Abstract
The mitochondrial protein frataxin is essential for cellular regulation of iron homeostasis. Although the exact function of frataxin is not yet clear, recent reports indicate the protein binds iron and can act as a mitochondrial iron chaperone to transport Fe(II) to ferrochelatase and ISU proteins within the heme and iron–sulfur cluster biosynthetic pathways, respectively. We have determined the solution structure of apo yeast frataxin to provide a structural basis of how frataxin binds and donates iron to the ferrochelatase. While the protein's α–β-sandwich structural motif is similar to that observed for human and bacterial frataxins, the yeast structure presented in this report includes the full N-terminus observed for the mature processed protein found within the mitochondrion. In addition, NMR spectroscopy was used to identify frataxin amino acids that are perturbed by the presence of iron. Conserved acidic residues in the helix 1–strand 1 protein region undergo amide chemical shift changes in the presence of Fe(II), indicating a possible iron-binding site on frataxin. NMR spectroscopy was further used to identify the intermolecular binding interface between ferrochelatase and frataxin. Ferrochelatase appears to bind to frataxin's helical plane in a manner that includes its iron-binding interface.
Frataxin, a mitochondrial protein known to participate in regulating cellular iron homeostasis (1-3), has recently been suggested to play a direct role in producing both heme and Fe–S clusters (4, 5). Reduced frataxin levels are the principal cause of the autosomal recessive neurodegenerative disorder Friedreich's ataxia, affecting one in 50 000 humans (6, 7). A cellular frataxin deficiency causes mitochondrial iron overload and impairment of both heme and Fe–S cluster biosynthesis (1, 2, 8). Cellular heme production is accomplished by the enzyme ferrochelatase, which utilizes iron and protoporphyrin as substrates to produce heme, and recent reports indicate frataxin binds to ferrochelatase with a nanomolar binding affinity (5, 9). Frataxin has also been shown to donate iron to ferrochelatase for the completion of in vitro heme synthesis (9, 10). These results suggest frataxin may act as an iron chaperone to deliver the Fe(II) required to complete cellular heme biosynthesis.
Frataxin has been shown to bind iron and partially protect metal against aerobic oxidation, suggesting in vivo protein could deliver the Fe(II) required by frataxin's protein binding partners to complete heme and iron–sulfur cluster biosynthesis (11-14). The presence of iron can induce aggregation in human and yeast frataxins (12, 13); however, this oligomerization is dependent on solution conditions (11, 15). Monomeric human and bacterial frataxins have been reported to bind up to six iron atoms, with dissociation constants of 55 and 3.8 μM, respectively (4, 14). In contrast, iron binding has only been observed in oligomeric forms of yeast frataxin (12, 13, 16). Mutational analyses implicate conserved acidic residues in frataxin's N-terminus in helping to mediate protein oligomerization (11); however, the specific amino acids that participate in iron binding have yet to be identified.
The structures of human and bacterial frataxins have previously been reported; however, the eukaryotic structures were determined for N-terminally truncated forms of the mature protein found within the mitochondria (17-19). The human structures presented in these reports represent the “mature” processed protein found within the mitochondria without the protein's 15 N-terminal residues, which were omitted because they were susceptible to proteolysis and assumed to be unstructured (18, 19). Both ortholog structures are defined by an α-β-sandwich motif, with two N- and C-terminal α-helices that construct the helical plane and five antiparallel β-strands that form the β-sheet plane. A sixth (and seventh in the human protein) β-strand intersects the helical and β-sheet planes.
In this report, we present the structure of a full-length mature frataxin and identify residues that construct frataxin's putative iron and ferrochelatase binding regions using NMR spectroscopy. These data describe the first structure of yeast frataxin from Saccharomyces cerevisiae and the first full-length mature eukaryotic frataxin structure. An iron titration was performed by NMR to determine which frataxin amino acids are affected by the presence of iron, and these data define what we believe is the iron binding region of the protein. Furthermore, NMR spectroscopy was also used to map the intermolecular interface where ferrochelatase binds to frataxin. When combined, these results present the first structural indication of how frataxin binds and delivers iron to ferrochelatase, providing direct insight into events that lead to cellular heme biosynthesis.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The coding sequence for the mature yeast frataxin, representing residues 52–174 of the complete open reading frame, was subcloned into the pET11a bacterial expression vector (Novagen). Unlabeled, 15N-labeled, and 15N- and 13C-labeled proteins were expressed in Escherichia coli BL21-(DE3) cells. Cells expressing unlabeled frataxin were grown in Luria-Bertani medium (Sigma), while the 15N-labeled protein and the 13C- and 15N-labeled protein were over-expressed in M9 minimal medium using 15N-labeled ammonium chloride (Isotec) and uniformly 13C-labeled glucose (Isotec), respectively. Cells were grown at 37 °C, induced at an OD600 of 0.4 with a final concentration of 1 mM IPTG for 6 h, lysed with a French press, and sonicated to liberate protein and disrupt DNA. The stepwise purification of the 123-amino acid protein (which includes an N-terminal methionine) involved two ammonium sulfate precipitation steps with a 40% cut to precipitate contaminating proteins and a 65% cut where frataxin precipitated. Following dialysis, the protein was subjected to anion exchange chromatography using a Pharmacia Q-Sepharose column equilibrated at pH 8.0 in 25 mM Tris buffer, 10 mM EDTA, and 5 mM β-mercaptoethanol (BME) and using a gradient to 1 M NaCl. The protein elutes at 600 mM NaCl. The final purification stage included a Pharmacia phenyl-Sepharose column with a decreasing 1 M ammonium sulfate gradient at pH 8.0 in 25 mM Tris buffer, 10 mM EDTA, and 5 mM BME. The protein elutes at 800 mM ammonium sulfate. An EDTA-containing protease inhibitor cocktail tablet (Roche) was added before the cells were lysed and before each purification step to prevent proteolysis of the protein's N-terminus. EDTA and salts were removed by dialysis prior to concentration of the protein. The protein was concentrated using Amicon cell concentrators (YM-10 membrane) and Millipore 10 kDa centricons; both membranes were washed extensively to remove glycerol and azide. Average yields per liter of growth medium were 70 and 30 mg under unlabeled and isotope-labeled conditions, respectively. The protein was characterized by N-terminal amino acid sequencing (NH3-M-E-S), amino acid analysis, and electrospray mass spectrometry (Mwcalc = 13 783 g/mol, Mwobs = 13 782 ± 2 g/mol). The protein was stored under an oxygen-depleted argon atmosphere at 4 °C in 25 mM sodium phosphate buffer at pH 7.0 and 2 mM DTT and used within 2 weeks of isolation to ensure protein integrity.
