Abstract
The aryl hydrocarbon receptor (AhR) is a ligand-dependent, basic helix–loop–helix Per-Arnt-Sim (PAS)-containing transcription factor that can bind and be activated by structurally diverse chemicals, including the toxic environmental contaminant 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD). Our previous three-dimensional homology model of the mouse AhR (mAhR) PAS B ligand binding domain allowed identification of the binding site and its experimental validation. We have extended this analysis by conducting comparative structural modeling studies of the ligand binding domains of six additional high-affinity mammalian AhRs. These results, coupled with site-directed mutagenesis and AhR functional analysis, have allowed detection of the “TCDD binding-fingerprint” of conserved residues within the ligand binding cavity necessary for high-affinity TCDD binding and TCDD-dependent AhR transformation DNA binding. The essential role of selected residues was further evaluated using molecular docking simulations of TCDD with both wild-type and mutant mAhRs. Taken together, our results dramatically improve our understanding of the molecular determinants of TCDD binding and provide a basis for future studies directed toward rationalizing the observed species differences in AhR sensitivity to TCDD and understanding the mechanistic basis for the dramatic diversity in AhR ligand structure.
The aryl hydrocarbon receptor (AhR)1 is a ligand-dependent transcription factor and a member of the basic helix–loop–helix Per-Arnt-Sim (bHLH/PAS) family of proteins (1–4). While the highest-affinity AhR ligands include persistent environmental contaminants such as 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) and related halogenated aromatic hydrocarbons (HAHs) and polycyclic aromatic hydrocarbons (PAHs) (5, 6), recently evidence has indicated that the AhR can be bound and activated by a structurally diverse range of chemicals, albeit lower-affinity ligands (5, 7–9). Mechanistically, ligand binding to the PAS B domain of the AhR in the cytosol leads to a conformational change (10) with exposure of an N-terminal nuclear localization sequence, followed by translocation of the liganded AhR protein complex into the nucleus(11). Dissociation of the AhR from its associated proteins [hsp90, XAP2, and p23 (12)] and its dimerization with ARNT (AhR nuclear trans-locator) convert the AhR into its high-affinity DNA binding form (13). Binding of the ligand–AhR–ARNT complex to its specific DNA recognition site, the dioxin responsive element (DRE), leads to an increased level of expression of CYP1A1 and other genes (2, 3, 14). The functional characteristics of AhRs are broadly conserved among vertebrate species, even though large species and strain differences in sensitivity to TCDD and other AhR ligands have been observed (15).
A molecular understanding of the events that result from ligand binding to the AhR requires detailed structural information about the PAS B domain. In fact, among the various domains of the AhR that exhibit distinct functional activities, PAS B is the one involved in ligand binding and also contains the primary hsp90 binding site (10, 16–18). Because of the lack of experimentally determined structures, the three-dimensional models of the mouse AhR (mAhR) PAS B domain that we previously developed by homology modeling techniques (19, 20) provided the first avenue for these studies. Our current model was built on the basis of the recently described NMR-determined structures of the PAS B domains of human hypoxia-inducible factor 2α (HIF-2α) (21) and the human ARNT protein (22). These are the best template structures available as they have the highest degrees of sequence identity and similarity with the AhR PASB domain among all the known PAS structures. In addition, both of these proteins also belong to the bHLH/PAS family of transcriptional factors and are functionally related to the AhR (4).
Examination of the resulting AhR PAS B homology model revealed the presence of a buried cavity within the core of the mAhR ligand binding domain (LBD), which was the putative ligand binding site. Site-directed mutagenesis experiments and functional analysis validated the proposed model and confirmed the identity of this cavity as the ligand binding pocket (20). The reported AhR LBD structures and structure-driven mutagenesis analysis also stimulated the studies of other authors, directed toward elucidating the determinants of ligand binding specificity as well as the differential responsiveness of AhRs of different species (23–25).
On the basis of the commonalities in the signaling pathways and the availability of TCDD binding affinity data and the sequences of various mammalian AhRs, here we have extended our structural modeling studies to the LBDs of six other mammalian AhRs. AhRs with relatively high binding affinities for TCDD (i.e., those with apparent dissociation constants of <5 nM) were selected for this analysis and included rat, hamster, rabbit, guinea pig, beluga whale, and seal AhRs (26–28). Structural modeling and comparison of these LBDs allowed further study of the AhR binding cavity characteristics that promote TCDD binding. Analysis of the importance of the conserved residues present in all the examined binding cavities performed by site-directed mutagenesis and AhR functional analysis allowed us to detect the “TCDD binding-fingerprint”, i.e., the group of residues in the AhR LBD whose physicochemical characteristics are needed for optimal TCDD binding. Further confirmation of the essential role of some of these residues in TCDD binding was obtained from a set of molecular docking simulations performed on the wild-type (wt) mAhR and mutant mAhRs that lacked both TCDD and TCDD-dependent DNA binding. These results allow for a complete understanding of the molecular determinants of TCDD binding.
MATERIALS AND METHODS
Homology Modeling
Homology modeling techniques were applied in predicting the LBD structures of the following mammalian AhRs: rat AhR (rtAhR) (gi 510269), hamster AhR (haAhR) (gi 9438131), rabbit AhR (rbAhR) (gi 25089567), guinea pig AhR (gpAhR) (gi 47571343), beluga whale AhR (bAhR) (gi 15625347), and seal AhR (sAhR) (gi 15528541).
The protocol previously proposed (20) for developing the model of the mouse AhR LBD, mAhR (gi 7304873), was also employed for the models of all the AhRs here presented. Three-dimensional models for the AhR LBDs were generated using MODELER version 8v1 (29–31), which implements an approach to comparative modeling by satisfying spatial restraints. The restraints, which were obtained empirically from a database of protein structure alignments, and CHARMM energy terms (32) were combined into an objective function. The resulting model was obtained by optimizing the objective function employing methods of conjugate gradients and molecular dynamics with simulated annealing.
Template identification by sequence similarity searches with the AhR LBD was performed independently for each target sequence using PSI-BLAST (33) against a database of known protein structures with default parameters. In all cases, the best hits corresponded to the NMR structures of hypoxia-inducible factor 2α (HIF-2α) and the AhR nuclear translocator (ARNT) PAS B domains. Therefore, these were chosen as template structures for homology modeling. Their coordinates were obtained from the Protein Data Bank (34): entries 1P97 for HIF-2α (21) and 1X0O for ARNT (22). The most representative structure within each of the NMR structure bundles was selected using NMRCLUST (35).
The templates were structurally aligned according to DALI-Lite (36, 37). The target–template sequence alignments were created with CLUSTALW (38), and the result was confirmed using the Align-2D command within MODELER (29–31). The quality of the models was assessed by the ProSA validation method (39) by using the web-based version (ProSA-web) (40). This method uses knowledge-based potentials of mean force to evaluate model accuracy and statistical significance, and it is widely employed in validating both experimentally determined protein structures and theoretical models. Secondary structures were attributed by DSSPcont (41). This has extended the discrete assignments of secondary structure in eight categories performed by DSSP (42), to a continuous assignment in the same categories. The continuum results are calculated by weighted averages over 10 discrete DSSP assignments with different hydrogen bond thresholds. Three-dimensional visualization and images of the AhR LBD structures were generated using PyMOL (43).
Analysis of Structural Cavities
Identification and characterization of surface pockets and internal cavities in the modeled structures were performed with the CASTp server (44). The program allows identification and calculation of the Connolly’s molecular surface and volume (45) for all pockets and cavities in a protein structure. It ranks the cavities by size, where the largest one is usually the binding site (46). The representations of the surface that includes the largest available cavity were produced with PyMOL (43).
Molecular Docking
AutoDock 4 (47) and its graphical front-end AutoDockTools (ADT) were used to set up and perform docking calculations. The mAhR LBD structure obtained by homology modeling was used as a starting structure for the wt mAhR. The single mutant models were built by substituting the mutated residue into the wild-type structure by PyMOL (43) and performing a short minimization (100 steps) with steepest descent with the GROMOS96 43a2 force field, as available in the GROMACS 3.2.1.2 package (48, 49). The structure of the 2,3,7,8-TCDD optimized with HF/6-31G* ab initio calculations (50) was employed for the ligand. In docking simulations, only polar hydrogens of each protein were considered and both ligand and protein structures were treated as rigid bodies.
Grid maps with0.375 Å spacing were defined for all the models to include the mAhR binding site, as defined by the CASTp program. Lamarckian genetic algorithms, as implemented in AutoDock, were employed to perform docking calculations. The maximum number of energy evaluations and generations was set to 25 million and 27000, respectively, and 100 runs were performed for each calculation, with a population of 150 individuals. Cluster analysis on the docked conformations was performed with AutoDock applying a root-mean-square deviation (RMSD) cutoff of 2 Å. All other parameters were default settings. For each of the docking cases, the lowest-energy docked conformation, according to the Autodock scoring function, was selected as the binding mode.
