Abstract
Purpose
To demonstrate an MRI method for directly visualizing amyloid-β (Aβ) plaques in the APP/PS1 transgenic (tg) mouse brain in vivo, and show that T1ρ relaxation rate increases progressively with Alzheimer’s disease (AD)-related pathology in the tg mouse brain.
Materials and Methods
We obtained in vivo MR images of a mouse model of AD (APP/PS1) that overexpresses human amyloid precursor protein, and measured T1ρ via quantitative relaxometric maps.
Results
A significant decrease in T1ρ was observed in the cortex and hippocampus of 12- and 18-month-old animals compared to their age-matched controls. There was also a correlation between changes in T1ρ and the age of the animals.
Conclusion
T1ρ relaxometry may be a sensitive method for noninvasively determining AD-related pathology in APP/PS1 mice.
Keywords: Alzheimer’s disease, amyloid-β, T1rho, spin-lock imaging, MRI
Alzheimer’s disease (AD) is the most common form of dementia in the elderly (1). Over 4 million people are affected by AD in the United States alone, and as the aging population increases, this number is expected to double by 2025 (2). The classic clinical symptoms of AD include memory loss and confusion. The neuropathological features are neurofibrillary tangles (NFTs) formed by paired helical filaments composed of tau protein, senile plaques (SPs) resulting from Aβ deposits, and neuron loss in the limbic and neocortical regions (3,4). Neuronal loss associated with early AD eventually leads to brain dysfunction and atrophy. Volumetric assessment methods are the current state of the art in magnetic resonance imaging (MRI) of AD in humans and in animal models of AD pathologies. These methods measure gross morphological changes and track disease progression, but do not provide information about biochemical changes. Conventional MRI of the brain, which relies on contrast generated by the variation of T1 and T2 or T2* relaxation times of water in tissue, has proven inadequate for observing authentic SPs and NFTs in vivo. This is primarily due to the high resolution and thin section (~10 µm) images required to visualize SPs, which currently are not achievable with MRI. However, interactions between macromolecules and bulk water, and changes in macromolecular content can be indirectly quantified by spatially mapping MR relaxation times. In these relaxometric maps, any change in relaxation times due to the presence of SPs will be reflected over an entire volume-averaged pixel, thereby providing an indirect method of detection without the need for very-high-resolution images.
Intriguing data suggesting that plaques can be detected in brain tissue specimens by T2*-weighted MRI have been reported (5); however, another study using similar methods failed to confirm this (6). Both studies were performed on formalin-fixed brain tissue specimens, and the MR characteristics of fixed tissue differ from those in vivo. T2-weighted MRI was employed to visualize plaques in a transgenic (tg) mouse model of AD ex vivo (7,8). Recent work has shown the possibility of detecting Aβ deposits in vivo in the APP/PS1 tg mouse model of AD using T2- and T2*-weighted MRI (9,10). The presence of iron in these plaques also exaggerates the actual size of plaques in T2*-weighted images, which makes the accurate quantification of plaque density particularly challenging (9). Also, although very-high-resolution T2*-weighted MRI (<100 µm) has been used to image plaque formations in human AD-affected tissue specimens (5), other experiments have contradicted such findings by demonstrating that plaques are not visible in T2* images (6). Previous studies have demonstrated the efficacy of gadolinium and magnetically labeled Aβ1–40 for visualizing Aβ deposits in an APP/PS1 tg model ex vivo (11) and in vivo (12). However, given the neurotoxicity of Aβ and the requirement for mannitol injection for the in vivo experiments, these methods may have limited utility in a clinical setting. Further, in vivo relaxometric measurements established differences in T2 on the order of a few milliseconds (13) between these mice and age-matched controls. Although these were meticulous experiments, Campeau et al (14) previously demonstrated that T2 did not significantly differ between AD patients and age-matched normal subjects, which suggests that T2-weighted MRI may have limited clinical utility.
An alternate contrast mechanism is T1ρ, or T1rho, the spin lattice relaxation time constant in the rotating frame, which determines the decay of the transverse magnetization in the presence of a “spin-lock” radiofrequency (RF) field. In biological tissues T1ρ is dependent on the macromolecular composition and provides contrast, unlike conventional T1/T2-based methods. While quantifying T2* with MRI can be prone to errors from susceptibility-induced signal losses, and T2-weighted MR signal is degraded by diffusion, T1ρ-weighted MR images are less prone to such losses and have a greater dynamic range of values as recently demonstrated in the human brain (15). Therefore, quantitative T1ρ MRI is an attractive alternative strategy for detecting and quantifying Aβ deposits, especially since it probes slow molecular interactions and the dynamics of water molecules in tissues based on variations in protein content (16).
