Abstract
Frataxin, a highly conserved protein found in prokaryotes and eukaryotes, is required for efficient regulation of cellular iron homeostasis. Humans with a frataxin deficiency have the cardio- and neurodegenerative disorder Friedreich’s ataxia, commonly resulting from a GAA trinucleotide repeat expansion in the frataxin gene. While frataxin’s specific function remains a point of controversy, the general consensus is that the protein assists in controlling cellular iron homeostasis by directly binding iron. This review focuses on the structural and biochemical aspects of iron binding by the frataxin orthologs and outlines molecular attributes that may help explain the protein’s role in different cellular pathways.
Keywords: Friedreich’s ataxia, heme biosynthesis, iron-sulfur cluster assembly, iron chaperone, Yfh1, CyaY, ferrochelatase, ISU, Aconitase
INTRODUCTION
Frataxin, a conserved protein found ubiquitously in prokaryotes and eukaryotes, is required for the cellular regulation of iron homeostasis. Although of extreme interest to many chemists and biologists alike, frataxin’s exact role in helping cells regulate iron chemistry and metal availability remains controversial at best. Frataxin has been proposed to participate in at least five different capacities: 1) as an iron chaperone during cellular heme and iron-sulfur (Fe-S) cluster production; 2) as an iron-storage protein during conditions of iron overload; 3) as an aid in the repair of oxidatively damaged aconitase Fe-S clusters; 4) as a factor that controls cellular oxidative stress by moderating the concentration of reactive oxygen species (ROS); and finally 5) as an active participant in pathways involving energy conversion and oxidative phosphorylation. While these functions are not necessarily exclusive, it seems extraordinary that one protein could directly control so many biological pathways within cells. Disruption of frataxin production does cause a general loss in the cellular control of iron bioavailability and reactivity; however, phenotypes are complicated, a fact that further inhibits definition of the protein’s exact function. The goal of this manuscript is to take a structural approach to help readers understand frataxin’s role during the regulation of cellular iron homeostasis. However, before we begin to draw structural correlations regarding frataxin’s function, it is beneficial to first look in detail at the iron chemistry the protein helps control.
Iron is extremely versatile in the chemistry it can perform, so it is no surprise that the metal is found ubiquitously in all living systems. Iron is found in the active sites of proteins involved in many diverse cellular functions, ranging from oxygen transport, oxidative metabolism, and electron transport to energy production. Iron’s ability to cycle between the ferrous (Fe[II]) and ferric (Fe[III]) oxidation states allows the metal to be extremely useful when performing redox chemistry, and “this chemical versatility is surely a reason why nature selected this element in so many life processes” (Cotton et al., 1988). Unfortunately iron’s flexibility toward performing redox chemistry means that the metal, if left unregulated, can perform deleterious reactions that elevate ROS concentrations, and these reactive oxygen species can damage cells by oxidatively attacking cellular membranes, proteins, and DNA alike. Ferrous iron will readily react with dioxygen to produce superoxide and ultimately hydroxyl radicals, as described by the Fenton reaction (Aisen et al., 2001; Kosman, 2003):
| (1) |
| (2) |
| (3) |
It is therefore beneficial to control the oxidation of ferrous iron as a means to prevent the damaging effects of hydroxyl radical production. Under normal physiological conditions (pH > 2), ferric iron undergoes stepwise hydrolysis leading to the formation of insoluble neutral species:
| (4) |
| (5) |
| (6) |
Organisms have therefore evolved protein-controlled mechanisms to modulate iron’s redox chemistry. These mechanisms allow cells to adapt quickly to enhancement or suppression of iron import, delivery of Fe for utilization, and finally the storage of this important element for future use. In humans, iron abstracted from diet is reduced and acidified by the low-pH environment of the stomach, and a portion is eventually transported for use in the production of heme and Fe-S clusters. A small percentage of the metal is also stored in a manner that can be easily mobilized. More than two thirds of the iron found in the body is incorporated into hemoglobin in developing erythroid precursor cells or in mature red blood cells (Andrews, 1999), highlighting the importance of heme Fe-prosthetic groups as components that help control local O2 concentration, availability, and reactivity. Owing to their remarkable structural plasticity and versatile chemical/electronic features, Fe-S clusters are additional prosthetic groups that help control cell and organism viability by participating in electron transfer events, substrate binding and activation, iron and sulfur storage, the regulation of gene expression, and in some cases enzyme activity (Johnson et al., 2005). The balance of iron in biological systems, however, is fragile, as both iron deficiency and overload are destructive to cells. Diseases related to the disruption in cellular iron homeostasis are, as a result, among the most common diseases found in humans.
Friedreich’s ataxia (FRDA), an autosomal recessive cardio- and neurodegenerative disorder that affects 1 in 50,000 humans, results from an inability to produce frataxin, and causes protein deficiency that leads to a disruption of cellular iron homeostasis (Delatycki et al., 2000). The majority of patients with FRDA (~96%) have extensive trinucleotide repeat expansions in the first intron of the gene encoding frataxin, while a small percentage of patients have frataxin point mutations (Campuzano et al., 1996). The expanded GAA repeat adopts abnormal DNA structures that impair frataxin transcription; the longer the repeat, the more profound the frataxin deficiency, the earlier the onset of the disorder, and the greater the intensity of the disease (Bidichandani et al., 1998; Ohshima et al., 1998; Pandolfo, 2002). In humans, frataxin is a 210-amino-acid protein that is nuclear encoded but has an N-terminal mitochondrial targeting sequence that is removed during processing (Campuzano et al., 1996; Gibson et al., 1996). Results from immunocytofluo-rescence and immunocytoelectron microscopy show processed frataxin is predominately located within the mitochondria and associated with mitochondrial membranes, crests and as free soluble protein within the matrix (Gibson et al., 1996). Frataxin mRNA is predominantly expressed in tissues with a high metabolic rate, including the liver, kidney, neurons and heart (Koutnikova et al., 1997). Phenotypes coupled with frataxin deficiency include mitochondrial iron accumulation (Babcock et al., 1997; Foury et al., 1997), disruption in both heme and Fe-S cluster production (Rotig et al., 1997; Stehling et al., 2004) and a progressive breakdown in cellular iron homeostasis (Babcock et al., 1997; Koutnikova et al., 1997). Mitochondrial iron accumulation diverts metal away from other cellular compartments and frataxin deficient cells accommodate for what they perceive as a general cellular iron deficiency by elevating mitochondrial iron import (Babcock et al., 1997). Boosting pathways that lead to increased mitochondrial iron import facilitates the problems associated with not being able to process the reactive iron that is already present, leading to further elevation in ROS levels, gradual cell damage and finally the loss of cell viability (Delatycki et al., 2000). FRDA patients therefore show a slow progression of muscular and neurological symptoms (loss of motor function, progressive limb and gait ataxia, etc.) linked to the disruption in proper iron regulation and the disorder is typically fatal due to complications resulting from cardiomyopathy (Orth et al., 2001).
A direct correlation between frataxin and cellular iron homeostasis is at present obvious, but early studies on the yeast frataxin homolog (Yfh1) provided the initial insight into what role(s) frataxin may play in helping cells maintain iron homeostasis (Babcock et al., 1997). Deletion of the frataxin gene results in the accumulation of mitochondrial iron deposits (Babcock et al., 1997; Foury et al., 1997) coupled with both aconitase and general Fe-S cluster protein deficiencies (Foury, 1999; Rotig et al., 1997). The presence of zinc suppresses iron accumulation phenotypes in yeast (Knight et al., 1998). ΔYfh1 cells were shown to be hypersensitive to H2O2, iron and copper levels (Babcock et al., 1997; Foury et al., 1997). Based on the observation that reintroduction of Yfh1 expression under depleted conditions can promote the recovery and export of these mitochondrial iron deposits, frataxin was originally proposed to directly control mitochondrial iron efflux (Radisky et al., 1999). Yfh1 was also shown to indirectly regulate mitochondrial iron uptake by interacting and partially controlling the activity of the yeast mitochondrial intermediate peptidase (YMIP), a metalloprotease required for maturation of ferrochelatase and other iron utilizing proteins (Branda, Yang et al., 1999).
In eukaryotes, frataxin is proteolytically processed during the production of the mature soluble protein found within the mitochondrial matrix. During translation, a frataxin precursor is produced which contains a N-terminal targeting sequence that directs the protein to the mitochondria and eventually into the matrix (Babcock et al., 1997; Knight et al., 1998). Processing of the yeast frataxin precursor protein takes place uniquely in two steps (Branda et al., 1999; Gordon et al., 2001). Typically, proteins targeted to the mitochondria are processed in a single step (Branda et al., 1999; Cavadini et al., 2000; Gordon et al., 2001). There are only a few additional proteins that show this variation; examples include the Rieske (Fe-S) containing proteins and Neurospora crassa ATPase subunit 9 (Branda et al., 1999; Cavadini et al., 2000; Gordon et al., 2001). Even within this category, only frataxin and ATPase subunit 9 use a single enzyme for processing during both steps (Cavadini et al., 2000). In the first step, the mitochondrial processing peptidase removes the N-terminal 20 residues (domain I) of the protein, generating a Yfh1 intermediate. This intermediate is then processed again by the peptidase to remove the next 31 amino acids (domain II), generating the 123 amino acid mature Yfh1 found within the matrix. The two processing steps are independent. Domain I is a typical mitochondrial targeting signal and it can be substituted by other mitochondrial targeting signals with similar cellular results. Domain II serves as a spacer. The first 20 amino acids of frataxin contain the 5 basic residues typically found in mitochondrial targeting signals. The N-terminal region of the mature protein is rich in acidic residues. A spacer separates basic residues in the targeting signal from acidic residues in the mature protein’s N-terminus. This prevents unwanted interactions that might hinder recognition of the target signal by the import proteins. Both domains are required for import into the mitochondria; however, protein with mutations blocking the first or second processing steps will still complement the Yfh1 deletion phenotype (Gordon et al., 2001).
N-terminal processing of human frataxin (HsFtx) is still a matter of debate. Cavadini and colleagues (2000) proposed a two-step processing of the N-terminus, whereas Gordon and coworkers could identify only a single processing event (Gordon et al., 2001). In the two-step processing proposal, the first cleavage occurs between residues 41 and 42, removing the first 41 residues of HsFtx to generate an intermediate. The second cleavage step occurs between residues 55 and 56, removing only the next 14 N-terminal residues. The kinetics of the first processing step were shown to be similar between the different species, however the second cleavage reaction rate is species specific. The second processing step (i.e., removing the 14 residues) is the rate-limiting step during HsFtx processing (Cavadini, Adamec et al., 2000). In contrast, Gordon and coworkers (2001) detected only a single processing event generating the 18 kDa mature human frataxin and similar results were observed with rat or yeast mitochondrial peptidase.
STRUCTURAL INSIGHTS INTO FRATAXIN’S FUNCTION
A detailed understanding of frataxin’s structure provides some insight into how the protein may function within its different proposed roles. Specific questions relevant to the molecular structure of frataxin include: Is there a patch of residues that could accommodate iron binding if frataxin is acting as a metal chaperone? Is there a favorable interface that could promote contacts between frataxin and protein partners coupled with heme biosynthesis, Fe-S cluster assembly, and aconitase Fe-S cluster repair? Are there molecular elements on frataxin’s surface that might promote protein aggregation during an iron storage function? And finally, does frataxin support an active site that could assist in the stabilization of the redox chemistry performed by the bound iron? We will attempt to address each question by looking closely at the structures of the different frataxin orthologs.
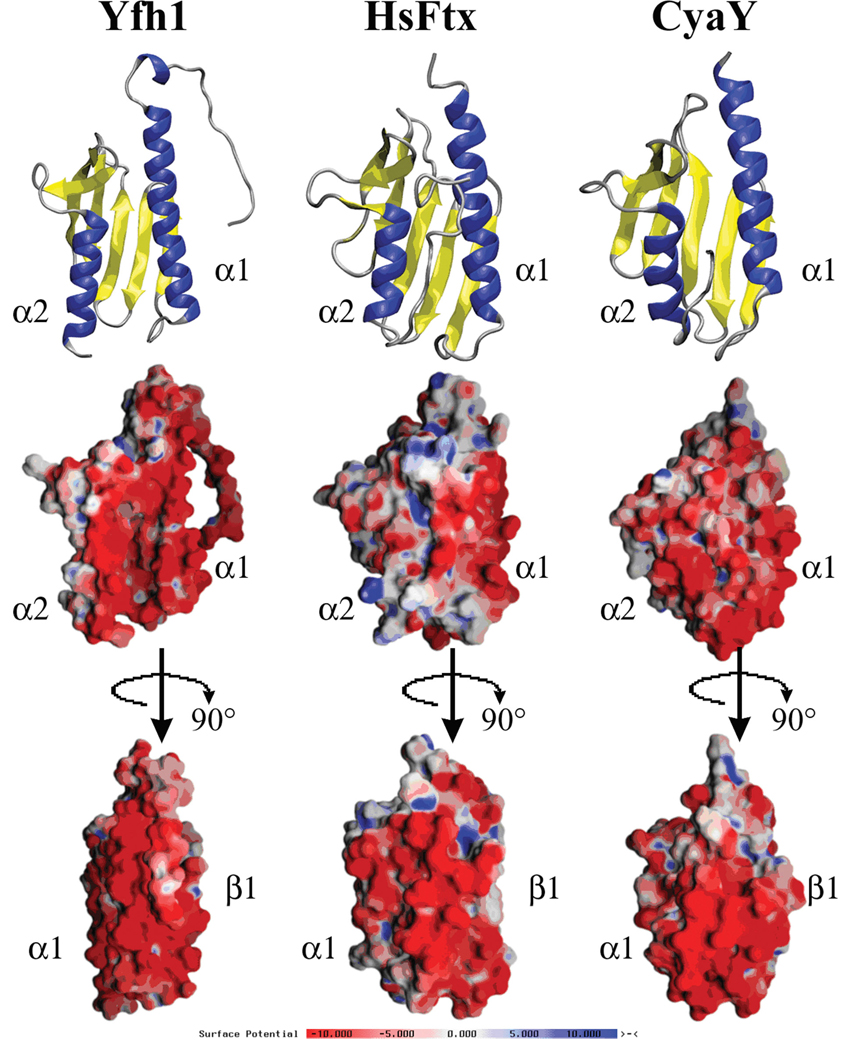
Solution and crystal structures have been reported for the yeast, human and bacterial (CyaY) frataxin orthologs (Cho et al., 2000; Dhe-Paganon et al., 2000; He et al., 2004; Lee et al., 2000; Musco et al., 2000; Nair et al., 2004). Frataxin has a unique fold that combines two terminal α-helices to make one plane, five antiparallel β-strands that construct the second plane of the protein and a sixth (and, in HsFtx, a seventh) β-strand that intersects the planes to give an overall planar α–β sandwich structure motif (Figure 1). The general shared frataxin topology is α1β1β2β3β4β5β6(β7)α2, strand seven is again only detected in HsFtx. In all three cases, the structures are extremely similar; RMSD’s of HsFtx versus CyaY and Yfh1 in regions of secondary structure are 1.34 Å and 0.65 Å, respectively. The overall structural dimensions of the orthologs are: 47 × 32 × 28 Å3 for HsFtx, 45 × 30 × 25 Å3 for CyaY and 47 × 29 × 23 Å3 for Yfh1. Mature HsFtx spans amino acids 75 to 210 of the total expressed sequence, however auto-degradation and proteolysis at the protein’s N-terminus prior to α1 residues have complicated structural studies of the full-length mature protein. Therefore, the structures solved of HsFtx begin at residue 88 in the crystal structure and at 90 in the solution structure (Dhe-Paganon et al., 2000; Musco et al., 2000). Circular dichroism data on full-length mature HsFtx indicate the N-terminal residues prior to residue 92 are predominately unstructured. Mature Yfh1 spans residues 52 to 174 of the expressed gene sequence. Unlike HsFtx, the 18 amino acids N-terminal to α1 in Yfh1 are stable and partially structured in a 310 helix observed between residues 65 and 68. Mature CyaY lacks any appreciable residues N-terminal to α1. In the case of HsFtx only, frataxin has a 17 amino acid C-terminal tail following α2. This C-terminal tail of HsFtx adopts a random coil structure that is tethered to the protein’s helical plane exposed surface.
