Summary
The proteasome is an abundant protease that is critically important for numerous cellular pathways. Proteasomes are activated in vitro by three known classes of proteins/complexes, including Blm10/PA200. Here we report a 3.4Å resolution crystal structure of a proteasome-Blm10 complex, which reveals that Blm10 surrounds the proteasome entry pore in the 1.2 MDa complex to form a largely closed dome that is expected to restrict access of potential substrates. This architecture, and the observation that Blm10 induces a disordered proteasome gate structure, challenges the assumption that Blm10 functions as an activator of proteolysis in vivo. The Blm10 C-terminus binds in the same manner as seen for 11S activators and inferred for 19S/PAN activators, and indicates a unified model for gate opening. We also demonstrate that Blm10 acts to maintain mitochondrial function. Consistent with the structural data, the C-terminal residues of Blm10 are needed for this activity.
Introduction
The bulk of proteolysis in the cytosol and nucleus of eukaryotes is performed by an ~700 kDa barrel-shaped protease called the proteasome (20S proteasome/core particle/CP), whose activity is important for protein quality control and the regulation of many biological pathways (Glickman and Ciechanover, 2002). Proteasomes comprise 28 protein subunits assembled into four heptameric rings, with outer rings composing α-subunits and inner rings β-subunits, to form a hollow complex that sequesters the proteolytic sites at the N-termini of β-subunits (Groll et al., 1997; Lowe et al., 1995; Seemuller et al., 1995). The seven distinct α-subunits (α1–7) and seven distinct β-subunits (β1–7) of eukaryotic proteasomes each occupy a unique position in their respective rings (Groll et al., 1997; Unno et al., 2002). Substrates enter the proteasome through a pore at the center of the α-subunit ring that is closed in the absence of an activator by interactions among the N-terminal peptides of α-subunits, with α2, α3, and α4 making the major contributions to closing the gate.
Three classes of activator facilitate substrate access to the proteasome interior by binding α-subunits. The 11S activators, PA28/REG/PA26, are heptameric rings that, as revealed by proteasome-PA26 crystal structures (Forster et al., 2005; Forster et al., 2003; Whitby et al., 2000), stimulate the hydrolysis of peptides by stabilizing an ordered open conformation of the entrance pore. The C-termini of 11S activators bind in pockets between proteasome α-subunits through main-chain to main-chain hydrogen bonds and a salt bridge between the C-terminal carboxylate and proteasome Lys66 (T.acidophilum proteasome numbering is used throughout). To open the gate, 11S activators utilize an internal “activation loop” (Zhang et al., 1998), which repositions the Pro17 reverse turns of proteasome α-subunits to destabilize the closed conformation and allow formation of a fully open conformation. Biochemical studies indicate that the unrelated PAN/19S activator induces the same open conformation as PA26 (Forster et al., 2003) and utilizes a similar mode of binding (Forster et al., 2005), although PAN/19S appears to lack an activation loop and achieves both binding and gate opening through interactions of C-terminal residues (Gillette et al., 2008; Rabl et al., 2008; Smith et al., 2007).
Unlike the oligomeric 11S and PAN/19S activators, which use multiple C-termini to bind in pockets between α-subunits, Blm10 (Fehlker et al., 2003; Iwanczyk et al., 2006; Schmidt et al., 2005), previously known as Blm3 (Doherty et al., 2004), and its mammalian homolog PA200 (Ustrell et al., 2002), are single chain proteins of 2143 residues (~250 kDa, S. cerevisiae sequence). Blm10 and PA200 are predominantly nuclear and stimulate the degradation of model peptides, although they do not appear to stimulate the degradation of proteins, recognize ubiquitin, or utilize ATP. The mouse PA200 knockout displays a defect in spermatogenesis (Khor et al., 2006) and roles in DNA repair and genomic stability have been proposed (Blickwedehl et al., 2008; Blickwedehl et al., 2007; Ustrell et al., 2002). Studies in yeast have produced inconsistent data that suggest roles in proteasome assembly/maturation (Fehlker et al., 2003; Marques et al., 2007) and proteasome inhibition (Lehmann et al., 2008), while early indications of bleomycin sensitivity were not supported by later studies, which found no role for Blm10 in the repair of DNA damage induced by bleomycin or other factors (Iwanczyk et al., 2006).
In order to better understand Blm10 mechanism, we have determined the crystal structure of a proteasome complex. The results challenge the model that Blm10 is a proteasome activator in vivo and also indicate that binding by 11S, 19S/PAN, and PA200/Blm10 is more similar than previously realized. We further report that yeast cells lacking Blm10 fail to maintain normal levels of mitochondrial function, and that this phenotype also results when just the C-terminal residues that make contacts between Blm10 and the proteasome are deleted.
