Abstract
The yeast HXT (glucose transporter gene) repressor Rgt1 lacks a dimerization domain and thus appears to bind as a monomer to its consensus binding site sequence (5′CGGANNA3′). The HXT1 promoter contains 8 Rgt1 binding sites, but its expression is not effectively repressed by Rgt1. In the present study, the Rgt1 binding sites in the HXT1 promoter were analyzed to examine how Rgt1 mediates transcriptional repression. I show that Rgt1 binds the HXT1 promoter, but does not significantly mediate repression. When engineered to be multimerized without the intervening sequences between the Rgt1 binding sites, however, 4 or more Rgt1 binding sites were required to provide sufficient Rgt1-dependent repression. These findings suggest that the intervening sequences between the Rgt1 binding sites are important for the regulation of Rgt1 function and that Rgt1 functions efficiently only through multiple binding sites.
Introduction
In the budding yeast S. cerevisiae, expression of the glucose transporter gene (HXT) is regulated by glucose; HXT expression is induced by high levels of glucose, and repressed when glucose levels are low [1, 2]. Repression of HXT expression appears to be mediated by the DNA-binding protein Rgt1, which binds and recruits the general corepressors Ssn6 and Tup1 to the HXT promoters [3–5]. Rgt1 binds DNA through its N-terminal domain containing a Zn2Cys6 binuclear cluster, one of the most common DNA-binding domains in the fungal DNA binding proteins [4, 5]. Many of the proteins containing this cluster, such as Gal4, Ppr1, and Put3, appear to bind as dimers to 2 ‘CGG’ sequences, separated by variable length spacers. Crystal structures of the proteins revealed that a leucine zipper-like coiled-coil motif mediates the homo-dimerization of each of the proteins, enabling them to specifically recognize their binding sites [6–7]. The Aspergillus nidulans transcriptional activator AlcR belongs to the zinc binuclear cluster protein family, but lacks the dimerization domain. Thus, AlcR binds DNA as a monomer to its single binding sites. AlcR, however, activates transcription only if multiple sites are present [8].
Like AlcR, Rgt1 lacks the coiled-coil dimerization domain and binds DNA motifs that contain a ‘CGG’ sequence, but not those containing a pair of these sequences [3, 4, 9]. These observations suggest that Rgt1 binds DNA as a monomer. Footprinting analysis of the upstream regions of HXT genes indicated that Rgt1 specifically recognizes the ‘CGGANNA’ sequence (N is any nucleoside) [5]. Rgt1 appears to bind cooperatively to multiple copies of the sequence, which provides a way for Rgt1 to limit its binding to the yeast genome. Different HXT promoters contain different numbers of Rgt1 binding sites; within 1000 bp upstream of the ATG start codon, the HXT1 promoter contains 8 sites, whereas the HXT2 and HXT4 promoters contain 3 sites. Moreover, in the absence of glucose, repression of the expression of the HXT2 and HXT4 genes is completely relieved by the deletion of RGT1, whereas HXT1 expression is still significantly repressed in an rgt1 mutant [10]. Therefore, Rgt1 functions differently at different promoter regions. Here, the DNA-binding properties of Rgt1 were examined to gain more insight into how Rgt1 functions through its binding sites.
Materials and Methods
DMS footprinting assay
The HXT1 upstream region from −400 to −550 was PCR-amplified and labeled with 32P as described previously [5]. For dimethylsulfate (DMS) protection footprinting [11], Rgt1 was incubated with 32P-labeled DNA fragments (2 × 105 cpm) in 20 μl of TGZD buffer (20 mM Tris-HCl, pH. 8.0, 75 mM KCl, 10 μM ZnCl2, 5% glycerol) containing 1μg of poly(dI-dC)-poly(dI-dC) at room temperature for 20 min. Rgt1-DNA complexes were incubated with 0.2% DMS (final concentration) at 20°C for 1 min. The methylated bases were cleaved by boiling DNA in PE buffer (10% piperidine and 10 mM EDTA) at 90°C for 30 min.
