Abstract
The first Swiss human embryonic stem cell (hESC) line, CH-ES1, has shown features of a malignant cell line. It originated from the only single blastomere that survived cryopreservation of an embryo, and it more closely resembles teratocarcinoma lines than other hESC lines with respect to its abnormal karyotype and its formation of invasive tumors when injected into SCID mice. The aim of this study was to characterize the molecular basis of the oncogenicity of CH-ES1 cells, we looked for abnormal chromosomal copy number (by array Comparative Genomic Hybridization, aCGH) and single nucleotide polymorphisms (SNPs). To see how unique these changes were, we compared these results to data collected from the 2102Ep teratocarcinoma line and four hESC lines (H1, HS293, HS401 and SIVF-02) which displayed normal G-banding result. We identified genomic gains and losses in CH-ES1, including gains in areas containing several oncogenes. These features are similar to those observed in teratocarcinomas, and this explains the high malignancy. The CH-ES1 line was trisomic for chromosomes 1, 9, 12, 17, 19, 20 and X. Also the karyotypically (based on G-banding) normal hESC lines were also found to have several genomic changes that involved genes with known roles in cancer. The largest changes were found in the H1 line at passage number 56, when large 5 Mb duplications in chromosomes 1q32.2 and 22q12.2 were detected, but the losses and gains were seen already at passage 22. These changes found in the other lines highlight the importance of assessing the acquisition of genetic changes by hESCs before their use in regenerative medicine applications. They also point to the possibility that the acquisition of genetic changes by ESCs in culture may be used to explore certain aspects of the mechanisms regulating oncogenesis.
Introduction
Human embryonic stem cells (hESCs) and human embryonal carcinoma cells (hECs) are two pluripotent cell types that share many characteristics [1] Human ECs are the malignant stem cells of teratocarcinomas, which are malignant tumors that have embryonal carcinoma components, and some may form teratocarcinomas when re-transplanted into an animal [2]. Both hECs and hESCs can differentiate into many cell types, but the differentiation potential of hECs is limited compared to that of hESC lines [1], [3], [4]. Before the clearly malignant line CH-ES1 [5] was developed, human ESC lines were reported to be benign; they form teratomas comprising differentiated tissue components of the three embryonic germ layers after injection into immune-incompetent mice, but they usually do not form teratocarcinomas. After culture adaptation, hESC lines can develop malignant features [6], but their ability to form tumors has not been analyzed in detail.
Human ESC lines have been most often derived from the inner cell mass of blastocyst-stage embryos [7] but they have also been derived from eight-cell stage morula embryos [8]. Klimanskaya et al. derived hESC lines from single isolated blastomeres at first by co-culture with other hESCs [9] but they were subsequently able to do so without such support [10]. These lines had normal karyotypes, and they formed teratomas when grown as xenografts. In another study, Van de Velde et al. [11] were able to obtain pluripotent cell lines from single blastomeres derived from four-cell stage embryos. These embryos had been established for this purpose and were of good quality. Nonetheless, the first line was karyotypically abnormal.
We established an hESC line from the only surviving blastomere of a four-cell stage embryo. This single cell survived freezing and thawing [5] and produced a cell line expressing the typical markers of hECs and hESCs. This line proved to be chromosomally very abnormal and was highly invasive when transplanted into SCID mice [5]. Hence, it has characteristics more similar to hECs than to hESCs.
In the present study, we have characterized the genomic changes that may explain the enhanced oncogenicity of the CH-ES1 teratocarcinoma-like hESC line relative to other pluripotent cell lines. We used both comparative genomic hybridization (aCGH) and single nucleotide polymorphism (SNP) genotyping to detect genomic changes in the CH-ES1 line, the 2102Ep teratocarcinoma line and four benign hESC lines (H1, HS293, HS401 and SIVF-02) originating from three different laboratories. In addition to finding extensive genomic abnormalities in the CH-ES1 line, we also found that the H1, HS293, HS401 and SIVF-02 lines share the general characteristics of hESCs that have been described by the International Stem Cell Initiative (ISCI) 1 [12]. We observed suggestive culture adaptation and growth advantages in these lines, as well as gains of known oncogenes and the possible deletion or loss of putative yet unrecognized tumor suppressor genes. The SNP arrays also revealed potentially tumorigenic changes in the karyotypically normal hESC lines.
Results
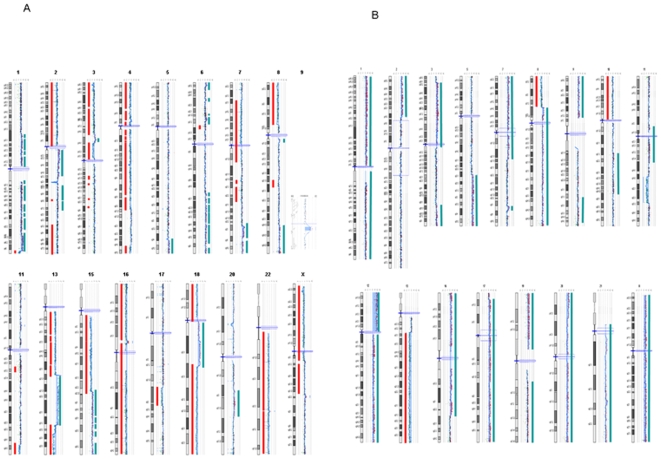
In the teratocarcinoma line 2102Ep and in the teratocarcinoma-like line CH-ES1, the chromosomal complement was highly aneuploid (Figure 1). CGH analysis of CH-ES1 confirmed a high level of genomic imbalances in agreement with earlier G-banding results. We performed a high resolution CGH analysis that revealed a higher frequency of genomic losses compared to gains in CH-ES1 (Table 1). The regions of reduced copy number ranged from 6 to 88 Mb and involved chromosomes 1q, 3p, 4p, 4q, 8p, 11q, 13q, 15q, 16, 17q, and 18p. Duplicated regions (0.4 to 60 Mb) were seen in chromosomes 2q, 5q, 6p, 6q, 7q, 8q, 9q, 13q, 15q, and 18q (Table 2). There were partial trisomies of chromosomes 1, 9, 12, 19, 20 and X, and a duplication of 17p13.2-qtel (3.674-tel). Only chromosome 14 was normal in the CGH assay. When compared to the 2102Ep teratocarcinoma line in the CGH assay, CH-ES1 displayed two common large aberrations, namely, duplication in 5q34 and a deletion of 13q32.1-q34 (Table 2).