In Vitro AhR Protein Expression and Mutagenesis
The pcDNA3/βAhR mouse AhR expression plasmid (51), obtained from O. Hankinson (University of California, Los Angeles, CA), and the pcDNA3.1/mβARNT mouse ARNT expression plasmid were used as templates for in vitro expression of the AhR and ARNT, respectively, using the TNT Quick coupled transcription/translation rabbit reticulocyte lysate system (Promega) as previously described (10). Mutation of selected amino acids within the AhR to alanine (F289A, Y316A, F318A, I319A, and F345A), phenylalanine (H285F, P291F, Y316F, and H320F), tyrosine (F289Y, F318Y, and I319Y), leucine (F345L), methionine (T283M), and glutamic acid (T283E) was carried out using the QuickChange mutagenesis technique (Stratagene). Translation grade L-[35S]methionine (>1175 Ci/mmol) was purchased from MP Biomedical (Solon, OH). 35S-radiolabeled wt and mutant mAhRs were synthesized in separate reactions in vitro, denatured, and subjected to SDS–polyacrylamide gel electrophoresis, and the level of expression of each protein was determined by phosphoimager analysis of the dried gel (see the Supporting Information).
DNA and Ligand Binding Analysis of Mutant AhRs
In vitro synthesized mouse ARNT and wt or mutant AhR were prepared as previously described (10). For gel retardation DNA binding analysis, in vitro synthesized mouse ARNT and wtAhR or mutant AhR (1.5 μL each) were combined with 7 μL of MEDG buffer [25 mM MOPES (pH 7.5), 1 mM EDTA, 1 mM DTT, and 10% (v/v) glycerol] and incubated in the presence of 20 nM TCDD or DMSO for 3 h at room temperature. These are conditions we have previously shown tobe sufficient for maximal formation of the wt or mutant AhR–Arnt–DNA complex (10). Aliquots of the incubation reaction mixture were mixed with [32P]DRE oligonucleotide and AhR transformation and DNA binding conducted by gel retardation analysis as described in detail previously (52). Protein–DNA complexes in the dried gels were visualized, and the amount of 32P-labeled DRE present in the TCDD-induced protein–DNA complex was quantitated using a Molecular Dynamics phosphoimager. The amount of specific TCDD-inducible AhR–Arnt–[32P]DRE complex was determined by quantitating the amount of radioactivity present in the TCDD-treated sample lane (i.e., the TCDD–AhR–DRE complex) and subtracting the amount of radioactivity present in the same position in the DMSO-treated sample lane. The DNA binding activities of all of the mutant AhRs were normalized to total DNA binding results obtained with the wtAhR that was included in each experiment, and results are expressed as a percentage of the amount of TCDD-inducible wtAhR DNA binding. For ligand binding analysis, in vitro synthesized AhR proteins diluted in MEDG buffer to 8 mg/mL protein were incubated with 4 nM [3H]TCDD [specific activity of 14.5 Ci/mmol, obtained from S. Safe (Texas A&M University, College Station, TX)] as described previously (53), and equivalent amounts of unprogrammed lysate were used as a nonspecific binding control. The hydroxyapatite binding assay (53) was used to measure the amount of [3H]TCDD binding in each sample, and the amount of specific binding was determined by subtracting the amount of [3H]TCDD bound to the unprogrammed lysate from the total amount of binding to lysate containing in vitro expressed AhR. The amount of specific binding of [3H]-TCDD to each mutant AhR was expressed as a percent of the total specific binding of [3H]TCDD to wtAhR. Values were expressed as the mean ± standard deviation of three to five individual analyses, and those values significantly different from the wtAhR positive control at p<0.05 were determined by the Student’s t test.
Transfection Analysis of Mutant AhRs
COS-1 cells were grown in α-minimal essential medium (α-MEM) (Gibco) supplemented with 10% fetal bovine serum (FBS) (Atlanta Biologicals). Cells were seeded into 24-well plates 24 h prior to transfection. Ninety minutes prior to transfection, the medium was replaced with α-MEM containing 2% FBS. Triplicate wells containing cells at ~60–65% confluence were cotransfected with Transfectol reagent according to the manufacturer’s protocol (Gene Choice, Frederick, MD), using 0.36 μg of the pGudLuc6.1 AhR-responsive luciferase reporter plasmid and 0.04 μg of pcDNA3/βAhR (containing wt or mutant AhRs) per well. The cells were maintained in α-MEM with 2% FBS during transfection, and after24 h, the culture medium was exchanged with fresh medium containing 2% FBS and 10 nM TCDD or 1% (v/v) DMSO (as the control). After 24 h, the cells were washed once with phosphate-buffered saline and lysed with 100 μL of passive lysis buffer (Promega, Madison, WI) and the plates shaken until complete cell lysis was achieved (15 min). Fifty microliters of the cell lysate was added to a well of a 96-well white microplate and luciferase activity determined using a Berthold microplate lumin-ometer with automatic injection of 50 μL of luciferase substrate and quantitation of light production after a 10 s read delay and integration times of 10 s.
RESULTS
Homology Modeling
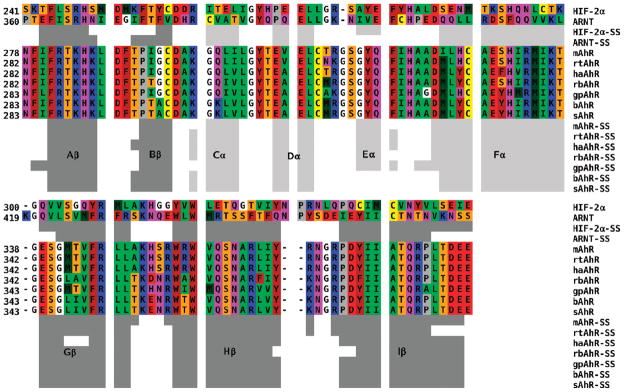
Recursive PSI-BLAST searches of the selected mammalian AhR LBD sequences in the regions corresponding to residues 230–421 of the mAhR were performed against the Protein Data Bank. The sequences that produced the most significant alignments were thoseofthe HIF-2α and ARNT PAS domains. This finding confirmed the previous choice of HIF-2α and ARNT as the optimal template structures (20) for modeling all of the AhR LBDs considered here. Both templates show a high degree of structural conservation of the typical PAS α- and β-fold. This includes a five-stranded β-sheet and a long central helix linked to a bulge of three small helices. Following the nomenclature generally adopted for PAS structures, the N-terminal β-strands are termed Aβ and Bβ, followed by the three small helices (Cα, Dα, and Eα), the long central helical connector (Fα), and the three C-terminal strands of the β-sheet (Gβ, Hβ, and Iβ). The secondary structure attribution obtained by the DSSPcont program for the two templates, labeled according this nomenclature, is shown in Figure 1.
Figure 1.
Multiple alignment of the selected mammalian AhRs against templates HIF-2α and ARNT (see the text). The coloring scheme is as follows: red for acidic, blue for basic, purple for polar, yellow for Cys, brown for aromatic, green for hydrophobic, orange for Ser and Thr, gray for Pro, and white for Gly. Secondary structures of both templates and AhR models attributed by the DSSPcont program: light gray bars for helices and dark gray bars for β-strands. The secondary structure elements are labeled according to the nomenclature generally adopted for the PAS structures.
The selected mammalian AhR sequences were first aligned internally and then aligned to the HIF-2α template using CLUSTALW, and the obtained alignment was in agreement with that generated using the Align-2D command within MODELER. A global alignment was subsequently generated by structurally aligning the ARNT and HIF-2α PAS B domains with DALI. The pairwise sequence identities and similarities between the target AhR LBDs and the two templates are presented in Table 1. The values indicate a medium to low degree of identity and similarity with both the template structures. Since the highest values are those obtained by comparison of the AhRs with HIF-2α, this template structure appears to be the most informative for modeling.
Table 1.
Sequence Identity and Similarity between the Ah Receptors and the PAS Domain Templates (110 aligned positions)
| HIF-2α |
ARNT |
|||
|---|---|---|---|---|
| AhR sequence | identity (%) | similarity (%) (BLOSUM 62) | identity (%) | similarity (%) (BLOSUM 62) |
| mAhR | 30.0 | 50.9 | 20.0 | 43.6 |
| rtAhR | 29.1 | 50.9 | 20.9 | 43.6 |
| haAhR | 29.1 | 49.1 | 19.1 | 42.7 |
| rbAhR | 28.2 | 48.2 | 20.0 | 44.6 |
| bAhR | 26.4 | 48.2 | 20.0 | 43.6 |
| sAhR | 26.4 | 48.2 | 20.0 | 43.6 |
Analysis of the obtained alignment, shown in Figure 1, reveals the presence of only a glycine insertion, in the DE loop, and a two-residue deletion, in the HI loop. A single residue deletion in both AhRs and HIF-2α with respect to ARNT is also present in the FG loops. On the basis of this limited number of insertions and deletions, no additional effort was needed to undertake loop modeling after the MODELER prediction cycle.
For each AhR sequence, 100 individual models were derived by MODELER from random generation of the starting structure and the model with the lowest value of the objective function was selected as the most representative. The quality of each final model was then evaluated by the MODELER’s ENERGY command. A limited number of violations of the MODELER stereochemical restraints were observed for all the models.
Additionally, the statistical significance of each modeled structure was assessed by calculation of the ProSA z-score. This measures the deviation of the total energy of each structure with respect to an energy distribution derived from random conformations (39). The z-score values obtained for the seven AhR models were between −3.71 and −4.27 and were within the ranges of scores typically found for native protein structures of similar size.