Results and Discussion
Blm10-proteasome structure determination
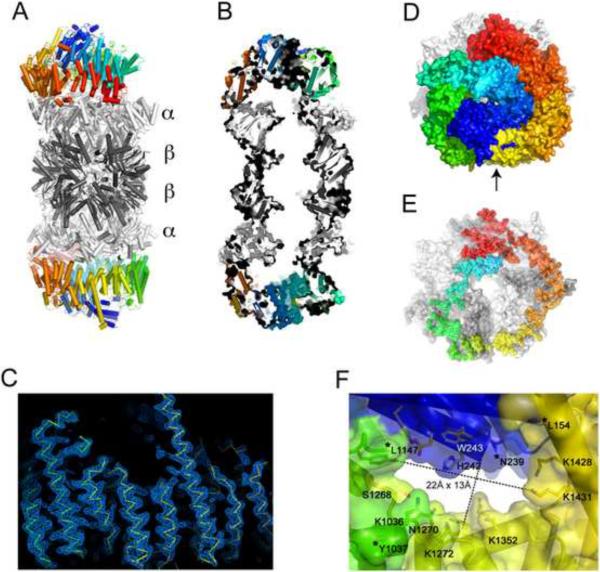
We determined a 3.4Å crystal structure of the S. cerevisiae proteasome capped on both ends by Blm10 (Figure 1, Figure S1) to an Rfree of 25% (Table 1). A variety of Blm10 constructs were screened and several crystal forms were obtained, with the best data collected from a construct that lacked the first 50 residues of Blm10, which are poorly conserved and predicted to be disordered. The ordered regions of Blm10 seen in the structure are residues 79–154, 239–1034, and 1147–2143 (C-terminus), consistent with proteolytic cleavage at residues ~154–239 and ~1043–1147 observed by SDS-PAGE and N-terminal sequencing upon storage at 4°C (Iwanczyk et al., 2006) and in crystals (data not shown). A number of observations argue that the crystal structure is not unduly influenced by lattice contacts (Figure S1A–D), including the very large Blm10-proteasome interface that includes all seven α-subunits and buries more than 10,000 Å2 of solvent accessible surface area (Figure 1E).
Figure 1. Structure of the Blm10:proteasome complex.
(A) Cartoon of the proteasome-Blm10 complex, side view. Proteasome white (α subunits) and gray (β subunits), Blm10 rainbow from N-terminus (blue) to C-terminus (red).
(B) Cutaway view with the molecular surface.
(C) Electron density for Blm10. All maps shown in this paper were phased on the proteasome molecular replacement model for which segments approaching Blm10 had been removed. The phases were refined by solvent flattening, histogram shifting, and four-fold non-crystallographic symmetry averaging.
(D) Top view, space-filling representation. The opening visible in the center of this view measures only ~6Å between atom centers. The largest opening, which is not visible in this orientation, is indicated with an arrow.
(E) Same as panel D, but only showing Blm10 residues that have at least one atom within 6 Å of the proteasome.
(F) Close-up view of the largest opening through the Blm10 dome. Orientation as indicated by the arrow in panel D. Asterisks denote the last ordered residues adjacent to disordered segments that have been omitted from the model.
Table 1.
Proteasome:Blm10 crystallographic data statistics.
| Crystallographic Data | |||||||
|---|---|---|---|---|---|---|---|
| ID/namea | c158/FL-1 | c164/FL-thim | c172/FL-PtCl4 | c280/NΔ50-1 | c290/NΔ50-MeHg | c292/NΔ50-PtCl4 | |
| Spacegroup | P212121 | P21 | P212121 | P21 | P21 | P21 | |
| Cell | a (Å) | 124.1 | 238.2 | 128 | 236.1 | 238.7 | 237.8 |
| b (Å) | 238.5 | 126.2 | 236.2 | 127.8 | 127.8 | 128.6 | |
| c (Å) | 489.8 | 528.7 | 515.1 | 532.3 | 535.6 | 537.1 | |
| β (°) | 102.5 | 102.8 | 102.5 | 102.8 | |||
| Resolution (Å)b | 50–3.9 (4.0–3.9) | 40–6.2 (6.4–6.2) | 50–6.0 (6.2–6.0) | 30–3.4 (3.5–3.4) | 30–6.8 (7.0–6.8) | 30–6.8 (7.0–6.8) | |
| Rmerge (%) | 20.9 (42.8) | 11.0 (43.6) | 18.2 (58.3) | 10.3 (31.4) | 9.6 (36.5) | 9.4 (35.9) | |
| I/σ(I) | 6.3 (2.2) | 8.6 (2.1) | 9.3 (2.4) | 10.3 (2.4) | 9.7 (2.0) | 9.4 (2.1) | |
| Completeness | 98.2 (97.3) | 99.6 (99.9) | 99.2 (97.7) | 98.6 (88.6) | 99.8 (99.6) | 99.8 (99.9) | |
| Refinement Statistics | |||||||
|---|---|---|---|---|---|---|---|
| Resolution (Å)b | 30–3.0 | ||||||
| # reflns used in refinementc | 490,961 | ||||||
| Rwork/Rfree (%) | 19.6/25.0 | ||||||
| Number of atomsc | 158,904 | ||||||
| <B> (Å2) | 104.3 | ||||||
| RMSD bond lengths (Å)d | 0.01 | ||||||
| RMSD bond Angles (°) | 1.291 | ||||||
Values in parentheses refer to the high-resolution shell.