Figure 1.
Top: ribbon diagram for yeast, human and bacterial frataxin. Middle: electropotential plots for proteins in same orientation. Bottom: electropotential plots for proteins rotated −90 degrees around the y-axis compared to top display. Structure figures made using solution structures of Yfh1 (PDB ID# 2GA5), HsFtx (PDB ID# 1LY7) and CyaY (PDB ID# 1SOY) frataxins.
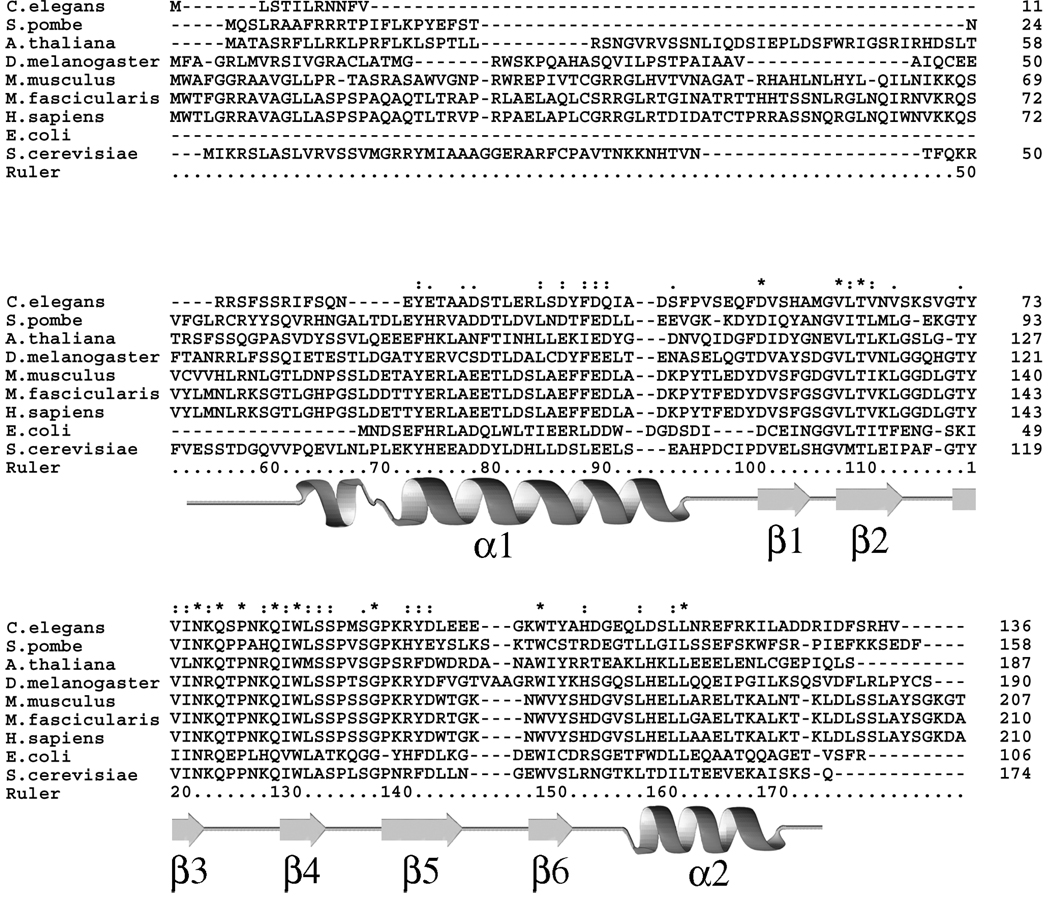
The strong structural similarity between frataxin orthologs results from the fact that these proteins share an extremely high degree of amino acid sequence similarity. Sequence identity of Yfh1 versus both CyaY and HsFtx are 28.1% and 37.8%, respectively, while respective sequence similarities are 59.8% and 65.0%. A large number of the highly conserved amino acids include a subset of Asp and Glu residues located in the N-terminal region of the protein. For Yfh1 this amounts to 9 of the first ~50 N-terminal residues in the mature protein constructed by conserved Asp and Glu residues (Figure 2). Conserved N-terminal acidic residues are located in the α1 and β1 secondary structural regions of frataxin. Electrostatic potential plots show these conserved acidic residues line the exposed surface of the α1 and β1 interfaces, generating a general negatively charged surface that covers roughly a quarter of frataxin’s total accessible surface (Dhe-Paganon et al., 2000). Carboxylate side chains from Asp and Glu residues often serve as ligands for bound metal in many iron-binding proteins, suggesting the possibility of these regions participating in metal binding. While HsFtx does not contain any cysteines, CyaY and Yfh1 contain poorly conserved cysteines at positions distant from the acidic patches.
Figure 2.
ClustalX alignment for a subset of characterized frataxin orthologs. Bottom three sequences represent structurally characterized frataxin orthologs. Secondary structural elements and ruler representing Yfh1 properties are given below the sequences.
Multiple factors have been shown to influence the stability of frataxin’s fold. The Pastore laboratory (MRC, London) performed extensive in vitro studies on the three characterized frataxins with the goal of defining factors that help stabilize each protein’s structure (Adinolfi et al., 2004; Adinolfi et al., 2002). Apo-bacterial, yeast and human frataxin are predominately stable as monomers at 1 mM protein concentrations, as confirmed by proton linewidths observed in 15N labeled protein 15N-HSQC specta (He et al., 2004; Mori, 1995; Musco et al., 1999; Musco et al., 2000; Nair et al., 2003; Nair et al., 2004). Apo-Yfh1 is however stable as a monomer for only up to 2 weeks after isolation, at which point it begins to self aggregate (Cook et al., 2006). Although CyaY, Yfh1 and HsFtx share a high degree of sequence homology and fold, the bacterial and human orthologs have melting points in the respective range of 54°C and 60°C (Adinolfi et al., 2004), while the yeast protein is less stable, with a melting point at 39°C (Adinolfi et al., 2004 and unpublished results from our laboratory). The type of buffer (organic vs. phosphate) has minimal effect on protein stability for these three frataxin orthologs, however the presence of salt generally increases the stability of frataxin’s fold (most pronounced in Yfh1). Lowering the pH raised the stability of Yfh1 but this had no effect on the helical fold of CyaY and HsFtx within the pH range of 6 to 8. The presence of iron at high metal to protein stoichiometries further increased the stability of all frataxin orthologs. Residues C-terminal to α2 in the protein’s structure appear to dramatically increase the general stability of frataxin. C-terminal CyaY truncation mutants matching the shorter Yfh1 construct showed a substantial (14°C) drop in the protein’s melting temperature. A C-terminal extension to Yfh1 caused a ~7°C increase in the protein’s melting temperature. Structural studies show the C-terminal extension in CyaY and HsFtx are unstructured but make surface contacts with α1 and α2 amino acids, most likely providing additional contacts that help stabilize the protein’s fold. Finally, the non-conserved N-terminal region of full-length HsFtx appears to be important for the partial apo-protein aggregation often found in the in vitro expressed/isolated protein (O’Neill et al., 2005).
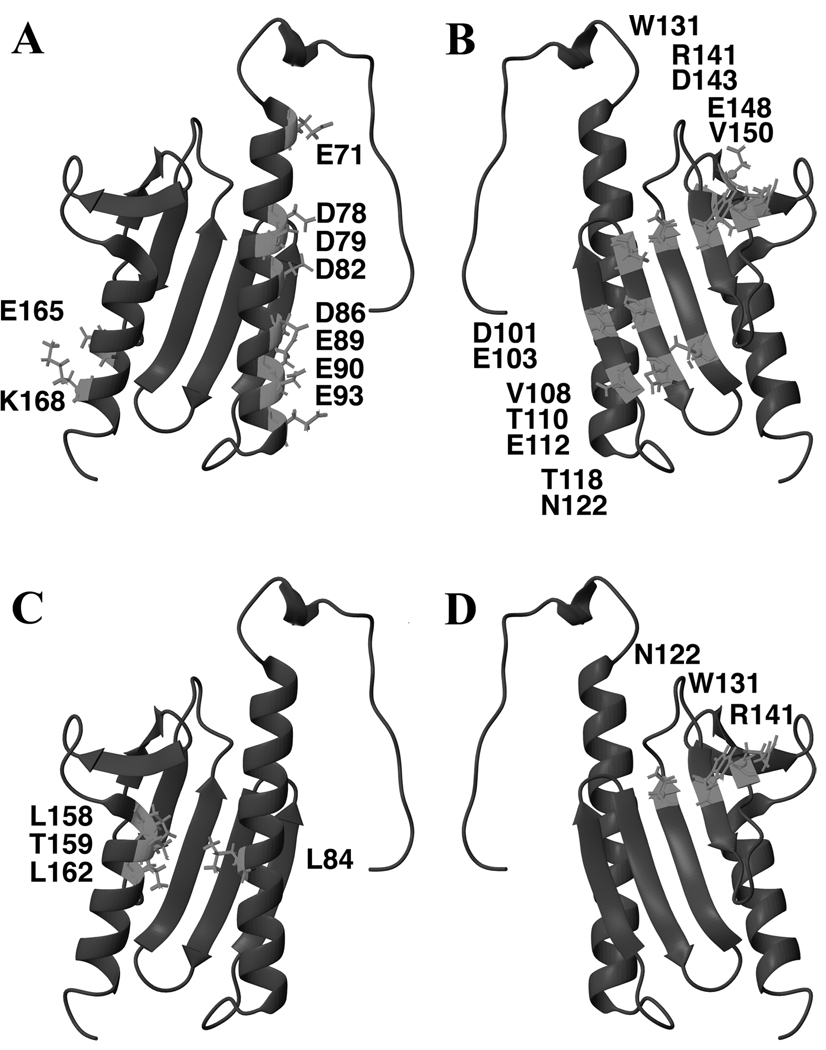
Close inspection of frataxin’s helical plane shows interesting structural attributes that may contribute to a better understanding of how the protein binds metal and also of mutations that cause FRDA (Delatycki et al., 2000). On Yfh1, α1 is lined with surface exposed acidic residues Glu71, Asp78, Asp79, Asp82, Asp86, Glu89, Glu90 and Glu93 (Figure 3A). The orientation and proximity of these conserved residues generates a conserved acidic patch on the protein in this region and these residues would be in a perfect position to contribute to cation binding. Frataxin helices α1 and α2 are parallel and oriented at an average adjacent backbone distance of ~11 Å, predominately due to interactions between the long side chains of hydrophobic and aromatic residues within this plane (in Yfh1, α1 residues in this region include Leu84, Leu88, and Leu91 and α2 residues include Leu162, Val166 and Ile170). Additional interactions between hydrophobic core amino acids on the β-sheet surface of the protein help secure the spatial orientation of α1 to α2. In case of the human frataxin, the C-terminal tail of HsFtx, including residues Thr196, Leu198, and Leu200, appear to further contribute to the stabilization of these inter-helical residue interactions (Cho et al., 2000; Dhe-Paganon et al., 2000; Lee et al., 2000; Musco et al., 2000; Nair et al., 2004). Surface exposed residues on the α2 surface of frataxin are less conserved; in Yfh1, Glu165, and Lys168 are fairly well conserved however the remaining surface residues are mixed. Yfh1 surface residues in the β6-loop (or in HsFtx β6-β7) region share highly conserved polar Arg153 and Asn154 residues. Disease-causing HsFtx helical plane point mutations are identified on the conserved Yfh1 sequence as α1 residue Leu84 mutation (to a Ser) and on α2, Leu158 mutation (to a His), Thr159 mutation (to an Arg) and Leu162 mutation (to an Arg) (Figure 3C) (Bartolo et al., 1998; Cossee et al., 1999; Filla, 1996; Zuhlke et al., 2004). Since these are all buried in the globular domain of the protein and play an important role in the packing of the two helices, mutating these residues most likely causes a disruption of elements that stabilize frataxin’s fold.
Figure 3.
Yfh1 residues that are highly conserved on the helical (A) and β-sheet (B) planes of the protein. Identity of HsFtx FRDA point mutations on the helical (C) and β-sheet (D) planes of Yfh1. Structure figures made using Yfh1 solution structure (PDB ID# 2GA5).
Numerous surface exposed residues on the β-sheet plane of frataxin are also highly conserved, and close inspection of this surface aids in the understanding of mutations that cause FRDA. Surface exposed residues on the Yfh1 β-sheet plane include Asp101, Glu103, Val108, Thr110, Glu112, Thr118, Asn122, Trp131, Arg141, Asp143, Glu148 and Val150, of which Asp101, Thr110, Asn122, Trp131, and Asp143 are fully conserved (Figure 3B). HsFtx amino acids, complementary in position to Yfh1 residues Asn122, Trp131 and Arg141, are surface exposed and mutated in a small set of FRDA patients (Figure 3D) (Forrest et al., 1998; Labuda et al., 1999; Zuhlke et al., 2004). Yfh1 β-sheet residues with side chains directed towards the hydrophobic core of frataxin include the conserved residues Ile121, Leu132 and Trp149. Along the β-sheet surface of HsFtx, Yfh1 residues at complementary positions of Ile130, Leu132 and Trp149 were found as point mutations in some FRDA patients, and mutating these residues would surely alter frataxin’s fold (Campuzano et al., 1996; Cossee et al., 1999). In addition, Gly107 in the loop region between Yfh1’s β1 and β2 is an additional point mutation found in a subset of FRDA patients (Bidichandani et al., 1997). Yfh1 residue Ile99 is the final HsFtx point mutation seen in a subset of FRDA patients, and this residue is located on the β-sheet plane of frataxin, connecting α1 to β1 (Cossee et al., 1999).