Chromatin Immunoprecipitation (ChIP)
ChIP assay was performed as described previously [5]. Briefly, yeast cells expressing Rgt1-HA [12] were treated with formaldehyde (1%, final concentration) and cell extracts were prepared by vortexing cell pellets with glass beads in lysis buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 1% Triton X-100, 0.1% sodium deoxycholate). After centrifugation, the cell lysates were sonicated five times with 10-s pulses using a microtip. Fragmented chromatins were precipitated with monoclonoal HA antibody. The cross-linking of the precipitated DNA to the protein was reversed by incubating them in elution buffer (50 mM Tris-HCl, pH 8.0, 1% SDS, 10 mM EDTA) at 65°C for 6 h.
Functional analysis of Rgt1 binding sites
Gene expression reporter plasmids containing Rgt1-binding sites were prepared by inserting various Rgt1 binding sites between the LEU2 upstream activation sequence and the TATA box of HIS3 fused to lacZ (pBM2832) [4]. Clusters of the Rgt1 binding sites in the HXT1 promoter were PCR-amplified. Multiple copies of Rgt1 binding sites without intervening sequences between the sites were prepared by annealing complementary oligonucleotides containing different numbers of Rgt1 binding site (CGAATCCGGAA AAACTACG).
Results and Discussion
Characterization of Rgt1 binding to the HXT1 promoter
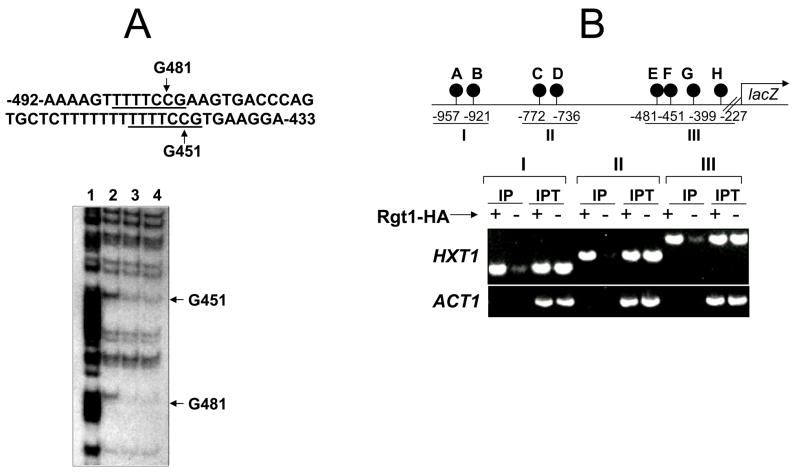
DMS is a small molecule that penetrates the protein-DNA complex and predominantly methylates the N-7 atoms of guanines in the major groove of B-DNA [11]. To probe the interactions between Rgt1 and the CGG triplet in the Rgt1 binding sites (5′CGGANNA3′), Rgt1-DNA complexes were treated with DMS and the resulting methylated bases were cleaved with piperidine. Footprinting analysis indicated that Rgt1 protects the central guanine residues corresponding to positions −451G and −481G (Fig. 1A), and that, like most other zinc cluster proteins, Rgt1 directly interacts with the CGG triplet. The HXT1 promoter contains 8 Rgt1 binding sites within 1000 bp upstream of the ATG start codon [5]. To detect Rgt1 binding to its sites in vivo, a chromatin immunoprecipitation assay of the rgt1 mutant expressing RGT1-HA [12] was performed using a monoclonal HA antibody. Rgt1 was shown to be associated with the 3 clusters of Rgt1 binding sites in the HXT1 promoter (I, II, and III; Fig.1B). There are 3 Rgt1 binding sites in the HXT2 (at −577, −430, and −393) and HXT4 (at −645, −424, and −295) promoters (numbers are the positions of the central G residues of the CGG triplet in relation to the ATG start codon). The individual Rgt1 binding sites in the HXT2 and HXT4 promoters could be amplified by polymerase chain reaction from chromatin immunoprecipitates, suggesting that Rgt1 can bind to its single binding sites (data not shown).
Fig. 1.
Rgt1 binds to the HXT1 promoter, but does not sufficiently mediate transcriptional repression.
A. Binding of Rgt1 to the HXT1 promoter in vitro. Top: The sequence of the HXT1 upstream region between −433 and −492. The Rgt1 binding sites (CGGANNA) were underlined. Bottom: In vitro methylation protection assay of the HXT1 upstream regulatory region (−400 to −550) containing two Rgt1 binding sites. The first 188 amino acids of Rgt1, encompassing the Zn2Cys6 DNA-binding domain in its N-terminus, fused to the glutathione-S-transferase was expressed in Escherichia coli BL21 (DE3) and was affinity-purified using the glutathione agarose beads [5]. Gel lanes: 1; A+G sequencing ladder, 2; without Rgt1, 3; 100 ng Rgt1, 4; 300 ng Rgt1.