Each data set was collected from a single crystal, which was given a unique identifier and a more descriptive name. Crystals were of full-length (FL) Blm10 or Blm10 missing the first 50 amino acid residues (Δ50). Mercury (Hg) or Platinum (Pt) heavy atom derivatives were prepared as described in Supplementary Experimental Procedures.
Resolution of a data set was formally defined as the Bragg spacing at which half of the measured reflections have an I/σ(I) value of at least 2.0, although the data were processed and used to smaller Bragg spacing. All crystallographic data values in this table refer to reflections within the formal resolution limits, whereas the refinement statistics refer to all data.
The total number of all 20S and Blm10 non-Hydrogen atoms in the asymmetric unit. No solvent molecules were included in the model.
RMSD denotes root mean square deviation from ideality.
Overall structure description
Blm10 encodes 32 HEAT repeat (HR)-like modules (Kajava et al., 2004), each comprising two helices joined by a turn, with adjacent repeats connected by a linker (Figure 1, Figure S1E). The first ordered Blm10 residue, Thr79, lies ~60Å above the proteasome surface and is followed by three short helices and loops before starting HR1 at His133. The following HEAT repeats continue almost to the C-terminus and spiral through a 1½ turn left-handed solenoid to form a dome that encloses a volume of ~110,000 Å3 above the proteasome. Whereas a standard HEAT repeat is composed of ~50 residues, the Blm10 HEAT repeats are highly variable. The length of helices ranges from 8 to 35 residues, turns range from 2 to 87 residues, and linkers range from 1 to 88 residues, with the longest linker, between HR21 and HR22, containing additional secondary structures (two strands and three helices).
Restricted opening through the Blm10 dome
The extensive Blm10 interface surrounds the proteasome entrance pore (Figure 1E). Consistent with the observation that Blm10/PA200 stimulates the hydrolysis of small peptides but not proteins, the largest opening through the Blm10 dome is only 13Å by 22Å when measured between atomic nuclei (Figure 1F). Moreover, access may be further restricted because segments of Blm10 that are not visible in the structure connect residues Leu154 to Asn239 and Tyr1037 to Leu1147, which are all adjacent to the mouth of the opening. A biological rationale for Blm10/PA200 to facilitate peptide hydrolysis in vivo is not obvious, and the structure is consistent with suggestions that Blm10 functions in proteasome assembly (Fehlker et al., 2003; Marques et al., 2007), as an adaptor (Rechsteiner and Hill, 2005), or as a physiological inhibitor (Lehmann et al., 2008). We cannot discount the possibilities that unfolded proteins might access the proteasome through this pore, perhaps with the assistance of an as yet unidentified ATPase, or that substrate proteins might be bound within the Blm10 dome prior to proteasome association.
The proteasome gate is disordered
The proteasome β-subunits do not move discernibly upon binding Blm10 (RMSD=0.4Å on all Cα atoms), while the α-subunits move somewhat toward the open conformation seen in complexes with PA26 to form a pore that is disordered rather than fully open or fully closed (Figure 2). This flexible conformation is expected to allow passage of small model substrates but to impede access of larger substrates (Benaroudj et al., 2003; Forster et al., 2003). Both the dome architecture and the proteasome pore conformation are therefore consistent with biochemical studies indicating that Blm10 and PA200 stimulate the hydrolysis of peptides but not proteins (Iwanczyk et al., 2006; Schmidt et al., 2005; Ustrell et al., 2002).