For overexpression and purification of ferrochelatase, the coding sequence for the mature processed ferrochelatase, including amino acids 32–393 of the open reading frame, was subcloned into a modified pET21b vector. The pET21b vector was modified by digestion with NdeI and SalI, filling in with Klenow, and religating, thereby removing the T7 tag. The ferrochelatase sequence was then inserted into the HindIII and XhoI sites, creating an expression plasmid with mature ferrochelatase and an appended His6 tag. The plasmid was transformed into E. coli strain BL21(DE3), and protein expression was induced with 1 mM IPTG for 4 h at 30 °C. Cells were lysed with a French press in 50 mM Tris-HCl (pH 8.0) and 50 mM NaCl and centrifuged at 9000g, and the supernatant was purified on a Ni–NTA column. The protein was eluted with imidazole, and 0.005% deoxycholate was added at this step to prevent protein aggregation. The eluted protein was concentrated using Amicon Ultrafilters, to approximately 20 mg/mL, in buffer consisting of 50 mM sodium phosphate (pH 7.0) and 0.0025% deoxycholate. The protein was highly active and able to support heme synthesis following purification and freezing.
NMR Spectroscopy
Sample Preparation
Frataxin samples used in the structure determination were prepared at a concentration of 1 mM in NMR buffer [25 mM sodium phosphate buffer (pH 7.0) and 2 mM DTT with 90% H2O and 10% D2O]. Deuterium-exchanged frataxin samples were prepared by washing the unlabeled protein within an Amicon concentrator with a 99.9% D2O solution buffered with deuterated 25 mM sodium phosphate (pD 7.4) and 2 mM DTT over an 8 day period (4 °C with six exchanges of solvent per day). NMR samples were degassed on a Schlenk line and flame-sealed under argon in 5 mm NMR tubes to prevent sample oxidation.
Data Collection and Assignments
Spectra used in the structural studies were recorded on a Varian INOVA 600 MHz NMR spectrometer equipped with a triple-resonance 1H, 13C, 15N Nalorac probe with z-axis pulsed field gradients. Spectra were collected at 30 °C and referenced using an external DSS control (20). Backbone atom assignments were made using a 1H–15N HSQC spectrum and standard triple-resonance experiments [HNCACB, CBCA(CO)NH, and HNCO]. Side chain aliphatic assignments were completed using 1H–13C HSQC, H(CCO)NH-TOCSY, C(CO)NH-TOCSY, HCCH-TOCSY, and 15N-filtered TOCSY experiments, while side chain aromatic assignments were made using two-dimensional (2D) 1H–1H TOCSY (deuterium exchange sample), 1H–13C HSQC, and 13C-filtered NOESY experiments (aromatic experiments centered in the carbon dimension at 125 ppm). TOCSY mixing times were between 40 and 60 ms, while a NOESY mixing time of 150 ms was used. Stereospecific assignments for 80 of the 180 β-methylene protons were made using 15N-filtered TOCSY and NOESY data. Interatomic structural NOE restraints were determined using three-dimensional (3D) versions of the 15N-filtered NOESY and 13C-filtered NOESY, while aromatic through-space correlations were determined using the 2D 1H–1H NOESY (deuterium exchange sample) and 3D 13C-filtered NOESY data. A complete list of NMR spectra, the collection parameters used during data collection, and references for each experiment are listed in Table 1.
Table 1.
Acquisition Parameters for NMR Experiments Performed on Apofrataxin
| no. of acquired data points (nucleus) |
spectral width (ppm) |
|||||||
|---|---|---|---|---|---|---|---|---|
| experiment | t1 | t2 | t3 | F1 | F2 | F3 | nta | ref |
| 2D 1H–15N HSQC | 128 (15N) | 2048 (1H) | 49.36 | 12.50 | 4 | 33 | ||
| 2D 1H–13C HSQC | 60 (13C) | 2048 (1H) | 79.58 | 12.50 | 64 | 33 | ||
| 3D HNCACB | 45 (13C) | 45 (15N) | 2048 (1H) | 60.01 | 41.13 | 12.50 | 32 | 34 |
| 3D CBCA(CO)NH | 40 (13C) | 48 (15N) | 2048 (1H) | 60.01 | 41.13 | 12.50 | 32 | 35 |
| 3D C(CO)NH | 128 (13C) | 48 (15N) | 2048 (1H) | 68.01 | 41.13 | 12.50 | 8 | 36 |
| 3D 15N TOCSY | 256 (1H) | 32 (15N) | 2048 (1H) | 12.50 | 41.13 | 12.50 | 8 | 37 |
| 3D H(CCO)NH | 120 (1H) | 40 (15N) | 2048 (1H) | 12.50 | 41.13 | 12.50 | 16 | 38 |
| 3D HCCH-TOCSY | 128 (1H) | 60 (15N) | 2048 (1H) | 12.50 | 39.78 | 12.50 | 8 | 39 |
| 3D HNCO | 64 (13C) | 40 (15N) | 2048 (1H) | 16.57 | 41.13 | 12.50 | 8 | 35 |
| 3D 15N NOESY | 32 (1H) | 256 (15N) | 2048 (1H) | 12.50 | 41.13 | 12.50 | 8 | 40 |
| 3D 13C NOESY | 128 (1H) | 64 (13C) | 2048 (1H) | 12.50 | 33.15 | 12.50 | 8 | 40 |
| 2D D2O NOESY | 512 (1H) | 4096 (1H) | 12.52 | 12.52 | 32 | 41 | ||
| 3D 13C NOESYb | 80 (1H) | 32 (13C) | 2048 (1H) | 12.50 | 29.83 | 12.50 | 16 | 40 |
| 2D D2O TOCSY | 512 (1H) | 4096 (1H) | 12.52 | 12.52 | 32 | 42 | ||
The value of nt refers to the number of transients in the acquisition dimension used in signal averaging the spectrum.
Carbon-filtered NOESY entered on the aromatic region.
Structure Determination
Frataxin structures were refined using torsion angle dynamics (TAD) within DYANA and regularized within CNS using a total of 1730 NOE inter-proton, 44 hydrogen bonding, and 144 ϕ/ψ dihedral angle restraints (21, 22). Backbone dihedral angle restraints were obtained on the basis of chemical shift values using TALOS (23). In DYANA, 200 structures were calculated using 16 000 cooling steps and starting from an annealing temperature of 9600 K. In CNS, 40 structures were further refined with 12 000 slow cooling steps starting from an annealing temperature of 25 K. Structures were analyzed using PROCHECK-NMR and displayed using MOLMOL (24, 25). The structural statistics for the protein are given in Table 2.
Table 2.