The development of novel diagnostic techniques for AD requires the study of an animal model in vivo before these techniques can be translated to clinical utility. For example, if the disease process can be detected in an in vivo animal model of AD with MRI, and this is validated by histological studies, a paradigm for visualizing the presence of AD pathology can be established. Transgenic (tg) mice (APP/PS1) expressing the pathogenic familial AD-associated form of amyloid precursor protein (APP-Swedish/Tg2576) and presenilin-1 (PS1, P246L/wt) were shown to develop a large number of fibrillary Aβ SP-like deposits in the brain, especially in the cerebral cortex and hippocampus (17,18).
In the present work we demonstrate an MRI method for directly visualizing Aβ plaques in the APP/PS1 tg mouse brain in vivo, and show that the T1ρ relaxation rate increases progressively with AD-related pathology in the tg mouse brain.
MATERIALS AND METHODS
Animal Preparation
The institutional animal care and use committee granted approval for all animal experiments discussed in this work. Two APP/PS1 tg mice and two age-matched controls in three age groups (6, 12, and 18 months) were imaged. The mice were initially anesthetized with intraperitoneal injection of 0.1 cc of ketamine/ acepromazine. The mice were then mounted on a bed and connected to an isoflurane (Abbott Labs, Abbott Park, IL, USA) inhalation anesthesia apparatus. A flow rate of 1 liter/minute with 1.5% isoflurane in air was maintained during the experiments. The bed was then placed into an in-house-built, 3-cm-diameter birdcage coil, and the entire apparatus was placed inside the center of the magnet bore. Throughout the experiments each animal’s heart rate was monitored by ECG, and body temperature was monitored by rectal thermometer.
Imaging Protocol
MRI was performed on a 4.7 Tesla horizontal-bore imaging spectrometer connected to a Varian console (Varian Inc., Palo Alto, CA, USA) with a 12-cm, actively-shielded gradient set capable of 25 G/cm amplitude (Magnex Scientific, Abingdon, UK). A two-dimensional (2D) gradient-echo localizer imaging sequence was run in three orthogonal planes through the center of the gradient sets to facilitate reproducible slice planning. The position of each slice relative to the rostral tip of the brain was noted, and extreme care was taken to ensure that the head and body of the animal lay flat on the bed to facilitate subsequent matching with histological sections.
High-Resolution MRI
Initially, two 18-month-old tg mice and two age-matched controls underwent high-resolution MRI. T1ρ-weighted images were acquired with a 2D gradient-echo-based T1ρ pulse sequence (19). Typical imaging parameters were TE/TR = 2 msec/500 msec, flip angle = 45°, FOV = 2 cm × 2 cm, thickness = 130 µm, matrix size = 256 × 256, in-plane pixel size = 78 µm × 78 µm. The time of spin-lock (TSL) was fixed at 15 msec, and the spin-lock field (γB1) was 500 Hz. The total imaging time was a little under three hours.
T1ρ Mapping
A series T1ρ-weighted images were acquired from each animal with a 2D spin-echo-based T1ρ pulse sequence (20) with six equally spaced TSLs from 10 to 60 msec. These images were 250 µm-thick pixel size was 156 µm × 156 µm and were used to generate T1ρ maps by fitting signal intensities as function of TSL by linear regression. In the fitting routine, pixels whose intensities correlated poorly (i.e., R2 < 0.95) with TSL were set to zero. This process guaranteed that noisy pixels were rejected from the maps. Other imaging parameters were TE/TR= 7 msec/2000 msec, FOV = 2 cm × 2 cm, and a spin-locking field of 500 Hz. The total imaging time was about three hours for an entire data set of six TSLs.