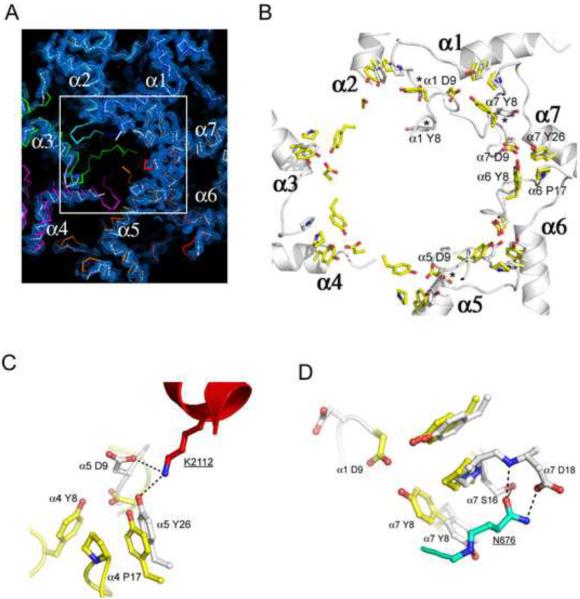
Figure 2. Proteasome conformational changes induced by Blm10.
(A) Top view of the proteasome pore region with electron density for the Blm10 complex. The absence of density for the N-terminal residues of proteasome α2, α3, and α4 indicates that they are disordered in the Blm10 complex (white), whereas they are ordered in the closed, unliganded conformation (colors) and in the fully open complex with PA26 (not shown in this panel).
(B) Open conformation seen in complexes with PA26 (yellow) and Blm10 (white). The stabilizing cluster residues (Tyr8, Asp9, Pro17, Tyr26; (Forster et al., 2003)) are labeled for the α6/α7 cluster, which is ordered in the unliganded proteasome (Groll et al., 1997) and in both the PA26 and Blm10 complexes shown here. Tyr8 and Asp9 residues are not ordered for α2, α3, or α4 in the Blm10 complex. Residues indicated with an asterisk are ordered in the Blm10 complex but are displaced from the open conformation seen with PA26. A version of this panel that also includes the closed conformation is shown in Figure S2.
(C) Contacts that stabilize α5Asp9 away from the open conformation.
(D) Contacts that stabilize α7Tyr8 away from the open conformation.
It is instructive to compare the proteasome complexes with Blm10 and PA26. The fully open conformation results from repositioning of the seven proteasome α-subunit Pro17 turns by the PA26 activation loop to form a wider, more circular arrangement, with the largest Pro17 Cα movement (3.6 Å) seen for α4 and the smallest Pro17 Cα movement (0.4 Å) seen for α1 (Forster et al., 2003). Repositioning of the Pro17 turns induces ordering of the Tyr8 and Asp9 residues of all seven proteasome α-subunits to form a continuous belt around the pore circumference that is stabilized by conserved clusters of Tyr8, Asp9, Pro17, and Tyr26 proteasome residues (Figure 2B) (notwithstanding the non-canonical α1/α2 cluster, (Forster et al., 2003)). Blm10 stabilizes the same Pro17 transition for proteasome α5 as seen with PA26, although it does so primarily by interactions of its C-terminal residues rather than by an internal activation loop. In contrast, the Pro17 turns of α2, α3, and α4 lack direct contacts with Blm10 and become disordered. Moreover, Blm10 blocks the fully open conformation by displacing α5Asp9 from a position where it could bind α4Tyr8 (Figure 2C) and by displacing α7Tyr8 from a position where it could bind α1Asp9 (Figure 2D). This explains why α1 and α4, and hence their contacting α2 and α3 subunits, do not form the same open conformation as seen with PA26.
Implications for binding and gate opening by 19S/PAN
The C-terminal residues of Blm10 bind in the pocket between proteasome α5 and α6 in a conformation that superimposes with the C-terminal residues of PA26 (Figure 3). PA26 is heptameric, and binds to all seven pockets of the seven-fold symmetric archaeal T. acidophilum proteasome and to four (α2/α3, α3/α4, α4/α5, α5/α6) pockets of the S. cerevisiae proteasome (Forster et al., 2005). Like PA26, the Blm10 C-terminal residues form β-sheet-like hydrogen bonds with the proteasome and the Blm10 C-terminal carboxylate forms a salt bridge with α6Lys66. Interestingly, biochemical (Forster et al., 2005; Smith et al., 2007) and electron microscopic (Rabl et al., 2008) data indicate that the C-termini of some of the 19S/RC activator ATPases and their archaeal homolog PAN also bind to the same site, presumably using the same interactions.