Structural Statistics for Apofrataxin
| (TAD)a | (CNS)a | |
|---|---|---|
| NOE distance restraintsb (Å) | 1730 | 1729 |
| intraresidue | 320 | 320 |
| sequential (|i − j| = 1) | 357 | 357 |
| medium-range (2 ≤ |i − j| ≤ 5) | 550 | 550 |
| long-range (|i − j| > 5) | 503 | 503 |
| hydrogen bonding restraintsc (Å) | 44 | 43 |
| dihedral angle restraints (TALOS) | 144 | 144 |
| stereospecific assignments (Hβ and Qγ1) | 100 | 100 |
| DYANA target function (Å4) | 1.20 | |
| CNS total energy | 132.97 ± 5.01 | |
| residual distance restraint violations | ||
| no. of violations >0.25 Å | 0 ± 0 | 0 ± 0 |
| sum of violations (Å or kcal/mol)d | 4.3 ± 0.5 | 15.88 ± 1.18 |
| maximum violation (Å) | 0.25 | |
| residual dihedral angle restraint violation | ||
| no. of violations >0.12° | 0 ± 0 | 0 ± 0 |
| sum of violations (deg or kcal/mol)d | 0.5 ± 0.1 | 61.97 ± 1.98 |
| maximum violation (deg) | 0.12 | |
| van der Waals violations | ||
| no. >0.3 Å | 1 | 1 |
| sum of violations (Å or kcal/mol)d | 5.2 ± 0.5 | 44.18 ± 2.66 |
| maximum violation (Å) | 0.37 | |
| Ramachandran statistics (%)e | ||
| favored | 74.6 | 75.3 |
| allowed | 19.2 | 21.6 |
| generously allowed | 4.5 | 2.4 |
| disallowed | 1.7 | 0.8 |
| rms deviations from average coordinates (Å)f | ||
| backbone | 0.67 ± 0.17 | 0.68 ± 0.10 |
| heavy atoms | 1.26 ± 0.18 | 1.24 ± 0.18 |
(TAD) is the ensemble of 20 lowest-penalty structures calculated using DYANA (21). (CNS) is the same ensemble after 2000 steps (15 ps each) of simulated annealing at 25 K, 2000 slow cooling steps to 0 K, and 10 000 steps of restrained Powell minimization in Cartesian space (anneal.inp protocol) (22).
Only meaningful and nonredundant restraints as determined by the DYANA CALIBA function.
Two upper limit distance restraints were used to define each hydrogen bond.
Violations from DYANA have units of angstroms, while violation energies from CNS are in kilocalories per mole.
Determined using PROCHECK-NMR (24).
The rmsds were calculated using MOLMOL (25) on the elements of secondary structure (residues 19–42, 50–55, 60–65, 69–74, 79–84, 88–94, 97–100, and 109–120).
Chemical Shift Perturbation Mapping Experiments
Amino Acids Affected by Iron
Chemical shift perturbation NMR experiments were performed to identify frataxin amino acids affected by the presence of iron. Data were collected on a Varian INOVA 720 MHz spectrometer equipped with a triple-resonance 1H, 13C, 15N Varian probe with z-axis pulsed field gradients. Using a 15N-labeled frataxin concentration of 333 μM, aqueous anaerobic ferrous ammonium sulfate (Sigma) in NMR buffer was added to independent protein samples to achieve Fe(II):protein stoichiometric ratios of 0.25, 0.50, 0.75, 1.00, 1.25, 1.50, 1.75, and 2.00. 15N-labeled frataxin in NMR buffer with 650 μM ammonium sulfate (Sigma) served as our apoprotein control. Samples were prepared at room temperature immediately before data collection, placed in the NMR spectrometer, and allowed to equilibrate to 30 °C for 30 min. Full 1H–15N HSQC spectra were collected at each titration point, and spectra were collected on reproducible independent samples. Amide chemical shifts were measured using XEASY (26), and normalized chemical shift changes (δ) resulting from the addition of Fe(II) to [15N]frataxin were measured and calculated using the equation δ = 25[(δHN)2 + (δN/5)2]0.5 (27).
Ferrochelatase Binding Interface
Amide chemical shift perturbation experiments were used to identify the intermolecular interface of frataxin that interacts with ferrochelatase. Reproducible, independent 15N-labeled frataxin samples were mixed with unlabeled ferrochelatase. The protein complex sample was prepared in NMR buffer with 0.005% deoxycholate, at a stoichiometric monomer ratio of 1:1 at a final individual protein concentration of 300 μM. 15N-labeled frataxin in NMR buffer with 0.005% deoxycholate served as our frataxin control. Because of the large size of the protein complex (yeast ferrochelatase is a dimer at ca. 89 kDa), the standard 1H–15N HSQC experiment gave amide line widths that were extremely broad and poorly resolved (data not shown). Therefore, we used a TROSY version of the 1H–15N HSQC experiment to collect spectra on the protein complex at a spectrometer frequency of 720 MHz. TROSY 1H–15N HSQC spectra were collected with eight transients, 2048 acquisition points, and 256 increments in the indirect dimension at 30 °C. Amide peak positions were measured using XEASY, and the normalized chemical shift changes (δ) were calculated in a manner identical to that described earlier.
RESULTS
Solution Structure of Frataxin
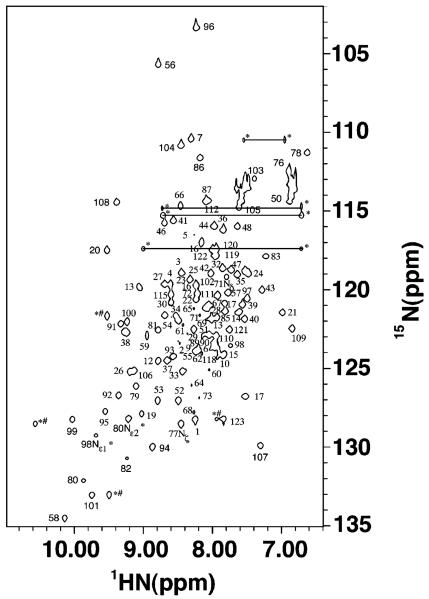
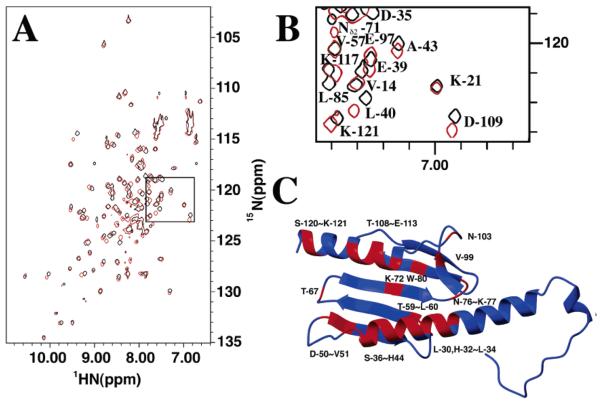
Apofrataxin is highly folded and a monomer in solution, as judged by the average 8 Hz amide proton half-height line widths observed within the 1H–15N HSQC spectrum (Figure 1) and by size exclusion chromatography. Nearly complete resonance and β-methylene stereospecific assignments were made for frataxin using 1H, 13C, 15N heteronuclear experiments. There is no indication within our data that there is more than a single protein conformation in solution, and our protein behaves like a well-folded homogeneous molecule for multiple weeks when NMR samples are stored under argon in sealed tubes (15). A total of 1730 NOE distance restraints, combined with 144 dihedral angle restraints (obtained from TALOS), 44 hydrogen bonding restraints (based on NOE patterns and chemical shift index analysis), and 80 Hβ stereospecific assignments (based on NOE patterns), were used to calculate 200 independent structures with the torsion angle dynamics simulated annealing algorithm within DYANA. The final ensemble of the 20 lowest-energy structures has heavy atom root-mean-square deviations in regions of secondary structure of 0.67 ± 0.17 Å (backbone) and 1.26 ± 0.18 Å (all) and a DYANA target function of 1.20 Å4. No distance violations were observed to be greater than 0.25 Å. According to PROCHECK-NMR analysis, the majority of the residues have ϕ and ψ angles in the favored region (74.6%), while only 1.7% of the residues (Thr-5 and Arg-102) are in the disallowed region. The 20 DYANA structures were then further refined using CNS (22). A comparison of the structural statistics for the 20 lowest-energy DYANA and CNS structures shows minimal differences between molecules calculated from each program (Table 2).