Histology
After the MR experiments the mice were subjected to histological analysis. The mice were lethally anesthetized and perfused intracardially with phosphate-buffered saline (PBS, 0.1 M, pH 7.4) followed by 10% neutral buffered formalin. Their brains were rapidly removed, immediately frozen on dry ice, and stored overnight at −80°. Blocks were cut by cryostat in a near serial array of 40-µm-thick coronal sections for analysis. Finally, we identified characteristic features of SPs in paraffin sections by subjecting them to histochemistry with Thioflavin-S and immunohistochemistry with anti-Aβ antibodies as described previously (21). Histologic slides were digitally scanned with a Nikon (Nikon Corp., Japan) slide scanner. Three consecutive immunostained sections were selected and manually overlaid by visual inspection of brain structures using Photo-Shop (Adobe Systems Inc., San Jose, CA, USA) software to generate a 120-µm-thick section for comparison with the corresponding MR image.
Data Processing
The MR images were transferred to a Macintosh G4 PowerBook computer (Apple Computers Inc., Cupertino, CA, USA). Basic image processing (3 × 3 smoothing filter, and contrast and brightness adjustments) was performed with custom-written software in the Interactive Data Language (RSI, Boulder, CO, USA). Signal-to-noise ratios (SNRs) were measured as the mean signal intensity in a 20 × 20 pixel region of interest (ROI) within the brain parenchyma in each image divided by the mean intensity of background pixels in a 20 × 20 pixel region outside the animal’s anatomy. An increase in plaque density is expected in older animals and in anatomical locations such as the hippocampi and cortex (22). Therefore, we chose ROIs for the measurement of the T1ρ values in these specific regions (Fig. 1). T1ρ values were recorded from 10 × 10 pixel ROIs, which were manually selected by a single user (A.B.) in the hippocampi, cortex, and thalami, as indicated in the figure. All 100 T1ρ values in each ROI were tabulated in an Excel spreadsheet (Microsoft Corp.). The data from both animals in each group were averaged at each location. A Student’s t-test was performed to determine the statistical significance of the difference between the data in each tg group and its corresponding age-matched control group. In a similar manner, the statistical significance of the inter-age difference among the three tg groups was also determined.
Figure 1.
Location of the ROIs used for all data analyses overlaid on a T1ρ-weighted image. Actual data were collected in the calculated T1ρ map, an example of which is shown in color in Fig. 3. The trapezoid represents the boundary of the measurements performed in the cortex, the solid squares are located in the hippocampi, and the dashed square is in the thalamus. The total number of distinct pixels and therefore T1ρ values in each location was 100.
RESULTS
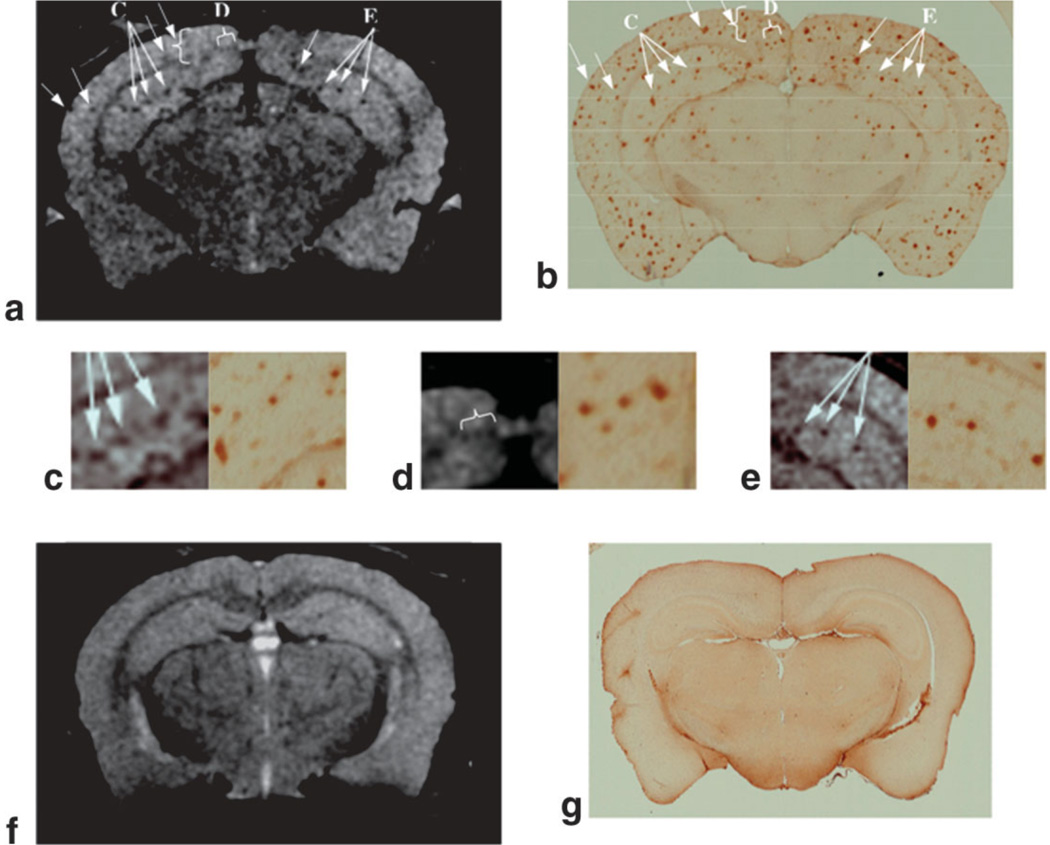