B. Binding of Rgt1 to the HXT1 promoter in vivo. The rgt1 mutant expressing HA-Rgt1 (+) or an empty plasmid (−) was grown to mid-log phase (OD600=1.0~1.2) in YP medium containing 2% galactose and subject to ChIP assay using the HA antibody. Rgt1 binding sites were PCR-amplified using different primer sets covering different regions of the promoter. IP; immunoprecipitated, IPT; input.
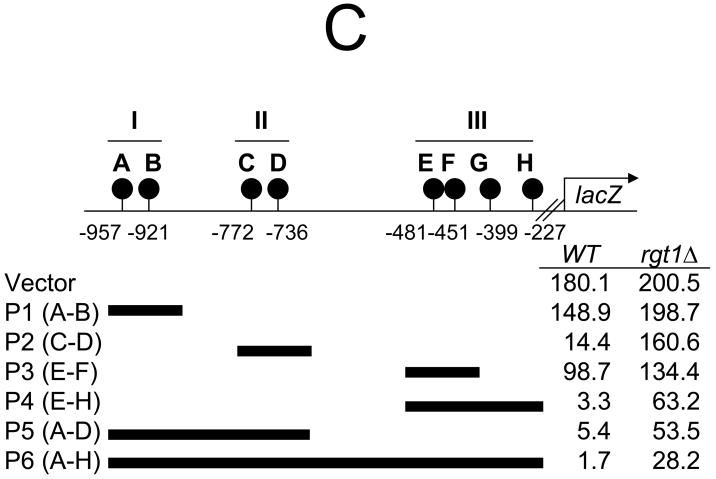
C. The upstream region of HXT1 was divided into three sections (cluster I, II and III) and inserted between the LEU2 upstream activation sequence and the TATA box of HIS3 fused to lacZ in pBM2832 [4]. β-Galactosidase activity was assayed using the yeast β-galactosidase assay kit (Pierce) and activities were presented in Miller units [5]. Yeast cells were grown to mid-log phase (OD600=1.0~1.2) in synthetic medium containing 2% galactose.
Functional analysis of the Rgt1 binding sites in the HXT1 promoter
To determine whether the Rgt1 binding sites in the HXT1 promoter are equally functional, the 3 clusters of Rgt1 binding sites were amplified by polymerase chain reaction and inserted into the promoter of the HIS3-lacZ reporter, and the constructs were expressed in wild-type and rgt1 mutant strains (Fig. 1C). Clusters I and II contained 2 Rgt1 binding sites that were 36 bp apart, but only binding to cluster II provided ~13-fold repression by Rgt1. This finding suggests that the intervening sequences between Rgt1 binding sites are involved in regulating HXT1 expression. When 4 Rgt1 binding sites were present, the reporter genes induced 33- (P5) and 54-fold (P4) repression by Rgt1, suggesting that Rgt1 binding on multiple sites synergistically mediates repression. Importantly, the full-length HXT1 promoter (P6) containing 8 Rgt1 binding sites induced about 100-fold repression, but Rgt1 appeared to mediate only 10% of the repression. These findings suggest that Rgt1 binds to the HXT1 promoter, but does not significantly mediate repression, and indicate that the intervening sequences between the Rgt1 binding sites have an important role in lowering the efficiency of Rgt1-dependent repression.