Figure 1.
1H–15N HSQC spectrum of 15N-labeled frataxin. The spectrum was collected at 30 °C on a Varian INOVA 600 MHz NMR spectrometer. Corresponding peak assignments are given adjacent to spectral features. Side chain amine signals are designated with asterisks and connected to partners by a line when applicable. Aliased peaks are designated with pound signs.
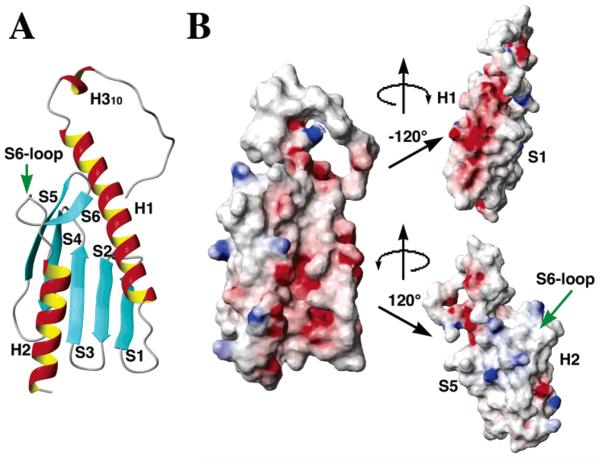
The solution structure of frataxin is consistent with an α-β-sandwich motif, with overall molecular dimensions, excluding the 18-amino acid N-terminus, of 47 Å × 29 Å × 23 Å (Figure 2A). The initial 11 N-terminal residues of frataxin are unstructured, while residues 12–15 form a 310-helix (defined as H310). The 18 N-terminal residues are dynamic in nature, as judged by the absence of long-range NOE patterns linking this region to the protein's globular domain. Within the globular domain of the protein, a helical plane is formed by two terminal α-helices [residues 19–42 (H1) an 109–120 (H2)] arranged in parallel fashion. A β-sheet plane is formed by five antiparallel β-strands of residues 50–55 (S1), 60–65 (S2), 69–74 (S3), 79–84 (S4), and 88–94 (S5). An additional β-strand, formed by residues 97–100 (S6), is connected to H2 by a loop of eight amino acids; β-strand 6 and loop residues will subsequently be called the protein's S6-loop domain.
Figure 2.
Ribbon drawing (A) and electrostatic surface plots (B) for frataxin at three different orientations around the vertical axis. (A) Labels for the different secondary structural elements are given adjacent to the corresponding helix or strand. (B) At the left is a surface electrostatic potential plot for frataxin at a molecular orientation identical to that shown in panel A. The surface was calculated using a probe radius of 1.4 Å and the potential surface displayed with a scale from −10 to 10 kbT using MOLMOL. At the right, the top and bottom panels correspond to a rotation of the central surface about the vertical axis by −120° and 120°, respectively. To orient the reader, secondary structural element landmarks have been labeled.
The electrostatic surface for frataxin, shown in three orientations in Figure 2B, reveals the helical plane, and the H1–S1 interface, are largely composed of exposed acidic surface residues. The pI for yeast frataxin (4.34) is partially the result of the 23 Asp and Glu residues centered about the H1–S1 and H2 structural elements. Surface-exposed acidic residues in the H1–S1 interface and in the H2 surface are highly conserved (11). The β-sheet surface of the protein is largely neutral in charge except for the presence of scattered surface basic residues.