Representative high-resolution (78 µm × 78 µm in-plane and 130-µm slice thickness) T1ρ-weighted MR images of the tg and control mice are shown in Fig. 2a and f, respectively. Several regions of hypointensity (some indicated by arrows) are evident in the brain of the tg animal (Fig. 2a) corresponding to the location of Aβ plaques in the histological section (Fig. 2b). This is also seen in the MR images in the middle row of the figure, where the regions indicated by the arrows in Fig. 2a and b are magnified alongside their corresponding histological images (Fig. 2c–e). Analysis of the images revealed that several large plaques were visible in the hippocampus and cortex by MRI. MR image pixels containing such plaques have reduced T1ρ values because they exhibit lower signal intensity in the T1ρ-weighted images. However, not all the plaques seen in the histological section could be matched with the MRI section, and some of the plaques visible on histology were too small to be detected in the T1ρ-weighted images.
Figure 2.
a: A T1ρ-weighted MR image of the APP/PS1 mouse brain shows hypointense regions, some of which are indicated by arrows. b: The corresponding histologic image created by combining five 10-µm-thick immunostained sections shows abundant Aβ deposits. c–e: The higher magnification views correspond to regions indicated by the arrows in part a to show identical clusters of Aβ deposits in the hippocampus and cortex in both MR images and histological sections. The T1ρ-weighted MR image of an age-matched control (f) and corresponding histology (g) failed to show SPs.
Typical in vivo T1ρ-weighted images that were used to generate T1ρ maps of the mouse brain are displayed in Fig. 3. The SNRs for the images displayed in this figure were 36:1 for the earliest TSL image (10 msec) and 16:1 for the longest TSL image (60 msec). These images provided T1ρ relaxometric maps (Fig. 4) in which anatomical structures, such as the hippocampi, corpus callosum, and ventricles, are readily discerned. In Fig. 4 a T1ρ map of an 18-month-old tg animal and its age-matched control are displayed above their corresponding Thioflavin-stained histologic images. The large diffused plaque aggregates, which are bright green under the fluorescein isothiocyanate filters, are located primarily in the cortex and hippocampi of the animals. Some of these aggregates (indicated with arrows) coincide with regions of decreased T1ρ values.
Figure 3.
Example of T1ρ images from a single data set obtained at different durations of the spin-lock pulse (TSL) that was used to calculate the T1ρ map (rightmost image, shown in color). In such maps, each pixel’s intensity is the actual T1ρ relaxation time constant of that pixel. Values above 100 msec, such as in fluid in the ventricles, are set to yellow, and pixels that did not correlate with TSL in the fitting routine, such as in the skull and background, were set to 0 (transparent).
Figure 4.
T1ρ color map overlaid on a grayscale T1ρ-weighted image (a) and corresponding Thioflavin-S stained histology section (b) of an 18-month-old tg mouse, and a T1ρ map of an age-matched control (c). During the MRI, identical slices were selected in both animals for meaningful comparisons. Although the histologic section is 40 µm thick and the MRI slice thickness is 250 µm, some of the pixels with low T1ρ values are in the same location as large Aβ deposits in the histology (indicated in a and b). The color bar-scale on the left indicates the range of T1ρ values.