Figure 1. Cartography of genetic aberrations which were found in the CH-ES1 and, 2102Ep cell lines.
A) Line CH-ES1, B) Line 2102Ep. Blue bars show duplicated regions and red ones show deleted regions.
Table 1. Genetic aberrations found in the H1, CH-ES1, and 2102Ep cell lines.
| Line | Number of genes | Status |
| 2102 Ep | 394 | Deleted |
| 7665 | Duplicated | |
| CH-ES1 | 4019 | Deleted |
| 1021 | Duplicated | |
| H1 | 71 | Deleted |
| 1471 | Duplicated |
Table 2. Summary of the genomic aberrations detected in CH-ES1 and 2102Ep cell lines by CGH-Array.
| 2102 Ep cells | CH-ES1 cells | ||||
| Cytoband | Aberration | Size (Mb) | Cytoband | Aberration | Size (Mb) |
| 1p36-p12 | gain | 124 | 1q44 | loss | 3.8 |
| 2p25-p16. | gain | 52 | 2q11-q21.2 | gain | 39 |
| 3p26-p11 | gain | 73 | 3p26-p16.1 | loss | 60 |
| 5q23.1-q35 | gain | 62 | 3p14.1-p12.1 | loss | 17 |
| 7p22-q21.13 | gain | 88 | 4p16-q31.21 | loss | 79 |
| 8p23-p12 | gain | 30 | 4q34.3-q35 | loss | 9 |
| 9p24-p12 | gain | 51 | 5q34 | gain | 13 |
| 11q14.3-q23.3 | loss | 23 | 6p25 | gain | 4 |
| 12p13-q24.3 | gain | 132 | 6p22.3 | gain | 2 |
| 13q12.3-q34 | loss | 84 | 6p21.31-p21.2 | gain | 4 |
| 16p13.3-q23.2 | gain | 72 | 6q21-q27 | gain | 60 |
| 17q13.2-q25 | gain | 78 | 7q33-q36 | gain | 25 |
| 19p11.3-p12 | gain | 28 | 8p23-p12 | loss | 35 |
| 20p13-q13.3 | gain | 62 | 8q24.12 9q21.31 | gain | 25 |
| 21q22.11 | gain | 1.4 | 11q24.3 | gain | 0.4 |
| Xp22.3-q28 | gain | 154 | 13q11q21.31 | loss | 6 |
| 13q21-q32 | loss | 42 | |||
| 13q32.1-q34 | gain | 33 | |||
| 15q11q22.32 | loss | 10 | |||
| 15q22.32-q26 | loss | 44 | |||
| 16p13-q24 | gain | 37 | |||
| 17q22q23.2 | loss | 88 | |||
| 18p11 | loss | 9 | |||
| 18q11-q12.3 | loss | 16 | |||
| 18q12.3-q23 | gain | 20 | |||
| loss | 39 | ||||
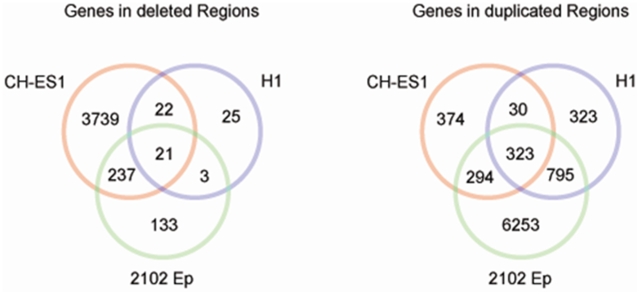
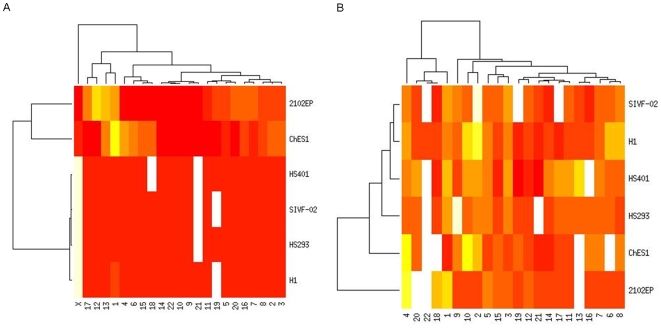
The CGH array showed extensive chromosomal changes in the teratocarcinoma line 2102Ep and in the malignant hESC line CH-ES1. There were also several visible changes in the H1 line at passage number 56, including partial 5 Mb duplications in 1q32.2 and 22q12.2, and these findings were confirmed by the SNP analysis. CGH analyses of the three other hESC lines did not identify any gains or deletions (Figure 2). A gene-level analysis revealed that, in CH-ES1, there were about 4019 hemizygously deleted genes and 1021 duplicated genes, whereas in 2102Ep, 394 genes were deleted and 7665 genes were duplicated. Similarly to other hESCs, control H1 cells showed about 71 deleted genes and 1471 duplicated genes (Table 1). When normal variations listed in the Genomic Variation databases [13] were excluded, 21 genes were deleted, and 323 were duplicated (Figure 2). The common deleted genes included BCL3, which is known to be mutated in B cell lymphomas (Table 3).
Figure 2. The number of genes in CH-ES1, 2102Ep and H1 lines in either deleted or duplicated regions.
A) deleted regions, B) duplicated regions.