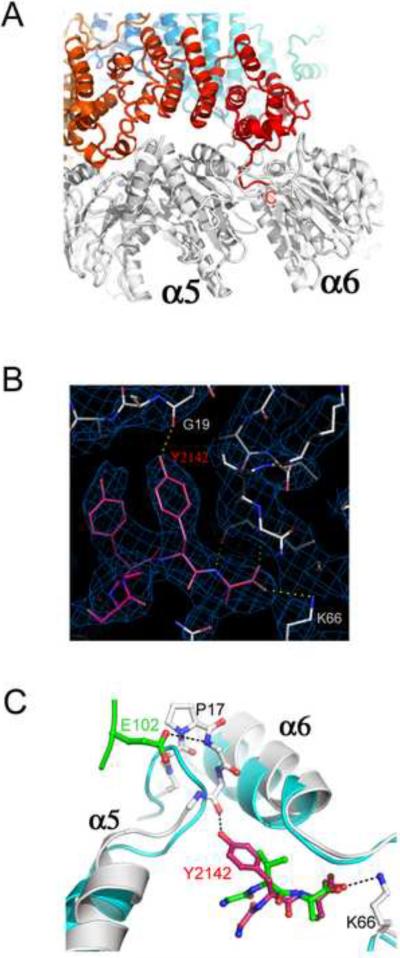
Figure 3. Interactions of the Blm10 C-terminal residues.
(A) Side view with Blm10 C-terminus labeled “C”.
(B) The electron density map is well defined for the Blm10 penultimate tyrosine (Tyr2142) and surrounding residues.
(C) The last three residues of PA26 (green) and Blm10 (red) are shown after overlap of the two complexes on surrounding proteasome residues. Unliganded proteasome (Groll et al., 1997), cyan. Blm10 Tyr2142 stabilizes the open position of α5 by hydrogen bonding with Gly19 O. PA26 stabilizes the same transition by hydrogen bonding interactions of its activation loop residue Glu102.
In contrast to the apparently shared mode of binding by 11S, Blm10/PA200, and 19S/PAN, an important difference is that peptides corresponding to the seven C-terminal residues of PAN and some 19S subunits are able to both bind proteasomes and stabilize the open gate conformation (Gillette et al., 2008; Rabl et al., 2008; Smith et al., 2007), whereas PA26/11S use their C-terminal sequences for binding but rely upon a distantly located activation loop to induce gate opening. Furthermore, a critical interaction for proteasome gate opening has been mapped to the penultimate tyrosine of PAN/19S ATPases, with some of the homologues containing a phenylalanine at this position (Gillette et al., 2008; Smith et al., 2007). Remarkably, the penultimate residue of Blm10, Tyr2142, is also invariably conserved as tyrosine or phenylalanine in an alignment of 46 sequences (Figure S1E). This residue possesses well-defined electron density in the Blm10:proteasome structure and the side chain reaches from the C-terminal binding site to hydrogen bond with the oxygen atom of α5Gly19 and stabilize the adjacent α5Pro17 reverse turn in the same open gate conformation as seen in proteasome complexes with PA26 (Figure 3C). Presumably, the penultimate tyrosine of PAN/19S subunits makes the same interactions as seen for the penultimate tyrosine of Blm10, with the monomeric Blm10 moving a single proteasome α-subunit Pro17 turn and disordering the gate, and the oligomeric PAN/19S moving multiple α-subunits to induce a fully open gate conformation. This model calls into question the proposal that PAN/19S induces gate opening without making direct contacts to the Pro17 turn (Rabl et al., 2008), and is consistent with a recent report that the penultimate tyrosine or phenylalanine of chimeric PA26 complexes designed to model the PAN/19S-proteasome interaction display equivalent contacts (Stadtmueller et al., 2009). Thus, while important questions remain, we favor the model that Blm10/PA200, 11S, and 19S/PAN all bind through their C-terminal residues and partially or fully open the proteasome gate by displacing one or multiple Pro17 turns, with Blm10 and PAN/19S using a penultimate tyrosine/phenylalanine to move the Pro17 turn and PA26/11S using an internal activation loop.
Blm10 is important for maintenance of mitochondrial function
Several genetic links have been reported between Blm10 and proteasome function, including synthetic growth defects in cells lacking both Blm10 and Ecm29 or Rpn4 (Schmidt et al., 2005), or lacking both Blm10 and the C-terminus of Pre4/β7 (Marques et al., 2007). We have been unable to confirm the interactions with Ecm29 (Iwanczyk et al., 2006) or Pre4, although we have observed genetic interactions between blm10 and rpn4 mutations (Figure S3A), supporting a role for Blm10 in a proteasome-related process.
We now report that deletion of Blm10 causes yeast cells to lose mitochondrial function at a high frequency (Figure 4). In the A364a genetic background, a strain with a deletion of the entire BLM10 gene yields about 9-fold more petite derivatives than a strain with normal Blm10. These cells are unable to grow on glycerol media because they lack the functional mitochondria required to metabolize this non-fermentable carbon source. This requirement is not specific to A364a cells, because the yield of petites is significantly elevated for blm10-Δ mutants in three other commonly used backgrounds (Figure 4B).