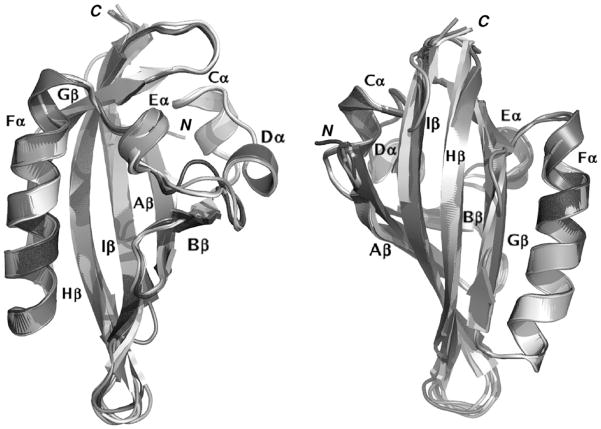
The final models for each of the seven AhR LBDs were structurally aligned with DALILite and visually compared. Snapshots of two orientations of the overlaid models are shown in cartoon style in Figure 2, with the secondary structure attribution obtained with DSSPcont. The visual comparison highlights that both the global fold and the secondary structure elements characteristic of the PAS domain structures are well-conserved in the seven mammalian AhR models.
Figure 2.
Cartoon representation of the modeled structures of the AhR LBDs from seven different species, structurally aligned with DALILite (36, 37) and shown in two different orientations.
A more detailed analysis (see also the aligned secondary structures in Figure 1) reveals a slight variability among the models that mainly involves the length and stereochemistry of the C-terminal β-strands and the arrangement of their connecting loops. In particular, the ordered structure of the Gβ strand of the templates appears twisted in all the models, with three central residues particularly strained. DSSPcont reports a 53% strand and a 47% loop attribution to these residues, except for the haAhR (68% loop). Moreover, the residues classified as a 310-helix in HIF-2α and as a helix turn in ARNT (Eα) are attributed as 310-helices in the mAhR, haAhR, and rbAhR, as α-helices in the rtAhR and gpAhR, and as helix turns in the bAhR and sAhR.
Analysis of Structural Cavities
The analysis of structural pockets and cavities, performed by the CASTp server, indicated the presence of a buried cavity in the core of all the modeled domains, delimited by the β-sheet and flanked by the helices and their connecting loops. All the cavity volumes fall in the range of 100–800 Å3 (see Table 2), a size commonly observed in protein binding pockets or cavities (46). Additionally, the range of values is comparable to the volume found for the mAhR cavity, whose role in TCDD binding was confirmed in our earlier mutagenesis experiments (20). Visual analysis of each molecular surface that includes the available volume indicates that the slight variability observed among the volumes for the different mammalian AhR cavities has to be attributed only to subtle differences in some internal side chain conformations. These were expected as a result of the modeling procedure using template structures with medium to low degrees of sequence identity with the target sequences. The marine mammalian AhRs, bAhR and sAhR, as a consequence of their sequence identity in the modeled region, present a common LBD fold and identical internal cavities. Their volumes are the smallest in this series of AhRs, but the shape of the available internal space resembles that of the central part of the larger cavities of the other mammalian AhRs.
Table 2.
Measures of the Area and Volume of the Main Cavity in the Connolly’s Molecular Surface, As Obtained by CASTp for the Ah Receptors
| AhR model | surface (Å2) | volume (Å3) |
|---|---|---|
| mAhR | 440 | 471 |
| rtAhR | 498 | 486 |
| haAhR | 423 | 442 |
| rbAhR | 400 | 419 |
| bAhR | 310 | 322 |
| sAhR | 310 | 322 |
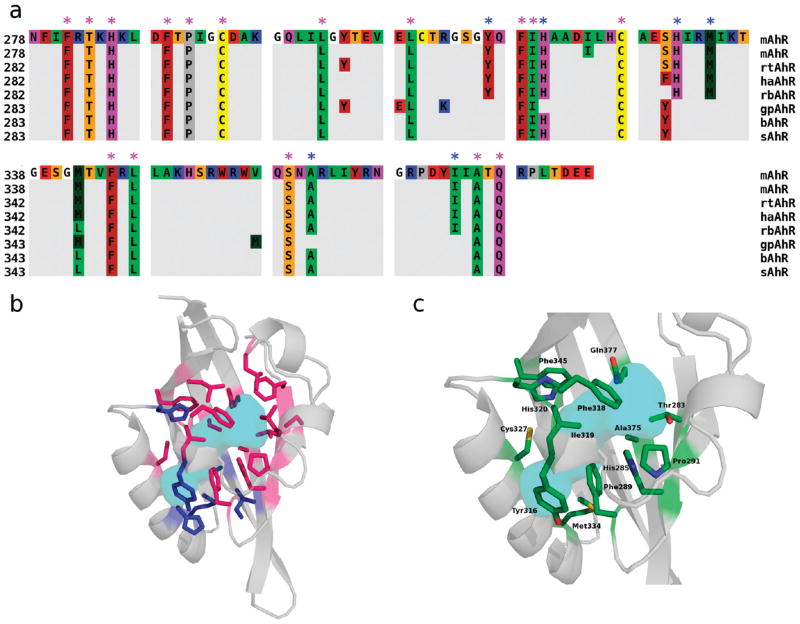
The detailed list of residues whose side chains contribute to the internal cavity surface was extracted from the CASTp output and compared. A multiple alignment highlighting these residues in each of the AhR LBDs is shown in Figure 3a. The location of boundary residues is consistent for all the AhR sequences in the alignment, providing further support that the modeled cavities are very similar. Moreover, a high degree of amino acid conservation of the common internal residues is observed, when compared to the mouse sequence, with only boundary residues at 330 and 342 showing a degree of variability.
Figure 3.
Identification of amino acids whose side chains contribute to the AhR ligand binding cavity. (a) Sequence alignment of the selected mammalian AhRs in which only the residues whose side chains are internal (pink asterisks) or borderline to the cavities (blue asterisks), as identified with CASTp, are shown. Seethe legend of Figure 1 for the residue color scheme. (b) Cartoon representation of the modeled mAhR LBD with conserved residues internal to all the AhR cavities (pink sticks) and borderline residues (blue sticks). The molecular surface that includes the consensus cavity (see the text), is colored cyan. (c) Cartoon representation of the modeled mAhR LBD with residues selected for mutagenesis and functional analysis (see Table 3) shown as sticks. Sticks are colored according to the atom type (green for carbon, red for oxygen, blue for nitrogen, and yellow for sulfur). The molecular surface that includes the consensus cavity is colored cyan.
Internal residues conserved in all modeled sequences are denoted with pink asterisks in Figure 3a. The cavity delimited by these residues (as calculated by CASTp) can be viewed as the internal space shared by all the domains, and as such, we will refer to it as the “consensus cavity”. As all these AhRs exhibit high binding affinities for TCDD, this common internal space represents the minimal space that is needed for high-affinity TCDD binding. Residues that are borderline to the cavities (i.e., those that are internal in some of the AhRs and external for others based on CASTp analysis) are denoted with blue asterisks in Figure 3a. Both internally conserved and borderline residues are also shown on the mAhR model structure in Figure 3b, along with the molecular surface including the consensus cavity.
On the basis of this structural analysis, the candidates for experimental analysis were selected among the internally conserved residues. Three of them were excluded because they are not conserved in other AhRs which can bind TCDD with high affinity: F281 and C294, which are Leu and Ile, respectively, in the zebrafish AhR2 (54), and S359, which is Ala in the chicken AhR (23). Moreover, L302, L309, and L347 were excluded because they lie relatively far from the cavity core (at the top of the cavity in Figure 3b). On the other hand, some borderline residues whose location with respect to the cavity core appeared to be particularly interesting (Y316, H320, and M334) were included in the analysis. The three- dimensional placement of the resulting group of 13 residues is shown in Figure 3c.
Mutagenesis and Ah Receptor Functional Analysis
Residues pointing into the proposed consensus cavity are expected to play key roles in the high-affinity binding of TCDD and other ligands. Given the above description and our previous results (20), the residues selected for mutation in this study were T283, H285, F289, P291, Y316, F318, I319, H320, and F345 (Figure 3c). To analyze the role of these targeted residues in both TCDD binding to and TCDD-dependent transformation and DNA binding of the AhR, we examined the effects of a series of amino acid substitutions in these residues on these AhR functionalities (Table 3). Since the side chains of these amino acids could play a role in ligand binding through steric effects on the size and shape of the cavity and/or their particular stereoelectronic characteristics useful in stabilizing ligand binding within the pocket, different types of amino acid substitutions in target residues were tested.
Table 3.