Mapping the Putative Iron-Binding Region of Frataxin
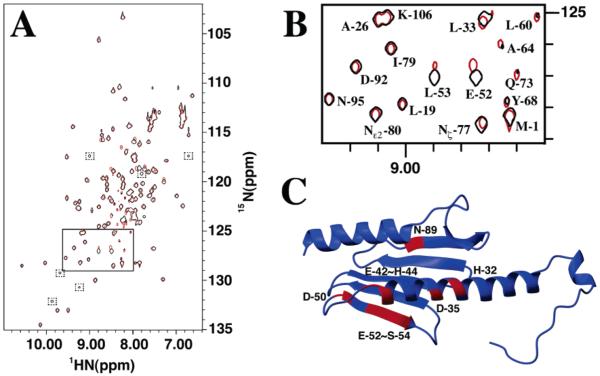
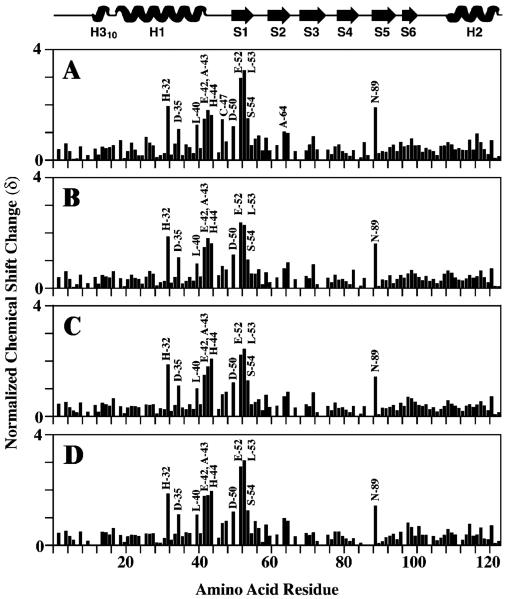
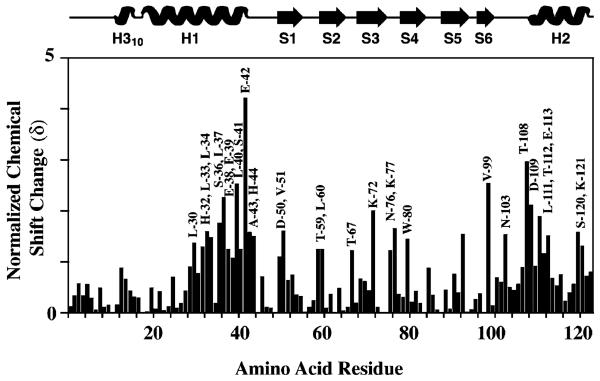
NMR spectroscopy was used to identify frataxin amino acids that are influenced by the presence of ferrous iron. A series of 1H–15N HSQC spectra were collected during the stepwise titration of anaerobic aqueous Fe(II) at concentrations between 0 and 2 equiv to a single equivalent of anaerobic 15N-labeled frataxin (Figure 3). The buffered Fe(II) solution was prepared under strict anaerobic conditions to prevent the oxidation of ferrous iron. Previous reports indicate that iron triggers oligomerization for yeast frataxin at these iron: protein stoichiometries in HEPES buffer (12); however, we saw no change in the overall amide proton line widths (8 Hz) at iron:protein stoichiometries in NMR buffer of up to 2:1, indicating under NMR buffer conditions frataxin remained a monomer in solution. At Fe:protein ratios above 2:1, all amide line widths were broadened, indicating that either the predominate protein had oligomerized or iron had bound nonselectively (data not shown). On the basis of the appearance of a single set of protein amide peaks at all iron concentrations, the Fe-bound frataxin complex appears to be in fast exchange between apo and metal-bound forms, and this is consistent with the weak iron binding affinity observed for the human and yeast proteins (Kd = 55 and 3.8 μM, respectively) (4). A comparison of backbone amide chemical shifts at intermediate Fe:protein ratios (between 0.25 and 2) revealed 10 frataxin amino acids underwent substantial normalized amide chemical shift changes (δ) in the range of 1–3 (Figure 4), while the remaining residues had an average δ value of 0.4. The 10 shifted residues include His-32, Asp-35, Glu-42, Ala-43, His-44, Asp-50, Glu-52, Leu-53, Ser-54, and Asn-89, and their position on the apofrataxin structure is shown in Figure 3C. If Asp-89 is excluded, the nine remaining amino acids are confined to the helix 1–strand 1 interface: H1 residues (His-32, Asp-35, and Glu-42), loop residues between H1 and S1 (Ala-43 and His-44), and S1 residues (Asp-50, Glu-52, Leu-53, and Ser-54). It is interesting that the average amide proton line widths for these 10 residues remained unchanged despite the presence of paramagnetic iron.
Figure 3.
NMR chemical shift mapping of frataxin residues that interact with iron. (A) 1H–15N HSQC spectra for frataxin in the presence (red) and absence (black) of iron. The iron spectrum was collected at a 2:1 iron:protein stoichiometric ratio. Peaks boxed with dashed lines are present only in the iron spectrum at low threshold levels. The region boxed with a solid line is expanded in Figure 4B. (B) Expansion of the region in the 15N HSQC spectrum for frataxin in the presence (red) and absence (black) of iron. (C) Residues identified on the apofrataxin structure that have normalized chemical shift (δ) values greater than 1 (colored red).
Figure 4.
Iron-induced amide chemical shift perturbations at varying iron:frataxin concentration ratios. Frataxin secondary structure information is given at the top. Perturbations in backbone 1H and 15N resonances, induced by the addition of Fe(NH4)2(SO4)2 to 15N-labeled apofrataxin, at the following iron:protein stoichiometries are plotted: (A) 2:1, (B) 1.5:1, (C) 1:1, and (D) 0.5:1.
Mapping the Ferrochelatase Binding Interface on Frataxin
NMR spectroscopy was used to map the ferrochelatase binding interface on frataxin. In the absence of metal, unlabeled ferrochelatase was added to 15N-labeled frataxin at a 1:1 monomeric protein stoichiometry. A TROSY version of the 1H–15N HSQC spectra was used to accommodate the large size of the protein complex, as ferrochelatase is a dimer and a 1:1 complex is ca. 120 kDa in size (28) (Figure 5). TROSY1H–15N HSQC spectra were collected on identical independent frataxin–ferrochelatase samples at 720 and 600 MHz. A total of 32 amino acids underwent normalized amide chemical shift changes (δ) within the range of 1–6 (Figure 6); these changes were corrected for the subtle change in frataxin amide chemical shifts due to the presence of a detergent (see the Supporting Information). Frataxin residues that were shifted in the presence of ferrochelatase included the iron-binding residues of the H1–S1 interface, as well as residues in both H2 and the S6-loop domains. The identity of these residues and their location on the apofrataxin structure are illustrated in Figure 5C and outlined in detail below: H1 residues (Leu-30, His-32, Leu-33, Leu-34, Ser-36, Leu-37, Glu-38, Glu-39, Leu-40, Ser-41, Glu-42, Ala-43, and His-44), S1 residues (Asp-50 and Val-51), the unstructured residue between S1 and S2 (Thr-59), an S2 residue (Leu-60), the unstructured residue between S2 and S3 (Thr-67), an S3 residue (Lys-72), the unstructured residues between S3 and S4 (Asn-76 and Lys-77), an S4 residue (Trp-80), an S6 residue (Val-99), loop residues between S6 and H2 (Asn-103 and Thr-108), H2 residues (Asp-109, Ile-110, Leu-111, Thr-112, Glu-113, and Ser-120), and a random coil residue (Lys-121).
Figure 5.
NMR chemical shift mapping of the ferrochelatase binding surface on frataxin. (A) Chemical shift-adjusted TROSY 1H–15N HSQC spectra for frataxin in the presence (red) and absence (black) of ferrochelatase at a 1:1 frataxin:ferrochelatase stoichiometry. (B) Expansion of the region identified in the spectra to the left. (C) Residues identified on the apofrataxin structure that have normalized chemical shift (δ) values greater than 1 (colored red).
Figure 6.
Identification of residues at yeast frataxin's ferrochelatase binding interface. Frataxin secondary structure information is given at the top. Perturbations in backbone 1H and 15N resonances were induced by the addition of ferrochelatase to 15N-labeled apofrataxin at ferrochelatase:protein concentration ratios of 1:1. Normalized chemical shift changes are plotted vs amino acid sequence number.