Figure 4. The C-terminal residues of Blm10 are important for its function.
(A) Isogenic strains were constructed in the A364a genetic background with the genotypes indicated (Table S1). Multiple independent colonies of each strain growing on glycerol medium to select for retention of mitochondrial function were used to inoculate rich medium containing glucose. Saturated cultures were diluted and plated on rich glucose medium, then mitochondrial function in clones was assessed using the tetrazolium staining method (Ogur et al., 1957). Results from multiple strains with the same genotype were combined (Table S1, total number indicated as N), with the average percentage yield of petite colonies plotted here. Error bars indicate the standard deviation of the measurements.
(B) As in panel A, except isogenic pairs from three other commonly used genetic backgrounds containing or lacking Blm10 were assayed by picking 120 colonies without regard to size, then replica plating to media containing glycerol or glucose to determine the number of petite colonies (Table S1). See also Figures S3.
Other mutations that cause defects in proteasome function have also been reported to cause increased levels of petite formation, although the mechanisms linking proteasomes to this phenotype remain under investigation and may be diverse. For example, loss of the 20S assembly chaperone Ump1 causes elevated levels of petite formation (Malc et al., 2009). This has been attributed to mitochondrial DNA damage due to increased production of reactive oxygen species (ROS) coupled with diminished levels of DNA repair (Malc et al., 2009). We therefore tested blm10-Δ deletion strains for increased ROS levels using a dihydrofluorescein diacetate assay (Malc et al., 2009) but found only a small, statistically insignificant increase (Figure S3B). Also unlike Ump1, deletion of Blm10 causes only a small increase in the yield of erythromycin resistant mutants (Figure S3B), which arise through mutation of a mitochondrial rDNA gene. Furthermore, deletion of Blm10 causes a small increase in the mRNA encoding the DNA repair protein Msh1 (David Stillman and Yaxin Yu, personal communication), whereas deletion of Ump1 is reported to show a 4-fold decrease (Malc et al., 2009). These data indicate that Blm10 and Ump1 employ distinct mechanisms for the maintenance of mitochondrial function.
Mutation of the 19S subunit Rpn11 also causes petite formation, although in this case the mechanism appears to involve mislocalization or aberrant persistence of factors that promote mitochondrial tubulation or that otherwise regulate fission and fusion, as these mutants display abnormal mitochondrial fragmentation (Rinaldi et al., 2008). The morphology of the mitochondria in blm10-Δ mutants appears to be normal, indicating a different mechanism for petite formation than that occurring in rpn11 mutants (not shown).
The C-terminus of Blm10 is important for its physiological function
To test the importance of the conserved C-terminus of Blm10, we deleted the last three codons (TyrTyrAla) in the normal genomic context. Consistent with the observation that these residues make intimate contact with the proteasome but do not contact other Blm10 residues, this mutant was as stable as the intact protein (Figure S3C) and localized to the nucleus in a manner indistinguishable from WT (data not shown). The truncated protein failed to maintain normal levels of mitochondrial function and the yield of petites was similar to that obtained with a complete deletion of Blm10 (Figure 4A). Other perturbations of the C-terminus, including deletion of just the last residue or mutation of the YYA sequence to AAA also caused severe impairment of Blm10 function (Figure 4A). These demonstrations that the C-terminus of Blm10 performs a physiologically important function are consistent with our structural finding that these residues play a specific role in proteasome binding and definition of the gate conformation.
Conclusions
We have verified that Blm10 functions in a proteasome-related process and demonstrated that it is required for the maintenance of mitochondrial function by a mechanism that is distinct from that of previously reported genes. We have also determined the structure of Blm10 in complex with the proteasome and found an unexpected similarity in binding by C-terminal residues, which indicates common modes of proteasome binding for Blm10, 11S, and 19S/PAN. Notably, the penultimate tyrosine residue makes specific interactions that suggest a refinement of current models for binding and gate opening by 19S/PAN activators. Consistent with the structure, genetic analysis indicates that the C-terminal residues of Blm10 are required for its physiological function. Despite these advances, a number of important questions remain. For example, we do not yet understand the role of Blm10 in maintaining mitochondrial vitality, although this phenotype provides both a conceptual rationale for the conservation of Blm10 among eukaryotes and an assay to probe the effect of mutating the BLM10 gene on a physiologically important function of Blm10. We also note that in view of the high abundance of mitochondria in spermatozoa, it is attractive to speculate that a role in maintaining mitochondrial function underlies the requirement of the mammalian Blm10 homolog PA200 for spermatogenesis (Khor et al., 2006). The structure appears somewhat at odds with Blm10's characterization as a proteasome activator in vitro, and seems more consistent with possible functions as an adaptor, assembly factor, or inhibitor. Thus, while fundamentally important questions of Blm10 biology remain to be answered, the structure and genetic demonstration of a role in maintaining mitochondrial function provide tools and insight that can guide future studies.