Effects of Mutagenesis of Individual Amino Acids within the mAhR LBD on [3H]TCDD Specific Binding and TCDD-Inducible AhR DNA Binding
| mutant | [3H]TCDD specific binding (% of that of wt mAhR) | TCDD-inducible DRE binding (% of that of wt mAhR) |
|---|---|---|
| wild-type AhR | 100 ±19a | 100 |
| T283E | 0 ± 1b | 6 ± 2b |
| T283M | 0 ±2b | −4 ± 3b |
| H285Ac | 0 ±1b | −2 ±4b |
| H285F | 7 ±2b | 32 ± 1b |
| F289A | −2 ± 3b | −3 ± 1b |
| F289Y | 4 ± 1b | 18 ± 3b |
| P291F | 26 ± 1b | 53 ± 7b |
| Y316A | 7 ± 3b | 4 ± 4b |
| Y316F | 45 ± 3b | 84 ± 4b |
| F318A | 20 ± 1b | 24 ± 4b |
| F318Y | 87 ± 2 | 86 ± 6 |
| I319A | 4 ± 2b | 1 ± 2b |
| I319Y | −1 ± 1b | 1 ± 2b |
| H320F | 28 ± 2b | 38 ± 2b |
| C327Ac | 59 ± 3b | 82 ± 11b |
| M334Ac | 57 ± 7b | 70 ± 9b |
| M334Ec | −16 ± 12b | 3 ±6b |
| F345A | 2 ± 2b | −3 ± 5b |
| F345L | 30 ± 2b | 51 ±8b |
| A375Lc | −2 ± 4b | 3 ± 8b |
| A375Vc | −6 ± 7b | 42 ± 12b |
| Q377Ac | 31 ± 4b | 52 ± 3b |
Values represent the mean ± standard deviation of three to five individual analyses.
Values are significantly different from that of wild-type mAhR at P<0.05 as determined by a Student’s t test.
From ref (20).
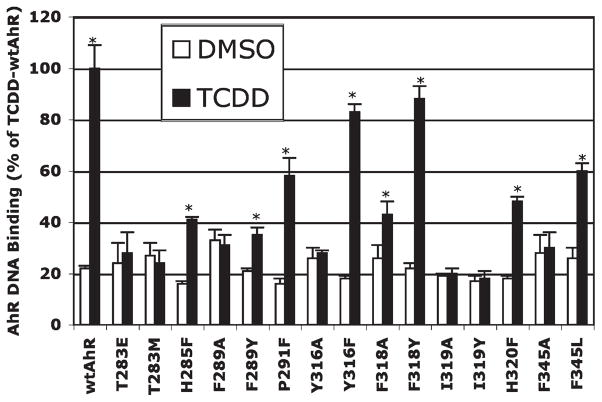
We have previously demonstrated that gel retardation analysis of in vitro synthesized wtAhR and ARNT incubated with TCDD results in the formation of an inducible TCDD–AhR–ARNT–DNA complex that is not observed in DNA binding reactions using unprogrammed lysate, or in vitro expressed AhR or ARNT alone (10, 20, 52). Utilizing this method, we examined the ability of each in vitro synthesized mutant AhR (in the presence of ARNT) to bind to [32P]DRE-containing DNA in a TCDD-inducible manner (using DMSO as the control), and the amount of protein–DNA complex was quantitated by phosphorimager analysis. The combined results of multiple gel retardation analysis of wt and mutant AhRs are shown in Figure 4 and Table 3. Given that the wt and mutant AhRs were synthesized in reticulocyte lysate at comparable levels [based on similar levels of expression of in vitro synthesized 35S-labeled wt and mutant AhRs (see Figure 1 of the Supporting Information)], the observed differences in DNA binding are not simply due to differences in the amount of AhR. Gel retardation analysis revealed that some mutations (Y316F and F318Y) had minimal effects on TCDD-dependent AhR DNA binding (reducing the level of DNA binding by <20%), some mutations reduced the level of DNA binding dramatically (H285F, F289Y, P291F, F318A, H320F, and F345L), and others eliminated DNA binding (T283E and -M, F289A, Y316A, I319A and -Y, and F345A). Mutations in the AhR LBD resulted in little change in the amount of background (i.e., ligand-independent) AhR DNA binding (Figure 4), with the exception of F289A, for which the background level of DNA binding was increased by ~50% over that of the wt AhR control (i.e., DMSO treatment). Mutation of F289 to tyrosine (another aromatic amino acid) did not result in increased in background DNA binding activity, but some ligand-dependent DNA binding of this mutant AhR was still retained (~20% of that of wt AhR).
Figure 4.
Effect of mutation of selected residues within the mAhR LBD on TCDD-dependent AhR DNA binding. In vitro expressed wild-type or mutant AhR and wt ARNT were incubated with DMSO (control) or TCDD, and formation of the AhR–ARNT–DRE complex was assessed by gel retardation analysis as described in Materials and Methods. Quantitation of the amount of AhR–ARNT–DRE complex was achieved by phosphoimager analysis, and the results are expressed as the mean ± standard deviation of multiple receptor preparations and gel retardation analyses (n≥3). The asterisk indicates that the values are significantly different from those of their respective DMSO control at p≤0.05 as determined by a Student’s t test. The amount of specific TCDD-inducible AhR–ARNT–DRE complex for each AhR determined after subtraction of the DMSO background is presented in Table 3.
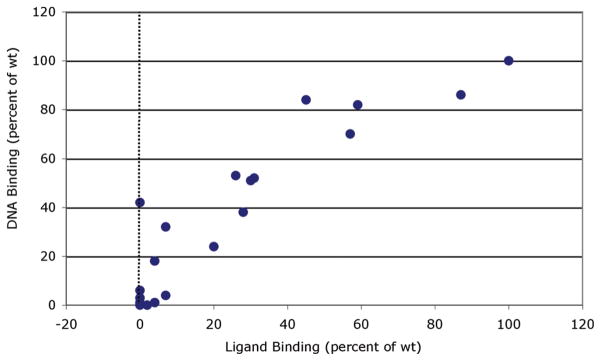
While gel retardation analyses demonstrated that a variety of mutations within the AhR LBD can adversely affect the ability of the AhR to transform and bind to DNA in a ligand (TCDD)-dependent manner, they do not address whether these mutations produce this effect by alterations in ligand binding and/or ligand-dependent transformation of the AhR. To address this, we examined the ability of [3H]TCDD to bind to each of the in vitro synthesized mutant mAhRs and compared these results with the ligand-dependent DNA binding activity of the same mutant AhRs (Figure 5 and Table 3). These results show a high degree of correlation between the ligand binding activity of various mutant AhRs and their ability to transform and bind to DNA in a ligand-dependent manner. Like the gel retardation assay results, these mutations produced a comparable range of effects on [3H]TCDD specific binding (Table 3). As expected, for those mutants that exhibited essentially no ligand-dependent DNA binding (T283E and -M, F289A, Y316A, I319A and -Y, and F345A), no [3H]TCDD specific binding was observed. These results are consistent with a role for these amino acids in high-affinity TCDD binding within the AhR LBD. The decrease in the level of binding of [3H]TCDD to mutants P291F, F318A, and H320F correlated well with the observed reduction in the level of TCDD-dependent DNA binding of these mutant AhRs, indicating a role for these residues in ligand binding and ligand-dependent transformation or DNA binding, consistent with the decreased response resulting from a reduction in ligand binding affinity.
Figure 5.
Correlation between [3H]TCDD ligand binding and TCDD-dependent DNA binding of mAhRs containing mutations of selected residues within the LBD. Ligand binding to in vitro expressed wild-type or mutant AhR incubated with 4 nM[3H]TCDD and unprogrammed reticulocyte lysate (nonspecific binding control) was assessed using hydroxyapatite binding, and DNA binding (formation of the AhR–ARNT–DRE complex) of in vitro expressed wild-type or mutant AhR and wt ARNT incubated with DMSO (control) or 20 nM TCDD was assessed by gel retardation analysis as described in Materials and Methods. The amount of [3H]TCDD specific binding to and AhR–ARNT–DRE complex formation of each mutant AhR was expressed relative to that of wild-type AhR (100%), and the specific values are listed in Table 3.
Two mutations (H285F and F289Y) exhibit interesting characteristics. While little [3H]TCDD specific binding to AhRs containing H285F and F289Y mutations was observed, the AhRs exhibited TCDD-dependent [32P]DRE-containing DNA binding (32 and 18% of that of wt AhR, respectively). Our previously reported results with the A375V mutation of the mAhR revealed a similar discrepancy in the functional analysis (i.e., no [3H]TCDD specific binding, but measurable TCDD-inducible DNA binding), and we subsequently demonstrated that this was due to a reduction in the affinity of TCDD binding (20). The dramatically greater specific activity of the 32P-labeled DNA (6000 Ci/mmol) compared to that of [3H]TCDD (14.5 Ci/mmol) accounted for our ability to detect TCDD-inducible DNA binding but not that of ligand binding (20). Increasing the concentration of [3H]TCDD in the binding reaction mixtures from 2 to 20 nM led to detectable [3H]TCDD specific binding to the mutant AhR. For the H285F and F289Y mutant mAhRs described here, increasing the concentration of available [3H]-TCDD in the binding reaction mixtureto20 nM led to an increase in [3H]TCDD specific binding to each receptor [from 141 ± 297 to 297 ± 91 dpm and from 46 ± 8 to 156 ± 53 dpm, respectively (data not shown)]. These results are consistent with the reduced level of TCDD binding to H285F and F289Y resulting from a decreased ligand (TCDD) binding affinity, similar to that previously observed for A375V (20).
