Abstract
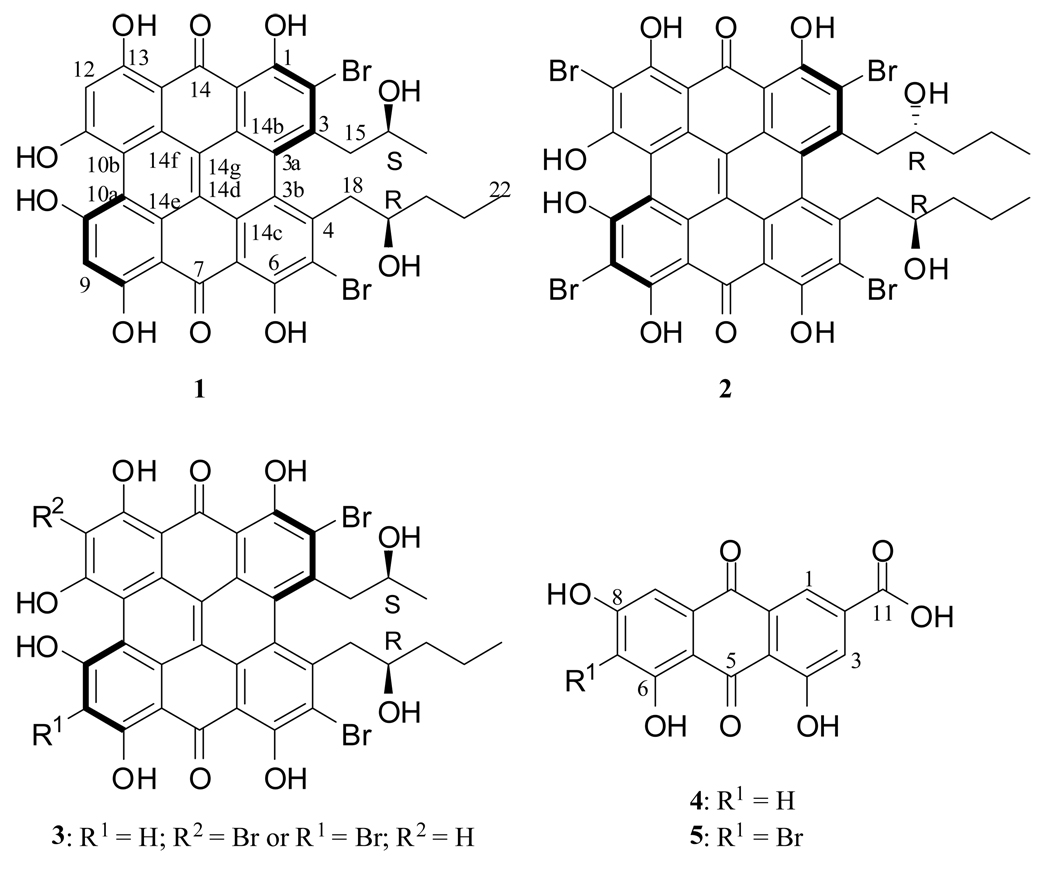
Bioactivity-guided fractionation of metabolites from the crinoid Holopus rangii led to the discovery of two new phenanthroperylenequinone derivatives, gymnochromes E (1) and F (2). Gymnochrome E showed cytotoxic activity toward the NCI/ADR-Res with an IC50 of 3.5 µM. It also inhibited histone deacetylase-1 with an IC50 of 3.3 µM. Gymnochrome F was a moderate inhibitor of Myeloid cell leukemia sequence 1 (MCL-1) binding to Bak. Two anthraquinone metabolites, emodic acid (4) and its new bromo derivative (5) were also isolated from the crinoid and show remarkable similarity to the phenanthroperylenequinone core, suggesting that these metabolites share the same polyketide biosynthetic pathway.