DISCUSSION
Structural Comparison of Frataxin Homologues
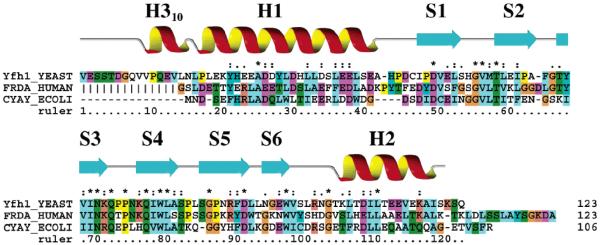
Our results confirm apofrataxin is a highly folded monomeric protein and a member of the α–β-sandwich structural motif family, with a structure very similar to those published for the human and bacterial orthologs (17-19). It is not surprising that the three characterized frataxins are structurally similar, given that their sequences are highly identical (37.8 and 28.1% between yeast and human and between yeast and E. coli, respectively) (29) and that the human protein functionally complements the yeast deletion mutant in S. cerevisiae (30). Although the yeast, human, and bacterial structures are similar, the published X-ray and NMR human structures were truncated N-terminally prior to helix 1, as these residues were susceptible to proteolysis. The configuration presented in this report includes the first full-length structure of a mature eukaryotic frataxin (Figure 7), and in yeast, the 18-amino acid N-terminus is marked by a 310-helix formed between residues 12 and 15.
Figure 7.
Alignment of structurally characterized frataxin orthologs. Multiple-sequence alignment of frataxin orthologs of known structure, displayed in ClustalX colors, corresponding to all residues in mature yeast frataxin (43, 44) as well as the structurally characterized regions of full-length bacterial and truncated mature human frataxin. Vertical lines in the human N-terminal sequence represent residues omitted from the mature human frataxin structures. The numbering refers to yeast frataxin. Secondary structural elements corresponding to frataxin are given directly above the sequences.
A common characteristic of all frataxin orthologs is their acidic nature (pI values for yeast, human, and E. coli of 4.34, 4.98, and 4.24, respectively), as a consequence of their conserved acidic residues. The helix 1–strand 1 surface of all structurally characterized frataxins is lined with the highly conserved exposed acidic residues (11). The side chains from conserved aspartic and glutamic acid residues give the H1–S1 region of frataxin a general negative charge that would be appropriate for binding positively charged ions such as aqueous Fe(II). The acidic surface constructed by H1–S1 residues extends to helix 2, making the helical plane of frataxin partially charged. There exists, however, a neutral patch of residues in the helical plane of frataxin constructed by S6-loop and H2 residues that would provide an appropriate binding surface for a neutrally surface charged protein binding partner.
Frataxin Residues Affected by the Presence of Iron
It was previously reported that under specific solution conditions (HEPES buffer and NaCl), in vitro yeast frataxin forms higher-order oligomers in the presence of high iron concentrations (12, 16). Mutational analyses of the bacterial frataxin homologue indicated that conserved acidic residues in the helix 1–strand 1 region are responsible for iron-induced protein oligomerization (11). On the basis of the amide spectral line widths observed in this report for frataxin in the presence of an up to 2:1 Fe:protein stoichiometry, the iron-bound protein remains in the monomeric state. At higher metal concentrations, amide line widths become very broad in the standard 15N HSQC spectrum, consistent with a substantial amount of protein existing in a higher-order oligomeric form or the general presence of abundant paramagnetic iron free in solution.
Chemical shift mapping experiments indicate acidic residues in frataxin's helix 1–strand 1 interface are affected by the presence of iron. It was recently shown that monomeric human and bacterial frataxin bind up to six iron atoms, although there is no information regarding which human frataxin amino acids participate in iron binding (4, 14). Common iron-binding ligands in proteins involve carboxylate oxygens, imidazole ring nitrogens, and sulfides from Asp/ Glu, His, and Cys residues, respectively (31). The majority of the 10 frataxin residues affected by the presence of iron contain oxygen- and nitrogen-based side chains that are suitable for serving as possible iron ligands (Asp, Glu, and His residues), while the single sulfur-containing amino acid in the protein (Cys-47) was unaffected by the presence of metal. The proximity of amino acids affected by the presence of iron suggests metal interacts with the protein at localized sites within the protein's H1–S1 region, and these may reflect possible iron binding sites utilized by frataxin when acting as an iron chaperone.
The observation that residues within this acidic H1–S1 region of the protein are well-conserved (Figure 7) suggests the frataxin residues we identified as being affected by the presence of iron may be important in metal regulation for all frataxin orthologs. Among the residues that were identified, Glu-42 is a conserved acidic residue in eukaryotes only, Glu-52 is a highly conserved acidic residue in eukaryotes and prokaryotes (although a serine in human frataxin), and His-44 is typically a Tyr in eukaryotes. Asp-50 is completely conserved, while Asp-35 is a conserved acidic residue across all prokaryotic and eukaryotic frataxins. His-32 is less well-conserved; however, this region typically has residues with potential side chain nitrogen (His, Lys, or Arg) or oxygen ligands (Ser, Thr, or Tyr) for binding iron (11).
Frataxin's Interface for Binding Ferrochelatase
Previous reports confirm frataxin forms a tight binding complex with ferrochelatase in solution. Recent in vitro data indicate frataxin binds to ferrochelatase with a nanomolar binding affinity (5, 9). Amide resonances for the 15N-labeled frataxin and ferrochelatase complex at a 1:1 monomer stoichiometry in a non-TROSY 1H–15N HSQC spectrum (data not shown) are extremely broad (spanning 40 Hz in the amide 1H dimension), indicating a large protein complex is formed in the absence of metal. It is reasonable to expect these broad line widths would be observed for the complex in a non-TROSY experiment given the large molecular size of the complex.
Residues within the helical plane and the H1–S1 interface construct the frataxin residues affected by binding the ferrochelatase. A significant number of frataxin residues involved in ferrochelatase binding overlap with what we believe is the Fe-binding surface of frataxin. It stands to reason that if frataxin donates metal to ferrochelatase, it would do so in a manner that involves the ferrochelatase overlapping frataxin's iron binding sites that we have identified.
It is interesting to note that residues in the S6-loop region of frataxin are also affected by ferrochelatase binding. Side chains for residues in the S6-loop region are predominately neutral in charge and provide a hydrophobic patch on frataxin on what is otherwise an overall acidic planar surface. Although the structure of yeast ferrochelatase has not been determined, the crystal structure of human ferrochelatase reveals a highly helical protein dimer whose surface is predominately hydrophobic (32). Sequence homologies suggest human and yeast ferrochelatases would have similar structures and similar surface charge distributions. Ferrochelatase is believed to associate with the mitochondrial inner membrane through a series of ridges at one end of the molecule. Attachment to the membrane through these ridges places the ferrochelatase substrate channels in a position to take up and release the poorly soluble porphyrin substrate and heme product through the mitochondrial membrane, respectively. The hydrophobic exterior of the protein is in stark contrast to the hydrophilic interior of the active sites, which are lined with conserved charged residues positioned well to receive positively charged iron. The S6-loop–H2 region in frataxin is the only predominately neutral region of the protein on the planar interface we identified that interacts with ferrochelatase. We therefore speculate that the S6-loop domain of frataxin may be the region that targets and facilitates binding of frataxin to the ferrochelatase.