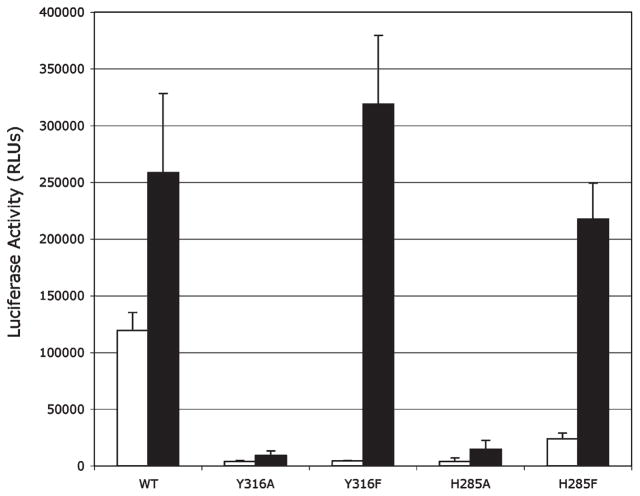
To confirm that the results of the in vitro mutagenesis and functional analysis are also observed in intact cells, the ability of selected mutant AhRs to stimulate TCDD-dependent gene expression was examined in transfection experiments. COS-1 cells were transiently cotransfected with an AhR-responsive luciferase reporter plasmid (pGudLuc6.1) and wild-type or mutant AhR expression vectors, and TCDD-inducible luciferase activity was determined. The AhRs selected for this analysis included those with substitutions at positions Y316 and H285 as mutation of these amino acids to alanine eliminated AhR functionality and their mutation to phenylalanine resulted in only a reduction in AhR functionality, compared to wtAhR (Table 3). Consistent with our in vitro analyses, AhRs containing Y316A and H285A mutations would not support TCDD-inducible reporter gene expression while those AhRs containing Y316F and H285F mutations allowed TCDD-dependent induction of gene expression (Figure 6). Interestingly, the background (DMSO) luciferase activity in cells transfected with the mutant AhRs was dramatically reduced compared to that of wt AhR. This is a function of the specific mutation, as background luciferase activity with AhR containing a F318Y mutation was not reduced compared to that of wtAhR (data not shown). We are currently examining the mechanism responsible for this reduction in constitutive AhR activity.
Figure 6.
Induction of the DRE-dependent reporter gene in COS-1 cells transiently transfected with an AhR-responsive luciferase reporter gene (pGudLuc.1) and wild-type or mutant AhR expression vectors. COS-1 cells (60–65% confluent) were cotransfected with the pGudLuc6.1 AhR-responsive luciferase reporter plasmid and wt or mutant AhR expression vectors (pcDNA3/βAhR) and incubated with DMSO (1%, v/v) or 10 nM TCDD for 24 h. Luciferase activity was measured as described in Materials and Methods, and the resulting values are expressed as the mean relative light units (RLUs) ± standard deviation of triplicate analyses.
Molecular Docking
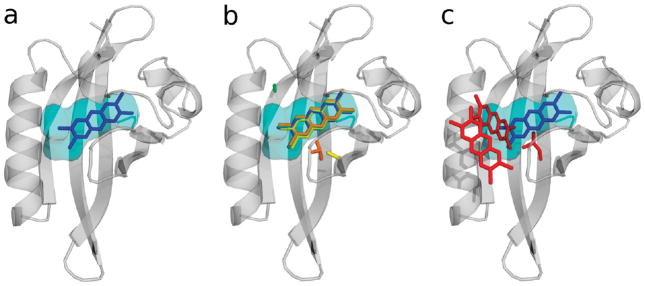
Molecular docking calculations were performed to simulate TCDD binding to the wild-type mAhR, by using AutoDock (see Materials and Methods). Despite the low atomic resolution of the homology model, it is conceivable that the binding site location as well as the relative positioning of the ligand can be reliably predicted by the docking algorithm.
A unique cluster of docked conformations was obtained, in agreement with a high binding specificity. The lowest-energy docked conformation is shown in Figure 6a, and the estimated free energy of binding (ΔG°binding) is reported in Table 4. The position of the TCDD was fully consistent with the location of the internal space available for ligand binding as described by the consensus cavity (Figures 3 and 7). On the basis of our mutagenesis results, all of the residues that appear to contribute to TCDD binding are in close contact with the docked TCDD molecule.
Table 4.
AutoDock-Estimated Free Energies of Binding (ΔG°binding) for the Lowest-Energy Docked Conformation of the Most Populated Clusters
| ΔG°binding (kcal/mol) | Na | |
|---|---|---|
| wild-type mAhR | −5.98 | 100 |
| H285A | −4.76 | 100 |
| F345A | −5.23 | 100 |
| A375 V | −3.69 | 98 |
| A375L | −3.95 | 31 |
| 0.63 | 63 |
N is the number of docked conformations in each cluster.
Figure 7.
Cartoon representation of the modeled mAhR LBD with (a) the lowest-energy docked conformation of TCDD (shown as blue sticks) and the consensus volume in the mammalian AhR cavities (see the text) (colored cyan). (b) Same as panel a, with the mutated residue and the lowest-energy docked conformation of TCDD in the mAhR mutants: H285A, yellow sticks; F345A, green sticks; A375V, orange sticks. (c) Same as panel a, with the mutated residue and the lowest-energy docked conformations of TCDD in the A375L mAhR mutant (red sticks).
It is known that the state-of-the-art docking methods do not lead to good estimates of the experimental binding free energies due to the employment of empirical scoring functions and the approximation of contributions like desolvation and entropy changes. Indeed, our calculations underestimate the experimental ΔG°binding by ~6 kcal/mol, leading to a TCDD dissociation constant (at 298 K) in the micromolar range instead of nanomolar range observed for wt AhRs (26). However, docking methods are able to correctly determine the relative order of magnitude in the ΔG°binding value for a series of docking experiments on similar systems. On this basis, some of the AhR mutants that exhibited a complete loss of both [3H]TCDD binding and TCDD-dependent DNA binding, H285A, F345A, and A375L (see Table 3), were subjected to molecular docking analysis, and the results were compared to those of the wild-type mAhR. The naturally occurring A375V mutant was included in the analysis for comparative purposes. Even though other mutants, such as F289A, Y316A, I319A, and I319Y, showed complete loss of TCDD and DRE binding activity (Table 3), they were not included in the analysis because of their location in flexible and exposed regions of the LBD. In cases such as these, more sophisticated docking approaches that take into account protein flexibility are required.
One or more clusters of docked conformations were obtained by AutoDock for the four selected mutant AhRs. The population of each cluster, along with the estimated free energy of binding for the lowest-energy docked conformation, is presented in Table 4, and the corresponding binding modes are shown in panels b and c of Figure 7.
In both of the binding modes obtained by docking of TCDD to A375L, the ligand is outside the modeled cavity, far from the TCDD binding conformation obtained for the wild-type mAhR, and shows unfavorable binding free energies. For the A375V mutant, a binding mode closer to the one in the wild-type AhR but with a difference of ~2 kcal/mol in ΔG°binding (3 orders of magnitude in the estimated dissociation constant) was predicted. Docking in the F345A and H285A mutants led to binding modes similar to that in the wild-type mAhR. However, it must be noted that their binding free energies differ from that of the wt mAhR by ~1 kcal/mol, and this corresponds to a difference of ~1 order of magnitude in the dissociation constants. Therefore, TCDD–receptor complexes formed with the A375V, F345A, and H285A mutants were remarkably less stable than that of the wt mAhR.
DISCUSSION
In a previous study (20), we reported that the mAhR PAS domain is characterized by an internal cavity with a volume falling in the range observed for binding cavities observed in proteins. It was also noted that the modeled conformation of the LBD that was developed could be viewed as being more appropriate for describing the ligand-bound form of mAhR, where the buried cavity can be made available for binding as a result of conformational changes in the flexible regions of the domain due to association of the unliganded AhR with hsp90. This information guided the first set of structure-driven experimental analyses that aimed to validate the proposed model (20).
The seven mammalian AhRs here presented are all characterized by high binding affinity values for the TCDD (Kd<5 nM) (26–28). Therefore, modeling and analysis of the three-dimensional features of their binding domains are useful in improving our understanding of the structural and chemical requirements for TCDD binding to the AhR and, upon validation by mutagenesis and functional analysis, for detecting the TCDD binding-fingerprint of critical residues in the AhR ligand binding pocket.
The modeled mammalian AhRs have a high degree of sequence identity and similarity, and as such, both the global fold and the secondary structure elements in the obtained models are very similar (Figures 1 and 2). They all maintain the well-conserved characteristics of the known PAS domain structures, with some minor differences in the arrangement of the Gβ strand and in the secondary structure attribution of residues located in the flexible region between the Dα and Fα helices. Therefore, modeling of this group of mammalian AhRs allowed us to confirm the structural characteristics of the mAhR LBD as the ones associated with high TCDD binding affinity. Moreover, on the basis of the modeled cavities of all these AhRs, it was possible to derive a common internal space, named the consensus cavity, that represents the steric characteristics of the internal cavity that are needed for optimal binding. More interestingly, the residues contributing to the internal cavity surfaces are remarkably conserved in AhRs of all species examined (Figure 3a). This confirms that the physicochemical characteristics of the ligand binding cavities are the same in mammalian AhRs with high TCDD affinity. Analysis of the conserved residues within the binding cavities (see Results and Figure 3b) allowed us to further define the molecular determinants of TCDD binding by focusing our mutagenesis experiments on a subgroup of 13 residues (Figure 3c). This choice was supported by previous results that identified a key role of four of these residues in TCDD binding to the mAhR (20).