The past decade has seen a dramatic increase of marine organism derived anticancer lead compounds entering human clinical trials1 resulting from recent technological advances in structure elucidation, organic synthesis, and biological assays. These lead compounds range in structural class from relatively simple linear peptides such as dolastatin 10,2 to more complex polyketides such as discodermolide,3 to very complex macrocyclic polyethers, such as halichondrin B.4 Equally diverse are the molecular modes of action by which these molecules impart their biological activity and the increasing number of compounds working through novel modes of action. As part of our on-going program to identify novel natural products with activity in target directed oncology assays, materials from the HBOI Peak Library (generated by reversed-phase medium pressure liquid chromatography) were assayed for their ability to inhibit the binding of MCL-1 to Bak using a FRET based assay.5 MCL-1 (an anti-apoptotic member of the BCL-2 family) binds Bak (a pro-apoptotic BCL-2 member) which upon release from MCL-1 regulates apoptosis. A fraction derived from the crinoid Holopus rangii inhibited the binding of MCL-1 to Bak with an IC50 of 10 µg/mL. NMR and MS analysis of the fraction suggested the presence of a series of highly unsaturated pigments. Bioassay- and spectroscopy-guided fractionation led to the isolation and characterization of two new members of the phenanthroperylenequinone family of natural products, which we have designated as Gymnochromes E (1) and F (2), the known isogymnochrome B (3), as well as two anthraquinones, emodic acid (4) and its 7-bromo derivative (5). Here we report the isolation and structure elucidation of compounds 1, 2 and 5 as well as their biological activities.
A sample of freeze-dried crinoid was cut into small pieces and crushed. The resultant powder was extracted successively with EtOH and EtOAc:EtOH 1:1. The combined extracts were concentrated by distillation under reduced pressure to give a dark green residue which was fractionated o n a C-18 stationary phase using vacuum column chromatography. Further purification using reversed-phase HPLC and monitoring by bioassay and mass spectrometry led to the isolation of 1 (3 mg), 2 (5 mg), 3 (1.5 mg), 4 (2.1 mg) and 5 (0.5 mg). The spectroscopic data observed for 3 was consistent with that reported for isogymnochrome B6 and has tentatively been assigned as isogymnochrome B. The spectroscopic data observed for 4 was identical to that reported for emodic acid,7 allowing for its identification.
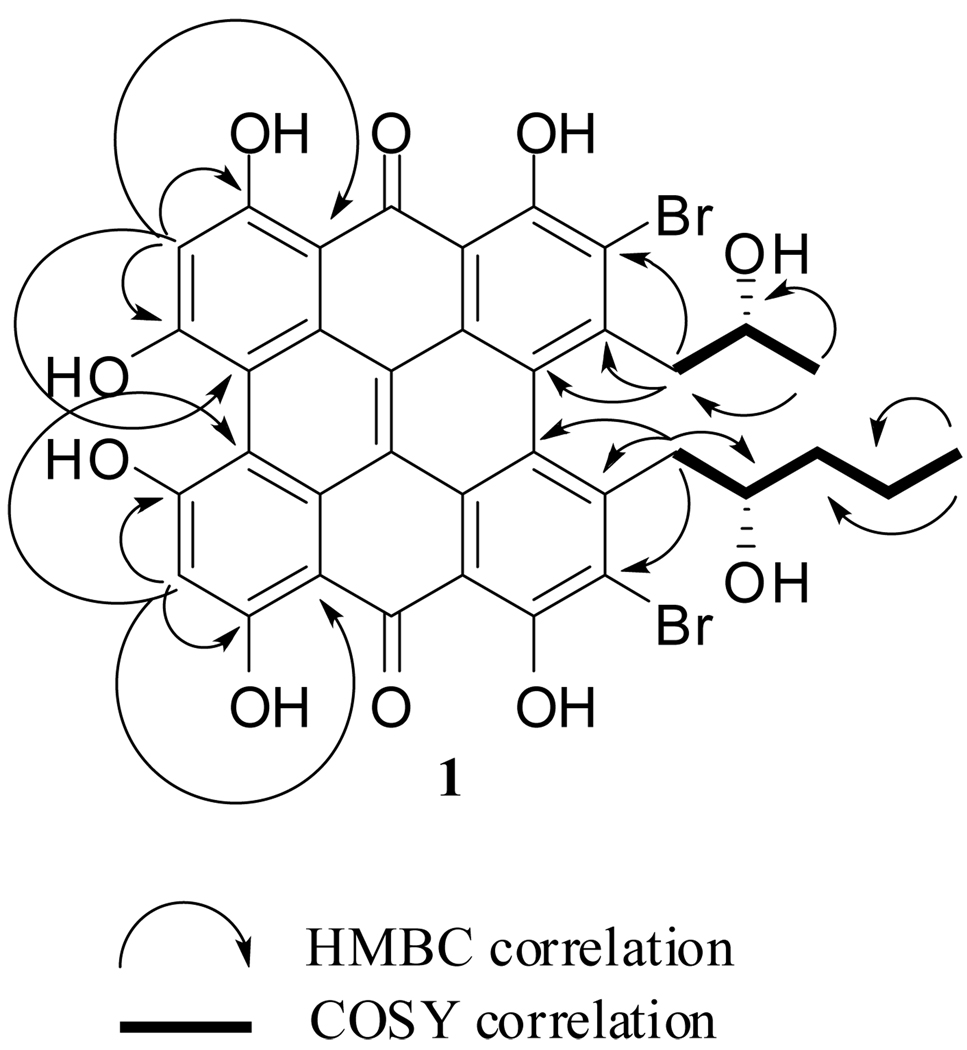