Rgt1 functions efficiently through its multiple binding sites
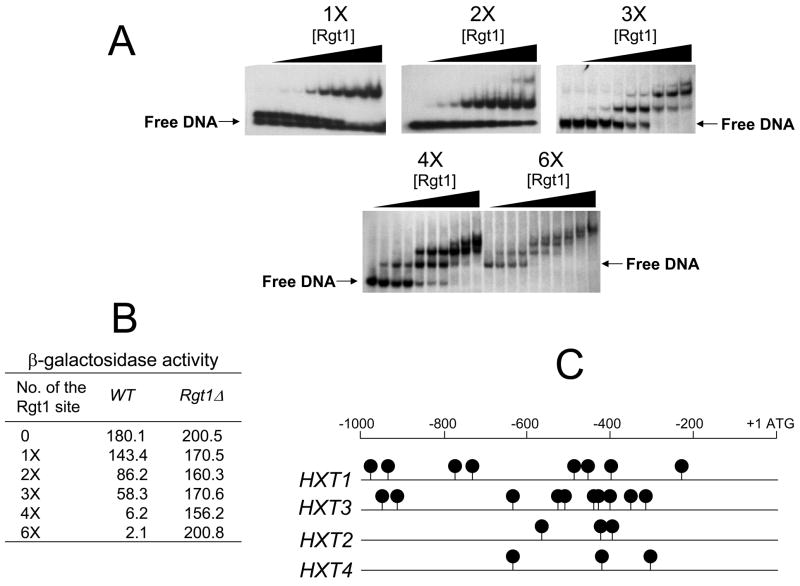
The questions of whether elimination of the intervening sequences between the Rgt1 binding sites increases the efficiency of Rgt1-dependent repression and the number of Rgt1 sites required to support sufficient Rgt1-dependent repression were examined next. To this end, complementary oligonucleotides containing different numbers of Rgt1 binding sites were inserted into the HIS3-lacZ promoter and the resulting constructs were expressed in yeast cells under the repressing condition. The activity of the synthetic DNA sequences was first tested by an electrophoretic mobility shift assay (EMSA) using the amino-terminal fragment of Rgt1 that contains a zinc binuclear cluster domain [5] (Fig. 2A). The results of the gel shift assay (EMSA) indicated that the truncated Rgt1 could bind to the DNA fragment containing an Rgt1-binding site, but bound with stronger affinity to the DNA probes that contained multiple binding sites. Further, the rate of migration of the DNA-Rgt1 complexes through polyacrylamide gel was progressively retarded with an increase in Rgt1 concentration, suggesting that the truncated Rgt1 fragment does not cooperatively bind DNA in vitro as reported previously [5]. A functional test of the synthetic, multimerized Rgt1 binding sites revealed that reporter genes containing 2 or 3 Rgt1 binding sites provided 2- to 3-fold repression, whereas those containing 4 and 6 sites induced about 30- and 90-fold repression, respectively (Fig. 2B). Rgt1 cooperatively binds to the cluster of 5 Rgt1 binding sites in the region between −290 to −400 in the HXT3 promoter in vivo, leading to an approximately 57-fold repression [5]. Therefore, the synergistic repression by RGt1 binding to multiple Rgt1 binding sites without intervening sequences is probably due to efficient recruitment of Rgt1 to the sites. These findings suggest that Rgt1 binds to single binding sites, but functions more efficiently through its binding to multiple sites, and that the DNA-binding properties of Rgt1 are similar to those of AlcR [8].
Fig. 2.
Rgt1 is efficiently recruited by artificially multimerized binding sites without intervening sequences and mediate synergistic repression of transcription.
A. Electromobility shift assay with the 32P-labelled oligonucleotides containing different numbers of Rgt1 binding sites (1x, 2x, 3x, 4x, and 6x) and the different amounts of N-terminal fragment of Rgt1 (0, 2.5, 5, 10, 20, 40, 80, 120, 250, and 500 ng per lane, from the left).
B. The complementary oligonucleotides containing different numbers of the Rgt1 binding sites were inserted between the LEU2 upstream activation sequence and the TATA box of HIS3 fused to lacZ [4]. β-Galactosidase activity was assayed as described in Fig. 1.
C. Different HXT promoters have different numbers of the Rgt1 binding sites. Black circles indicate positions of the Rgt1 consensus binding site sequence (5′CGGANNA3′).
Yeast HXT promoters regulated by Rgt1 have different numbers of Rgt1 binding sites, and different effects on Rgt1-dependent repression. For example, the HXT2 and HXT4 promoters contain 3 Rgt1 binding sites spaced 100~200 bp apart (Fig. 2C), and repression is completely relieved in an rgt1 mutant [4]. Rgt1-dependent repression appears to be more effective in the HXT2 and HXT4 promoters with fewer Rgt1 binding sites than in the HXT1 promoter with more Rgt1 binding sites. Therefore, Rgt1 functions differently at different promoters, perhaps due to different architecture of the Rgt1 binding sites in the promoters.