Therefore, site-directed mutagenesis experiments on the selected set of residues (Table 3) were designed for the mAhR to shed light on three main aspects: the side chains’ steric characteristics that are consistent with the internal space needed for TCDD binding, the involvement of aromatic interactions in the molecular association with the TCDD, and the role of the polar residues that are present in the mainly hydrophobic core of the cavity. Accordingly, more than one type of amino acid substitution was tested in some of the positions. The effects of point mutations observed on both [3H]TCDD binding and TCDD-dependent DNA binding (see Table 3) of the mAhR provide insights in all of these aspects.
With regard to the role of side chain steric characteristics, the adverse effects observed when TCDD binds to mutants with increasing steric hindrance in the central 375 position (A375V and A375L) (20) suggest that the internal space available in the high-affinity AhRs is necessary for accommodating the TCDD molecule within the LBD cavity. This was confirmed by the comparison of the TCDD binding mode in the wild-type mAhR model to those in the A375L and A375V mutants, obtained by docking calculations. In fact, in the docking simulation on the wild-type mAhR, the TCDD steric requirements appear to perfectly fit both the volume and the shape of the consensus cavity (Figure 7a). In contrast, the binding cavity of the A375L mutant appeared to make it inaccessible to TCDD (Figure 7c). In fact, the single mutation in that central zone of the cavity not only reduces its internal volume but also causes a break in the accessible space for TCDD. The cavity of the A375V mutant AhR, despite being available for ligand binding (Figure 7b), showed a drastic reduction in the estimated binding free energy for TCDD (Table 4).
Similar observations emerged from a recent study (25) which demonstrated that the A361V and A361L mutants essentially eliminated TCDD binding and TCDD-induced DRE binding in mAhR. This residue lies on the strand adjacent to A375 and is conserved in all seven mammalian AhRs examined here.
More in-depth insights into steric effects were provided by two additional mutants: P291F and I319Y. These were designed after we had observed the presence of phenylalanine and tyrosine residue in the corresponding positions of HIF-2α. Analysis of the NMR structureof this domain (21) indicated the presence of only small internal cavities, with a total volume of <100 Å3, and the side chains mentioned above contribute greatly to a reduction in the size of the internal space. Interestingly, very recent protein crystallization studies reported the occurrence of a larger internal cavity in the HIF-2α PAS domain as well as the ability of a small molecule to bind within this cavity (55). As Scheuermann et al. stated, the significance of this observation remains to be determined as there is no current evidence that HIF-2α actually requires a cofactor or involves a ligand-dependent mechanism of activation in vivo, and these studies used a non-natural small molecule ligand during HIF-2α protein crystallization. However, this observation suggests that most PAS domains have a putative ligand binding function; specific side chain arrangements and different degrees of structural flexibility decide if this is also exploited in the cellular environment. The CASTp analysis of the mAhR models generated by the in silico mutagenesis of these two residues (P291 and I319) revealed a reduction in the cavity volume from 470 Å3 for the wild-type AhR to ~350 Å3 for the P291F mutant and to ~290 Å3 for the I319Y mutant. The P291F mutation reduced AhR ligand binding by 75% and decreased the ligand-dependent DNA binding by 50%; the I319Y substitution completely eliminated both TCDD and DNA binding (Table 3). Also the double mutant P291F/I319Y completely abolished ligand and DNA binding (data not shown). These findings confirm our hypothesis about the need for all of the available space within the modeled cavity for the accession and binding of ligand (TCDD). On the other hand, results with the I319A mutation suggest that the hydrophobic stabilization of the long isoleucine side chain in this position is essential for TCDD binding. This is in agreement with the results of other investigators who reported that alanine substitution for isoleucine at amino acid 319 resulted in a dramatic reduction in the magnitude of induction of luciferase activity by the mAhR (24). Moreover, mutagenesis experiments in the corresponding position of the tern AhR (i.e., amino acid 325) indicated that a single V325I substitution increased the TCDD binding and transactivation abilities of this AhR to those of the more responsive chicken AhR, which like the high-affinity mAhR, has an isoleucine in this position (23).
The role of aromatic interactions in TCDD binding stabilization was confirmed by a number of mutations, with single amino acid substitutions causing a complete (F289A, Y316A, and F345A) or partial loss (F318A and F345L) of [3H]TCDD and [32P]DRE DNA binding activity (Table 3). TCDD docking calculations on the F345A mutant (Figure 7b and Table 4) showed a reduction in the estimated binding free energy with respect to that of the wild-type mAhR, the difference of ~1 kcal/mol being consistent with the destabilization due to the loss of intermolecular interactions between aromatic rings. The results with the F289Y mutation indicated that a hydrophobic aromatic functionality is specifically required at that position; indeed, substitution with the polar aromatic tyrosine essentially abolished TCDD binding. In contrast, the Y316F and F318Y mutants, which slightly decreased (Y316F) and substantially maintained (F318Y) the binding properties of the wild-type AhR, respectively, highlight the requirement of a general aromatic residue at those positions. Similarly, mutagenesis of Y320 in the rat AhR (corresponding to Y316 in the mouse AhR) to phenylalanine maintained the binding activity of the AhR (56). In addition, loss of induction of AhR-dependent luciferase reporter gene activity was observed with AhRs containing F318A and F318L mutations as well as a reduction in the overall induction by AhRs containing F318Y and F318W substitutions (24). The remaining aromatic residue belonging to the list of residues in the cavity surface is F281, which was excluded from our analysis because it is not conserved in high-affinity zebrafish AhR2 (where it is a leucine) (54). Previous mutagenesis confirmed that this position was not crucial for TCDD binding to the AhR, because its substitution with tryptophan did not affect TCDD specific binding or ligand-dependent DNA binding (25).
A specific point should be made on the role of histidines in AhR ligand binding. The complete elimination of TCDD binding caused by the H285A substitution suggested the possible role of this residue in stabilizing the TCDD association by aromatic interaction. This hypothesis was supported by molecular docking results on the H285A mutant, which showed a reduction of more than 1 kcal/mol in binding free energy, corresponding to an ~1 order of magnitude change in the dissociation constant, with respect to that of the wild-type mAhR. Additionally, we previously hypothesized that the polarity of the imidazole ring of this residue was involved in electrostatic stabilization of the AhR–TCDD complex (20) because of the electronic polarization of the ligand induced by the four lateral withdrawing chlorine atoms (50, 57). This hypothesis was confirmed by our additional substitution at this position. The H285F substitution abolished TCDD binding and considerably weakened TCDD-dependent DNA binding, thus supporting the specific role of the polar aromatic histidine at this position. Interestingly, a similar conclusion can be drawn from the mutagenesis of the other histidine (H320) that lies at the boundary of the cavity on the opposite side with respect to H285 (Figure 3c). Substitution of histidine at amino acid 320 with phenylalanine resulted in an ~70% reduction in [3H]TCDD specific binding and a 60% reduction in the DRE binding (Table 3). These results are consistent with a reduction in the induction of AhR-dependent luciferase activity by TCDD observed in cells transfected with an AhR containing the H320A substitution (24).
Finally, our previous results based on the substitutions of other polar residues C327, M334, and Q377, showed a reduction in TCDD binding to all the mutants obtained by alanine substitution and a complete loss of binding activity with an M334E mutation (20). These findings lead us to hypothesize that additional stabilization of the AhR–TCDD interaction could result from a network of weak interactions, like the sulfur–arene interactions with the methionine and cysteine side chains and the N–H/π hydrogen bonds with glutamine observed in other proteins (58), and that the introduction of the negatively charged Glu residue in this environment would be expected to strongly perturb the existing intermolecular interaction.
More dramatic effects on TCDD and DNA binding were observed for the T283M and T283E mutations. Substitution of this threonine, polarity and availability of which as a hydrogen bond donor may further stabilize binding to TCDD, with both a methionine and the charged residue glutamic acid completely abolished binding. These findings point out a key role for this residue.
The global picture that emerges from the mutagenesis analysis (see Figure 3c as a reference) is that: (1) the binding cavity, in its ligand-bound form, provides sufficient internal space to accommodate a molecule with the size of the TCDD but easily loses its binding activity as a consequence of point mutations that restrict the available space, (2) the surface of the cavity is characterized by a considerable aromatic character (six of the 13 internal residues here considered) that is essential in stabilizing the TCDD binding, presumably by π–π interactions, and (3) the surface also includes a histidine and a threonine residue whose polarity seems to be necessary for specific binding to the TCDD and a group of additional polar residues at both sides of the cavity surface that contribute to TCDD stabilization probably through a network of weak interactions.