Although frataxin’s fold is fairly unique, an α–β sandwich motif structure is seen in a surprising number of proteins that share similar functions to those proposed for frataxin. The α–β sandwich motif can be easily recognized in the ferredoxin-like fold of some cytosolic copper chaperones. Although human HAH1 (Anastassopoulou et al., 2004), yeast ATX1 (Arnesano et al., 2001), and BsCopZ (Banci et al., 2003) (PDB ID’s: 1TL5, 1FES and 1P8G) are smaller in their overall size compared to frataxin (ca. 18Å × 22Å × 30 Å), and their helices are tilted at 45° to each other, the helices and beta strands still form a general two-layer structure that is similar to the α–β sandwich motif found in frataxin. The copper binding domains of these proteins involve a conserved CXXC motif in a loop region; this binding site is surrounded by hydrophobic residues that are essential for binding to partner proteins but distinct in position from the acidic patch on frataxin’s α1/β1 surface (i.e., the proposed metal binding region of frataxin). Recent crystallographic data on complex I of the Thermus thermophilus mitochondrial respiratory chain revealed a previously unknown protein Nqo15 that is also a member of this superfamily (Sazanov et al., 2006). Nqo15 binds in close contact with four other domains to help stabilize the larger complex. This protein is constructed by 129 amino acids, held together in an α–β sandwich motif fold. Despite a low sequence homology (average of 12%), the crystal structures superimpose well with RMSD values of 2.5 Å and 3.3 Å for CyaY and HsFtx, respectively. This domain binds to the complex through its β-sheet surface in a manner that creates a hydrophilic channel connecting the solvent with the N3 subdomain. The N-terminal β1 and loop connecting β1 and β2 contain several surface exposed histidine residues. Based on the fold similarity between frataxin and Nqo15, and a recent publication from the Cowan laboratory (Ohio State University) that proposed frataxin’s β-sheet plane may be an essential interface when interacting with ISU proteins during Fe-S cluster assembly (Yoon et al., 2003), the authors of the Nqo15 structure proposed that the conserved β-sheet plane histidine residues could serve as an iron binding site during iron storage and delivery by the protein when reconstitution of the Fe-S cluster in subdomain N3 is required.
FRATAXIN’S IRON BINDING ABILITY
Given the spatial arrangement of conserved acidic residues on the α1/β1 regions of frataxin (Figure 1), and the fact that Asp and Glu carboxylate side chains are chemically suited as ligands for binding metals, it is not surprising to learn that frataxin is an iron binding protein. However, the Isaya laboratory (Mayo Clinic) was the first to identify that frataxin directly binds iron; this iron binding ability was first determined in the yeast system (Adamec et al., 2000). The iron binding ability of additional frataxin orthologs has since been confirmed for both the human and bacterial proteins (Adinolfi et al., 2002; Bou-Abdallah et al., 2004; Cavadini et al., 2002; Cook et al., 2006; Yoon et al., 2003; Yoon et al., 2004). While it was initially recognized that iron binding in the yeast system induces frataxin to oligomerize under unique solution conditions, numerous subsequent reports suggest that oligomerization is not essential when the protein functions in an iron chaperone capacity (i.e., during heme and Fe-S cluster assembly), but it may be important when the protein participates in helping control ROS production under iron overloading conditions. In an attempt to deconvolute the published reports concerning the iron binding ability of frataxin in its different oligomeric states and to determine how these relate to the different proposed functions of frataxin, we will selectively discuss, in the following text, issues concerning: first, the iron binding ability of monomeric frataxin; second, frataxin’s ability to form iron loaded oligomers; and finally the protein’s ability to regulate the redox chemistry performed by iron bound to the protein. Each of these points is summarized below.
Frataxin Will Bind Iron as a Stable Protein Monomer
Metal to protein stoichiometries, measured for monomeric bacterial, yeast and N-terminally truncated human frataxin show these proteins will tightly bind 2, 2 and 7 Fe(II) atoms, respectively (Bou-Abdallah et al., 2004; Cook et al., 2006; Yoon et al., 2003). Iron dissociation constants, measured for CyaY, Yfh1 and N-terminally truncated HsFtx give average Kd values of 3.8, 3.0 and 55.0 µM, respectively (Bou-Abdallah et al., 2004; Cook et al., 2006; Yoon et al., 2003). The presence of magnesium or calcium salts, at physiologically relevant concentrations found within the mitochondria, stabilize CyaY and Yfh1 in the iron bound monomeric state against oligomerization, while a large portion of bacterially overexpressed HsFtx is stable as an iron-loaded monomer regardless of the solution conditions (Adinolfi et al., 2002; Cook et al., 2006). N-terminally truncated HsFtx was shown to bind ~6 Fe(III) atoms at a Kd of 10.2 µM (Yoon et al., 2003). In the presence of oxidants, monomeric CyaY can bind up to 6 Fe(III) atoms and will interact with up to 25 Fe(III) atoms/monomer (Bou-Abdallah et al., 2004). Ferric iron binding studies for Yfh1 are linked only to protein homooligomerization (Adamec et al., 2000).
NMR spectroscopy provided a powerful tool for identifying frataxin amino acids directly affected by the presence of iron. Backbone amide resonances for the three characterized frataxins are generally well dispersed, hence providing a fingerprint for the majority of protein amino acids (He et al., 2004; Musco et al., 1999; Nair et al., 2003). Subtle perturbations in the amide resonances and general signal intensity due to the addition of iron therefore serve as markers for residues affected by the presence of paramagnetic iron. Ferrous iron titrations performed anaerobically with the 15N-labeled frataxin orthologs show distinct alterations in α1 and β1 amino acid NMR resonances and, in the case of the CyaY and HsFtx, these perturbations are dependent on metal to protein stoichiometry. General trends in amide resonance perturbations upon addition of iron can be divided into two categories: amide resonances that were significantly line-broadened (often beyond detection), and amide chemical shifts that were shifted in the presence of metal. Bacterial amide resonances broadened upon addition of a single Fe(II) atom include α1 residues Arg20, Leu21, Asp22, and Asp23 (Table 1), while human resonances include α1 residues Asp112, Leu113, Asp115, and Val125 (Nair et al., 2004). Additional bacterial resonances broadened upon the addition of a second Fe(II) atom include α1 residues Ser4, Glu5, Phe6, His7, Glu19 and Trp24, and β1 residues Asp31 and Cys32; human resonances include α1 residues Asp104, Ala107, Glu108, Phe110, Ala114, Thr119, Phe120, Asp122, and Asp124. Addition of up to six Fe(II) atoms to CyaY and HsFtx caused a general broadening of amide resonances throughout the α1 and β1 regions. A number of additional CyaY and HsFtx resonances in the α1 and β1 regions underwent chemical shift perturbations upon the addition of ferrous metal. Yfh1 resonances broadened under similar buffer conditions by addition of up to two iron atoms include α1 residues Asp79, Asp82, His83, and Asp86, the β1 residue Glu103 and the β2 residue Glu112, while amide resonances for additional α1, β1 and β2 residues undergo chemical shift perturbations in the presence of iron (Cook et al., 2006). Ferric iron titrations were performed on CyaY with similar results to those obtained from ferrous iron binding. CyaY residues significantly broadened at a 1:1 Fe(III) to protein stoichiometry include α1 residues Arg20, Asp22 and Asp23. In the presence of a 2:1 ratio, additional broadened resonances include α1 residue Asp29, as well as β1 residues Ile30, Asp31, Cys32, Glu33, and Ile34. Again, addition of up to six iron atoms causes a general broadening of α1 and β1 resonances. Finally binding studies performed on CyaY with the non-paramagnetic Ca(II) ions showed general chemical shift perturbations in only the α1 and β1 regions (Nair et al., 2004). In summary, these NMR titrations paint a picture of frataxin’s metal binding residues located predominately in the conserved acidic α1 and β1 regions of frataxin (Table 1).
TABLE 1.
Comparison of the iron interaction with Yfh1, HsFtx and CyaY residues in α1 thru β2 secondary structural regions*.
| Yfh1 |
HsFtx |
CyaY |
Structure Consensus |
|||
|---|---|---|---|---|---|---|
| Sequence | Fe Effect | Sequence | Fe Effect | Sequence | Fe Effect | |
| Glu71 | Thr93 | Ser4 | br2 | |||
| Lys72 | Thr94 | Glu5 | br2 | |||
| Tyr73 | Tyr95 | Phe6 | br2 | α1 | ||
| His74 | Glu96 | His7 | br2 | α1 | ||
| Glu75 | sh2 | Arg97 | Arg8 | α1 | ||
| Glu76 | sh2 | Leu98 | Leu9 | α1 | ||
| Ala77 | Ala99 | Ala10 | α1 | |||
| Asp78 | sh2 | Glu100 | Asp11 | α1 | ||
| Asp79 | br2 | Glu101 | Gln12 | α1 | ||
| Tyr80 | Thr102 | Leu13 | α1 | |||
| Leu81 | Leu103 | Trp14 | α1 | |||
| Asp82 | br2 | Asp104 | br2 | Leu15 | α1 | |
| His83 | br2 | Ser105 | Thr16 | α1 | ||
| Leu84 | Leu106 | Ile17 | α1 | |||
| Leu85 | Ala107 | br2 | Glu18 | α1 | ||
| Asp86 | br2 | Glu108 | br2 | Glu19 | br2 | α1 |
| Ser87 | sh2 | Phe109 | Arg20 | br1 | α1 | |
| Leu88 | Phe110 | br2 | Leu21 | br1 | α1 | |
| Glu89 | sh2 | Glu111 | Asp22 | br1 | α1 | |
| Glu90 | sh2 | Asp112 | br1 | Asp23 | br1 | α1 |
| Leu91 | Leu113 | br1 | Trp24 | br2 | ||
| Ser92 | sh2 | Ala114 | br2 | Asp25 | ||
| Glu93 | sh2 | Asp115 | br1 | Gly26 | ||
| Ala94 | Lys116 | — | ||||
| — | Pro117 | — | ||||
| His95 | Tyr118 | — | ||||
| Pro96 | Thr119 | br2 | — | |||
| Asp97 | Phe120 | br2 | Asp27 | |||
| Cys98 | Glu121 | Ser28 | ||||
| Ile99 | Asp122 | br2 | Asp29 | |||
| Pro100 | Tyr123 | Ile30 | ||||
| Asp101 | Asp124 | br2 | Asp31 | br2 | β1 | |
| Val102 | Val125 | br1 | Cys32 | br2 | β1 | |
| Glu103 | br2 | Ser126 | Glu33 | β1 | ||
| Leu104 | Phe127 | Ile34 | β1 | |||
| Ser105 | sh2 | Gly128 | Asn35 | β1 | ||
| His106 | Ser129 | Gly36 | ||||
| Gly107 | Gly130 | Gly37 | ||||
| Val108 | Val131 | Val38 | β2 | |||
| Met109 | Leu132 | Leu39 | β2 | |||
| Thr110 | Thr133 | Thr40 | β2 | |||
| Leu111 | sh2 | Val134 | Ile41 | β2 | ||
| Glu112 | br2 | Lys135 | Thr42 | β2 | ||
| Ile1113 | Leu136 | Phe43 | β2 | |||
The table represents a stoichiometry of between 1 to 2 Fe(II) per protein monomer. br1 and br2 (broadened and shifted at 1:1 and 2:1 respective Fe(II) to protein ratios) and sh2 (shifted at a 2:1 Fe(II) to protein). The final column contains the ortholog consensus secondary structure. The dashed line represents gaps in sequence homology (see Figure 2 for clarification).
Structural studies directed at characterizing the metal-ligand coordination geometry and electronic properties of bound iron provide additional insight into the metal binding ability of the monomeric frataxin orthologs. X-ray absorption spectroscopy (XAS) provides a powerful tool for characterizing structural and electronic properties of metals bound to proteins in solution (Teo, 1986). This technique has been used to characterize iron bound to Yfh1 and HsFtx monomers. Iron bound to monomeric Yfh1 and HsFtx is stable in the ferrous state when samples are prepared anaerobically or in the presence of the reducing agent dithionite (Bencze et al., 2006; Cook et al., 2006). Analysis of the iron 1s→3d transitions in the x-ray absorption near edge structure (XANES) portion of the XAS spectra are consistent with ferrous iron existing in the high-spin state and coordinated in a highly centrosymmetric metal-ligand coordination geometry when bound to monomeric HsFtx and Yfh1 (Bencze et al., 2006; Cook et al., 2006). Structural analysis of the Fe(II)-ligand coordination geometry for monomeric Yfh1 and HsFtx from the extended x-ray absorption fine structure (EXAFS) portion of the XAS spectrum are consistent with iron bound in a highly symmetric six-coordinate ligand environment. Ligands coordinating Fe(II) are exclusively oxygen and/or nitrogen based, in agreement with α1 and β1 Asp, Glu and His residues identified in the multiple NMR titrations as residues that possibly interact with iron. Additional ligands to the protein bound metal surely also come from water or hydroxide ions. There is no evidence for any metal–metal interaction for the ferrous iron bound to monomeric frataxin, even when up to two metals are bound in both Yfh1 and HsFtx.
Frataxin Can Form Stable Iron Loaded Homooligomers
Frataxin’s ability to homooligomerize, forming aggregates with metal binding abilities similar to that of ferritin, suggests frataxin may act as an iron storage protein. Under elevated metal-to-protein stoichiometries, in the presence of oxygen and the absence of salt, Yfh1 will form a 48-multimeric homooligomer that can bind up to 50 iron atoms per protein monomer (Gakh et al., 2002). CyaY aggregates of a similar size and iron loading capacity will also form under low-salt, aerobic, and iron-overloaded solution conditions (Adinolfi et al., 2002). Assembly in both systems is predominately averted under high concentrations of other divalent metal ions or in the presence of physiological salt concentrations (Adamec et al., 2000; Adinolfi et al., 2002; Cook et al., 2006), indicating CyaY and Yfh1 assembly is iron specific and will proceed under low salt but molecular oxygen (O2) dependent conditions. Monomeric HsFtx does not self-assemble even at high iron concentrations, however a ~59-mer HsFtx homooligomer is isolated as part of the Escherchia coli overexpressed protein and this oligomer can bind approximately 10 iron atoms under aerobic and salt-free conditions (Cavadini et al., 2002). The bulk iron bound in Yfh1 and HsFtx assemblies is predominately in the high-spin Fe(III) state (Nichol et al., 2003).