Compound 1 was isolated as a dark-brown oil. The IR spectrum of 1 shows an absorption band observed at 1634 cm−1 characteristic of the carbonyl of a hydrogen-bonded quinone. The UV-vis spectrum of 1 showed maxima (λmax) at 311, 372, 524 and 596 nm that are similar to those of the well-known compound hypericin8, 9 suggesting that 1 is a phenanthroperylenequinone derivative. The electrospray mass spectrum (ESIMS) of 1 detected in negative mode showed a complex multiplet of three peaks at m/z 775, 777 and 779, suggesting that 1 is dibrominated. The 1H NMR spectrum of 1 recorded in DMSO-d6 showed the presence of two methyl resonances [δH −0.09 (H3-22), 0.94 (H3-17)] as well as an aromatic proton resonance at δH 6.59 (H-9 and H-12) that were also suggestive that 1 is a dibromophenanthroperylenequinone derivative. In addition to the signals attributable to the dibromohydroxy phenanthroperylenequinone skeleton, analyses of the 13C and HSQC spectra of 1 revealed the presence of two different aliphatic side chains. The first one consisted of three methylene carbons, an oxymethine carbon and a methyl carbon. It was unambiguously identified as a 2-hydroxypentyl moiety based upon correlations observed in the COSY spectrum that revealed the sequential connectivity of H2-18 → H-19 → H2-20 → H2-21 → H3-22. For the second aliphatic side chain a methylene carbon, an oxymethine carbon and a methyl carbon were identified. The COSY spectrum clearly established the connectivity of H2-15 → H-16 → H3-17 and assigned the presence of a 2-hyroxypropyl moiety in 1. This assignment was further supported by the HMBC spectrum which showed correlations between the methyl protons H3-17 (δ0.94, d, J = 6.1) and both C-15 and C-16; as well as correlations between the methylene protons H2-15 (H-15a, δH 3.47 dd, J = 8.2, 13.0; H-15b, δH 3.64 dd, J = 13.0, 2.0) and the methyl carbon C-17 (δC 24.8). The two aliphatic fragments were connected to the dibromohydroxy phenanthroperylenequinone moiety with the aid of diagnostic correlations observed in the HMBC spectrum (Figure 1). In particular through three-bond connectivity observed between H2-18 (H-18a, δH 3.56 dd, J = 13.0, 8.9; H-18b δH 3.73 dd, J = 13.0, 4.8) with C-5 (δc 115.4) and C-3b (δc 122.0) as well as connectivity between H2-15 with C-3a (δc 122.5) and C-2 (δc 115.0). Based upon this data the structure of 1 was assigned as 1,6,8,10,11,13- hexahydroxy-2,5-dibromo-3-(2-hydroxypropyl)-4-(2-hydroxypentyl)-phenanthroperylene-7,14-quinone, which we are designating gymnochrome E. Further support for this assignment came from the ESI-MS/MS analysis of 1, which showed a fragment ion at m/z 645, representing the cleavage of the side chains (see Supporting Information). Gymnochrome E represents the 9 or 12 debromo derivative of isogymnochrome B (3).