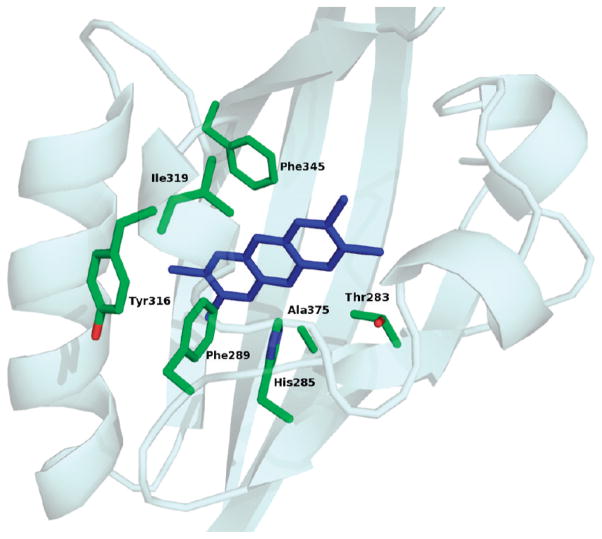
Moreover, it should be noted that data derived from functional analysis on the mAhR mutants highlighted a group of mutants that caused the complete loss of both TCDD and TCDD-dependent DNA binding. By excluding mutants with the negatively charged Glu residue that are expected to strongly perturb the intermolecular interactions within the cavity, we found the only mutants that produced such dramatic effects were T283M, H285A, F289A, Y316A, I319A and -Y, F345A, and A375L (Table 3). Therefore, on the basis of our experimental analysis, these seven residues (see Figure 8) were identified as the key determinants for high-affinity TCDD binding, and they can be regarded as the TCDD binding-fingerprint for the mammalian AhRs. The A375 and I319 residues play a critical role in molecular recognition of TCDD due to their steric characteristics, F289, Y316, and F345 as a consequence of aromatic interactions, and T283 and H285 for electrostatic stabilization.
Figure 8.
TCDD binding-fingerprint of conserved residues within the AhR ligand binding cavity necessary for optimal high-affinity TCDD binding. The modeled mAhR LBD is shown as cartoons; the selected residues are shown as sticks, colored according to atom type (green for carbon, red for oxygen, and blue for nitrogen), and the lowest-energy docked conformation of TCDD is shown as blue sticks.
In conclusion, comparative structural modeling studies with AhRs from seven species have highlighted the fact that the physicochemical characteristics of the ligand binding cavities are conserved in mammalian AhRs with high TCDD affinity. This evidence, coupled with site-directed mutagenesis experiments, has allowed determination of the TCDD binding-fingerprint of conserved residues within the AhR ligand binding cavity necessary for optimal high-affinity TCDD binding. These findings were supported by the results of the first AhR molecular docking simulations for TCDD within the AhR binding pocket. These results, when combined with our previous analysis (20), have greatly improved our understanding of the molecular determinants of TCDD binding within the PAS B ligand binding pocket and provide a basis for future studies directed toward rationalizing the observed species differences in AhR sensitivity to TCDD. The application of docking approaches for other classes of AhR ligands will also provide an avenue for mechanistic explanations into the observed structural diversity of AhR ligands.
Supplementary Material
Acknowledgments
We thank Dr. Steve Safe (Texas A&M University) for the [3H] TCDD and TCDF and Dr. Oliver Hankinson (University of California, Los Angeles) and Dr. James P. Whitlock, Jr., for mAhR and mARNT expression vectors.
Footnotes
Abbreviations: AhR, aryl hydrocarbon receptor; bHLH, basic helix–loop–helix; PAS, Per-Arnt-Sim; LBD, ligand binding domain; HAH, halogenated aromatic hydrocarbon; PAH, polycyclic aromatic hydrocarbon; TCDD, 2,3,7,8-tetrachlorodibenzo-p-dioxin; hsp90, heat shock protein of 90 kDa; XAP2, X-associated protein 2; ARNT, AhR nuclear translocator; HIF-2α, hypoxia-inducible factor 2α; DRE, dioxin responsive element; mAhR, mouse AhR; rtAhR, rat AhR; haAhR, hamster AhR; rbAhR, rabbit AhR; gpAhR, guinea pig AhR; bAhR, beluga whale AhR; sAhR, seal AhR.
This research was supported by the National Institutes of Environmental Health Sciences (Grants ES07685 and ES012498) and the California Agricultural Experiment Station.
SUPPORTING INFORMATION AVAILABLE
Expression levels of in vitro synthesized wild-type and mutant AhRs. 35S-Labeled wild-type (wt) and mutant AhRs (specific mutation indicated) were synthesized in vitro, denatured, and resolved by SDS–polyacrylamide gel electrophoresis and phosphoimager analysis of the dried gels as described in Materials and Methods (Figure 1). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Schmidt JV, Bradfield CA. Ah receptor signaling pathways. Annu Rev Cell Dev Biol. 1996;12:55–89. doi: 10.1146/annurev.cellbio.12.1.55. [DOI] [PubMed] [Google Scholar]
- 2.Denison MS, Elferink CF, Phelan D. The Ah receptor signal transduction pathway. In: Denison MS, Helferich WG, editors. Toxicant-Receptor Interactions in the Modulation of Signal Transduction and Gene Expression. Taylor and Francis; Philadelphia: 1998. pp. 3–33. [Google Scholar]
- 3.Ma Q. Induction of CYP1A1. The AhR/DRE paradigm: Transcription, receptor regulation, and expanding biological roles. Curr Drug Metab. 2001;2:149–164. doi: 10.2174/1389200013338603. [DOI] [PubMed] [Google Scholar]
- 4.Kewley RJ, Whitelaw ML, Chapman-Smith A. The mammalian basic helix-loop-helix/PAS family of transcriptional regulators. Int J Biochem Cell Biol. 2004;36:189–204. doi: 10.1016/s1357-2725(03)00211-5. [DOI] [PubMed] [Google Scholar]
- 5.Denison MS, Seidel SD, Rogers WJ, Ziccardi M, Winter GM, Heath-Pagliuso S. Natural and synthetic ligands for the Ah receptor. In: Puga A, Wallace KB, editors. Molecular Biology Approaches to Toxicology. Taylor and Francis; Philadelphia: 1998. pp. 393–410. [Google Scholar]
- 6.Safe S. Polychlorinated biphenyls (PCBs), dibenzo-p-dioxins (PCDDs), dibenzofurans (PCDFs), and related compounds: Environmental and mechanistic considerations which support the development of toxic equivalency factors (TEFs) Crit Rev Toxicol. 1990;21:51–88. doi: 10.3109/10408449009089873. [DOI] [PubMed] [Google Scholar]
- 7.Denison MS, Heath-Pagliuso S. The Ah receptor: A regulator of the biochemical and toxicological actions of structurally diverse chemicals. Bull Environ Contam Toxicol. 1998;61:557–568. doi: 10.1007/pl00002973. [DOI] [PubMed] [Google Scholar]
- 8.Denison MS, Pandini A, Nagy SR, Baldwin EP, Bonati L. Ligand binding and activation of the Ah receptor. Chem-Biol Interact. 2002;141:3–24. doi: 10.1016/s0009-2797(02)00063-7. [DOI] [PubMed] [Google Scholar]
- 9.Denison MS, Nagy SR. Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu Rev Pharmacol Toxicol. 2003;43:309–334. doi: 10.1146/annurev.pharmtox.43.100901.135828. [DOI] [PubMed] [Google Scholar]
- 10.Soshilov AA, Denison MS. Role of the Per/Arnt/Sim Domains in Ligand-dependent Transformation of the Aryl Hydrocarbon Receptor. J Biol Chem. 2008;283:32995–33005. doi: 10.1074/jbc.M802414200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hord NG, Perdew GH. Physiochemical and immunochemical analysis of aryl hydrocarbon receptor nuclear translocator: Characterization of two monoclonal antibodies to the aryl hydrocarbon receptor nuclear translocator. Mol Pharmacol. 1994;46:618–624. [PubMed] [Google Scholar]
- 12.Petrulis JR, Perdew GH. The role of chaperone proteins in the aryl hydrocarbon receptor core complex. Chem-Biol Interact. 2002;141:25–40. doi: 10.1016/s0009-2797(02)00064-9. [DOI] [PubMed] [Google Scholar]
- 13.Hankinson O. The aryl hydrocarbon receptor complex. Annu Rev Pharmacol Toxicol. 1995;35:307–340. doi: 10.1146/annurev.pa.35.040195.001515. [DOI] [PubMed] [Google Scholar]
- 14.Whitlock JP., Jr Induction of cytochrome P4501A1. Annu Rev Pharmacol Toxicol. 1999;39:103–125. doi: 10.1146/annurev.pharmtox.39.1.103. [DOI] [PubMed] [Google Scholar]
- 15.Hahn ME. Aryl hydrocarbon receptors: Diversity and evolution. Chem-Biol Interact. 2002;141:131–160. doi: 10.1016/s0009-2797(02)00070-4. [DOI] [PubMed] [Google Scholar]
- 16.Coumailleau P, Poellinger L, Gustafsson JA, Whitelaw ML. Definition of a minimal domain of the dioxin receptor that is associated with Hsp90 and maintains wild type ligand binding affinity and specificity. J Biol Chem. 1995;270:25291–25300. doi: 10.1074/jbc.270.42.25291. [DOI] [PubMed] [Google Scholar]
- 17.Perdew GH, Bradfield CA. Mapping the 90 kDa heat shock protein binding region of the Ah receptor. Biochem Mol Biol. 1996;39:589–593. doi: 10.1080/15216549600201651. [DOI] [PubMed] [Google Scholar]
- 18.McGuire J, Okamoto K, Whitelaw ML, Tanaka H, Poellinger L. Definition of a dioxin receptor mutant that is a constitutive activator of transcription: Delineation of overlapping repression and ligand binding functions within the PAS domain. J Biol Chem. 2001;276:41841–41849. doi: 10.1074/jbc.M105607200. [DOI] [PubMed] [Google Scholar]