Yfh1 homooligomers have a unique morphology that highly resemble those seen from the iron storage protein ferritin, suggesting iron storage may be an important function of frataxin (Adamec et al., 2000; Gakh et al., 2002). Unlike ferritin, which forms spherical aggregates in the absence of metal (Theil, 1973), Yfh1 will only self-assemble in the presence of Fe, O2, and low salt (Adinolfi et al., 2002; Park et al., 2002). Stoichiometric and higher Fe(II):protein ratios drive Yfh1 to progressively self-assemble into larger homooligomers beginning from a protein trimer repeating unit, directing next to a hexamer, then to a 12-mer, 24-mer and finally resulting in the 48-mer spherical protein aggregate that resembles ferritin (Adamec et al., 2000). The 48-mer Yfh1 aggregate stores bulk metal, of greater than 2000 iron atoms, predominately as ferrihydrite (Nichol et al., 2003). The ~59-mer HsFtx homooligomers, isolated from a portion of the protein overexpressed in E. coli, will store ~600 iron atoms as ferrihydrite in a rod-shaped polymeric protein structural morphology (Cavadini et al., 2002). Non-covalent protein subunit interactions, mediated by the non-conserved N-terminal region of human frataxin, were shown to be important for assembly (O’Neill, Gakh et al., 2005). Iron does not induce the assembly of the monomeric truncated portion of overexpressed HsFtx isolated from E. coli (Adinolfi et al., 2002). CyaY can also form aggregates in the presence of iron and like Yfh1, aggregation is closely controlled by the presence of salt (Adinolfi et al., 2002). Analytical ultracentrifugation studies show that CyaY tetramers can be formed when Fe(II) is added anaerobically; different protein aggregates are formed upon oxidation of the bound Fe(II) (Bou-Abdallah et al., 2004). These in vitro assembly results are intriguing as they suggest protein aggregation may be an important property of the protein, in that it may help regulate cellular iron homeostasis under iron overloaded cellular conditions.
Residues in the α1/β1 region are essential for controlling frataxin homooligomerization. Specific conserved acidic residues in the α1/β1 region of Yfh1 and CyaY are required for assembly. Mutating CyaY α1 residues Glu18, Glu19, and Asp22, along with β1 residue Glu33 abolishes the iron induced oligomerization behavior (Adinolfi et al., 2002). Mutating conserved Yfh1 acidic residues Asp86, Glu90 and Asp93 abolishes the iron induced oligomerization behavior of the protein, surprisingly with no real in vivo phenotypes under normal growth conditions (Aloria et al., 2004). Additional Yfh1 mutational studies indicate Asp93 is critical for protein self-assembly (Gakh et al., 2006). Unlike CyaY and Yfh1, a portion of the full-length mature HsFtx (residues 56 to 210) will self-assemble in the absence of metal; N-terminally truncated HsFtx lacking the initial 22 residues does not self assemble in the absence of iron (Cavadini et al., 2002; O’Neill, Gakh et al., 2005).
XAS results show iron bound to Yfh1 and HsFtx spherical aggregates is predominately stable in the ferric state when samples are prepared aerobically at high metal to protein ratios (Nichol et al., 2003). Analysis of the 1s→3d transitions in the XANES spectra indicate ferric iron is coordinated in a highly centrosymmetric metal-ligand structural environment. Structural analysis of ferric iron coordinated to Yfh1 and HsFtx aggregates are consistent with bulk metal being bound as ferrihydrite, a biomineral composed of ferric oxide/hydroxide octahedra. The ligands coordinating the metal come predominately from complexed mono- and bidentate oxygen ligands that hold the multinuclear Fe cluster together and not from protein based ligands. These structural studies were essential for the early identification of how frataxin oligomers store bulk metal, hence participating as an iron storage capacity, maintaining metal in an inert form to shield the cell against unwanted iron redox chemistry.
Functional correlations between frataxin oligomers and ferritin are obvious. Mitochondrial ferritin (MtF) is an additional iron storage protein found in eukaryotes. Mature MtF is a 22 kDa protein encoded by an intronless gene on 5q23.1 chromosome in humans. MtF shares a 79% sequence identity with H-ferritin and the proteins possess ferroxidase activity. Its expression is restricted to tissues with high number of mitochondria (i.e., the testis) unlike frataxin, which is ubiquitous in its expression. MtF expression is not correlated to tissues like liver, which are involved in iron storage. MtF expression is observed in iron-loaded erythroblasts from patients with sideroblastic anemia, but not in normal erythroblasts, suggesting it may be induced only under conditions of stress in some cells. Consistent with these data, reports show that MtF expression in normal cells results in cytoplasmic and mitochondrial iron deprivation and a decrease in enzymatic activity of Fe-S cluster containing enzymes like aconitase (Levi et al., 2004). Correlating these findings with what is known about iron storage role of frataxin, it is possible that frataxin may be the iron storage molecule under low-salt but normal iron level conditions whereas both frataxin and mitochondrial ferritin scavenge iron to protect the cell from oxidative damage under iron overloading conditions.
Frataxin Controls Iron’s Redox Chemistry
As previously outlined, iron is extremely reactive toward oxygen-based redox chemistry, so it is not surprising that a breakdown in cellular iron homeostasis resulting from a frataxin deficiency causes elevated oxidative stress to affected cells. Controlling the ability of iron to perform redox chemistry appears to be an additional function of frataxin, suggesting this protein may directly participate in controlling cellular oxidative stress by reducing ROS production. In vivo reports indicate frataxin deficiency leads to oxidative damage in humans (Emond et al., 2000; Schultz et al., 2000), mice (Ristow et al., 2003; Thierbach et al., 2005), yeast (Karthikeyan et al., 2003), and Caenorhabditis elegans (Vazquez-Manrique et al., 2006). Mitochondrial iron overload, resulting from a frataxin deficiency, leads to oxidative damage in mitochondrial and nuclear DNA as well as to Fe-S clusters in mitochondrial aconitase and other respiratory enzymes (Babcock et al., 1997; Cavadini et al., 2000; Foury, 1999; Karthikeyan et al., 2002). Interestingly, frataxin affords protection for DNA against iron-induced oxidative damage (Gakh et al., 2006; O’Neill et al., 2005), presumably by directly binding iron and modulating its oxidation chemistry (Bou-Abdallah et al., 2004).
In numerous biological systems, protein-controlled ferroxidase centers direct the controlled oxidation of ferrous iron. These centers are common among proteins within different metabolic pathways that involve iron. One of many examples of ferroxidase centers is found in the iron storage protein ferritin (Theil, 1987; Theil, 2003). Mammals contain two types of ferritin subunits, the heavy (H) chain and the light (L) chain, which differ in size. Ferritin binds ferrous iron via Glu and His residues on the H chain and metal is ultimately oxidized at the protein’s ferroxidase center to form an oxygen bridged diferric center. Metal is then moved from the ferroxidase center to the nucleation site for storage as inert ferric oxide cores.
Recent reports have shown that frataxin will also perform ferroxidase chemistry (Park et al., 2002). In Yfh1 at sub-stoichiometric metal to protein levels, coupled with ferrous iron oxidation, H2O2 is most likely generated through O2 consumption during the ferroxidase reaction. The presence of hydrogen peroxide is only minimally detected under these conditions, and therefore it has been proposed that H2O2 immediately reacts with protein to attenuate all Fenton chemistry. Ferroxidation is progressively overcome by slower auto-oxidation at stoichiometric and higher iron to protein concentrations in Yfh1, and metal oxidation leads to progressive assembly of higher order protein oligomers. Therefore, it has been suggested that frataxin’s ability to initially perform ferroxidase chemistry, retaining a portion of bound metal in a bioavailable form, and eventually storing iron in a less accessible form, allows this protein to serve both as an iron chaperone and an iron storage protein (Park et al., 2003). Formation of the ferroxidase center in yeast frataxin is correlated to α1 residues Asp79 and Asp82; yeast mutants targeting these residues have reduced ferroxidase activity and assemble at a slower rate than wild type protein (Gakh et al., 2006). A series of frataxin α1/β1 residues were implicated as participating in the iron mineralization chemistry observed under iron overloaded conditions, possibly by acting as iron binding amino acids in the yeast frataxin oligomers. Yfh1 residues α1 Asp86, Glu89, Glu90, and Glu93, and β1 residues Asp101 and Glu103 are generally important in promoting the iron binding and metal sequestering properties of frataxin oligomers (Gakh et al., 2006).
Ferroxidase centers were also detected in HsFtx and CyaY aggregates (O’Neill et al., 2005). The oligomeric form of iron loaded HsFtx showed distinct protection of DNA against oxidative damage in the presence of H2O2 (O’Neill et al., 2005). Iron oxidation studies on CyaY monomers showed no apparent ferroxidase activity in the presence of molecular oxygen; rather, O2 acted as a rather poor oxidant of Fe(II) bound to the protein (Bou-Abdallah et al., 2004). The oxidation of Fe(II) bound to CyaY was accelerated in the presence of H2O2 and as a result there was an increase in the protein’s ability to bind additional metals (Bou-Abdallah et al., 2004). Unpublished results from our laboratory for Yfh1 monomers showed similar results to those published for CyaY monomers, with O2-induced Fe(II) oxidation rates on the minutes time scale (50% conversion in ~10 minutes at 100 µm protein/iron concentrations). In these published studies, during the reduction of O2 to 2H2O, the presence of highly reactive H2O2 or hydroxyl radicals was minimal.
FRATAXIN’S ROLE IN CELLULAR HEME BIOSYNTHESIS
Cells often utilize heme prosthetic groups when helping control normal cellular pathways. Given that frataxin is required for the production of heme, it is not a far stretch to suggest this protein could directly participate in heme bioassembly. Heme, a ubiquitous iron-containing tetrapyrrole ring system, is involved in multiple aspects of cellular metabolism. The Fe-prosthetic groups are used by globins, cytochromes and several additional enzymes (Ponka, 1999) that play key roles in the sensing and/or utilization of molecular oxygen in all living organisms (Andrew et al., 1990; Padmanaban et al., 1989; Zhu et al., 1999). Heme centers help drive cellular energy generation during respiration by storing and transporting oxygen (Atamna, 2004). Heme groups can also participate in both catalytic and regulatory functions within cells (Zhu et al., 2002). This prosthetic group can function as an effector center that regulates several biological processes encompassing transcription, translation, protein translocation and erythroid differentiation (Padmanaban et al., 1989). They function as ligands for transcription factors within prokaryotes (Monson et al., 1992) and in yeast (Creusot et al., 1988; Fytlovich et al., 1993; Pfeifer et al., 1989; Zhang et al., 1998). In higher eukaryotes, heme helps control the activity of specific transcription factors (Ogawa et al., 2001; Sassa et al., 1996) and proteins in several biochemical pathways by binding to the short protein sequence labeled the “heme regulatory motif” (Zhang et al., 1995). Heme regulation is essential during erythroid differentiation. In mammalian erythroid cells, heme initiates changes in key factors controlling numerous activities ranging from cell cycle and Ras signaling to chromatin structure, splicing, and protein folding (Zhu et al., 1999). Thus the multifunctional roles of heme suggests fluctuations in its concentration help control several key aspects of cellular metabolism (Atamna, 2004). In order to better understand the role frataxin plays in heme bioassembly, we present a brief description of the assembly pathway as it applies to our protein.
The heme biosynthetic pathway was outlined in detail during the 1950’s and 60’s (Labbe et al., 1999). Cellular heme biosynthesis occurs in eight sequential steps, four of which (step 1 and steps 6 to 8) occur within the mitochondria, while the other steps occur within the cytoplasm (Tait, 1978). The enzyme ferrochelatase catalyses the final step in the heme biosynthetic pathway. Ferrous iron is inserted into porphyrin by ferrochelatase to produce a functional heme prosthetic group (Taketani, 2005). Ferrochelatase is associated with the inner mitochondrial membrane in eukaryotes; in prokaryotes, ferrochelatase is found within the cytoplasm (Bacillus subtillus) or associated to the cytoplasmic membrane (Dailey et al., 2000). To this date, structures of apo- and metal-loaded bacterial (Bacillus subtilis), yeast (Saccharomyces cerevisiae), and human ferrochelatases have been solved (Al-Karadaghi et al., 1997; Karlberg et al., 2002; Wu et al., 2001). While the functional bacterial protein is monomeric, human and yeast ferrochelatase are homodimers (Karlberg et al., 2002). Mammalian, yeast (Schizosaccharomyces pombe) and some bacterial ferrochelatases are metalloenzymes that contain [2Fe-2S] prosthetic groups, which are required for activity (Ferreira, 1999). All three orthologs have a conserved overall fold of two Rossmann-type domains with a four-stranded parallel β-sheet that is flanked by α-helices. These domains contribute to the formation of the porphyrin-binding cleft. The differences between the eukaryotic and prokaryotic structures are mostly limited to the region in the eukaryotic ferrochelatase suggested to be involved in interactions with membranes and to the C-terminus, which contains the Fe-S cluster in the human but not the S. cerevisiae and B. subtilis enzymes (Karlberg et al., 2002). The localization of ferrochelatase to the matrix side of the inner mitochondrial membrane ensures uptake of the poorly soluble porphyrin and heme release (Ferreira, 1999; Taketani, 2005). The hydrophobic exterior of ferrochelatase is in stark contrast to the hydrophilic interior of the active sites that are lined with conserved charged residues well positioned to receive the positively charged iron. The porphyrin-binding cleft of ferrochelatase exhibits a high degree of conservation from prokaryotes to higher eukaryotes (Al-Karadaghi et al., 2006).
Ferrochelatase can chelate various divalent metal ions besides Fe2+, most prominent being Zn2+ (Dailey, 2003). A general mechanism for the metallation of the tetrapyrrole, proposed in 1974, suggested that deformation of the porphyrin ring after an outer-sphere complex formation between the metal ion and porphyrin would generate an appropriate configuration for metal insertion (Hambright et al., 1974). More recent studies have looked into mechanisms that endow ferrochelatase assisted ferrous ion metallation of porphyrin. Quantum and molecular mechanical calculations showed ferrochelatases induce a thermodynamically favorable distortion of free base protoporphyrin IX (Sigfridsson et al., 2003). Al-Karadaghi and colleagues (2006) recently proposed that the degree of distortion imposed by ferrochelatase modulates the type of metal ion that is inserted, and this in turn is determined by subtle structural changes in the highly conserved active sites in the porphyrin binding cleft of ferrochelatase (Al-Karadaghi et al., 2006).
Ferrochelatase requires ferrous iron as a substrate to complete heme assembly (Taketani, 2005) and the cellular mechanism that makes ferrous iron available, while also limiting the toxic effects of the metal, most likely involves the presence of a ferrous iron chaperone. Recent reports indicate that frataxin is the elusive iron chaperone that delivers the ferrous iron to ferrochelatase, hence promoting heme biosynthesis in a direct manner. In frataxin deleted yeast strains, Foury and colleagues (1997) observed a decrease in cytochrome c oxidase activity. Later Dancis and coworkers, working with frataxin null yeast strains, observed a severe deficiency of cytochromes b, c, and (a+a3) (Lesuisse et al., 2003). Using a yeast strain with a single copy of Yfh1, placed under the control of a regulatable promoter to prevent rho minus conversion and secondary nuclear mutations that might mask the defect, the Dancis laboratory (University of Pennsylvania, Philadelphia) observed the recovery of cytochromes in general and cytochrome c in particular upon re-induction of frataxin expression (Lesuisse et al., 2003). Although frataxin deleted cells have reduced levels of ferrochelatase, it was shown that cellular heme production is modulated by the insertion of zinc instead of iron into the porphyrin ring (Lesuisse et al., 2003). Furthermore, increasing ferrochelatase levels using a multicopy plasmid did not correct the heme synthesis defect (Lesuisse et al., 2003). These experiments proved that mitochondrial iron located in yeast frataxin deficient cells is not available for heme synthesis. Surface plasmon resonance studies showed, in the absence of iron, that recombinant Yfh1 interacts with yeast ferrochelatase with a high affinity (Kd = 40 nM) (Lesuisse et al., 2003). Working toward obtaining a global result of frataxin deficiency, Schoenfeld and colleagues (2005) found that frataxin deficiency leads to the down regulation of mitochondrial transcripts and a kinetic inhibition of the heme pathway. An additional significant observation from their studies was a large increase in zinc chelatase activity of ferrochelatase (Schoenfeld et al., 2005), consistent with earlier studies by Lesuisse and coworkers (2003). This growing evidence implicates frataxin in determining the specificity of metal for the ferrochelatase by directly binding to the protein partner.