Figure 1.
Key HMBC and COSY correlations observed for Gymnochrome E (1)
Compound 2 was isolated as dark-brown oil. The UV-vis spectrum as well as the ESI-MS/MS fragmentation pattern suggested that 2 is a homologue of 1. The electrospray mass spectrum (ESIMS) of 2 indicated in negative mode a complex multiplet of five peaks at m/z 959, 961, 963, 965, and 967, suggesting that 2 is tetra brominated. The lack of an aromatic proton signal in the 1H NMR spectrum of 2 suggested that the aromatic part of 2 was fully substituted. Furthermore, the 1H NMR spectrum of 2 revealed the presence of a side chain, which could be ascribed to a 2-hydroxypentyl-side chain, based upon the correlations observed in the HMBC and COSY spectra. The structure of 2 was thus elucidated as 1,6,8,10,11,13-hexahydroxy-2,5,9,12-tetrabromo-3,4-(2-hydroxypentyl)-phenanthroperylene-7,14-quinone, which we have designated as gymnochrome F. Compound 2 was reported previously as the solvolysis product of its disulfated derivative isogymnochrome D.6 As it was possible that compound 2 was an artifact from our isolation procedure in which H2O-TFA (0.05%) was used as the elution solvent during HPLC, the crude MeOH extract of the crinoid was analyzed by ESI-MS. The detection of an ion with m/z 963 (negative ionization mode) in the mass spectrum strongly supported the natural origin of 2.
The configuration of the chiral centers in the side chains have been tentatively assigned using arguments similar to those described in detail by De Riccardis et al.6 Briefly, it has been determined that the orientation of the side chain in perylenequinones is regulated both by the helicity of the ring system and the configurations of the chiral centers in the side chain. When the helicity of the ring and the configuration of the side chain chiral carbon are M and R respectively, then the side chain is oriented above the aromatic ring system and is in the shielding zone of the aromatic ring system. When the helicity of the ring and the configuration of the side chain chiral carbon are M and S respectively, then the side chain is directed away from the aromatic ring and is outside the shielding zone of the aromatic ring system. Therefore if one knows the helicity of the ring system, one can assign configuration to the chiral center of the side chain based upon the observation of shielding (or lack thereof) in the 1H NMR data observed for the protons in the side chain.