- 19.Procopio M, Lahm A, Tramontano A, Bonati L, Pitea D. A model for recognition of polychlorinated dibenzo-p-dioxins by the aryl hydrocarbon receptor. Eur J Biochem. 2002;269:13–18. doi: 10.1046/j.0014-2956.2002.02619.x. [DOI] [PubMed] [Google Scholar]
- 20.Pandini A, Denison MS, Song Y, Soshilov AA, Bonati L. Structural and functional characterization of the aryl hydrocarbon receptor ligand binding domain by homology modeling and mutational analysis. Biochemistry. 2007;46:696–708. doi: 10.1021/bi061460t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Erbel PJ, Card PB, Karakuzu O, Bruick RK, Gardner KH. Structural basis for PAS domain heterodimerization in the basic helix–loop–helix-PAS transcription factor hypoxia-inducible factor. Proc Natl Acad Sci USA. 2003;100:15504–15509. doi: 10.1073/pnas.2533374100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Card PB, Erbel PJ, Gardner KH. Structural basis of Arnt PAS-B dimerization: Use of a common β-sheet interface for hetero- and homodimerization. J Mol Biol. 2005;353:664–678. doi: 10.1016/j.jmb.2005.08.043. [DOI] [PubMed] [Google Scholar]
- 23.Karchner SI, Franks DG, Kennedy SW, Hahn ME. The molecular basis for differential dioxin sensitivity in birds: Role of the aryl hydrocarbon receptor. Proc Natl Acad Sci USA. 2006;103:6252–6257. doi: 10.1073/pnas.0509950103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goryo K, Suzuki A, Del Carpio CA, Siizaki K, Kuriyama E, Mikami Y, Kinoshita K, Yasumoto K, Rannug A, Miyamoto A, Fujii-Kuriyama Y, Sogawa K. Identification of amino acid residues in the Ah receptor involved in ligand binding. Biochem Biophys Res Commun. 2007;354:396–402. doi: 10.1016/j.bbrc.2006.12.227. [DOI] [PubMed] [Google Scholar]
- 25.Henry EC, Gasiewicz TA. Molecular determinants of species-specific agonist and antagonist activity of a substituted flavone towards the aryl hydrocarbon receptor. Arch Biochem Biophys. 2008;472:77–88. doi: 10.1016/j.abb.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Denison MS, Wilkinson CF, Okey AB. Ah receptor for 2,3,7,8-tetrachlorodibenzo-p-dioxin: Comparative studies in mammalian and nonmammalian species. Chemosphere. 1986;15:1665–1672. [Google Scholar]
- 27.Jensen BA, Hahn ME. cDNA cloning and characterization of a high affinity aryl hydrocarbon receptor in a cetacean, the beluga, Delphinapterus leucas. Toxicol Sci. 2001;64:41–56. doi: 10.1093/toxsci/64.1.41. [DOI] [PubMed] [Google Scholar]
- 28.Kim EY, Hahn ME. cDNA cloning and characterization of an aryl hydrocarbon receptor from the harbor seal (Phoca vitulina): A biomarker of dioxin susceptibility? Aquat Toxicol. 2002;58:57–73. doi: 10.1016/s0166-445x(01)00221-1. [DOI] [PubMed] [Google Scholar]
- 29.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 30.Marti-Renom MA, Stuart A, Fiser A, Sánchez R, Melo F, Sali A. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 31.Fiser A, Do RK, Sali A. Modeling of loops in protein structures. Protein Sci. 2000;9:1753–1773. doi: 10.1110/ps.9.9.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacKerell AD, Jr, Bashford D, Bellott M, Dunbrack RL, Jr, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, III, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 33.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kelley LA, Gardner SP, Sutcliffe MJ. An automated approach for clustering an ensemble of NMR-derived protein structures into conformationally related subfamilies. Protein Eng. 1996;9:1063–1065. doi: 10.1093/protein/9.11.1063. [DOI] [PubMed] [Google Scholar]
- 36.Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 37.Holm L, Park J. DaliLite workbench for protein structure comparison. Bioinformatics. 2000;16:566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- 38.Thompson JD, Higgins DG, Gibson TJ. CLUS-TAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sippl MJ. Recognition of errors in three-dimensional structures of proteins. Proteins. 1993;17:355–362. doi: 10.1002/prot.340170404. [DOI] [PubMed] [Google Scholar]
- 40.Wiederstein M, Sippl MJ. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35:W407–W410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andersen CA, Palmer AG, Brunak S, Rost B. Continuum secondary structure captures protein flexibility. Structure. 2002;10:175–184. doi: 10.1016/s0969-2126(02)00700-1. [DOI] [PubMed] [Google Scholar]
- 42.Kabsch W, Sander C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 43.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
- 44.Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J. CASTp: Computed atas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 2006;34:W116–W118. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Connolly ML. Analytical molecular surface calculation. J Appl Crystallogr. 1983;16:548–558. [Google Scholar]
- 46.Liang J, Edelsbrunner H, Woodward C. Anatomy of protein pockets and cavities: Measurement of binding site geometry and implications for ligand design. Protein Sci. 1998;7:1884–1897. doi: 10.1002/pro.5560070905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huey R, Morris GM, Olson AJ, Goodsell DS. A semiempirical free energy force field with charge-based desolvation. J Comput Chem. 2007;28:1145–1152. doi: 10.1002/jcc.20634. [DOI] [PubMed] [Google Scholar]
- 48.Berendsen HJC, van der Spoel D, van Drunen R. GROMACS: A message-passing parallel molecular dynamics implementation. Comput Phys Commun. 1995;91:43–56. [Google Scholar]
- 49.Lindahl E, Hess B, van der Spoel D. GROMACS 3.0: A package for molecular simulation and trajectory analysis. J Mol Model. 2001;7:306–317. [Google Scholar]
- 50.Fraschini E, Bonati L, Pitea D. Molecular polarizability as a tool for understanding the binding properties of polychlorinated dibenzo-p-dioxins: Definition of a reliable computational procedure. J Phys Chem. 1996;100:10564–10569. [Google Scholar]
- 51.Fukunaga BN, Hankinson O. Identification of a novel domain in the aryl hydrocarbon receptor required for DNA binding. J Biol Chem. 1996;271:3743–3749. doi: 10.1074/jbc.271.7.3743. [DOI] [PubMed] [Google Scholar]
- 52.Rushing SR, Denison MS. The Silencing Mediator of Retinoic Acid and Thyroid Hormone Receptors (SMRT) Can Interact with the Ah Receptor but Fails to Repress Ah receptor-dependent Gene Expression. Arch Biochem Biophys. 2002;403:189–201. doi: 10.1016/s0003-9861(02)00233-3. [DOI] [PubMed] [Google Scholar]
- 53.Denison MS, Rogers JM, Rushing SR, Jones CL, Tetangco SC, Heath-Pagliuso S. Analysis of the Ah Receptor Signal Transduction Pathway. In: Maines M, Costa LG, Reed DJ, Sassa S, Sipes IG, editors. Current Protocols in Toxicology. John Wiley and Sons; New York: 2002. pp. 4.8.1–4.8.45. [DOI] [PubMed] [Google Scholar]
- 54.Hahn ME, Karchner SI, Evans BR, Frank DG, Merson RR, Lapseritis JM. Unexpected diversity of aryl hydrocarbon receptors in non-mammalian vertebrates: Insights from comparative genomics. J Exp Zool, Part A. 2006;305:693–706. doi: 10.1002/jez.a.323. [DOI] [PubMed] [Google Scholar]
- 55.Scheuermann TH, Tomchick DR, Machius M, Guo Y, Bruick RK, Gardner KH. Artificial ligand binding within the HIF2α transcription factor. Proc Natl Acad Sci USA. 2009;106:450–455. doi: 10.1073/pnas.0808092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Backlund M, Ingelman-Sundberg M. Different Structural Requirements of the Ligand Binding Domain of the Aryl Hydrocarbon Receptor for High- and Low-Affinity Ligand Binding and Receptor Activation. Mol Pharmacol. 2004;65:416–425. doi: 10.1124/mol.65.2.416. [DOI] [PubMed] [Google Scholar]
- 57.Bonati L, Fraschini E, Lasagni M, Palma Modoni E, Pitea D. A hypothesis on the mechanism of PCDD biological activity based on molecular electrostatic potential modeling. Part 2. THEOCHEM. 1995;340:83–95. [Google Scholar]
- 58.Meyer EA, Castellano RK, Diederich F. Interactions with aromatic rings in chemical and biological recognition. Angew Chem, Int Ed. 2003;42:1210–1250. doi: 10.1002/anie.200390319. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.