Experimental Procedures
See supplemental data for a more complete description of the methods. Double capped S. cerevisiae proteasome:Blm10 and Δ50Blm10 complexes were prepared largely as described (Iwanczyk et al., 2006). Protein was concentrated to 20–25 mg/ml for crystallization by vapor diffusion. Diffraction data were collected at 100K at the National Synchrotron Light Source beamline X29 and were phased by molecular replacement using the unliganded proteasome (Groll et al., 1997) (pdb code 1ryp) as the search model.
Supplementary Material
Acknowledgments
We thank Charisse Kettelkamp and Hua Xin for technical assistance, Pavel Afonine for running Phenix calculations, Heidi Schubert for assistance and advice on aspects of the crystallography, Daniel Finley and David Legett for providing the yeast strain sDL135 that expresses the protein A-tagged Pre1 proteasome subunit, David Stillman and Yaxin Yu for MSH1 mRNA data, and Martin Rechsteiner and Beth Stadtmueller for critical comments on the manuscript. We gratefully acknowledge the DNA Synthesis and Sequencing Core Facilities at the University of Utah. X-ray diffraction data for this study were measured at beamline X29 of the National Synchrotron Light Source (NSLS). Financial support for NSLS comes principally from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the US Department of Energy, and from the National Center for Research Resources of the National Institutes of Health. This work was supported by National Institutes of Health Grant RO1 GM059135. Coordinates and diffraction data have been deposited at the Protein Data Bank with accession code 3L5Q.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Data Supplemental Data include three additional figures, one table, and experimental procedures, and can be found with this article online.
References
- Benaroudj N, Zwickl P, Seemuller E, Baumeister W, Goldberg AL. ATP hydrolysis by the proteasome regulatory complex PAN serves multiple functions in protein degradation. Mol Cell. 2003;11:69–78. doi: 10.1016/s1097-2765(02)00775-x. [DOI] [PubMed] [Google Scholar]
- Blickwedehl J, Agarwal M, Seong C, Pandita RK, Melendy T, Sung P, Pandita TK, Bangia N. Role for proteasome activator PA200 and postglutamyl proteasome activity in genomic stability. Proc Natl Acad Sci U S A. 2008;105:16165–16170. doi: 10.1073/pnas.0803145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blickwedehl J, McEvoy S, Wong I, Kousis P, Clements J, Elliott R, Cresswell P, Liang P, Bangia N. Proteasomes and proteasome activator 200 kDa (PA200) accumulate on chromatin in response to ionizing radiation. Radiat Res. 2007;167:663–674. doi: 10.1667/RR0690.1. [DOI] [PubMed] [Google Scholar]
- Doherty K, Pramanik A, Pride L, Lukose J, Moore CW. Expression of the expanded YFL007w ORF and assignment of the gene name BLM10. Yeast. 2004;21:1021–1023. doi: 10.1002/yea.1146. [DOI] [PubMed] [Google Scholar]
- Fehlker M, Wendler P, Lehmann A, Enenkel C. Blm3 is part of nascent proteasomes and is involved in a late stage of nuclear proteasome assembly. EMBO Rep. 2003;4:959–963. doi: 10.1038/sj.embor.embor938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster A, Masters EI, Whitby FG, Robinson H, Hill CP. The 1.9 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions. Mol Cell. 2005;18:589–599. doi: 10.1016/j.molcel.2005.04.016. [DOI] [PubMed] [Google Scholar]
- Forster A, Whitby FG, Hill CP. The pore of activated 20S proteasomes has an ordered 7-fold symmetric conformation. EMBO J. 2003;22:4356–4364. doi: 10.1093/emboj/cdg436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillette TG, Kumar B, Thompson D, Slaughter CA, DeMartino GN. Differential roles of the COOH termini of AAA subunits of PA700 (19 S regulator) in asymmetric assembly and activation of the 26 S proteasome. J Biol Chem. 2008;283:31813–31822. doi: 10.1074/jbc.M805935200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik HD, Huber R. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- Iwanczyk J, Sadre-Bazzaz K, Ferrell K, Kondrashkina E, Formosa T, Hill CP, Ortega J. Structure of the Blm10-20 S proteasome complex by cryo-electron microscopy. Insights into the mechanism of activation of mature yeast proteasomes. J Mol Biol. 2006;363:648–659. doi: 10.1016/j.jmb.2006.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajava AV, Gorbea C, Ortega J, Rechsteiner M, Steven AC. New HEAT-like repeat motifs in proteins regulating proteasome structure and function. J Struct Biol. 2004;146:425–430. doi: 10.1016/j.jsb.2004.01.013. [DOI] [PubMed] [Google Scholar]