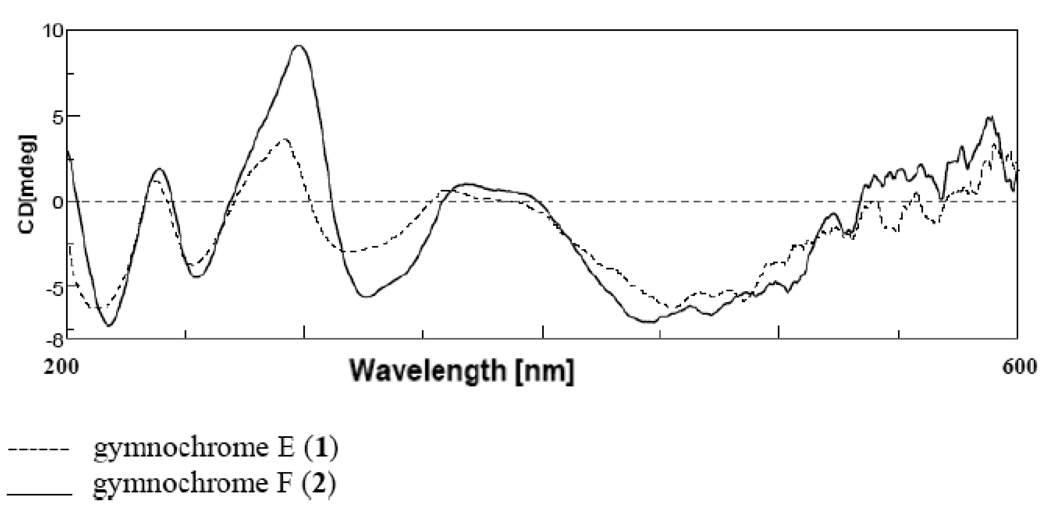
Compounds 1 and 2 have CD spectra (Figure 2) that are virtually superimposable with that of isogymnochrome D which has been reported previously to have M axial chirality.6 For compound 1, the propyl side chain methyl (H3-17) of compound 1 is not shielded (δH 0.94) which argues that it is directed away from the aromatic ring and that C-16 has S configuration. The 1H resonance of the methyl group of the pentyl moiety (H3-22) is shielded (δH −0.09) and therefore lies above the aromatic ring which suggests an R configuration for the C-19 chiral center. In compound 2 which has C2 symmetry, the H3-19 methyl resonance is shielded suggesting that it is oriented above the ring and therefore C-16 has been assigned R configuration. Additional data to confirm this assignment is the close agreement in the 1H NMR data observed for 2 with that reported for the desulfated isogymnochrome D produced during the course of the assignment of the structure of isogymnochrome D.6 Gymnochrome F is therefore the desulfated form of isogymnochrome D.
Figure 2.
CD spectra of 1 and 2.
Compound 5 was obtained as a yellow oil. The HR-ESIMS suggested a formula of C15H7BrO7 for 5. The UV-vis spectrum of 5 was similar to that observed for 4, suggesting that they belong to the same family of natural products. The primary difference in the 1H NMR spectra was the lack of the proton resonance observed at δH 6.51 (H-7) in 5, which suggested that 5 is the 7-bromo derivative of 4. Thus 5 was identified as 7-bromoemodic acid. The co-occurrences of 1–5 in the same organism strongly suggest a biogenetic relationship. Biosynthetic studies on the hypericin class of compounds indicates that the biosynthesis of the phenathroperylenequinones and related natural products involves two consecutive reaction cascades, consisting of the biosynthesis of the two anthraquinones, followed by their condensation.10
Gymnochrome E (1) inhibited the proliferation of the NCI/ADR-Res (multi drug resistant ovarian cancer cell line) with an IC50 value of 3.5 µM and did not show significant inhibitory activity at a concentration of 6.4 µM against the PANC-1 pancreatic carcinoma or DLD-1 human colorectal adenocarcinoma cell lines. Gymnochrome E also inhibited histone deacetylase-1 (HDAC-1) with an IC50 of 10.9 µM. Gymnochrome F (2) did not show significant inhibitory activity at a concentration of 5.2 µM against the PANC-1, NCI/ADR-Res or DLD-1 tumor cell lines but was a moderate inhibitor of MCL-1 binding to Bak with an IC50 of 3.3 µM. Gymnochrome E (1) exhibited minimum inhibition concentrations (MICs) of 25 µg/mL against both Staphylococcus aureus and methicillin-resistant S. aureus (MRSA) while gymnochrome F (2) exhibited MICs of 12.5 µg/mL against S. aureus and MRSA. Methicillin-resistant S. aureus has emerged as a serious threat due to its resistance to penicillin-class antibiotics. Compounds 1 and 2 did not show activity against either Pseudomonas aeruginosa or Candida albicans at the concentrations tested. Emodic acid (4) and its 7-bromoderivative (5) showed no significant activity in any of the assays.