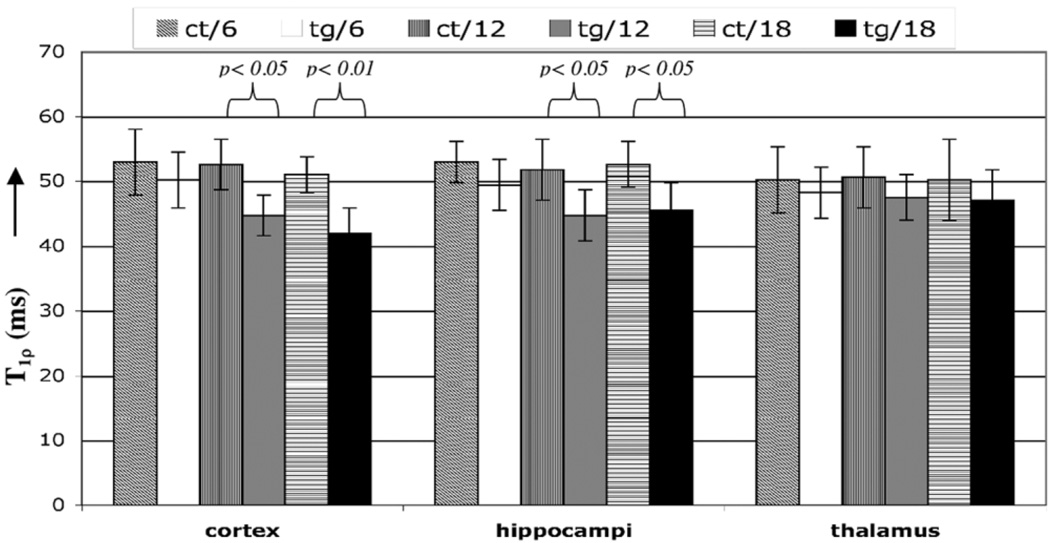
The results of T1ρ measurements are summarized in Table 1 and displayed in Fig. 5. At 6 months, when the tg animals had few plaques, they did not show any significant decrease in relaxation times. However, T1ρ was reduced in the cortex of the tg animals compared to the control animals at both 12 months (P < 0.05) and 18 months (P < 0.01) of age, when plaques are abundant in mice (18). A similar trend was observed in the hippocampus of the 12- and 18-month-old tg animals compared to their age-matched controls, but the difference was smaller (P < 0.05 in both cases). The inter-age difference in relaxation times among the three age groups of tg animals is tabulated in Table 2. Statistically significant (P < 0.05) differences were observed between the 12-month-old animals that exhibited an 11% decreased T1ρ in the cortex than the 6-month-old tg animals and 10% decrease in the hippocampus. The 18-month-old animal had 16% and 8% lower T1ρ in the cortex and hippocampus, respectively.
Table 1.
Summary of the Results of T1ρ Measurements
| T1 ρ (msec) | ||||||
|---|---|---|---|---|---|---|
| 6 month | 12 month | 18 month | ||||
| Control (N = 2) | tg (N = 2) | Control (N = 2) | tg (N = 2) | Control (N = 2) | tg (N = 2) | |
| Cortex | 53.1 ± 5.1 | 50.2 ± 4.3 | 52.6 ± 3.9 | 44.7 ± 3.1 | 51 ± 2.8 | 41.9 ± 3.8 |
| Hippocampus | 52.9 ± 3.1 | 49.4 ± 3.7 | 51.8 ± 4.7 | 44.7 ± 3.9 | 52.5 ± 3.5 | 45.5 ± 4.3 |
| Thalamus | 50.2 ± 5.1 | 48.2 ± 3.9 | 50.6 ± 4.3 | 47.4 ± 3.5 | 50.2 ± 6.3 | 47.1 ± 4.7 |
Figure 5.
T1ρ values averaged from ROIs in both animals, in the locations indicated. The maximum difference was observed between the 18-month-old tg animals and controls (9 msec), and was statistically significant (P < 0.01). The difference between the 12-month-old tg mice and controls was significant in both the cortex and hippocampus. The thalamus had reduced T1ρ values (~2 msec) in all age groups, but the difference was not significant.
Table 2.