- Khor B, Bredemeyer AL, Huang CY, Turnbull IR, Evans R, Maggi LB, Jr., White JM, Walker LM, Carnes K, Hess RA, et al. Proteasome activator PA200 is required for normal spermatogenesis. Mol Cell Biol. 2006;26:2999–3007. doi: 10.1128/MCB.26.8.2999-3007.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann A, Jechow K, Enenkel C. Blm10 binds to pre-activated proteasome core particles with open gate conformation. EMBO Rep. 2008;9:1237–1243. doi: 10.1038/embor.2008.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- Malc E, Dzierzbicki P, Kaniak A, Skoneczna A, Ciesla Z. Inactivation of the 20S proteasome maturase, Ump1p, leads to the instability of mtDNA in Saccharomyces cerevisiae. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 2009 doi: 10.1016/j.mrfmmm.2009.05.008. [DOI] [PubMed] [Google Scholar]
- Marques AJ, Glanemann C, Ramos PC, Dohmen RJ. The C-terminal extension of the beta7 subunit and activator complexes stabilize nascent 20 S proteasomes and promote their maturation. J Biol Chem. 2007;282:34869–34876. doi: 10.1074/jbc.M705836200. [DOI] [PubMed] [Google Scholar]
- Ogur M, St. John R, Nagai S. Tetrazolium overlay technique for population studies of respiration deficiency in yeast. Science. 1957;125:928–929. doi: 10.1126/science.125.3254.928. [DOI] [PubMed] [Google Scholar]
- Rabl J, Smith DM, Yu Y, Chang SC, Goldberg AL, Cheng Y. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol Cell. 2008;30:360–368. doi: 10.1016/j.molcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechsteiner M, Hill CP. Mobilizing the proteolytic machine: cell biological roles of proteasome activators and inhibitors. Trends Cell Biol. 2005;15:27–33. doi: 10.1016/j.tcb.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Rinaldi T, Hofmann L, Gambadoro A, Cossard R, Livnat-Levanon N, Glickman MH, Frontali L, Delahodde A. Dissection of the carboxyl-terminal domain of the proteasomal subunit Rpn11 in maintenance of mitochondrial structure and function. Mol Biol Cell. 2008;19:1022–1031. doi: 10.1091/mbc.E07-07-0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M, Haas W, Crosas B, Santamaria PG, Gygi SP, Walz T, Finley D. The HEAT repeat protein Blm10 regulates the yeast proteasome by capping the core particle. Nat Struct Mol Biol. 2005;12:294–303. doi: 10.1038/nsmb914. [DOI] [PubMed] [Google Scholar]
- Seemuller E, Lupas A, Stock D, Lowe J, Huber R, Baumeister W. Proteasome from Thermoplasma acidophilum: a threonine protease. Science. 1995;268:579–582. doi: 10.1126/science.7725107. [DOI] [PubMed] [Google Scholar]
- Smith DM, Chang SC, Park S, Finley D, Cheng Y, Goldberg AL. Docking of the proteasomal ATPases' carboxyl termini in the 20S proteasome's alpha ring opens the gate for substrate entry. Mol Cell. 2007;27:731–744. doi: 10.1016/j.molcel.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtmueller BM, Ferrell K, Whitby FG, Heroux A, Robinson H, Myszka DG, Hill CP. Structural models for interactions between the 20S proteasome and its PAN/19S activators. J Biol Chem. 2009 doi: 10.1074/jbc.C109.070425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unno M, Mizushima T, Morimoto Y, Tomisugi Y, Tanaka K, Yasuoka N, Tsukihara T. The structure of the mammalian 20S proteasome at 2.75 A resolution. Structure. 2002;10:609–618. doi: 10.1016/s0969-2126(02)00748-7. [DOI] [PubMed] [Google Scholar]
- Ustrell V, Hoffman L, Pratt G, Rechsteiner M. PA200, a nuclear proteasome activator involved in DNA repair. EMBO J. 2002;21:3516–3525. doi: 10.1093/emboj/cdf333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitby FG, Masters EI, Kramer L, Knowlton JR, Yao Y, Wang CC, Hill CP. Structural basis for the activation of 20S proteasomes by 11S regulators. Nature. 2000;408:115–120. doi: 10.1038/35040607. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Clawson A, Realini C, Jensen CC, Knowlton JR, Hill CP, Rechsteiner M. Identification of an activation region in the proteasome activator REGalpha. Proc Natl Acad Sci U S A. 1998;95:2807–2811. doi: 10.1073/pnas.95.6.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.