The presence of frataxin in vitro has been shown to stimulate heme development under controlled conditions. In vitro ferrochelatase activity assays, in the presence of citrate, showed that monomeric human frataxin can deliver the ferrous iron required for heme synthesis; an optimal ferrochelatase activity was observed at a stoichiometric ratio of one frataxin monomer per ferrochelatase dimer (Yoon et al., 2004). The Cowan laboratory, using isothermal titration calorimetry and fluorescence quenching experiments, were able to quantitatively investigate complex formation between human frataxin and ferrochelatase. The Kd for the interaction between HsFtx and human ferrochelatase in the presence of iron was 17 nM; no interaction was detected for the human proteins in the absence of iron. In the yeast system, the Isaya laboratory showed ferrous iron associated with Yfh1 oligomers was available to yeast ferrochelatase for stimulation of in vitro heme synthesis (Park et al., 2003). Metal transfer was shown to occur in the presence of Fe(II) specific chelators, suggesting again a strong intermolecular interaction is formed at the ferrous iron site in the frataxin-ferrochelatase protein complex (Park et al., 2003).
Spectroscopic analysis of the yeast frataxin/ferrochelatase complex has been useful in helping to identify where and how the proteins interact. Our laboratory has shown that solution titrations, adding unlabeled yeast ferrochelatase into 15N-labeled Yfh1, allowed us to probe by NMR which frataxin amino acids underwent amide chemical shift perturbations as a result of complex formation (He et al., 2004). Frataxin chemical shift perturbations were localized predominately on the helical plane of the protein. Specific Yfh1 residues affected by ferrochelatase binding included surface exposed α1 residues His83, Glu89, Glu90, Glu93 and His95, in the metal binding region of the protein (of which all Glu’s are conserved), and β6-loop-α2 residues Val150, Asn154, Thr159, Asp160, Thr163, Glu164, Ser171, and Lys172 (of which Val150, Asn154, Thr163, and Glu164 are conserved in their physical properties). Yfh1 residues in the β6-loop region provide a hydrophobic patch on the protein and thus it is tempting to speculate that this domain might help target and facilitate the binding of frataxin to the hydrophobic exterior of ferrochelatase (He et al., 2004). Complementary experiments were also performed on the human proteins by our laboratory in collaboration with the Cowan laboratory, with similar results identifying the helical residues perturbed upon complex formation (Bencze et al., 2006). These results suggest that the helical plane of frataxin, including the α1 iron-binding residues, is responsible for generating a favorable interface when frataxin binds to ferrochelatase.
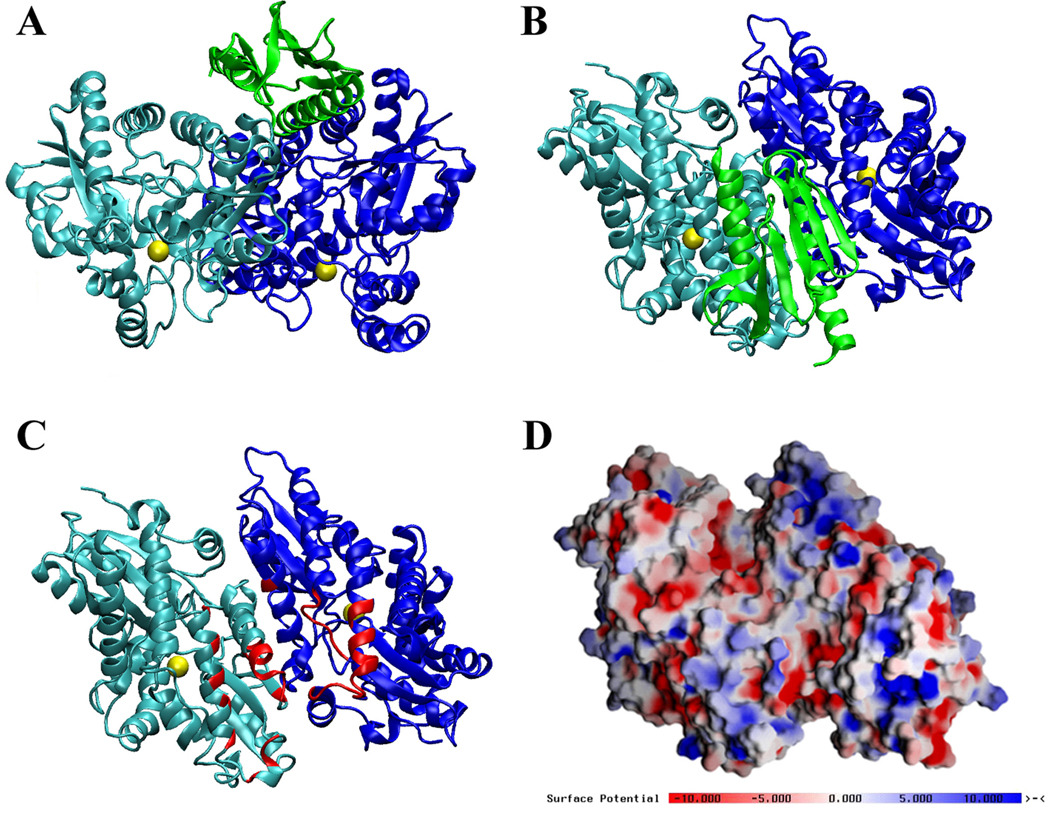
Recently we performed modeling studies to predict how frataxin may interact with ferrochelatase. Using the Yfh1 solution structure (He et al., 2004) and the Co2+ bound yeast ferrochelatase crystal structure (Karlberg et al., 2002), we were able to generate a plausible model for the interaction between these two proteins (Figure 4) in an orientation that may suggest how iron is delivered to ferrochelatase during heme biosynthesis. We generated a large collection of complexes using the automatic docking program ZDOCK; however, we applied strict inclusion criteria that match published biophysical data when selecting appropriate docking interactions. The selection criteria utilized were: a) Yfh1 residues identified in our NMR titration as perturbed by the presence of ferrochelatase should be located at the interface, b) the location of the metal binding sites from both the human and yeast metal loaded ferrochelatase structures should face each other, and c) excluding the region of ferrochelatase that docks to the membrane (Wu et al., 2001). In our docking model, the Yfh1 monomer interacts with the yeast ferrochelatase dimer through highly conserved frataxin helices α1/α2 and the highly conserved ferrochelatase metal docking site including β4-loop-α6, α8 and α11 amino acids. Human ferrochelatase binds cobalt at this docking site using residues His231 and Asp383, located opposite to the membrane binding loops near a groove lined by arginines and lysines (Wu et al., 2001). These Lys and Arg residues could provide complementary electrostatic interactions to the acidic helices found in frataxin (Figure 1). In our docking model, the Yfh1 iron-binding α1 unit docks in close proximity to the ferrochelatase cobalt binding site found in the human ortholog; in yeast, two glutamate side chains are at this site and these could function during iron binding by the ferrochelatase. Iron bound to ferrochelatase would then be free to translocate towards the heme assembly active site (i.e., the position of bound Co2+ in the yeast ferrochelatase structure), as has been proposed by Wu and colleagues (2001). These data are also consistent with the published ratio of 1:1 frataxin monomer/ferrochelatase dimer binding interactions seen in both the yeast and human systems (Lesuisse et al., 2003; Yoon et al., 2004). A recent report postulated protoporphyrin might help modulate the frataxin-ferrochelatase interaction, with porphyrin stabilizing the interaction (Al-Karadaghi et al., 2006). Porphyrin was, however, not added to our docking simulation. Given that the entry site for porphyrin is at the ferrochelatase surface opposite to the frataxin binding site, one would expect no direct effect of porphyrin binding on frataxin-ferrochelatase affinity. However, the lowest energy normal mode explored by free ferrochelatase, and the mode most likely to be physiologically relevant, shows that opening the porphyrin binding site partially closes the frataxin binding site. These results suggest a possible indirect effect on frataxin binding by ferrochelatase and vice versa. Current studies are underway in our laboratory to determine if iron or porphyrin alters the binding affinity between the wild type yeast frataxin and ferrochelatase proteins, and if mutating surface contact residues between the partners affects complex formation and metal transfer.
Figure 4.
Lowest energy simulation of Yfh1 monomer docked to the metal loaded yeast ferrochelatase. (A)Side view of single Yfh1 monomer (green) docked to yeast ferrochelatase dimer (dark and light blue). Co2+ is bound in the yeast ferrochelatase structure in the assembly active site close to the four membrane attachment lips at the bottom of the figure. (B) Side view (90°—Horizontal rotation of Figure 4A) showing monomeric Yfh1 interacts with both units in the ferrochelatase dimer. (C) Ferrochelatase side view with Yfh1 structure removed to show the residues that directly interact with frataxin (in red). (D) Electrostatic potential plots (calculated and vendered with Grasp) of ferrochelatase, side view. Figures A, B, and C prepared using VMD (Humphry et al., 1996). Docking simulations were performed using ZDock (Chen et al., 2003) using the Yfh1 structure simulations (PDB ID# 2GA5) and Co2+ loaded yeast ferrochelatase structure (PDB ID# 1L8X).
FRATAXIN AND Fe-S CLUSTER ASSEMBLY
Mounting evidence implicates frataxin as having a direct role in Fe-S cluster biosynthesis. Fe-S clusters are among the most complex and ancient prosthetic groups found in biology. They play essential roles in cellular processes ranging from electron transport, catalysis, gene regulation, and iron uptake (Chen et al., 2004). As an illustration of their importance, there are three separate pathways in bacteria for the production of Fe-S clusters. The first pathway identified involves the nitrogen fixation machinery that assembles the Fe-S clusters required for the maturation of nitrogenase (Frazzon et al., 2002; Rees et al., 2000). For most cellular needs, Fe-S clusters are provided by the second pathway, involving the ISC-assembly machinery (Kispal et al., 1999; Zheng et al., 1998). In some bacteria, a minor contributor under stressed conditions is the third pathway, the sulfur mobilization (SUF) machinery (Takahashi et al., 2002). Many bacteria contain more than one of these pathways, and interestingly they appear to be somewhat interchangeable (Ali et al., 2004; Takahashi et al., 2002). The mitochondria of eukaryotes however utilize only ISC orthologs for Fe-S cluster assembly and numerous reports outline the different characteristics of this highly important pathway (Johnson et al., 2005; Lill et al., 2005; Mansy et al., 2004). A brief outline of the proteins within the yeast ISC-assembly machinery is given below as a basis for introducing frataxin’s role in Fe-S cluster assembly.
Within the ISC-assembly pathway, yeast Fe-S cluster biosynthesis begins with the liberation of sulfur by the cysteine desulfurase, Nfs1 (Kispal et al., 1999). A disulfide bond is formed between Nfs1 and free mitochondrial cysteine, leading to bond cleavage and alanine release (Lill et al., 2005). Arh1, a ferredoxin reductase, and Yah1, a ferredoxin, are both required for efficient ISC assembly (Muhlenhoff et al., 2003), and it has been proposed they provide the electrons necessary for sulfur liberation (Lill et al., 2005). Next, sulfur is transferred to the cluster assembly scaffold proteins, a ~28 kDa protein dimer constructed of Isu1 (and/or) 2, usually via heterotetrameric complexes of the scaffold and the desulfurase (Garland et al., 1999; Gerber et al., 2003; Lill et al., 2005; Schilke, 1999). The recently identified protein Isd11 is required to both stabilize and act as an adaptor between Nfs1, and the scaffold proteins, and this helps promote sulfur release (Adam et al., 2006; Muhlenhoff et al., 2003; Wiedemann et al., 2006).
A general property of the scaffold proteins is that they are dynamic in nature, hence providing a flexible platform on which to construct Fe-S clusters while retaining the ability to release the prosthetic group for transfer to future acceptor proteins (Adinolfi et al., 2004; Bertini et al., 2003; Mansy et al., 2004). While the structure of the yeast ortholog has not yet been solved, structures for Isu orthologs are consistent with having a β-sheet surface connected to a globular helical protein arrangement (Bertini et al., 2003; Liu et al., 2005; Ramelot et al., 2004). The structure of Zn2+ bound to the Streptococcus pyogenes and Haemophilus influenzae orthologs shows that divalent metal is ligated by conserved cysteine residues in a surface exposed region of the protein monomer (Liu et al., 2005; Ramelot et al., 2004).
Frataxin is required for the in vivo production of Fe-S clusters and is believed to play a direct role in their assembly. A Fe-S cluster deficiency was identified in FRDA patients early in the recognition of the disorder, suggesting the need for frataxin in Fe-S cluster assembly (Rotig et al., 1997). This idea was supported by knockout mice showing similar disease phenotypes (Puccio et al., 2001) and more recently in the bacterial system (VivasC et al., 2006). The majority of the direct frataxin in vivo Fe-S cluster correlations come again however from studies in the yeast system (Chen et al., 2002; Duby et al., 2002; Muhlenhoff et al., 2002). Suppressing Yfh1 expression results in respiratory deficiency, mitochondrial iron accumulation and reduced Fe-S enzyme activity while non Fe-S containing enzymes remained active (Chen et al., 2002). Additional studies suggest that frataxin may not be essential for Fe-S cluster assembly, but it does improve the efficiency of the assembly process (Duby et al., 2002). A direct interaction between frataxin and the assembly apparatus proteins has been detected (Gerber et al., 2003; Muhlenhoff et al., 2003; Ramazzotti et al., 2004), and the requirement of frataxin for the maturation of Fe-S clusters in yeast has been confirmed (Stehling et al., 2004). Taken together, these data suggest frataxin plays a direct role in the cellular assembly of Fe-S clusters.
Recent in vitro studies show that frataxin binds to the ISU scaffold protein with high affinity and stimulates the production of Fe-S clusters, indicating that frataxin may be acting as the iron chaperone that delivers the Fe(II) required for bioassembly. Binding between in vitro human frataxin and the ISU dimer has been reported to occur with an affinity in the nanomolar range (Yoon et al., 2003). Binding of monomeric HsFtx with the human ISU dimer was dependent on the presence of iron, in close correlation with yeast studies showing that the interaction between Yfh1 and Isu1/Nfs1 is also iron dependent (Gerber et al., 2003). In vitro activity assays for the yeast and human protein systems show that the presence of frataxin stimulates Fe-S cluster assembly (Muhlenhoff, Richhardt, Gerber et al., 2002; Yoon et al., 2003). Based on the fact that frataxin is an iron-binding protein, that it binds tightly to the ISU complex and finally that it stimulates Fe-S cluster assembly, it seems highly probable that frataxin can act as the iron chaperone during Fe-S cluster bioassembly.