Interage Difference Among the Three Age Groups of tg Animals
| ΔT1ρ (msec) | ||
|---|---|---|
| 6 month tg | 12 month tg | |
| 12 month tg | 5.5 msec, P < 0.05 (cortex) | |
| 4.7 msec, P < 0.05 (hippocampus) | ||
| 18 month tg | 8.3 msec, P < 0.01 (cortex) | 2.8 msec, P < 0.05 (cortex) |
| 3.9 msec, P < 0.05 (hippocampus) | 0.8, not significant (hippocampus) | |
DISCUSSION
In conclusion, analyses of T1ρ values in this study showed that the difference in mean T1ρ values between 18-month-old tg mice and their age-matched controls was greater than the T2 difference (3 msec) reported in a similar tg animal model (13). The lower dynamic range of spin-spin relaxation time could also explain why no difference was observed between AD-affected and normal human brain tissue specimens in T2*-weighted images (6). T1ρ relaxation occurs in the presence of a spin-lock pulse that minimizes the effects of diffusion and susceptibility on the signal, which would explain why T1ρ values are greater than the reported T2 values in the same animal. Further, at the spin-lock amplitude of 500 Hz used in the current experiments, the T1ρ signal was sensitive to relaxation processes that occur in the milliseconds range (e.g., chemical exchange-mediated dipolar relaxation). The greater distribution of T1ρ values further implies that T1ρ-weighted MRI can be used to distinguish tissues with similar T2. Incidentally, the increased distribution of T1ρ values compared to T2 was also observed in the human brain (15).
The observed reduction in T1ρ in the older tg animals may result not only from increased Aβ deposition, as demonstrated in Fig. 4a, but also from decreased blood flow and blood volume, which are associated with Aβ pathology (23,24). More studies are needed to conclusively determine the relevant relaxation processes and the degrees to which these factors affect the T1ρ relaxation time. To this end, arterial spin-labeled (25) and spin-lock (26) MRI methods can be incorporated into the current protocol to quantify blood flow and perfusion.
The SNR of the images used to calculate T1ρ maps has to be high to reduce the error in the measurement of T1ρ via curve-fitting methods. Since noisier data values are introduced into the fitting procedure, T1ρ maps appear more pixelated because identical tissues would erroneously exhibit different T1ρ values. In the data presented, the longest-TSL image had an SNR of 16:1; however, higher SNR can be achieved by the use of surface coils for T1ρ-weighted MRI (27), which is currently being developed for application in the tg animal model.
The MRI slice thickness is three times that of the histologic image, and the MR image’s in-plane resolution is 78 µm × 78 µm. These factors lead to partial voluming and cause mismatching of some large plaques and inadequate resolution to visualize smaller plaques. However, this resolution cannot be achieved on clinical MRI scanners in a practical imaging time. Therefore, it is difficult to translate this technique to direct visualization of plaques in human patients. However, as the T1ρ-weighted image of the tg mouse brain demonstrates, the presence of plaques reduces T1ρ relaxation times and could therefore be indirectly quantified even with lower-resolution T1ρ maps.
The results from this study will form the basis for extending this strategy to a large cohort of animals to study disease progression or response to therapeutic intervention, and eventually to similar measurements in humans in vivo. However, extending T1ρ mapping to clinical studies would require additional considerations, such as patient comfort and involuntary motion, which places constraints on the total imaging time. Furthermore, for clinical imaging the imaging protocol must not exceed FDA-mandated energy deposition levels (measured as the specific absorption rate (SAR)) (28). A protocol for mapping T1ρ in the human brain images with submillimeter resolution in-plane and 2-mm slice thickness can be achieved in two minutes for each TSL by combining a fast spin-echo T1ρ sequence with a low-SAR sequence (15,29). By employing such a protocol, the total MRI time for five different TSL images that will be needed to calculate T1ρ maps is ~10 minutes.
An accurate comparison of measurements from histology and MRI necessitates a comparison of data generated with different orientations, slice thicknesses, and resolutions. Ideally, the same slice section thicknesses and orientations should be obtained by both methods for comparison. In the current experiments we achieved this by taking extreme care in aligning the animals inside the scanner and measuring the location of the imaged slice for subsequent histology. However, we will explore the application of several image registration methods in future studies to enable more accurate and reproducible comparisons of MRI and histology. Nonetheless, T1ρ MRI holds promise for imaging AD to confirm the diagnosis of this disorder and perhaps detect it in its earliest stages without the need to inject tracers or contrast agents.
ACKNOWLEDGMENTS
We thank Professor John S. Leigh for his encouragement and support. This work was performed at the MMRRCC, an NIH regional resource (NIH RR02305), and the Center for Neurodegenerative Disease Research, an NIH-funded Alzheimer’s Disease Center.