Table 3. Deleted genes in common between H1, CH-ES1, and 2102Ep cell lines.
| Input ID | Entrez Gene ID | Symbol | Name |
| ALDH16A1 | 126133 | ALDH16A1 | aldehyde dehydrogenase 16 family, member A1 |
| APOC1 | 341 | APOC1 | apolipoprotein C-I |
| APOC2 | 344 | APOC2 | apolipoprotein C-II |
| APOC4 | 346 | APOC4 | apolipoprotein C-IV |
| APOE | 348 | APOE | apolipoprotein E |
| BCAM | 4059 | BCAM | basal cell adhesion molecule (Lutheran blood group) |
| BCL3 | 602 | BCL3 | B-cell CLL/lymphoma 3 |
| CBLC | 23624 | CBLC | Cas-Br-M (murine) ecotropic retroviral transforming sequence c |
| CCDC155 | |||
| CLPTM1 | 1209 | CLPTM1 | cleft lip and palate associated transmembrane protein 1 |
| FCGRT | 2217 | FCGRT | Fc fragment of IgG, receptor, transporter, alpha |
| FLT3LG | 2323 | FLT3LG | fms-related tyrosine kinase 3 ligand |
| PIH1D1 | |||
| PTH2 | |||
| PVRL2 | 5819 | PVRL2 | poliovirus receptor-related 2 (herpesvirus entry mediator B) |
| RCN3 | 57333 | RCN3 | reticulocalbin 3, EF-hand calcium binding domain |
| RPL13A | 23521 | RPL13A | ribosomal protein L13a |
| RPS11 | 6205 | RPS11 | ribosomal protein S11 |
| SLC17A7 | 57030 | SLC17A7 | solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 7 |
| TOMM40 | 10452 | TOMM40 | translocase of outer mitochondrial membrane 40 homolog (yeast) |
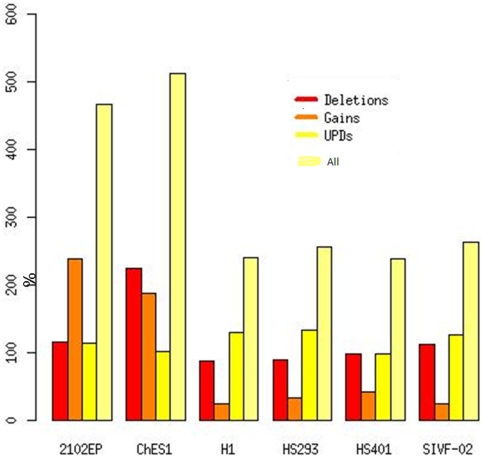
Further analyses using Affymetrix 6.0 SNP arrays confirmed all the changes observed by CGH and also identified an additional 1275 copy number variant sites. Of these, about 33% were shorter than 43 kb; the median resolution was 44 kb for the Agilent CGH arrays. About 60% of the CNVs were detected in the CH-ES1 and 2102Ep stem cell lines, consistent with their abnormal behavior (Figure 3).
Figure 3. The number of aberrations per cell line detected by Affymetrix SNP6.0 arrays.
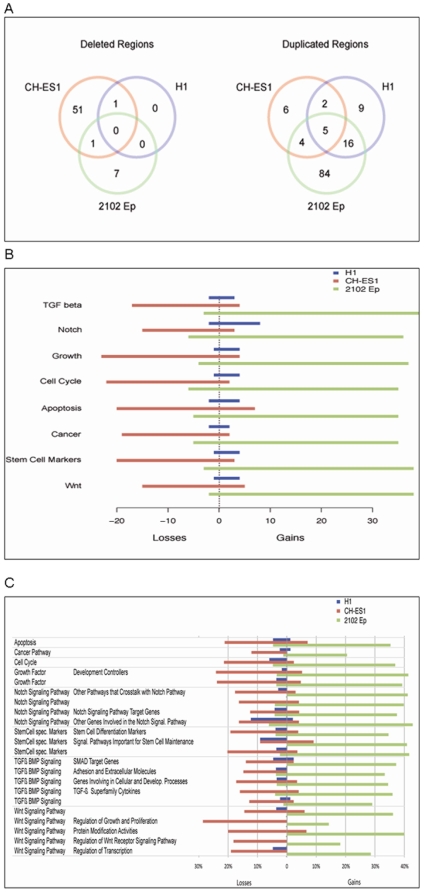
After assignment of the identified genes to KEGG pathways, we found that there were no common pathways among the 21 gene deletions shared by the H1, CH-ES1 and 2102Ep cells. However, there were five pathways that were altered among the 323 duplicated genes shared between these cell lines, including MAPK signaling, axon guidance, natural killer cell-mediated cytotoxicity, tight junction and Fc epsilon RI signaling pathways (Figure 4A). Almost all of these pathways were altered by gene deletions in CH-ES1 and gene duplications in 2102Ep (Figure 4B). However, we did not find any significantly predominant type of mutation in H1 (Figure 4C).
Figure 4. Pathway analyses of gained and lost genes in the analysed cell lines.
A) The number of pathways which were statistically significantly enriched per line involving deleted (left panel) and duplicated (right panel) genes. B) The number of lost and gained genes per cell line in summary pathways. C) The percentage of genes altered per pathway by deletion or duplication. Only pathways with copy number variations are shown.
Out of the 1275 CNVs detected by the SNP arrays, 165 were not previously reported in the database of Genomic Variants, suggesting that these are unique to the stem cell lines analyzed and may be pertinent to their specific behavior (Figure 5A). The median length of these mutations was approximately 28 kb and the total average genome coverage was 4.2 Mb per cell line. Further annotation using the ENTREZ gene database [14] mapped 181 genes to these unique CNVs. Out of these, 85 were found to be expressed in normal stem cell lines.
Figure 5. The number of CNVs and UPDs per chromosome and cell line as detected by Affymetrix 6.0 arrays.
CH-ES1 and 2102Ep are the only female lines. A) The numer of CNVs, B) The number of UPDs.
We matched this list of genes with the OMIM disease database [15] and found that 27 were previously implicated in a range of disorders encompassing different forms of cancer (CYLD, NOD2, SLC19A1, COL18A1), cardiovascular (ADRA1B, NEBL, NRG1, ZFPM2, UMOD) and psychiatric disorders (GABA3, DAOA, NRG1, KMO, CTNNA3, ZDHHC17, PPP2RB2, OPRD1). The primers for validation are given in Table S1.
We further detected 1269 LOH sites (Figure 5B). We matched these with the CNVs and further annotated them according to the Toronto Database of Variation. The number of annotated copy number neutral LOHs (or uniparental disomies (UPDs)) was 211, with a median length of 235 kb and coverage of 10 Mb per cell line. In total, 363 genes reported in the ENTREZ database [14] were located in the UPD regions. Out of these, 128 were found to be expressed in normal hESCs. Again, annotation to OMIM [15] revealed that several cancer-related genes were involved in these LOH regions (MLH1, ZMAT3, ADCY7, PIK3CA).
