Google Scholar]
- 90.Huber T, Botelho AV, Beyer K, Brown MF. Membrane model for the GPCR prototype rhodopsin: Hydrophobic interface and dynamical structure. Biophys J. 2004;86:2078–2100. doi: 10.1016/S0006-3495(04)74268-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fishkin N, Berova N, Nakanishi K. Primary events in dim light vision: A chemical and spectroscopic approach toward understanding protein/chromophore interactions in rhodopsin. Chem Rec. 2004;4:120–135. doi: 10.1002/tcr.20000. [DOI] [PubMed] [Google Scholar]
- 92.Fujimoto Y, Fishkin N, Pescitelli G, Decatur J, Berova N, Nakanishi K. Solution and biologically relevant conformations of enantiomeric 11-cis-locked cyclopropyl retinals. J Am Chem Soc. 2002;124:7294–7302. doi: 10.1021/ja020083e. [DOI] [PubMed] [Google Scholar]
- 93.Jäger S, Lewis JW, Zvyaga TA, Szundi I, Sakmar TP, Kliger DS. Chromophore structural changes in rhodopsin from nanoseconds to microseconds following pigment photolysis. Proc Natl Acad Sci USA. 1997;94:8557–8562. doi: 10.1073/pnas.94.16.8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fujimoto Y, Ishihara J, Maki S, Fujioka N, Wang T, Furuta T, Fishkin N, Borhan B, Berova N, Nakanishi K. On the bioactive conformation of the rhodopsin chromophore: Absolute sense of twist around the 6-s-cis bond. Chem Eur J. 2001;7:4198–4204. doi: 10.1002/1521-3765(20011001)7:19<4198::aid-chem4198>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 95.Sugihara M, Hufen J, Buss V. Origin and consequences of steric strain in the rhodopsin binding pocket. Biochemistry. 2006;45:801–810. doi: 10.1021/bi0515624. [DOI] [PubMed] [Google Scholar]
- 96.Gascon JA, V, Batista S. QMMM study of energy storage and molecular rearrangements due to the primary event in vision. Biophys J. 2004;87:2931–2941. doi: 10.1529/biophysj.104.048264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Crocker E, Eilers M, Ahuja S, Hornak V, Hirshfeld A, Sheves M, Smith SO. Location of Trp265 in meta-rhodopsin II: Implications for the activation mechanism of the visual receptor rhodopsin. J Mol Biol. 2006;357:163–172. doi: 10.1016/j.jmb.2005.12.046. [DOI] [PubMed] [Google Scholar]
- 98.Park JH, Scheerer P, Hofmann KP, Choe HW, Ernst OP. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008;454:183–188. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- 99.Ruprecht JJ, Mielke T, Vogel R, Villa C, Schertler GFX. Electron crystallography reveals the structure of metarhodopsin I. EMBO J. 2004;23:3609–3620. doi: 10.1038/sj.emboj.7600374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chabre M, Breton J. The orientation of the chromophore of vertebrate rhodopsin in the “meta” intermediate states and the reversibility of the meta II-meta III transition. Vision Res. 1979;19:1005–1018. doi: 10.1016/0042-6989(79)90226-8. [DOI] [PubMed] [Google Scholar]
- 101.Yan ECY, Kazmi MA, Ganim Z, Hou JM, Pan D, Chang BSW, Sakmar TP, Mathies RA. Retinal counterion switch in the photoactivation of the G protein-coupled receptor rhodopsin. Proc Natl Acad Sci USA. 2003;100:9262–9267. doi: 10.1073/pnas.1531970100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lüdeke S, Beck R, Yan ECY, Sakmar TP, Siebert F, Vogel R. The role of Glu181 in the photoactivation of rhodopsin. J Mol Biol. 2005;353:345–356. doi: 10.1016/j.jmb.2005.08.039. [DOI] [PubMed] [Google Scholar]
- 103.Martínez-Mayorga K, Pitman MC, Grossfield A, Feller SE, Brown MF. Retinal counterion switch mechanism in vision evaluated by molecular simulations. J Am Chem Soc. 2006;128:16502–16503. doi: 10.1021/ja0671971. [DOI] [PubMed] [Google Scholar]
- 104.Huber T, Rajamoorthi K, Kurze VF, Beyer K, Brown MF. Structure of docosahexaenoic acid-containing phospholipid bilayers as studied by 2H NMR and molecular dynamics simulations. J Am Chem Soc. 2002;124:298–309. doi: 10.1021/ja011383j. [DOI] [PubMed] [Google Scholar]
- 105.Crozier PS, Stevens MJ, Forrest LR, Woolf TB. Molecular dynamics simulation of dark-adapted rhodopsin in an explicit membrane bilayer: Coupling between local retinal and larger scale conformational change. J Mol Biol. 2003;333:493–514. doi: 10.1016/j.jmb.2003.08.045. [DOI] [PubMed] [Google Scholar]
- 106.Lau PW, Grossfield A, Feller SE, Pitman MC, Brown MF. Dynamic structure of retinylidene ligand of rhodopsin probed by molecular simulations. J Mol Biol. 2007;372:906–917. doi: 10.1016/j.jmb.2007.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Isin B, Schulten K, Tajkhorshid E, Bahar I. Mechanism of signal propagation upon retinal isomerization: Insights from molecular dynamics simulations of rhodopsin restrained by normal modes. Biophys J. 2008;95:789–803. doi: 10.1529/biophysj.107.120691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Henzler-Wildman KA, Thai V, Lei M, Ott M, Wolf-Watz M, Fenn T, Pozharski E, Wilson MA, Petsko GA, Karplus M, Hübner CG, Kern D. Intrinsic motions along an enzymatic reaction trajectory. Nature. 2007;450:838–844. doi: 10.1038/nature06410. [DOI] [PubMed] [Google Scholar]