Biological Implications
These results provide the first complete description of the molecular structure for a full-length eukaryotic frataxin, as well as the first structural report for yeast frataxin that is not based on modeling arguments. Furthermore, we have shown that monomeric yeast frataxin interacts with ferrous iron, and identified what we believe are residues that interact with iron. Finally, we now have structural data that indicate how frataxin and ferrochelatase interact in solution. Mutational experiments both in vivo and in vitro are in progress in our labs to test the physiological relevance of the frataxin residues identified at both the iron and ferrochelatase binding interfaces. We believe that these data provide insight into how yeast frataxin binds and interacts with both ferrous iron and its protein-binding partner ferrochelatase during heme biosynthesis.
Supplementary Material
ACKNOWLEDGMENT
Yfh1 cDNA was kindly supplied by Dr. Jerry Kaplan (University of Utah, Salt Lake City, UT). NMR spectra used in determining the solution structure of frataxin were collected at the Biomolecular NMR Facility (University of Utah, Salt Lake City, UT) partially with time generously contributed by Dr. Wesley I. Sundquist. High-field NMR data were collected at 720 MHz on the National High Field Magnet Laboratory (NHMFL, Tallahassee, FL).
Footnotes
This work is supported by the American Heart Association, Midwest Affiliate (Grant 0130527Z to T.L.S.) and by the National Institutes of Health (Grant DK53953 to A.D.).
NMR assignments, atomic coordinates, and chemical shift data for frataxin have been deposited (PDB entry 1XAQ, BMRB entry 11688).
SUPPORTING INFORMATION AVAILABLE
Normalized chemical shift changes for all yeast frataxin amino acids in the presence of deoxycholate. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Babcock M, de Silva D, Oaks R, Davis-Kaplan S, Jiralerspong S, Montermini L, Pandolfo M, Kaplan J. Regulation of Mitochondrial Iron Accumulation by Yfh1p, a Putative Homolog of Frataxin. Science. 1997;276:1709–1712. doi: 10.1126/science.276.5319.1709. [DOI] [PubMed] [Google Scholar]
- 2.Foury F, Cazzalini O. Deletion of the yeast homologue of the human gene associated with Friedreich's ataxia elicits iron accumulation in mitochondria. FEBS Lett. 1997;411:373–377. doi: 10.1016/s0014-5793(97)00734-5. [DOI] [PubMed] [Google Scholar]
- 3.Koutnikova H, Campuzano V, Foury F, Dolle P, Cazalini O, Koenig M. Studies of human, mouse and yeast homologues indicate a mitochondrial function of frataxin. Nat. Genet. 1997;16:345–351. doi: 10.1038/ng0897-345. [DOI] [PubMed] [Google Scholar]
- 4.Yoon T, Cowan JA. Iron–sulfur cluster biosynthesis. Characterization of frataxin as an iron donor for assembly of [2Fe-2S] clusters in ISU-type proteins. J. Am. Chem. Soc. 2003;125:6078–6084. doi: 10.1021/ja027967i. [DOI] [PubMed] [Google Scholar]
- 5.Lesuisse E, Santos R, Matzanke BF, Knight SA, Camadro JM, Dancis A. Iron use for haeme synthesis is under control of the yeast frataxin homologue (Yfh1) Hum. Mol. Genet. 2003;12:879–889. doi: 10.1093/hmg/ddg096. [DOI] [PubMed] [Google Scholar]
- 6.Puccio H, Koenig M. Recent advances in the molecular pathogenesis of Friedreich ataxia. Hum. Mol. Genet. 2000;9:887–892. doi: 10.1093/hmg/9.6.887. [DOI] [PubMed] [Google Scholar]
- 7.Campuzano V, Montermini L, Molto MD, Pianese L, Cossee M, Cavalcanti F, Monros E, Rodius F, Duclos F, Monticelli A, Zara F, Canizares J, Koutnikova H, Bidichandani SI, Gellera C, Brice A, Trouillas P, De Michele G, Filla A, De Frutos R, Palau F, Patel PI, Di Donato S, Mandel JL, Cocozza S, Koenig M, Pandolfo M. Freidreich's Ataxia: Autosomal Recessive Disease Caused by an Intronic GAA Triplet Repeat Expansion. Science. 1996;271:1423–1427. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- 8.Rotig A, Lonlay PD, Chretien D, Foury F, Koenig M, Sidi D, Munnich A, Rustin P. Aconitase and mitochondrial iron-sulphur protein deficiency in Friedreich's ataxia. Nat. Genet. 1997;17:215–217. doi: 10.1038/ng1097-215. [DOI] [PubMed] [Google Scholar]
- 9.Yoon T, Cowan JA. Frataxin-mediated iron delivery to ferrochelatase in the final step of heme biosynthesis. J. Biol. Chem. 2004;279:25943–25946. doi: 10.1074/jbc.C400107200. [DOI] [PubMed] [Google Scholar]
- 10.Park S, Gakh O, O'Neill HA, Mangravita A, Nichol H, Ferreira GC, Isaya G. Yeast frataxin sequentially chaperones and stores iron by coupling protein assembly with iron oxidation. J. Biol. Chem. 2003;278:31340–31351. doi: 10.1074/jbc.M303158200. [DOI] [PubMed] [Google Scholar]
- 11.Adinolfi S, Trifuoggi M, Politou AS, Martin S, Pastore A. A structural approach to understanding the iron-binding properties of phylogenetically different frataxins. Hum. Mol. Genet. 2002;11:1865–1877. doi: 10.1093/hmg/11.16.1865. [DOI] [PubMed] [Google Scholar]
- 12.Adamec J, Rusnak F, Owen WG, Naylor S, Benson LM, Gacy AM, Isaya G. Iron-Dependent Self-Assembly of Recombinant Yeast Frataxin: Implications for Friedreich's Ataxia. Am. J. Hum. Genet. 2000;67:549–562. doi: 10.1086/303056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cavadini P, O'Neill HA, Benada O, Isaya G. Assembly and iron-binding properties of human frataxin, the protein deficient in Friedreich ataxia. Hum. Mol. Genet. 2002;11:217–227. doi: 10.1093/hmg/11.3.217. [DOI] [PubMed] [Google Scholar]
- 14.Bou-Abdallah F, Adinolfi S, Pastore A, Laue TM, Chasteen DN. Iron binding and oxidation kinetics in frataxin CyaY of Escherichia coli. J. Mol. Biol. 2004;341:605–615. doi: 10.1016/j.jmb.2004.05.072. [DOI] [PubMed] [Google Scholar]