Even the smaller abnormalities involved potential oncogenes, as illustrated in Figure 4. Both openly malignant lines (2102Ep and CH-ES1) had multiple large deletions and insertions involving genes participating in the cell cycle, apoptosis, growth regulation and oncogenesis (Figure 4, Table S2, Table S3). These included, for example, MYC, BRCA2, p53, and others the numerous deleted and duplicated genes according to the pathways they reperesent are listed in detail in (Table S3). The results regarding genomic structure indicate the high instability of CH-ES1 and 2102Ep cells. Remarkably, several numerical abnormalities were observed in the HS401, HS293 and SIVF-02 hESC lines, as illustrated in Figure 5. Two genes were also verified for copy number variation on DNA level by quantitave real-time PCR, namely loss of GRB10 in HS293 and loss of MLLT1 in HS401 (Table 4)
Table 4. Primers used in real-time PCR.
| Gene | Forward primer (5′-3′) | Reverse primer (5′-3′) | Size(bp) |
| GRB10-A | AGGTGCTGGGTAGCATGTTC | GGCTACAACACCCCACTGAC | 130 |
| GRB10-B | TGTAGGGCCTCCAGAATTGA | TTTCCATTGAGCATCAAAACAG | 138 |
| MLLT1-A | CGTCCAGGTGAGGTTAGAGC | CCAGAAGACCACCTTCTCCA | 145 |
| MLLT1-B | CTGACAGCGGCAGATGTTTA | GAGAAGAAAACGCGATCCTG | 107 |
| HEM3 * | TGCACGGCAGCTTAACGAT | AGGCAAGGCAGTCATCAAGG | 202 |
It is noteworthy that alterations of several potential oncogenes (such as RAB6A) were also seen in these hESC lines, which induced only benign tumors in the mouse teratoma assay.
Validation of mRNA expression by quantitative real time PCR of a given gene with an increased copy number or a deletion, also showed corresponding modification of its mRNA level (Table 5). Totally 11 genes that were either duplicated or deleted in the different hESC lines were tested for their mRNA expression. The increased copy numbers and deletions were seen in the hESC line, H1, already at earliest available passage 22 as revealed by the validation assay. The only cell line that showed altered mRNA expression from what was expected was the highly malignant CH-ES1 with high level of genomic imbalances.
Table 5. Primers used in real-time quantitative PCR.
| Gene | Forward primer (5′-3′) | Reverse primer (5′-3′) | Size(bp) |
| MLLT1 | CAGCAAGCCTGAGAAGATCC | TTGAAGTGGCCAGTCTCCTC | 150 |
| PK1A | TCCTGGTTTCCTCTGCAAGT | CGTTGTGCATCTTCTTCACC | 97 |
| GRB10 | GAAGCAGTACAACGCCCCTA | CTCTGCACAGAGCAACCTCA | 94 |
| FGFR1 | TCCGTCAATGTTTCAGATGC | TTCCATCTTTTCTGGGGATG | 144 |
| EHMT1 | AGAGGACAGCAGGACTTCCA | TTCCGAACTCAGGTCAGACTC | 142 |
| RAB6A | TTGCTGACAAGAGGCAAGTG | CAAAGCTGCTGCTACACGTC | 134 |
| MAP3K15 | AGGGCGATAATGTTCTGGTG | TCTCAGGTGCCATGTACTGC | 135 |
| FAM49B | CATATTCTCCCACCCAGCAT | TGGCAGGATTTGTCATCTTG | 107 |
| JAK1 | AACTGAAGTGGACCCCACAC | CACCTGCTCCCCTGTATTGT | 130 |
| FH | TCGATTTTTGGGTTCTGGTC | CCATGGTCATTGCTTCACAC | 122 |
| PTPN13 | ACCTCCACCTGGTGTGCTAC | ATCTGAGCTGGTGCTTTGCT | 133 |
| GAPDH* | GCAGCCCTGGTGACCAG | GGGAAGGTGAAGGTCGGA | 62 |
Discussion
The hESC line CH-ES1 showed many characteristics typical of a teratocarcinoma-derived EC cell line. Spontaneous teratocarcinomas generally arise from primordial germ cells, typically in the testis, but also occasionally in the ovary or at non-gonad sites. Experimental teratocarcinomas may also be derived from ectopically transplanted embryos [12]. A single blastomere of a four-cell stage human embryo could therefore also form a teratocarcinoma. It is likely that the blastomere cell that gave rise to the CH-ES1 line had an abnormal genetic constitution, which is very common in human pre-implantation embryos [16], [17].
Human EC cells commonly have nearly triploid genomes and DNA content with gross chromosomal changes and a large number of variations [3]. It has been suggested that such tumor cells originate from a tetraploid derivative of primordial germ cells. These cells subsequently lose and rearrange their chromosomes to first generate a seminoma and then the more malignant and pluripotent EC cells, which stabilize at an approximately 3n DNA content [18], [19]. It is thus tempting to speculate that the blastomere that gave rise to CH-ES1 may have been tetraploid and that subsequent chromosomal loss resulted in an EC-like phenotype by a mechanism comparable to that by which EC cells arise.
Our results emphasize the importance of not only cytogenetic testing but also more detailed genetic testing of hESC lines by microarray methods before their clinical application in regenerative medicine. A large proportion of early human embryos are chromosomally abnormal, particularly those with poor morphology or developmental delays. The embryos donated for research are often of poor quality, but reported chromosomal abnormalities in hESC lines are not common, at least in early passages. For instance, all 30 hESC lines derived in our laboratory at Karolinska Institutet display karyotypically normal G-banding patterns [20]. It may be that genetically abnormal embryos do not form hESC lines as easily as normal ones. It is unlikely that the abnormalities in CH-ES1 would have been caused by the derivation process itself or by early culture, because we used identical conditions to those used to produce the 30 chromosomally normal hESC lines [20. Instead, derivation from a single blastomere may play a role, since Geens et al. [11], who succeeded in deriving hESC lines from embryos that were established for the study of early development, obtained a cytogenetically abnormal line.