FRATAXIN ROLE IN ADDITIONAL PATHWAYS
Numerous reports have linked frataxin deficiency with an enhanced reduction in aconitase activity (Chen et al., 2002; Foury, 1999; Rotig et al., 1997). Additional reports indicate frataxin may also deliver Fe(II) to aconitase for the use in repairing oxidatively damaged [4Fe-4S]2+ clusters converted to the inactive [3Fe-4S]+ form (Bulteau et al., 2004). Damage to aconitase, a Krebs-cycle enzyme that converts citrate to isocitrate (Beinert et al., 1996), has shown to be a marker for cellular oxidative damage (Bulteau et al., 2003). Reduced aconitase activity leads to increased transcription of the iron-import mechanism and this leads to further mitochondrial iron overload in FRDA patients (Chen et al., 2004). Frataxin interacts in a citrate dependent manner with oxidatively damaged aconitase, promoting aconitase enzyme reactivation suggesting frataxin delivers the Fe(II) required to reactivate the damaged iron-deficient Fe-S center (Bulteau et al., 2004). Furthermore, aconitase appears to directly associate with frataxin under high concentrations of ROS, possibly as a means to further protect aconitase against [4Fe-4S]2+ cluster disassembly, irreversible inactivation, and potential degradation (Bulteau et al., 2005). ΔYfh1 yeast cells have low manganese superoxide-dismutase activity that can be recovered by supplementing Mn or limiting Fe in the media, suggesting a correlation between cellular iron overload and reduced SOD activity as a cause of oxidative stress in FRDA cells (Irazusta et al., 2006). Finally, the overexpression of frataxin in human cell lines leads to increased mitochondrial oxidative metabolism, as shown by an increased aconitase activity, mitochondrial membrane potential, cellular respiration, and ATP content (Schulz et al., 2006). These data suggest that frataxin provides a direct line of defense against cellular oxidative stress by controlling aconitase activity and iron import, as well as a possible additional indirect correlation between the partial regulation of the enzymatic activity of other proteins that control ROS activity.
OPEN QUESTIONS REGARDING FRATAXIN
Frataxin has been shown to play an important role in regulating cellular iron homeostasis, although the exact function(s) of the protein continues to remain controversial. While numerous groups have implicated frataxin as a protein that directs the utility of bioavailable iron, many key questions remain unanswered. If frataxin is an iron chaperone, what drives this chaperone to deliver iron specifically to multiple different protein partners (i.e., ferrochelatase, ISU scaffold or aconitase) when members of the copper chaperone family are highly specific with regard to their protein partners (Rosenzweig et al., 2000)? Certainly, additional structural data of frataxin in complex with each protein partner will help provide a better understanding of how and why these proteins interact within the different pathways. How can frataxin participate in two counterproductive roles, providing bioavailable and labile Fe(II,) when acting as an iron chaperone, while at other times retaining iron as inert Fe(III) when acting in an iron storage capacity, to help prevent toxicity? Structural data regarding iron-loaded frataxin oligomers will surely help address this issue of how and why these assemblies form, and identification of conditions when these assemblies can be observed in vivo will provide additional insight into their physiological relevance. Finally, what is the dominant function of frataxin? Recently frataxin has been shown to interact with the succinate dehydrogenase complex subunits, implying an additional possible role for frataxin in mitochondrial electron transport (Gonzalez-Cabo et al., 2005). Frataxin has also been shown to be a key activator of mitochondrial energy conversion and oxidative phosphorylation, suggesting yet another role for the protein in energy production (Ristow et al., 2000). Finally, CyaY has recently been shown to interact with the cysteine desulfurase IscS to promote both ferric iron reduction and Fe-S cluster assembly, suggesting iron delivery by frataxin may only occur when coupled with iron reduction (Layer et al., 2006). In vivo mutational studies that split the individual phenotypes for each pathway will provide biological relevance to help understand frataxin’s role in each of the many functions proposed for this important protein. Addressing these and other equally important issues regarding how frataxin functions will surely help in developing improved treatment strategies to control iron regulation in disorders like Friedreich’s ataxia.
ACKNOWLEDGMENTS
T. L. Stemmler acknowledges financial support from the American Heart Association (0130527Z) and the National Institutes of Health (DK068139).
Contributor Information
Krisztina Z. Bencze, Department of Biochemistry and Molecular Biology, Wayne State University, School of Medicine, Detroit, Michigan, USA
Kalyan C. Kondapalli, Department of Biochemistry and Molecular Biology, Wayne State University, School of Medicine, Detroit, Michigan, USA
Jeremy D. Cook, Department of Biochemistry and Molecular Biology, Wayne State University, School of Medicine, Detroit, Michigan, USA
Stephen McMahon, Department of Biochemistry and Molecular Biology, Wayne State University, School of Medicine, Detroit, Michigan, USA.
César Millán-Pacheco, Facultad de Ciencias, Universidad Autonoma del Estado de Morelos, Cuernavaca, Morelos, Mexico.
Nina Pastor, Facultad de Ciencias, Universidad Autonoma del Estado de Morelos, Cuernavaca, Morelos, Mexico.
Timothy L. Stemmler, Department of Biochemistry and Molecular Biology, Wayne State University, School of Medicine, Detroit, Michigan, USA
REFERENCES
- Adam AC, Bornhovd C, Prokisch H, Neupert W, Hell K. The Nfs1 interacting protein Isd11 has an essential role in Fe/S cluster biogenesis in mitochondria. EMBO J. 2006;25:174. doi: 10.1038/sj.emboj.7600905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamec J, Rusnak F, Owen WG, Naylor S, Benson LM, Gacy AM, Isaya G. Iron-dependent self-assembly of recombinant yeast frataxin: implications for Friedreich ataxia. Am J Hum Genet. 2000;67:549. doi: 10.1086/303056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adinolfi S, Trifuoggi M, Politou AS, Martin S, Pastore A. A structural approach to understanding the iron-binding properties of phylogenetically different frataxins. Hum. Mol. Genet. 2002;11:1865. doi: 10.1093/hmg/11.16.1865. [DOI] [PubMed] [Google Scholar]
- Adinolfi S, Nair M, Politou A, Bayer E, Martin S, Temussi P, Pastore A. The factors governing the thermal stability of frataxin orthologues: how to increase a protein’s stability. Biochemistry. 2004;43:6511. doi: 10.1021/bi036049+. [DOI] [PubMed] [Google Scholar]
- Adinolfi S, Rizzo F, Masino L, Nair M, Martin SR, Pastore A, Temussi PA. Bacterial IscU is a well folded and functional single domain protein. Eur J Biochem. 2004;271:2093. doi: 10.1111/j.1432-1033.2004.04112.x. [DOI] [PubMed] [Google Scholar]
- Aisen P, Enns C, Wessling-Resnick M. Chemistry and biology of eukaryotic iron metabolism. Int J Biochem Cell Biol. 2001;33:940. doi: 10.1016/s1357-2725(01)00063-2. [DOI] [PubMed] [Google Scholar]
- Al-Karadaghi S, Hansson M, Nikonov S, Jonsson B, Hederstedt L. Crystal structure of ferrochelatase: the terminal enzyme in heme biosynthesis. Structure. 1997;5:1501. doi: 10.1016/s0969-2126(97)00299-2. [DOI] [PubMed] [Google Scholar]
- Al-Karadaghi S, Franco R, Hansson M, Shelnutt JA, Isaya G, Ferreira GC. Chelatases: distort to select? Trends Biochem Sci. 2006;31:135. doi: 10.1016/j.tibs.2006.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali V, Shigeta Y, Tokumoto U, Takahashi Y, Nozaki T. An intestinal parasitic protist, Entamoeba histolytica, possesses a non-redundant nitrogen fixation-like system for iron-sulfur cluster assembly under anaerobic conditions. J Biol Chem. 2004;279:16863. doi: 10.1074/jbc.M313314200. [DOI] [PubMed] [Google Scholar]
- Aloria K, Schilke B, Andrew A, Craig EA. Iron-induced oligomerization of yeast frataxin homologue Yfh1 is dispensable in vivo. EMBO Rep. 2004;5:1096. doi: 10.1038/sj.embor.7400272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastassopoulou I, Banci L, Bertini I, Cantini F, Katsari E, Rosato A. Solution structure of the apo and copper(I)-loaded human metallochaperone HAH1. Biochemistry. 2004;43:13046. doi: 10.1021/bi0487591. [DOI] [PubMed] [Google Scholar]
- Andrew TL, Riley PG, Dailey HA. Regulation of heme biosynthesis in higher animals. In: Dailey HA, editor. Biosynthesis of Heme and Cholorophylls. New York: Green Pub. Associates and Wiley-Interscience; 1990. p. 183. [Google Scholar]
- Andrews NC. Disorders of iron metabolism. N Engl J Med. 1999;341:1986. doi: 10.1056/NEJM199912233412607. [DOI] [PubMed] [Google Scholar]
- Arnesano F, Banci L, Bertini I, Huffman DL, O’Halloran TV. Solution structure of the Cu(I) and apo forms of the yeast metallochaperone, Atx1. Biochemistry. 2001;40:1528. doi: 10.1021/bi0014711. [DOI] [PubMed] [Google Scholar]
- Atamna H. Heme, iron, and the mitochondrial decay of ageing. Ageing Res Rev. 2004;3:303. doi: 10.1016/j.arr.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Babcock M, de Silva D, Oaks R, Davis-Kaplan S, Jiralerspong S, Montermini L, Pandolfo M, Kaplan J. Regulation of mitochondrial iron accumulation by Yfh1p, a putative homolog of frataxin. Science. 1997;276:1709. doi: 10.1126/science.276.5319.1709. [DOI] [PubMed] [Google Scholar]
- Banci L, Bertini I. Solution structure of apo CopZ from Bacillus subtilis: further analysis of the changes associated with the presence of copper. Biochemistry. 2003;42:13422. doi: 10.1021/bi0353326. [DOI] [PubMed] [Google Scholar]
- Bartolo C, Mendell JR, Prior TW. Identification of a missense mutation in a Friedreich’s ataxia patient: implications for diagnosis and carrier studies. Am J Med Genet. 1998;79:396. [PubMed] [Google Scholar]
- Beinert H, Kennedy MC, Stout CD. Aconitase as ironminus signsulfur protein, enzyme, and iron-regulatory protein. Chem Rev. 1996;96:2335. doi: 10.1021/cr950040z. [DOI] [PubMed] [Google Scholar]
- Bencze KZ, Yoon T, Millán-Pacheco C, Pastor N, Cowan JA, Stemmler TL. Human frataxin: Iron structure and ferrochelatase binding surface. J Am Chem Soc. 2006 doi: 10.1039/b703195e. (Submitted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertini I, Cowan JA, Del Bianco C, Luchinat C, Mansy SS. Thermotoga maritima IscU. Structural characterization and dynamics of a new class of metallochaperone. J Mol Biol. 2003;331:907. doi: 10.1016/s0022-2836(03)00768-x. [DOI] [PubMed] [Google Scholar]
- Bidichandani SI, Ashizawa T, Patel PI. Atypical Friedreich ataxia caused by compound heterozygosity for a novel missense mutation and the GAA triplet-repeat expansion. Am J Hum Genet. 1997;60:1251. [PMC free article] [PubMed] [Google Scholar]
- Bidichandani SI, Ashizawa T, Patel PI. The GAA triplet-repeat expansion in Friedreich ataxia interferes with transcription and may be associated with an unusual DNA structure. Am J Hum Genet. 1998;62:111. doi: 10.1086/301680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bou-Abdallah F, Adinolfi S, Pastore A, Laue TM, Dennis Chasteen N. Iron binding and oxidation kinetics in frataxin CyaY of Escherichia coli. J Mol Biol. 2004;341:605. doi: 10.1016/j.jmb.2004.05.072. [DOI] [PubMed] [Google Scholar]
- Branda SS, Yang ZY, Chew A, Isaya G. Mitochondrial intermediate peptidase and the yeast frataxin homolog together maintain mitochondrial iron homeostasis in Saccharomyces cerevisiae. Hum Mol Genet. 1999;8:1099. doi: 10.1093/hmg/8.6.1099. [DOI] [PubMed] [Google Scholar]
- Branda SS, Cavadini P, Adamec J, Kalousek F, Taroni F, Isaya G. Yeast and human frataxin are processed to mature form in two sequential steps by the mitochondrial processing peptidase. J Biol Chem. 1999;274:22763. doi: 10.1074/jbc.274.32.22763. [DOI] [PubMed] [Google Scholar]
- Bulteau AL, Ikeda-Saito M, Szweda LI. Redox-dependent modulation of aconitase activity in intact mitochondria. Biochemistry. 2003;42:14846. doi: 10.1021/bi0353979. [DOI] [PubMed] [Google Scholar]
- Bulteau AL, Lundberg KC, Ikeda-Saito M, Isaya G, Szweda LI. Reversible redox-dependent modulation of mitochondrial aconitase and proteolytic activity during in vivo cardiac ischemia/reperfusion. Proc Natl Acad Sci U S A. 2005;102:5987. doi: 10.1073/pnas.0501519102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulteau AL, O’Neill HA, Kennedy MC, Ikeda-Saito M, Isaya G, Szweda LI. Frataxin acts as an iron chaperone protein to modulate mitochondrial aconitase activity. Science. 2004;305:242. doi: 10.1126/science.1098991. [DOI] [PubMed] [Google Scholar]
- Campuzano V, Montermini L, Molto MD, Pianese L, Cossee M, Cavalcanti F, Monros E, Rodius F, Duclos F, Monticelli A, et al. Friedreich’s ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271:1423. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- Cavadini P, Gellera C, Patel PI, Isaya G. Human frataxin maintains mitochondrial iron homeostasis in Saccharomyces cerevisiae. Hum Mol Genet. 2000;9:2523. doi: 10.1093/hmg/9.17.2523. [DOI] [PubMed] [Google Scholar]