Experimental Section
General Experimental Procedures
The UV spectra were collected on a Hitachi U-3010 spectrophotometer. The circular dichroism (CD) spectra were collected on a JASCO J-815 circular dichroism system. The IR spectra were collected on a Midac M-1200 with Galactic GRAMS/386 software. NMR data were collected on a JEOL ECA-600 spectrometer operating at 600.17 MHz for 1H; 150.9 MHz for 13C. The edited-g-HSQC spectrum was optimized for 140 Hz; the g-HMBC spectrum was optimized for 8 Hz; chemical shifts were referenced to solvent, (CD3OD δH observed at 3.31 ppm and δC observed at 49.0 ppm; for DMSO-d6 δH observed at 2.46 and δc observed at 40.0 ppm). The ESIMS were measured using a Finnigan LTQ mass spectrometer and the HRESIMS were measured using a Kratos MS50TC mass spectrometer. HPLC was performed using a Hitachi LaChrom binary gradient HPLC system equipped with a diode array detector (L-7455) monitoring at 230 nm; semi-preparative C18 column (218TP1010, Grace Vydac). All solvents used were HPLC grade. Simulated 13C-NMR spectra were calculated for comparison to the observed spectra using ACD 13C predictor version 11.
Biological Material
The specimen used in this study was identified as Holopus rangii d’Orbigni, 1837 (phylum Echinodermata, class Crinoidea, order Cyrtocrinida, family Holopodidae). Taxonomy was confirmed by Dr. David Pawson, Senior Research Scientist, National Museum of Natural History, Smithsonian Institution. A museum voucher specimen is cataloged in the Harbor Branch Oceanographic Museum (HBOI Sample ID 8-V-00-1-002; Catalog No. 070:00047). H. rangii is a stalked crinoid with an enlarged root that typically attaches to a hard substrate. Four arms attach to a thick pentagonal plate and each arm divides in two, forming eight arms which are conical, tuberculated on the outer surface, and almost twice as long as the foot. The arms stretch out to feed, but retract into a tight ball when disturbed or to prevent predation. The live animal is dark green and remains green in alcohol. The museum specimen is approximately 4 cm in length. The samples were collected with the Johnson-Sea-Link submersible (dive number JSL II-3203) from a depth of 358 m on a rocky slope on the south coast of Curacao (12° 04.34’N, 68° 53.62’W), May 8, 2000.