There are several possible explanations for the malignancy of the CH-ES1 and the teratocarcinoma lines, including partial triploidy [21]. Many of the trisomies that have been identified in cancers and culture-adapted cells [4], [17], [18] were also seen in CH-ES1 cells, such as trisomy of chromosomes 1, 12, and X and a duplication of 17p13.2-qtel(3.674-tel). In addition, there were trisomies of chromosomes 9, 19, 20 and 21. In fact, only chromosome 14 was normal in the CGH assays of the CH-ES1 line. It is not difficult to understand why this particular cell line is particularly malignant and invasive. According to the CGH analysis, the changes observed in H1 (the oldest hESC line, which was at passage number 56 at the time of analysis) may have been there from the beginning. However, it is also possible that these changes arose during culture adaptation. We do not presently have CGH or SNP array data from earlier passages or from other laboratories.
Culture adaptation of hESCs and accumulation of chromosomal changes during long term culture occur as a result of the successive increase of selective growth advantages provided by certain abnormalities in the cells [4], [21], [22]. Furthermore, smaller changes than can be seen by G-banding have been described, and these may offer growth advantages similar to those that occur in cancer. Impaired imprinting and aberrations in mitochondrial DNA have been described [23], and impaired X-chromosome activation occurs during culture adaptation [24]. Culture adaptation has also been described in teratocarcinoma lines [3]. It is possible that least some of the aberrations in the studied lines are caused by culture adaptation. The aneuploidic increases in copy number of genes that may promote tumor formation, including ARHGAP26, GRB10, DDHD2, FGFR1, CTNNA3, PTPN1 and MLLT1 in the apparently stable and karyotypically normal lines HS293 and HS401, are cause for concern. Among the altered genes, ARHGAP26 and MLLT1 have been associated with leukemia-specific translocations, DDHD2, FGFR1 and PTPN1 have tumor-promoting potential in breast cancer, and CTNNA3 may promote tumor formation in urothelial cancer. Furthermore, a copy of the GRB10 gene, which acts as a growth inhibitor, was lost from HS293, supporting the idea of an acquired growth advantage. Such losses or gains of these potential growth- or cancer-promoting genes may increase the likelihood of malignant transformation with the accumulation of later mutations.
The SNP analysis was made using different passage levels to see what possible changes the lines displaying normal G-banding finding contain. In the quantitative PCR analysis of RNA expression, eleven genes were analysed for elevated or decreased expression according to the losses and gains in the different cell lineages. All the findings of the PCR validation were consistent with the SNP array results, but the malignant CH-ES-1 behaved differently.
Translocated genes may come under the influence of different promoters and enhancers disturbing and altering their gene expression. This is a possible explanation to decreased PTPN13 mRNA expression although the gene was shown to be duplicated, and an increased mRNA expression of FH although loss of a gene copy in the teratocarcinoma-like CH-ES1
Long term testing in immune-suppressed animals is neither an adequate nor a sufficient model to study cancer transformation of hESC lines. It will be difficult to exclude the possibility that cells carrying copy number alterations of growth-promoting or tumor suppressor genes have malignant potential by studying them in model organisms. In xeno-models, all tumorigenic cells are more likely to be rejected than in transplantation between individuals of the same species [4], [25]. Immunosuppression of the recipient makes the problem of possible tumor formation even more serious. The only way to avoid such risks is to use cells at the earliest possible passage number to decrease the likelihood of such changes.
According to the CGH and SNP array results, the profiles were consistent among all six cell lines studied. However, as expected, the higher resolution offered by the SNP arrays revealed 1275 additional changes smaller than 43 kb (the threshold of CGH resolution). In addition, SNP arrays can identify copy number neutral aberrations showing LOH, such as gene conversions and uniparental isodisomies. Altogether, we found 211 segments with a median length of 235 kb and coverage of 10 Mb per genome that showed LOH, and these have not been previously recorded as CNVs. In total, these regions contained 363 ENTREZ [14] genes, of which 128 were found to be expressed in normal hESCs. We conclude that the increased resolution offered by the SNP arrays is required for assessing potentially harmful alterations in hESC lines.
In conclusion, the first teratocarcinoma-like hESC line derived from a single blastomere showed many features typical of malignant cells, such as trisomies, duplications, deletions, and increased copy numbers of oncogenes, explaining its malignancy. In addition, benign and cytogenetically normal hESC lines also displayed many potentially tumorigenic genomic alterations, which may be due to the derivation method or to the prolonged culture conditions. Hence, at a minimum, SNP-profiling of the hESC lines before their use in regenerative medicine is important.
Materials and Methods
The lines HS293 and HS401 were previously derived from fresh poor quality embryos that had been donated for research after informed consent in the Fertility Unit of the Karolinska University Hospital, Huddinge, Sweden, as described [26], [27]. They were derived using postnatal human skin fibroblasts as feeder cells and Knockout Serum Replacement (SR, Invitrogen)-containing medium. The Ethics Board of the Karolinska Institutet approved the derivation and research use of these lines. At the time of DNA extraction, HS293 was at passage number 47, and HS401 was at passage number 25. The lines have been karyotyped several times after derivation, and they were found repeatedly to be cytogenetically normal. After injection into SCID mice, they formed benign teratomas containing differentiated tissue components of the three germ layers.
The line CH-ES1 was derived under the same culture conditions as the lines produced at Karolinska Institutet using postnatal skin fibroblasts and SR-containing medium [5]. The derivation of this line was accomplished under the ethics permission and license of Swiss authorities. Surprisingly, the first karyotype performed at passage number three by G-banding showed many substantial chromosomal aberrations. Moreover, when CH-ES1 cells were injected into mice, they induced highly invasive tumors with clearly malignant cell composition [5]. At the time of DNA extraction, CH-ES1 was at passage number 19.
The clonal subline 2102Ep, an hEC line derived from a testicular teratocarcinoma, was maintained by one of us (PWA) in Sheffield as previously described [28]; DNA was extracted from the clone at passage number 40.