- Cavadini P, O’Neill HA, Benada O, Isaya G. Assembly and iron-binding properties of human frataxin, the protein deficient in Friedreich ataxia. Hum Mol Genet. 2002;11:217. doi: 10.1093/hmg/11.3.217. [DOI] [PubMed] [Google Scholar]
- Cavadini P, Adamec J, Taroni F, Gakh O, Isaya G. Two-step processing of human frataxin by mitochondrial processing peptidase. Precursor and intermediate forms are cleaved at different rates. J Biol Chem. 2000;275:41469. doi: 10.1074/jbc.M006539200. [DOI] [PubMed] [Google Scholar]
- Chen OS, Hemenway S, Kaplan J. Inhibition of Fe-S cluster biosynthesis decreases mitochondrial iron export: evidence that Yfh1p affects Fe-S cluster synthesis. Proc Natl Acad Sci U S A. 2002;99:12321. doi: 10.1073/pnas.192449599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen OS, Crisp RJ, Valachovic M, Bard M, Winge DR, Kaplan J. Transcription of the yeast iron regulon does not respond directly to iron but rather to iron-sulfur cluster biosynthesis. J Biol Chem. 2004;279:29513. doi: 10.1074/jbc.M403209200. [DOI] [PubMed] [Google Scholar]
- Chen R, Li L, Weng Z. ZDOCK: an initial-stage protein-docking algorithm. Proteins. 2003;52:80. doi: 10.1002/prot.10389. [DOI] [PubMed] [Google Scholar]
- Cho SJ, Lee MG, Yang JK, Lee JY, Song HK, Suh SW. Crystal structure of Escherichia coli CyaY protein reveals a previously unidentified fold for the evolutionarily conserved frataxin family. Proc Natl Acad Sci U S A. 2000;97:8932. doi: 10.1073/pnas.160270897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook JD, Bencze KZ, Jankovic AD, Crater AK, Busch CN, Bradley PB, Stemmler AJ, Spaller MR, Stemmler TL. Monomeric yeast frataxin is an iron binding protein. Biochemistry. 2006;45:7767. doi: 10.1021/bi060424r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossee M, Durr A, Schmitt M, Dahl N, Trouillas P, Allinson P, Kostrzewa M, Nivelon-Chevallier A, Gustavson KH, Kohlschutter A. Friedreich’s ataxia: point mutations and clinical presentation of compound heterozygotes. Ann Neurol. 1999;45:200. doi: 10.1002/1531-8249(199902)45:2<200::aid-ana10>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Cotton FA, Wilkinson G. Advanced Inorganic Chemistry. New York: John Wiley & Sons; 1988. [Google Scholar]
- Creusot F, Verdiere J, Gaisne M, Slonimski PP. CYP1 (HAP1) regulator of oxygen-dependent gene expression in yeast. I. Overall organization of the protein sequence displays several novel structural domains. J Mol Biol. 1988;204(2):263. doi: 10.1016/0022-2836(88)90574-8. [DOI] [PubMed] [Google Scholar]
- Dailey HA, Dailey TA, Wu CK, Medlock AE, Wang KF, Rose JP, Wang BC. Ferrochelatase at the millennium: structures, mechanisms and [2Fe-2S] clusters. Cell Mol Life Sci. 2000;57:1909. doi: 10.1007/PL00000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dailey HA, T A. Ferrochelatase. In: Kadish KM, Saith KM, Guilard R, editors. The Porphyrin handbook. Vol. 12. USA: Elsevier Science; 2003. p. 93. [Google Scholar]
- Delatycki MB, Williamson R, Forrest SM. Friedreich ataxia: an overview. J Med Genet. 2000;37:1. doi: 10.1136/jmg.37.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhe-Paganon S, Shigeta R, Chi YI, Ristow M, Shoelson SE. Crystal structure of human frataxin. J Biol Chem. 2000;275:30753. doi: 10.1074/jbc.C000407200. [DOI] [PubMed] [Google Scholar]
- Duby G, Foury F, Ramazzotti A, Herrmann J, Lutz T. A non-essential function for yeast frataxin in iron-sulfur cluster assembly. Hum Mol Genet. 2002;11:2635. doi: 10.1093/hmg/11.21.2635. [DOI] [PubMed] [Google Scholar]
- Emond M, Lepage G, Vanasse M, Pandolfo M. Increased levels of plasma malondialdehyde in Friedreich ataxia. Neurology. 2000;55:1752. doi: 10.1212/wnl.55.11.1752. [DOI] [PubMed] [Google Scholar]
- Ferreira GC. Ferrochelatase. Int J Biochem Cell Biol. 1999;31:995. doi: 10.1016/s1357-2725(99)00066-7. [DOI] [PubMed] [Google Scholar]
- Filla A, De Michele G, Cavalcanti F, Pianese L, Monticelli A. The relationship between trinucleotide (GAA) repeat length and clinical features in Friedreich ataxia. Am J Hum Genetics. 1996;59:554. [PMC free article] [PubMed] [Google Scholar]
- Forrest SM, Knight M, Delatycki MB, Paris D, Williamson R, King J, Yeung L, Nassif N, Nicholson GA. The correlation of clinical phenotype in Friedreich ataxia with the site of point mutations in the FRDA gene. Neurogenetics. 1998;1:253. doi: 10.1007/s100480050037. [DOI] [PubMed] [Google Scholar]
- Foury F. Low iron concentration and aconitase deficiency in a yeast frataxin homologue deficient strain. FEBS Letters. 1999;456:281. doi: 10.1016/s0014-5793(99)00961-8. [DOI] [PubMed] [Google Scholar]
- Foury F, Cazzalini O. Deletion of the yeast homologue of the human gene associated with Friedreich’s ataxia elicits iron accumulation in mitochondria. FEBS Lett. 1997;411:373. doi: 10.1016/s0014-5793(97)00734-5. [DOI] [PubMed] [Google Scholar]
- Frazzon J, Dean DR. Biosynthesis of the nitrogenase iron-molybdenum-cofactor from Azotobacter vinelandii. Met Ions Biol Syst. 2002;39:163. [PubMed] [Google Scholar]
- Fytlovich S, Gervais M, Agrimonti C, Guiard B. Evidence for an interaction between the CYP1(HAP1) activator and a cellular factor during heme-dependent transcriptional regulation in the yeast Saccharomyces cerevisiae. Embo J. 1993;12:1209. doi: 10.1002/j.1460-2075.1993.tb05762.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gakh O, Adamec J, Gacy AM, Twesten RD, Owen WG, Isaya G. Physical evidence that yeast frataxin is an iron storage protein. Biochemistry. 2002;41:6798. doi: 10.1021/bi025566+. [DOI] [PubMed] [Google Scholar]
- Gakh O, Park S, Liu G, Macomber L, Imlay JA, Ferreira GC, Isaya G. Mitochondrial iron detoxification is a primary function of frataxin that limits oxidative damage and preserves cell longevity. Hum Mol Genet. 2006;15:467. doi: 10.1093/hmg/ddi461. [DOI] [PubMed] [Google Scholar]
- Garland SA, Hoff K, Vickery LE, Culotta VC. Saccharomyces cerevisiae ISU1 and ISU2: members of a well-conserved gene family for iron-sulfur cluster assembly. J Mol Biol. 1999;294:897. doi: 10.1006/jmbi.1999.3294. [DOI] [PubMed] [Google Scholar]
- Gerber J, Muhlenhoff U, Lill R. An interaction between frataxin and Isu1/Nfs1 that is crucial for Fe/S cluster synthesis on Isu1. EMBO Rep. 2003;4:906. doi: 10.1038/sj.embor.embor918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson TJ, Koonin EV, Musco G, Pastore A, Bork P. Friedreich’s ataxia protein: phylogenetic evidence for mitochondrial dysfunction. Trends Neurosci. 1996;19:465. doi: 10.1016/S0166-2236(96)20054-2. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Cabo P, Vazquez-Manrique RP, Garcia-Gimeno MA, Sanz P, Palau F. Frataxin interacts functionally with mitochondrial electron transport chain proteins. Hum Mol Genet. 2005;14:2091. doi: 10.1093/hmg/ddi214. [DOI] [PubMed] [Google Scholar]
- Gordon DM, Wang J, Amutha B, Pain D. Self-association and precursor protein binding of Saccharomyces cerevisiae Tom40p, the core component of the protein translocation channel of the mitochondrial outer membrane. Biochem J. 2001;356:207. doi: 10.1042/0264-6021:3560207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon DM, Kogan M, Knight SA, Dancis A, Pain D. Distinct roles for two N-terminal cleaved domains in mitochondrial import of the yeast frataxin homolog, Yfh1p. Hum Mol Genet. 2001;10:259. doi: 10.1093/hmg/10.3.259. [DOI] [PubMed] [Google Scholar]
- Hambright P, Chock PB. Metal-porphyrin interactions. 3. A dissociative-interchange mechanism for metal ion incorporation into porphyrin molecules. J Am Chem Soc. 1974;96:3123. doi: 10.1021/ja00817a018. [DOI] [PubMed] [Google Scholar]
- He Y, Alam SL, Proteasa SV, Zhang Y, Lesuisse E, Dancis A, Stemmler TL. Yeast frataxin solution structure, iron binding, and ferrochelatase interaction. Biochemistry. 2004;43:16254. doi: 10.1021/bi0488193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphry W, Dalke A, Schulten K. VMD—visual molecular dynamics. J Molec Graphics. 1996;14:33. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Irazusta V, Cabiscol E, Reverter-Branchat G, Ros J, Tamarit J. Manganese is the link between frataxin and iron-sulfur deficiency in the yeast model of Friedreich’s ataxia. J Biol Chem. 2006;281:12227. doi: 10.1074/jbc.M511649200. [DOI] [PubMed] [Google Scholar]
- Johnson DC, Dean DR, Smith AD, Johnson MK. Structure, function, and formation of biological iron-sulfur clusters. Annu Rev Biochem. 2005;74:247. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- Karlberg T, Lecerof D, Gora M, Silvegren G, Labbe-Bois R, Hansson M, Al-Karadaghi S. Metal binding to Saccharomyces cerevisiae ferrochelatase. Biochemistry. 2002;41:13499. doi: 10.1021/bi0260785. [DOI] [PubMed] [Google Scholar]
- Karthikeyan G, Lewis LK, Resnick MA. The mitochondrial protein frataxin prevents nuclear damage. Hum Mol Genet. 2002;11:1351. doi: 10.1093/hmg/11.11.1351. [DOI] [PubMed] [Google Scholar]
- Karthikeyan G, Santos JH, Graziewicz MA, Copeland WC, Isaya G, Van Houten B, Resnick MA. Reduction in frataxin causes progressive accumulation of mitochondrial damage. Hum Mol Genet. 2003;12:3331. doi: 10.1093/hmg/ddg349. [DOI] [PubMed] [Google Scholar]
- Kispal G, Csere P, Prohl C, Lill R. The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. Embo J. 1999;18:3981. doi: 10.1093/emboj/18.14.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SA, Sepuri NB, Pain D, Dancis A. Mt-Hsp70 homolog, Ssc2p, required for maturation of yeast frataxin and mitochondrial iron homeostasis. J Biol Chem. 1998;273:18389. doi: 10.1074/jbc.273.29.18389. [DOI] [PubMed] [Google Scholar]
- Kosman DJ. Molecular mechanisms of iron uptake in fungi. Mol Microbiol. 2003;47:1185. doi: 10.1046/j.1365-2958.2003.03368.x. [DOI] [PubMed] [Google Scholar]
- Koutnikova H, Campuzano V, Foury F, Dolle P, Cazzalini O, Koenig M. Studies of human, mouse and yeast homologues indicate a mitochondrial function for frataxin. Nat Genet. 1997;16:345. doi: 10.1038/ng0897-345. [DOI] [PubMed] [Google Scholar]
- Labbe RF, Vreman HJ, Stevenson DK. Zinc protoporphyrin: a metabolite with a mission. Clin Chem. 1999;45:2060. [PubMed] [Google Scholar]
- Labuda M, Poirier J, Pandolfo M. A missense mutation (W155R) in an American patient with Friedreich ataxia. Human Mutation. 1999;13:506. [Google Scholar]
- Layer G, Ollagnier de Choudens S, Sanakis Y, Fontecave M. Iron-sulfur cluster biosynthesis: Characterization of Escherichia coli cyay as an iron donor for the assembly of [2Fe-2S] clusters in the scaffold ISCU. J Biol Chem. 2006;281:16256. doi: 10.1074/jbc.M513569200. [DOI] [PubMed] [Google Scholar]
- Lee MG, Cho SJ, Yang JK, Song HK, Suh SW. Crystallization and preliminary X-ray crystallographic analysis of Escherichia coli CyaY, a structural homologue of human frataxin. Acta Crystallogr D Biol Crystallogr. 2000;56(Pt 7):920. doi: 10.1107/s0907444900005916. [DOI] [PubMed] [Google Scholar]
- Lesuisse E, Santos R, Matzanke BF, Knight SA, Camadro JM, Dancis A. Iron use for haeme synthesis is under control of the yeast frataxin homologue (Yfh1) Hum Mol Genet. 2003;12:879. doi: 10.1093/hmg/ddg096. [DOI] [PubMed] [Google Scholar]
- Levi S, Arosio P. Mitochondrial ferritin. Int J Biochem Cell Biol. 2004;36:1887. doi: 10.1016/j.biocel.2003.10.020. [DOI] [PubMed] [Google Scholar]
- Lill R, Muhlenhoff U. Iron-sulfur-protein biogenesis in eukaryotes. Trends Biochem Sci. 2005;30:133. doi: 10.1016/j.tibs.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Liu J, Oganesyan N, Shin DH, Jancarik J, Yokota H, Kim R, Kim SH. Structural characterization of an iron-sulfur cluster assembly protein IscU in a zinc-bound form. Proteins. 2005;59:875. doi: 10.1002/prot.20421. [DOI] [PubMed] [Google Scholar]
- Mansy SS, Cowan JA. Iron-sulfur cluster biosynthesis: toward an understanding of cellular machinery and molecular mechanism. Acc Chem Res. 2004;37:719. doi: 10.1021/ar0301781. [DOI] [PubMed] [Google Scholar]
- Monson EK, Weinstein M, Ditta GS, Helinski DR. The FixL protein of Rhizobium meliloti can be separated into a heme-binding oxygen-sensing domain and a functional C-terminal kinase domain. Proc Natl Acad Sci U S A. 1992;89:4280. doi: 10.1073/pnas.89.10.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S, Abeygunawardana C, O’Neil Johnson M, van Zijl PC. Improved sensitivity of HSQC spectra of exchanging protons at short interscan delays using a new fast HSQC (FHSQC) detection scheme that avoids water saturation. J Magn Res B. 1995;108:94. doi: 10.1006/jmrb.1995.1109. [DOI] [PubMed] [Google Scholar]
- Muhlenhoff U, Richhardt N, Gerber J, Lill R. Characterization of iron-sulfur protein assembly in isolated mitochondria. A requirement for ATP, NADH, and reduced iron. J Biol Chem. 2002;277:29810. doi: 10.1074/jbc.M204675200. [DOI] [PubMed] [Google Scholar]