- 15.Adinolfi S, Nair M, Politou A, Bayer E, Martin S, Temussi P, Pastore A. The Factors Governing the Thermal Stability of Frataxin Orthologues: How To Increase a Protein's Stability. Biochemistry. 2004;43:6511–6518. doi: 10.1021/bi036049+. [DOI] [PubMed] [Google Scholar]
- 16.Gakh O, Adamec J, Gacy AM, Twesten RD, Owen WG, Isaya G. Physical evidence that yeast frataxin is an iron storage protein. Biochemistry. 2002;41:6798–6804. doi: 10.1021/bi025566+. [DOI] [PubMed] [Google Scholar]
- 17.Cho SJ, Lee MG, Yang JK, Song HK, Suh SW. Crystal structure of E. coli CyaY protein reveals a previously unidentified fold for the evolutionarily conserved frataxin family. Proc. Natl. Acad. Sci. U.S.A. 2000;97:8932–8937. doi: 10.1073/pnas.160270897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dhe-Paganon S, Shigeta R, Chi YI, Ristow M, Shoelson SE. Crystal structure of human frataxin. J. Biol. Chem. 2000;275:30753–30756. doi: 10.1074/jbc.C000407200. [DOI] [PubMed] [Google Scholar]
- 19.Musco G, Stier G, Kolmerer B, Adinolfi S, Martin S, Frenkiel T, Gibson T, Pastore A. Towards a Structural Understanding of Friedreich's Ataxia: The Solution Structure of Frataxin. Structure. 2000;8:695–707. doi: 10.1016/s0969-2126(00)00158-1. [DOI] [PubMed] [Google Scholar]
- 20.Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, Oldfield L, Markley JL, Sykes BD. 1H, 13C, and 15N chemical shift referencing in biomolecular NMR. J. Biomol. NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- 21.Guntert P, Mumenthaler C, Wuthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 22.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D54. 1998;(Part 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 23.Cornilescu G, Delagio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 24.Laskowski RA, Rullmann JAC, MacArthur MW, Kaptein R, Thorton JM. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 25.Koradi R, Billeter M, Wuthrich K. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 26.Bartels CH, Xia T-H, Billeter M, Güntert P, Wüthrich K. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J. Biomol. NMR. 1995;5:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 27.Grzesiek S, Bax A, Clore GM, Gronenborn AM, Hu JS, Kaufman J, Palmer I, Stahl SJ, Wingfield PT. The solution structure of HIV-1 Nef reveals an unexpected fold and permits delineation of the binding surface for the SH3 domain of Hck tyrosine protein kinase. Nat. Struct. Biol. 1996;3:340–345. doi: 10.1038/nsb0496-340. [DOI] [PubMed] [Google Scholar]
- 28.Salzmann M, Pervushin K, Wider G, Senn H, Wuthrich K. TROSY in triple-resonance experiments. Proc. Natl. Acad. Sci. U.S.A. 1998;95:13585–13590. doi: 10.1073/pnas.95.23.13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Accelrys. Accelrys Inc.; San Diego, CA: 2002. [Google Scholar]
- 30.Cavadini P, Gellera C, Patel PI, Isaya G. Human frataxin maintains mitochondrial iron homeostasis in Saccaromyces cerevisiae. Hum. Mol. Genet. 2000;9:2523–2530. doi: 10.1093/hmg/9.17.2523. [DOI] [PubMed] [Google Scholar]
- 31.Lippard SJ, Berg JM. Principles of Bioinorganic Chemistry. University Science Books, Mill; Valley, CA: 1994. [Google Scholar]
- 32.Wu CK, Dailey HA, Rose JP, Burden A, Sellers VM, Wang BC. The 2.0 Å structure of human ferrochelatase, the terminal enzyme of heme biosynthesis. Nat. Struct. Biol. 2001;8:156–160. doi: 10.1038/84152. [DOI] [PubMed] [Google Scholar]
- 33.Bodenhausen G, Ruben DJ. Natural abundance nitrogen-15 NMR by enhanced heteronuclear spectroscopy. Chem. Phys. Lett. 1980;69:185–188. [Google Scholar]
- 34.Wittekind M, Meuller L. HNCACB, A High-Sensitivity 3D NMR Experiment to Correlate Amide-Proton and Nitrogen Resonances with the α- and β-Carbon Resonances in Proteins. J. Magn. Reson., Ser. B. 1993;101:201–205. [Google Scholar]
- 35.Kay LE, Ikura M, Tschudin R, Bax A. Three-dimensional triple-resonance NMR spectroscopy of isotopically enriched proteins. J. Magn. Reson. 1990;89:496–514. doi: 10.1016/j.jmr.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 36.Muhandiram DR, Kay LE. Gradient-enhanced triple-resonance three-dimensional NMR experiments with improved sensitivity. J. Magn. Reson., Ser. B. 1994;103:203–216. [Google Scholar]
- 37.Talluri S, Wagner G. An optimized 3D NOESYHSQC. J. Magn. Reson., Ser. B. 1996;112:200–205. doi: 10.1006/jmrb.1996.0132. [DOI] [PubMed] [Google Scholar]
- 38.Sattler M, Schleucher J, Griesinger C. Prog. NMR Spectrosc. 1999;34:93–158. [Google Scholar]
- 39.Kay LE, Xu GY, Singer AU, Muhandiram DR, Forman-Kay JD. A gradient-enhanced HCCH_TOCSY experiment for side chain 1H and 13C correlations in H2O samples of proteins. J. Magn. Reson., Ser. B. 1993;101:333–337. [Google Scholar]
- 40.Wider G, Neri D, Otting G, Wuthrich K. A heteronuclear three-dimensional NMR experiment for measurements of small heteronuclear coupling constants in biological macromolecules. J. Magn. Reson. 1989;85:426–431. [Google Scholar]
- 41.Wider G, Macura S, Kumar A, Ernst RR, Wuthrich K. Homonuclear two-dimensional 1H NMR of proteins. Experimental procedures. J. Magn. Reson. 1984;56:207–234. [Google Scholar]
- 42.Bax A, Davis DG. MLEV-17-based two-dimensional homonuclear magnetization transfer spectroscopy. J. Magn. Reson. 1985;65:355–360. [Google Scholar]
- 43.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gordon DM, Kogan M, Knight SA, Dancis A, Pain D. Distinct roles for two N-terminal cleaved domains in mitochondrial import of the yeast frataxin homolog, Yfh1p. Hum. Mol. Genet. 2001;10:259–269. doi: 10.1093/hmg/10.3.259. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.