The hESC lines H1 from WiCell Research Institute (Madison, WI), SIVF02 (non-GMP line, a kind gift of Sydney IVF, Australia), CH-ES1 [5], HS293 and HS401 were maintained in DMEM/F-12 medium supplemented with 20% serum replacement, L-glutamine, non-essential amino acids, and 4 ng/ml human basic fibroblast growth factor. All hESC lines were cultured on irradiated human foreskin fibroblasts and passaged mechanically. The fibroblast feeders were cultured in DMEM (Invitrogen) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (both from Invitrogen). Cells were mitotically inactivated by irradiation at 35 Gy before seeding on a gelatin-coated 6-well plate at 3.5×105 cells/plate. The hESC culture medium was changed daily.
Prior to DNA extraction for SNP analysis, cells were cultured for at least four passages under feeder-free culture conditions on Matrigel Growth Factor Reduced (Becton Dickinson AG, Basel, Switzerland) coated 6-well plates with feeder-conditioned medium (CM). Matrigel was diluted 1∶30 with DMEM/F12 and 0.5 ml of the dilution was added to cover each well of a 6-well plate and allowed to gel for 1 h at 37°C. Plates were immediately used after the coating procedure. CM was prepared by incubating stem cell media overnight on irradiated feeder cells plated at the same density for hESC culture. CM was harvested after 24 h and supplemented with 20 ng/mL bFGF immediately before use with hESC cultures. This procedure was repeated for one week before discarding the feeder cells.
Array CGH
DNA was extracted from cells using the QUIamp DNA extract kit (Qiagen Germantown, MD) following standard protocols. The same DNA samples were used for both SNP arrays and array-CGH.
Array-CGH was performed using the Agilent Human Genome CGH Microarray Kit 44B (Agilent Technologies, Santa Clara, California, USA). This platform is a high-resolution 60-mer oligonucleotide-based microarray that allows genome-wide surveys and molecular profiling of genomic aberrations with a resolution of ∼75 kb. Labeling and hybridization were performed following the protocols provided by Agilent. Briefly, 500 ng of purified DNA from a patient and a control (Promega Corporation, Madison, Wisconsin, USA) was double-digested with RSAI and AluI for two hours at 37°C. After twenty minutes at 65°C, each digested sample was labeled by the Agilent random primers labeling kit for two hours using Cy5-dUTP for the patient DNA and Cy3-dUTP for the control DNA. Labeled products were purified on columns and prepared according to the Agilent protocol. After probe denaturation and pre-annealing with 5 µl of Cot-1 DNA, hybridization was performed at 65°C with rotation for 40 hours. After two washing steps the arrays were analyzed with the Agilent scanner and Feature Extraction software (v9.1.3). A graphical overview was obtained using the CGH analytics software (v3.4.27).
The identification of aberrant chromosomal regions was performed manually using CGH-Analytics software (v3.4.27) (Agilent Technologies) according to the UCSC Genome Bioinformatics, (2010) [29] http://genome.ucsc.edu) and the Database of Genomic Variants 2010) [13] (http://projects.tcag.ca/variation/) on the Human March 2006 assembly.
Associations between genomic instability and Pathways, Gene Ontology and manually assembled gene lists were tested with R/bioconductor [30] and Webgestalt [31]. Losses and gains were considered separately, and enrichment was assessed with hypergeometric tests corrected for multiple testing using False Discovery Rate (FDR).
SNP Arrays
The genotyping to detect both copy-number variations and loss of heterozygosity (LOH) without loss of chromosomal material was performed using the Affymetrix Genome-Wide Human SNP Array 6.0 (San Diego, CA). Labeling and hybridization were performed following the protocols provided by the manufacturer. The CRMA method [32] from the Aroma Affymetrix package [33] was used to asses total CNV.
As a CNV neutral reference group, we used data from a set of 20 arrays of nonmalignant blood cell DNA samples that had been previously hybridized in the same laboratory (JK).
To separate signal from noise, we considered only CNVs with intensities larger than one standard deviation of the raw copy number signal across all of the stem cell arrays. Moreover, we required CNVs to be tagged by at least four consecutive probes. Further testing by qPCR of CNVs close to the cut off confirmed the adequacy of this choice.
LOH was estimated using genotyping calls from Affymetrix proprietary software Genotyping Console (birdseed method) and a Hidden Markov Chain Method (HMCM) as implemented in the software dChip. LOH and CNVs were compared in order to determine Uniparental Disomy (UPD [34] or copy number neutral LOH.
Verification of copy number variation with quantitative real-time PCR
Two selected variations in the hESC lines, the deletions of the GRB10 gene in HS293 and the MLLT1 gene in HS401, were verified by designing PCR amplicons within the deleted segments. A copy number neutral amplicon, HEM3, was used as a reference [35].
Two amplicons per gene were designed in the Primer Express v2.0 program (table 6). qRT-PCR analyses were performed in 20 µl volumes with 1 x Fast SYBR Green PCR Master Mix (Applied Biosystems), 10 ng genomic DNA and optimized primer concentrations: for HEM3 600 nmol/L, and for GRB10 and MLLT1 400 nmol/L. Each amplicon was quantified in triplicate using the Fast SYBR program (95°C for 20 s, followed by 40 cycles of 95°C for 3 s and 60°C for 30 s) on a 7500 Real-time PCR machine (Applied Biosystems). Relative copy number estimates were derived through ΔΔ Ct calculations for the copy number neutral amplicon, the HEM3 control gene. Three laboratory control DNA samples were used as standards for analyzing relative copy number.
Table 6. Summary of selected genes showing gains or losses in the different cell lineages and their corresponding mRNA expression.