Conclusion
The separation of multimers of potential binding sites by intervening sequences is a common feature of regulatory proteins in natural promoters [13]. The HXT1 promoter contains 8 binding sites for the HXT repressor Rgt1, which does not significantly mediate the repression of HXT1 expression. The intervening sequences between the Rgt1 binding sites in the HXT1 promoter, however, likely play a regulatory role in Rgt1-dependent repression. A yet unidentified regulatory protein(s) may bind to the intervening sequences to influence the Rgt1-dependent repression. This view is supported by the finding that Rgt1 is efficiently recruited to multiple copies of the Rgt1 binding sites without intervening sequences and mediates synergistic repression of transcription.
Acknowledgments
I thank Mark Johnston and Özcan Sabire for providing plasmids. I also thank David JouandotII for technical assistance. This work was supported by NIH Grant RR016476-06 (MS INBRE program to Glen Shearer).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Özcan S, Johnston M. Function and regulation of yeast hexose transporters. Microbiol Mol Biol Rev. 1999;63:554–569. doi: 10.1128/mmbr.63.3.554-569.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnston M, Kim JH. Glucose as a hormone: receptor-mediated glucose sensing in the yeast Saccharomyces cerevisiae. Biochem Soc Trans. 2005;33:247–252. doi: 10.1042/BST0330247. [DOI] [PubMed] [Google Scholar]
- 3.Özcan S, Leong T, Johnston M. Rgt1p of Saccharomyces cerevisiae, a key regulator of glucose-induced genes, is both an activator and a repressor of transcription. Mol Cell Biol. 1996;16:6419–6426. doi: 10.1128/mcb.16.11.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Özcan S, Johnston M. Two different repressors collaborate to restrict expression of the yeast glucose transporter genes HXT2 and HXT4 to low levels of glucose. Mol Cell Biol. 1996;16:5536–5545. doi: 10.1128/mcb.16.10.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim JH, Polish J, Johnston M. Specificity and regulation of DNA binding by the yeast glucose transporter gene repressor Rgt1. Mol Cell Biol. 2003;23:5208–5216. doi: 10.1128/MCB.23.15.5208-5216.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marmorstein R, Carey M, Ptashne M, Harrison SC. DNA recognition by GAL4: structure of a protein-DNA complex. Nature. 1992;356:408–414. doi: 10.1038/356408a0. [DOI] [PubMed] [Google Scholar]
- 7.Swaminathan K, Flynn P, Reece RJ, Marmorstein R. Crystal structure of a PUT3-DNA complex reveals a novel mechanism for DNA recognition by a protein containing a Zn2Cys6 binuclear cluster. Nat Struct Biol. 1997;4:751–759. doi: 10.1038/nsb0997-751. [DOI] [PubMed] [Google Scholar]
- 8.Felenbok B, Flipphi M, Nikolaev I. Ethanol catabolism in Aspergillus nidulans: a model system for studying gene regulation. Prog Nucleic Acid Res Mol Biol. 2001;69:149–204. doi: 10.1016/s0079-6603(01)69047-0. [DOI] [PubMed] [Google Scholar]
- 9.Kim JH. Immobilized DNA-binding assay, an approach for in vitro DNA-binding assay. Anal Biochem. 2004;334:401–402. doi: 10.1016/j.ab.2004.06.045. [DOI] [PubMed] [Google Scholar]
- 10.Özcan S, Johnston M. Three different regulatory mechanisms enable yeast hexose transporter (HXT) genes to be induced by different levels of glucose. Mol Cell Biol. 1995;15:1564–1572. doi: 10.1128/mcb.15.3.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maxam AM, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 12.Mosley AL, Lakshmanan J, Aryal BK, Özcan S. Glucose-mediated phosphorylation converts the transcription factor Rgt1 from a repressor to an activator. J Biol Chem. 2003;278:10322–10327. doi: 10.1074/jbc.M212802200. [DOI] [PubMed] [Google Scholar]
- 13.Ligr M, Siddharthan R, Cross FR, Siggia ED. Gene expression from random libraries of yeast promoters. Genetics. 2006;172:2113–2122. doi: 10.1534/genetics.105.052688. [DOI] [PMC free article] [PubMed] [Google Scholar]