REFERENCES
- 1.Dawbarn D, Allen SJ. Neurobiology of Alzheimer’s disease. New York: Oxford University Press; 2001. p. 409. xvi. [Google Scholar]
- 2.Prince M. The need for research on dementia in developing countries. Trop Med Int Health. 1997;2:993–1000. doi: 10.1046/j.1365-3156.1997.d01-156.x. [DOI] [PubMed] [Google Scholar]
- 3.Selkoe DJ. Translating cell biology into therapeutic advances in Alzheimer’s disease. Nature. 1999;399 Suppl 6738:A23–A31. doi: 10.1038/399a023. [DOI] [PubMed] [Google Scholar]
- 4.Lee VM, Goedert M, Trojanowski JQ. Neurodegenerative tauopathies. Annu Rev Neurosci. 2001;24:1121–1159. doi: 10.1146/annurev.neuro.24.1.1121. [DOI] [PubMed] [Google Scholar]
- 5.Benveniste H, Einstein G, Kim KR, Hulette C, Johnson GA. Detection of neuritic plaques in Alzheimer’s disease by magnetic resonance microscopy. Proc Natl Acad Sci USA. 1999;96:14079–14084. doi: 10.1073/pnas.96.24.14079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dhenain M, Privat N, Duyckaerts C, Jacobs RE. Senile plaques do not induce susceptibility effects in T2*-weighted MR microscopic images. NMR Biomed. 2002;15:197–203. doi: 10.1002/nbm.760. [DOI] [PubMed] [Google Scholar]
- 7.Zhang J, Yarowsky P, Gordon MN, et al. Detection of amyloid plaques in mouse models of Alzheimer’s disease by magnetic resonance imaging. Magn Reson Med. 2004;51:452–457. doi: 10.1002/mrm.10730. [DOI] [PubMed] [Google Scholar]
- 8.Lee SP, Falangola MF, Nixon RA, Duff K, Helpern JA. Visualization of beta-amyloid plaques in a transgenic mouse model of Alzheimer’s disease using MR microscopy without contrast reagents. Magn Reson Med. 2004;52:538–544. doi: 10.1002/mrm.20196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jack CR, Jr, Garwood M, Wengenack TM, et al. In vivo visualization of Alzheimer’s amyloid plaques by magnetic resonance imaging in transgenic mice without a contrast agent. Magn Reson Med. 2004;52:1263–1271. doi: 10.1002/mrm.20266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vanhoutte G, Dewachter I, Borghgraef P, Van Leuven F, Van der Linden A. Noninvasive in vivo MRI detection of neuritic plaques associated with iron in APP[V717I] transgenic mice, a model for Alzheimer’s disease. Magn Reson Med. 2005;53:607–613. doi: 10.1002/mrm.20385. [DOI] [PubMed] [Google Scholar]
- 11.Poduslo JF, Wengenack TM, Curran GL, et al. Molecular targeting of Alzheimer’s amyloid plaques for contrast-enhanced magnetic resonance imaging. Neurobiol Dis. 2002;11:315–329. doi: 10.1006/nbdi.2002.0550. [DOI] [PubMed] [Google Scholar]
- 12.Wadghiri YZ, Sigurdsson EM, Sadowski M, et al. Detection of Alzheimer’s amyloid in transgenic mice using magnetic resonance microimaging. Magn Reson Med. 2003;50:293–302. doi: 10.1002/mrm.10529. [DOI] [PubMed] [Google Scholar]
- 13.Helpern JA, Lee SP, Falangola MF, et al. MRI assessment of neuropathology in a transgenic mouse model of Alzheimer’s disease. Magn Reson Med. 2004;51:794–798. doi: 10.1002/mrm.20038. [DOI] [PubMed] [Google Scholar]
- 14.Campeau NG, Petersen RC, Felmlee JP, O’Brien PC, Jack CR., Jr Hippocampal transverse relaxation times in patients with Alzheimer disease. Radiology. 1997;205:197–201. doi: 10.1148/radiology.205.1.9314985. [DOI] [PubMed] [Google Scholar]
- 15.Borthakur A, Wheaton AJ, Gougoutas AJ, et al. In vivo measurement of T1rho dispersion in the human brain at 1.5 tesla. J Magn Reson Imaging. 2004;19:403–409. doi: 10.1002/jmri.20016. [DOI] [PubMed] [Google Scholar]