| Gene | Sample | Chr | Aberration | mRNA expression# |
| MLLT1 | HS401 | 19 | loss | 2-fold decrease |
| PK1A | HS401 | 8 | gain | 2-fold increase |
| GRB10 | HS293 | 7 | loss | 2-fold decrease |
| FGFR1 | HS203 | 8 | gain | 2-fold increase |
| EHMT1 | H1 | 9 | loss | 3 - 4-fold decrease* |
| RAB6A | H1 | 11 | gain | 1.3-2-fold increase* |
| MAP3K15 | SIVF-02 | X | loss | 4-fold decrease |
| FAM49B | SIVF-02 | 8 | gain | 2-fold increase |
| JAK1 | CH-ES1 | 1 | loss | 3-fold decrease |
| FH | CH-ES1 | 1 | loss | 1.5-fold increase |
| PTPN13 | CH-ES1 | 4 | gain | 3-fold decrease |
RNA extraction and quantitative real time polymerase chain reaction
Total RNA was extracted from the different cell lineages using the Trizol reagent according to the manufacturer's instruction (Invitrogen). By the time of RNA extraction, HS401 was at passage 35, HS293 at passage 54, H1 was analysed from three different passage levels 22, 33 and 69, SIVF-02 at passage 45 and CH-ES 1 at passage 14. cDNA was synthesized from 1 µg of total RNA with the SuperScript II First-Strand synthesis system (Life Technologies). Quantitative RT-PCR measurements of individual cDNAs were performed in a final volume of 10 µl using SYBR green PCR master mix (Applied Biosystems) to measure duplex DNA formation with the 7500 Real-time PCR machine (Appplied Biosystems). Gene-specific primers were designed using the Primer3 software [35] with standard selection criteria in order to amplify approximately 90–150 bp long PCR fragments (table 5). Real-time PCR primers were used at a final concentration of 100 nM. Melting curve analysis and agarose gel electrophoresis was performed to monitor production of the appropriate PCR product. Each PCR reaction was performed in triplicates with negative controls. The results were normalized to endogenous GAPDH and PSMB mRNA levels.
Bioinformatics
Putative phenotypically neutral CNV and UPD sites were purged by comparing the detected changes to the polymorphisms and aberrations from the database of Genomic Variants November 2008 Assembly (hg18) [13] (http://projects.tcag.ca/variation/). The remaining sites were annotated with the genes from the ENTREZ database (http://www.ncbi.nlm.nih.gov/sites/entrez) and the genes were further annotated to the Human disease database David, Bioinformatics Resources. NIH (2009) [36] (http://david.abcc.ncifcrf.gov/) and OMIM. NIH (2009) [15] (http://www.ncbi.nlm.nih.gov/OMIM).
Expression microarrays
Microarray data on Affymetrix HGU133plus2 chips (San Diego, CA) that had been hybridized with normal stem cell lines (HS237 and HS181) in a previous experiment were used to evaluate gene activity. Presence calls from the Affymetrix MAS5 algorithm where used to establish whether a gene was expressed in normal stem cell lines. As the hybridization was performed in two technical replicates and genes could be interrogated by several probe sets, we designated a gene as expressed when it was present at least half of the time it was interrogated.
Supporting Information
Primers used in real-time quantitative PCR.
(0.03 MB DOC)
Altered pathways in the studied lines.
(0.09 MB DOC)
Deleted (DEL) and duplicated (DUPL) genes in H1, CH-ES1, and 2102 Ep cell lines related to several signaling pathways.
(0.07 MB DOC)
Acknowledgments
We are grateful to Ingrid Wagner, Sandrine Vianin, and Ami Stromberg for their technical support. We also thank Professors Paul Bischof, and Jean-Bernard Dubuisson for their scientific support and fruitful discussions. We also thank Teneille Ludwig for sending us passage 22 H1 cells for the q-PCR validation analysis.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the Swiss National Foundation, Medical Research Council UK, Roche Research Foundation, the Ernst & Lucie Schmidheiny Foundation, the Geneva University Hospitals, the Geneva Faculty of Medicine, Genico, the EU for ESTOOLS, the Swedish Research Council and RD grants from Stockholm County and Karolinska Institutet. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Solter D. From teratocarcinomas to embryonic stem cells and beyond: a history of embryonic stem cell research. Nat Rev Genet. 2006;7:319–327. doi: 10.1038/nrg1827. [DOI] [PubMed] [Google Scholar]
- 2.Damjanov I. The road from teratocarcinoma to human embryonic stem cells. Stem Cell Rev. 2005;1:273–276. doi: 10.1385/SCR:1:3:273. [DOI] [PubMed] [Google Scholar]
- 3.Andrews PW, Matin MM, Bahrami AR, Damjanov I, Gokhale P, et al. Embryonic stem (ES) cells and embryonal carcinoma (EC) cells: opposite sides of the same coin. Biochem Soc Trans. 2005;33:1526–1530. doi: 10.1042/BST0331526. [DOI] [PubMed] [Google Scholar]
- 4.Blum B, Benvenisty N. The tumorigenicity of human embryonic stem cells. Adv Cancer Res. 2008;100:133–158. doi: 10.1016/S0065-230X(08)00005-5. [DOI] [PubMed] [Google Scholar]
- 5.Feki A, Bosman A, Dubuisson JB, Irion O, Dahoun S, et al. Derivation of the first Swiss human embryonic stem cell line from a single blastomere of an arrested four-cell-stage embryo. Swiss Medical Weekly. 2008;138:540–543. doi: 10.4414/smw.2008.12385. [DOI] [PubMed] [Google Scholar]
- 6.Baker DE, Harrison NJ, Maltby E, Smith K, Moore HD, et al. Adaptation to culture of human embryonic stem cells and oncogenesis in vivo. Nat Biotechno. 2007;25:207–215. doi: 10.1038/nbt1285. [DOI] [PubMed] [Google Scholar]
- 7.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz M A, Swiergiel JJ, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 8.Strelchenko N, Verlinsky O, Kukharenko V, Verlinsky Y. Morula-derived human embryonic stem cells. Reprod Biomed Online. 2004;9:623–629. doi: 10.1016/s1472-6483(10)61772-5. [DOI] [PubMed] [Google Scholar]
- 9.Klimanskaya I, Chung Y, Becker S, Lu SJ, Lanza R. Human embryonic stem cell lines derived from single blastomeres. Nature. 2006;444:481–485. doi: 10.1038/nature05142. [DOI] [PubMed] [Google Scholar]
- 10.Chung Y, Klimanskaya I, Becker S, Li T, Maserati M, et al. Human embryonic stem cell lines generated without embryo destruction. Cell Stem Cell. 2008;2:113–117. doi: 10.1016/j.stem.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 11.Geens M, Mateizel I, Sermon K, De Rycke M, Spits C, et al. Human embryonic stem cell lines derived from single blastomeres of two 4-cell stage embryos. Hum Reprod. 2009;24:2709–2717. doi: 10.1093/humrep/dep262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adewumi O, Aflatoonian B, Ahrlund-Richter L, Amit M, Andrews PW, et al. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat Biotechnol. 2007;25:803–816. doi: 10.1038/nbt1318. [DOI] [PubMed] [Google Scholar]
- 13.Database of Genomic Variants. 2010. http://projects.tcag.ca/variation.