- Muhlenhoff U, Gerber J, Richhardt N, Lill R. Components involved in assembly and dislocation of iron-sulfur clusters on the scaffold protein Isu1p. EMBO J. 2003;22:4815. doi: 10.1093/emboj/cdg446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlenhoff U, Richhardt N, Ristow M, Kispal G, Lill R. The yeast frataxin homolog Yfh1p plays a specific role in the maturation of cellular Fe/S proteins. Hum Mol Genet. 2002;11:2025. doi: 10.1093/hmg/11.17.2025. [DOI] [PubMed] [Google Scholar]
- Musco G, Stier G, Kolmerer B, Adinolfi S, Martin S, Frenkiel T, Gibson T, Pastore A. Towards a structural understanding of Friedreich’s ataxia: the solution structure of frataxin. Structure. 2000;8:695. doi: 10.1016/s0969-2126(00)00158-1. [DOI] [PubMed] [Google Scholar]
- Musco G, de Tommasi T, Stier G, Kolmerer B, Bottomley M, Adinolfi S, Muskett FW, Gibson TJ, Frenkiel TA, Pastore A. Assignment of the 1H, 15N, and 13C resonances of the C-terminal domain of frataxin, the protein responsible for Friedreich ataxia. J Biomol NMR. 1999;15:87. doi: 10.1023/a:1008398832619. [DOI] [PubMed] [Google Scholar]
- Nair M, Adinolfi S, Kelly G, Frenkiel TA, Pastore A. NMR assignment of the 1H, 15N and 13C resonances of the E. coli frataxin orthologue, CyaY. J Biomol NMR. 2003;27:403. doi: 10.1023/a:1025845513018. [DOI] [PubMed] [Google Scholar]
- Nair M, Adinolfi S, Pastore C, Kelly G, Temussi P, Pastore A. Solution structure of the bacterial frataxin ortholog, CyaY: mapping the iron binding sites. Structure (Camb) 2004;12:2037. doi: 10.1016/j.str.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Nichol H, Gakh O, O’Neill HA, Pickering IJ, Isaya G, George GN. Structure of frataxin iron cores: an X-ray absorption spectroscopic study. Biochemistry. 2003;42:5971. doi: 10.1021/bi027021l. [DOI] [PubMed] [Google Scholar]
- O’Neill HA, Gakh O, Isaya G. Supramolecular assemblies of human frataxin are formed via subunit-subunit interactions mediated by a non-conserved amino-terminal region. J Mol Biol. 2005;345:433. doi: 10.1016/j.jmb.2004.10.074. [DOI] [PubMed] [Google Scholar]
- O’Neill HA, Gakh O, Park S, Cui J, Mooney SM, Sampson M, Ferreira GC, Isaya G. Assembly of human frataxin is a mechanism for detoxifying redox-active iron. Biochemistry. 2005;44:537. doi: 10.1021/bi048459j. [DOI] [PubMed] [Google Scholar]
- Ogawa K, Sun J, Taketani S, Nakajima O, Nishitani C, Sassa S, Hayashi N, Yamamoto M, Shibahara S, Fujita H. Heme mediates derepression of Maf recognition element through direct binding to transcription repressor Bach1. EMBO J. 2001;20:2835. doi: 10.1093/emboj/20.11.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima K, Montermini L, Wells RD, Pandolfo M. Inhibitory effects of expanded GAA.TTC triplet repeats from intron I of the Friedreich ataxia gene on transcription and replication in vivo. J Biol Chem. 1998;273:14588. doi: 10.1074/jbc.273.23.14588. [DOI] [PubMed] [Google Scholar]
- Orth M, Schapira AH. Mitochondria and degenerative disorders. Am J Med Genet. 2001;106:27. doi: 10.1002/ajmg.1425. [DOI] [PubMed] [Google Scholar]
- Padmanaban G, Venkateswar V, Rangarajan PN. Haem as a multifunctional regulator. Trends Biochem Sci. 1989;14:492. doi: 10.1016/0968-0004(89)90182-5. [DOI] [PubMed] [Google Scholar]
- Pandolfo M. Iron metabolism and mitochondrial abnormalities in Friedreich ataxia. Blood Cells Mol Dis. 2002;29:536. doi: 10.1006/bcmd.2002.0591. [DOI] [PubMed] [Google Scholar]
- Park S, Gakh O, Mooney SM, Isaya G. The ferroxidase activity of yeast frataxin. J Biol Chem. 2002;277:38589. doi: 10.1074/jbc.M206711200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Gakh O, O’Neill HA, Mangravita A, Nichol H, Ferreira GC, Isaya G. Yeast frataxin sequentially chaperones and stores iron by coupling protein assembly with iron oxidation. J Biol Chem. 2003;278:31340. doi: 10.1074/jbc.M303158200. [DOI] [PubMed] [Google Scholar]
- Pfeifer K, Kim KS, Kogan S, Guarente L. Functional dissection and sequence of yeast HAP1 activator. Cell. 1989;56:291. doi: 10.1016/0092-8674(89)90903-3. [DOI] [PubMed] [Google Scholar]
- Ponka P. Cell biology of heme. Am J Med Sci. 1999;318:241. doi: 10.1097/00000441-199910000-00004. [DOI] [PubMed] [Google Scholar]
- Puccio H, Simon D, Cossee M, Criqui-Filipe P, Tiziano F, Melki J, Hindelang C, Matyas R, Rustin P, Koenig M. Mouse models for Friedreich ataxia exhibit cardiomyopathy, sensory nerve defect and Fe-S enzyme deficiency followed by intramitochondrial iron deposits. Nat Genet. 2001;27:181. doi: 10.1038/84818. [DOI] [PubMed] [Google Scholar]
- Radisky DC, Babcock MC, Kaplan J. The yeast frataxin homologue mediates mitochondrial iron efflux: evidence for a mitochondrial iron cycle. J Biol Chem. 1999;274:4497. doi: 10.1074/jbc.274.8.4497. [DOI] [PubMed] [Google Scholar]
- Ramazzotti A, Vanmansart V, Foury F. Mitochondrial functional interactions between frataxin and Isu1p, the iron-sulfur cluster scaffold protein, in Saccharomyces cerevisiae. FEBS Lett. 2004;557:215. doi: 10.1016/s0014-5793(03)01498-4. [DOI] [PubMed] [Google Scholar]
- Ramelot TA, Cort JR, Goldsmith-Fischman S, Kornhaber GJ, Xiao R, Shastry R, Acton TB, Honig B, Montelione GT, Kennedy MA. Solution NMR structure of the iron-sulfur cluster assembly protein U (IscU) with zinc bound at the active site. J Mol Biol. 2004;344:567. doi: 10.1016/j.jmb.2004.08.038. [DOI] [PubMed] [Google Scholar]
- Rees DC, Howard JB. Nitrogenase: standing at the crossroads. Curr Opin Chem Biol. 2000;4:559. doi: 10.1016/s1367-5931(00)00132-0. [DOI] [PubMed] [Google Scholar]
- Ristow M, Pfister MF, Yee AJ, Schubert M, Michael L, Zhang CY, Ueki K, Michael MD, Lowell BB, Kahn CR. Frataxin activates mitochondrial energy conversion and oxidative phosphorylation. Proc Natl Acad Sci U S A. 2000;97:12239. doi: 10.1073/pnas.220403797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristow M, Mulder H, Pomplun D, Schulz TJ, Muller-Schmehl K, Krause A, Fex M, Puccio H, Muller J, Isken F, et al. Frataxin deficiency in pancreatic islets causes diabetes due to loss of beta cell mass. J Clin Invest. 2003;112:527. doi: 10.1172/JCI18107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenzweig AC, O’Halloran TV. Structure and chemistry of the copper chaperone proteins. Curr Opin Chem Biol. 2000;4:140. doi: 10.1016/s1367-5931(99)00066-6. [DOI] [PubMed] [Google Scholar]
- Rotig A, de Lonlay P, Chretien D, Foury F, Koenig M, Sidi D, Munnich A, Rustin P. Aconitase and mitochondrial iron-sulphur protein deficiency in Friedreich ataxia. Nat Genet. 1997;17:215. doi: 10.1038/ng1097-215. [DOI] [PubMed] [Google Scholar]
- Sassa S, Nagai T. The role of heme in gene expression. Int J Hematol. 1996;63:167. doi: 10.1016/0925-5710(96)00449-5. [DOI] [PubMed] [Google Scholar]
- Sazanov LA, Hinchliffe P. Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus. Science. 2006;311:1430. doi: 10.1126/science.1123809. [DOI] [PubMed] [Google Scholar]
- Schilke B, Voisine C, Beinert H, Craig E. Evidence for a conserved system for iron metabolism in the mitochondria of Saccharomydes cervisiae. Proc Natl Acad Sci USA. 1999;96:10206. doi: 10.1073/pnas.96.18.10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfeld RA, Napoli E, Wong A, Zhan S, Reutenauer L, Morin D, Buckpitt AR, Taroni F, Lonnerdal B, Ristow M, et al. Frataxin deficiency alters heme pathway transcripts and decreases mitochondrial heme metabolites in mammalian cells. Hum Mol Genet. 2005;14:3787. doi: 10.1093/hmg/ddi393. [DOI] [PubMed] [Google Scholar]
- Schultz JB, Dehmer T, Schols L, Mende H, Hardt C, Vorgerd M, Burk K, Matson W, Dichgans J, Beal MF. Oxidative stress in patients with Friedreich’s ataxia. Neurology. 2000;55:1719. doi: 10.1212/wnl.55.11.1719. [DOI] [PubMed] [Google Scholar]
- Schulz TJ, Thierbach R, Voigt A, Drewes G, Mietzner B, Steinberg P, Pfeiffer AF, Ristow M. Induction of oxidative metabolism by mitochondrial frataxin inhibits cancer growth: Otto Warburg revisited. J Biol Chem. 2006;281:977. doi: 10.1074/jbc.M511064200. [DOI] [PubMed] [Google Scholar]
- Sigfridsson E, Ryde U. The importance of porphyrin distortions for the ferrochelatase reaction. J Biol Inorg Chem. 2003;8:273. doi: 10.1007/s00775-002-0413-8. [DOI] [PubMed] [Google Scholar]
- Stehling O, Elsasser HP, Bruckel B, Muhlenhoff U, Lill R. Iron-sulfur protein maturation in human cells: evidence for a function of frataxin. Hum Mol Genet. 2004;13:3007. doi: 10.1093/hmg/ddh324. [DOI] [PubMed] [Google Scholar]
- Tait GH. The Biosynthesis and degradation of heme. In: De Matteis F, Aldridge NW, editors. Heme and Hemoproteins. Berlin: Springer-Verlag; 1978. pp. 1–35. [Google Scholar]
- Takahashi Y, Tokumoto U. A third bacterial system for the assembly of iron-sulfur clusters with homologs in archaea and plastids. J Biol Chem. 2002;277:28380. doi: 10.1074/jbc.C200365200. [DOI] [PubMed] [Google Scholar]
- Taketani S. Aquisition, mobilization and utilization of cellular iron and heme: endless findings and growing evidence of tight regulation. Tohoku J Exp Med. 2005;205:297. doi: 10.1620/tjem.205.297. [DOI] [PubMed] [Google Scholar]
- Teo BK. EXAFS: Basic Principles and Data Analysis. New York: Springer-Verlag; 1986. [Google Scholar]
- Theil EC. Amphibian red blood cell ferritin. J Biol Chem. 1973;248:622. [PubMed] [Google Scholar]
- Theil EC. Ferritin: structure, gene regulation, and cellular function in animals, plants, and microorganisms. Annu Rev Biochem. 1987;56:289. doi: 10.1146/annurev.bi.56.070187.001445. [DOI] [PubMed] [Google Scholar]
- Theil EC. Ferritin: at the crossroads of iron and oxygen metabolism. J Nutr. 2003;133:1549S. doi: 10.1093/jn/133.5.1549S. [DOI] [PubMed] [Google Scholar]
- Thierbach R, Schulz TJ, Isken F, Voigt A, Mietzner B, Drewes G, von Kleist-Retzow JC, Wiesner RJ, Magnuson MA, Puccio H. Targeted disruption of hepatic frataxin expression causes impaired mitochondrial function, decreased life span and tumor growth in mice. Hum Mol Genet. 2005;14:3857. doi: 10.1093/hmg/ddi410. [DOI] [PubMed] [Google Scholar]
- Vazquez-Manrique RP, Gonzalez-Cabo P, Ros S, Aziz H, Baylis HA, Palau F. Reduction of Caenorhabditis elegans frataxin increases sensitivity to oxidative stress, reduces lifespan, and causes lethality in a mitochondrial complex II mutant. Faseb J. 2006;20:172. doi: 10.1096/fj.05-4212fje. [DOI] [PubMed] [Google Scholar]
- Vivas E, Skovran E, Downs DM. Salmonella enterica strains lacking the frataxin homolog CyaY show defects in Fe-S cluster metabolism in vivo. J Bacteriol. 2006;188:1175. doi: 10.1128/JB.188.3.1175-1179.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann N, Urzica E, Guiard B, Muller H, Lohaus C, Meyer HE, Ryan MT, Meisinger C, Muhlenhoff U, Lill R. Essential role of Isd11 in mitochondrial iron-sulfur cluster synthesis on Isu scaffold proteins. EMBO J. 2006;25:184. doi: 10.1038/sj.emboj.7600906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CK, Dailey HA, Rose JP, Burden A, Sellers VM, Wang BC. The 2.0 A structure of human ferrochelatase, the terminal enzyme of heme biosynthesis. Nat Struct Biol. 2001;8:156. doi: 10.1038/84152. [DOI] [PubMed] [Google Scholar]
- Yoon T, Cowan JA. Iron-sulfur cluster biosynthesis. Characterization of frataxin as an iron donor for assembly of [2Fe-2S] clusters in ISU-type proteins. J Am Chem Soc. 2003;125:6078. doi: 10.1021/ja027967i. [DOI] [PubMed] [Google Scholar]
- Yoon T, Cowan JA. Frataxin-mediated iron delivery to Ferrochelatase in the final step of Heme biosynthesis. J Biol Chem. 2004;279:25943. doi: 10.1074/jbc.C400107200. [DOI] [PubMed] [Google Scholar]
- Zhang L, Guarente L. Heme binds to a short sequence that serves a regulatory function in diverse proteins. EMBO J. 1995;14:313. doi: 10.1002/j.1460-2075.1995.tb07005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Hach A, Wang C. Molecular mechanism governing heme signaling in yeast: a higher-order complex mediates heme regulation of the transcriptional activator HAP1. Mol Cell Biol. 1998;18:3819. doi: 10.1128/mcb.18.7.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Cash VL, Flint DH, Dean DR. Assembly of iron-sulfur clusters. Identification of an iscSUA-hscBA-fdx gene cluster from Azotobacter vinelandii. J Biol Chem. 1998;273:13264. doi: 10.1074/jbc.273.21.13264. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Hon T, Zhang L. Heme initiates changes in the expression of a wide array of genes during the early erythroid differentiation stage. Biochem Biophys Res Commun. 1999;258:87. doi: 10.1006/bbrc.1999.0586. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Hon T, Ye W, Zhang L. Heme deficiency interferes with the Ras-mitogen-activated protein kinase signaling pathway and expression of a subset of neuronal genes. Cell Growth Differ. 2002;13:431. [PubMed] [Google Scholar]
- Zuhlke CH, Dalski A, Habeck M, Straube K, Hedrich K, Hoeltzen-bein M, Konstanzer A, Hellenbroich Y, Schwinger E. Extension of the mutation spectrum in Friedreich’s ataxia: detection of an exon deletion and novel missense mutations. Eur J Hum Genet. 2004;12:979. doi: 10.1038/sj.ejhg.5201257. [DOI] [PubMed] [Google Scholar]