- 16.Akella SV, Reddy Regatte R, Gougoutas AJ, et al. Proteoglycan induced changes in T1ρ-relaxation of articular cartilage at 4T. Magn Reson Med. 2001;46:419–423. doi: 10.1002/mrm.1208. [DOI] [PubMed] [Google Scholar]
- 17.Jaffar S, Counts SE, Ma SY, et al. Neuropathology of mice carrying mutant APP(swe) and/or PS1(M146L) transgenes: alterations in the p75(NTR) cholinergic basal forebrain septohippocampal pathway. Exp Neurol. 2001;170:227–243. doi: 10.1006/exnr.2001.7710. [DOI] [PubMed] [Google Scholar]
- 18.Flood DG, Reaume AG, Dorfman KS, et al. FAD mutant PS-1 gene-targeted mice: increased A beta 42 and A beta deposition without APP overproduction. Neurobiol Aging. 2002;23:335–348. doi: 10.1016/s0197-4580(01)00330-x. [DOI] [PubMed] [Google Scholar]
- 19.Borthakur A, Wheaton AJ, Charagundla SR, et al. Three-dimensional T1ρ-weighted MRI at 1.5 Tesla. J Magn Reson Imaging. 2003;17:730–736. doi: 10.1002/jmri.10296. [DOI] [PubMed] [Google Scholar]
- 20.Wheaton AJ, Borthakur A, Charagundla SR, Reddy R. Pulse sequence for multislice T1rho-weighted MRI. Magn Reson Med. 2004;51:362–369. doi: 10.1002/mrm.10705. [DOI] [PubMed] [Google Scholar]
- 21.Skovronsky DM, Zhang B, Kung MP, Kung HF, Trojanowski JQ, Lee VM. In vivo detection of amyloid plaques in a mouse model of Alzheimer’s disease. Proc Natl Acad Sci USA. 2000;97:7609–7614. doi: 10.1073/pnas.97.13.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reilly JF, Games D, Rydel RE, et al. Amyloid deposition in the hippocampus and entorhinal cortex: quantitative analysis of a transgenic mouse model. Proc Natl Acad Sci USA. 2003;100:4837–4842. doi: 10.1073/pnas.0330745100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang F, Eckman C, Younkin S, Hsiao KK, Iadecola C. Increased susceptibility to ischemic brain damage in transgenic mice overexpressing the amyloid precursor protein. J Neurosci. 1997;17:7655–7661. doi: 10.1523/JNEUROSCI.17-20-07655.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niwa K, Kazama K, Younkin SG, Carlson GA, Iadecola C. Alterations in cerebral blood flow and glucose utilization in mice overexpressing the amyloid precursor protein. Neurobiol Dis. 2002;9:61–68. doi: 10.1006/nbdi.2001.0460. [DOI] [PubMed] [Google Scholar]
- 25.Zhang W, Williams DS, Detre JA, Koretsky AP. Measurement of brain perfusion by volume-localized NMR spectroscopy using inversion of arterial water spins: accounting for transit time and cross-relaxation. Magn Reson Med. 1992;25:362–371. doi: 10.1002/mrm.1910250216. [DOI] [PubMed] [Google Scholar]
- 26.Dixon WT, Oshinski JN, Trudeau JD, Arnold BC, Pettigrew RI. Myocardial suppression in vivo by spin locking with composite pulses. Magn Reson Med. 1996;36:90–94. doi: 10.1002/mrm.1910360116. [DOI] [PubMed] [Google Scholar]
- 27.Borthakur A, Charagundla SR, Wheaton A, Reddy R. T1ρ-weighted MRI using a surface coil to transmit spin-lock pulses. J Magn Reson. 2004;167:306–316. doi: 10.1016/j.jmr.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 28.Guidance for industry and FDA staff-criteria for significant risk investigations of magnetic resonance diagnostic devices. Guidance for industry and FDA staff—criteria for significant risk investigations of magnetic resonance diagnostic devices. [Accessed 2003 September 25];U.S. Food and Drug Administration. 2003 July 14; http://www.fda.gov/cdrh/ode/guidance/793/html.
- 29.Wheaton AJ, Borthakur A, Corbo M, Charagundla SR, Reddy R. Method for reduced SAR T1rho-weighted MRI. Magn Reson Med. 2004;51:1096–1102. doi: 10.1002/mrm.20141. [DOI] [PubMed] [Google Scholar]