- 14.NCBI website. 2009. ( http://www.ncbi.nlm.nih.gov/sites/entrez)
- 15.OMIM database, NIH. 2009. ( http://www.ncbi.nlm.nih.gov/OMIM)
- 16.Ambartsumyan G, Clark AT. Aneuploidy and early human embryo development. Hum Mol Genet. 2008;17:R10–15. doi: 10.1093/hmg/ddn170. [DOI] [PubMed] [Google Scholar]
- 17.Hardarson T, Caisander G, Sjogren A, Hanson, C, Hamberger L, Lundin K. A morphological and chromosomal study of blastocysts developing from morphologically suboptimal human pre-embryos compared with control blastocysts. Hum Reprod. 2003;18:399–407. doi: 10.1093/humrep/deg092. [DOI] [PubMed] [Google Scholar]
- 18.Oliver RT. Clues from natural history and results of treatment supporting the monoclonal origin of germ cell tumours. Cancer Surv. 1990;9:333–368. [PubMed] [Google Scholar]
- 19.Oosterhuis JW, Looijenga LH. Current views on the pathogenesis of testicular germ cell tumours and perspectives for future research: highlights of the 5th Copenhagen Workshop on Carcinoma in situ and Cancer of the Testis. APMIS. 2003;111:280–289. doi: 10.1034/j.1600-0463.2003.1110131.x. [DOI] [PubMed] [Google Scholar]
- 20.Strom S, Inzunza J, Grinnemo KH, Holmberg K, Matilainen E, et al. Mechanical isolation of the inner cell mass is effective in derivation of new human embryonic stem cell lines. Hum Reprod. 2007;22:3051–3058. doi: 10.1093/humrep/dem335. [DOI] [PubMed] [Google Scholar]
- 21.Harrison NJ, Baker D, Andrews PW. Culture adaptation of embryonic stem cells echoes germ cell malignancy. Int J Androl. 2007;30:275–281. doi: 10.1111/j.1365-2605.2007.00762.x. [DOI] [PubMed] [Google Scholar]
- 22.Draper JS, Moore HD, Ruban LN, Gokhale PJ, Andrews PW. Culture and characterization of human embryonic stem cells. Stem Cells Dev. 2004;13:325–336. doi: 10.1089/scd.2004.13.325. [DOI] [PubMed] [Google Scholar]
- 23.Maitra A, Arking DE, Shivapurkar N, Ikeda M, Stastny V, et al. Genomic alterations in cultured human embryonic stem cells. Nat Genet. 2005;37:1099–1103. doi: 10.1038/ng1631. [DOI] [PubMed] [Google Scholar]
- 24.Enver T, Soneji S, Joshi C, Brown J, Iborra F, et al. Cellular differentiation hierarchies in normal and culture-adapted human embryonic stem cells. Hum Mol Genet. 2005;14:3129–3140. doi: 10.1093/hmg/ddi345. [DOI] [PubMed] [Google Scholar]
- 25.Erdo F, Buhrle C, Blunk J, Hoehn M, Xia Y, et al. Host-dependent tumorigenesis of embryonic stem cell transplantation in experimental stroke. J Cereb Blood Flow Metab. 2003;23:780–785. doi: 10.1097/01.WCB.0000071886.63724.FB. [DOI] [PubMed] [Google Scholar]
- 26.Hovatta O, Mikkola M, Gertow K, Stromberg AM, Inzunza J, et al. A culture system using human foreskin fibroblasts as feeder cells allows production of human embryonic stem cells. Hum Reprod. 2003;18:1404–1409. doi: 10.1093/humrep/deg290. [DOI] [PubMed] [Google Scholar]
- 27.Inzunza J, Gertow K, Stromberg AM, Matilainen E, Blennow E, et al. Derivation of human embryonic stem cell lines in serum replacement medium using postnatal human fibroblasts as feeder cells. Stem Cells. 2005;23:544–549. doi: 10.1634/stemcells.2004-0201. [DOI] [PubMed] [Google Scholar]
- 28.Andrews PW, Goodfellow PN, Shevinsky LH, Bronson DL, Knowles BB. Cell-surface antigens of a clonal human embryonal carcinoma cell line: morphological and antigenic differentiation in culture. Int J Cancer. 1982;29:523–531. doi: 10.1002/ijc.2910290507. [DOI] [PubMed] [Google Scholar]
- 29.USCS Genome Bioinformatics. 2010. http://genome.ucsc.edu.
- 30.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bengtsson H, Wirapati P, Speed TP. A single-array preprocessing method for estimating full-resolution raw copy numbers from all Affymetrix genotyping arrays including GenomeWide SNP 5 &6. Bioinformatics. 2009;25:2149–2156. doi: 10.1093/bioinformatics/btp371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bengtsson H, Irizarry R, Carvalho B, Speed TP. Estimation and assessment of raw copy numbers at the single locus level. Bioinformatics. 2008;24:759–67. doi: 10.1093/bioinformatics/btn016. [DOI] [PubMed] [Google Scholar]
- 34.Bruce S, Leinonen R, Lindgren CM, Kivinen K, Dahlman-Wright K, et al. Global analysis of uniparental disomy using high density genotyping arrays. J Med Genet. 2005;42:847–51. doi: 10.1136/jmg.2005.032367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weksberg R, Hughes S, Moldovan L, Bassett AS, Chow EW, et al. A method for accurate detection of genomic microdeletions using real-time quantitative PCR. BMC Genomics. 2005;6:180. doi: 10.1186/1471-2164-6-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Human disease database David, Bioinformatics Resources. NIH. 2009. http://david.abcc.ncifcrf.gov/) and OMIM. NIH.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used in real-time quantitative PCR.
(0.03 MB DOC)
Altered pathways in the studied lines.
(0.09 MB DOC)
Deleted (DEL) and duplicated (DUPL) genes in H1, CH-ES1, and 2102 Ep cell lines related to several signaling pathways.
(0.07 MB DOC)