Extraction and Isolation
152 g of the frozen crinoid Holopus rangii was freeze dried and then extracted exhaustively by macerating with EtOH and EtOAc:EtOH 1:1 v/v using a Waring blender (3×400 mL). The combined filtered extracts were concentrated by distillation under reduced pressure. The resulting dark-brown oil (3.97 g) was separated under vacuum-column chromatographic conditions on a reversed-phase C18 stationary phase. A 150 mL buchner funnel fitted with a medium porosity fritted glass disc was used as the column. The stationary phase was packed to a total height of 4 cm. The crude extract was absorbed onto a portion of the C18 stationary phase and applied as slurry to the column. Fractions were eluted using the following elution series: Fraction 1: H2O (150 mL), Fraction 2: ACN: H2O 8:2 (150 mL), Fraction 3: IPA (isopropanol) (150 mL), Fraction 4: ACN: H2O:TFA 8:2:0.1 (150 mL) and fraction 5: CH2Cl2:MeOH 1:1 (150 mL). Active components were isolated on the basis of bioassays and spectroscopic analysis. Final purification was achieved by semi-preparative HPLC on fraction 2 using a Vydac C18 Protein and Peptide Column (10 mm×250 mm, 10 µm particle size), flow rate 2.5 mL/min; eluent: H2O containing 0.05% TFA-ACN 2:3 (v/v); monitored by UV at 230 nm, yielding 1 (3 mg), 2 (5 mg), 3 (1.5 mg), 4 (2.1 mg) and 5 (0.5 mg)
Gymnochrome E (1)
dark-brown oil; UV (MeOH) λmax (log ε): 293 (4.32), 311 (4.24), 372 (4.01), 524 (4.08) and 596 (4.32) nm; CD (MeOH, c = 10 µg/mL) reported in figure 2; IR (Film) 3388, 2928, 1634, 1609, 1574, 1458, 1245, 1114 and 1032 cm−1; 1H NMR (DMSO-d6) see table 1; 13C NMR (DMSO-d6) see table 1; ESI-MS/MS (m/z): 718, and 645 [M - H] −; HRESIMS: m/z 776.9966 [M + H]+ (calcd for C36H27 Br2O10, 776.9965).
Table 1.
NMR Spectroscopic Data for Gymnochrome E (1) and Gymnochrome F (2).a
| No | 1 | 2 | |||||
|---|---|---|---|---|---|---|---|
| δCb,e | δH (J in Hz)b | HMBCc | δCd,e | δH (J in Hz)d | HMBC | ||
| 1, 6 | 158.9 | 158.9 | 159.6 | ||||
| 2, 5 | 115.0 | 115.4 | 116.4 | ||||
| 3, 4 | 145.6 | 143.7 | 144.0 | ||||
| 3a, 3b | 122.5 | 122.0 | 121.5 | ||||
| 6a, 14a | 109.5 | 109.4 | 109.2 | ||||
| 7, 14 | 183.0 | 183.0 | 185.0 | ||||
| 7a, 13a | 102.5 | 102.5 | 103.0 | ||||
| 8, 13 | 168.9 | 168.9 | 163.7 | ||||
| 9, 12 | 106.3 | 106.2 | 6.59, s | 7a, 8, 10, 10a | 102.0 | ||
| 10b, 11, 13, 13a | |||||||
| 10, 11 | 175.7 | 175.7 | 168.3 | ||||
| 10a, 10b | 120.3 | 120.3 | 120.7 | ||||
| 14b, 14c | 127.5 | 127.6 | 125.1 | ||||
| 14d, 14g | 121.7 | 121.6 | 121.5 | ||||
| 14e, 14f | 124.7 | 124.9 | 125.6 | ||||
| 15 | 45.7 | 3.47, dd (13.0, 8.2) | 2, 3, 3a, 17 | 45.7 | 3.85, dd (13.7, 6.8) | 2, 3, 3a, 16, 17 | |
| 3.64, dd (13.0, 2.0) | 2, 3, 3a, 17 | 3.98, dd (13.7, 5.5) | 2, 3, 3a, 16, 17 | ||||
| 16 | 67.5 | 3.42, brs | 70.8 | 3.54, brs | |||
| 17 | 24.8 | 0.94, d (6.1) | 15, 16 | 37.0 | 0.43, brs | ||
| 0.51, brs | |||||||
| 18 | 45.4 | 3.56, dd (13.0, 8.9) | 3b, 4, 5, 19, 20 | 17.5 | 0.70, brs | ||
| 3.73, dd (13.0, 4.8) | 3b, 4, 5, 19, 20 | 0.85, brs | |||||
| 19 | 69.5 | 3.29, brs | 12.3 | 0.22, t (6.8) | 17, 18 | ||
| 20 | 37.1 | 0.03, m | |||||
| 0.27, m | |||||||
| 21 | 17.3 | 0.58, m | |||||
| 0.81, m | |||||||
| 22 | 13.6 | −0.09, t (6.8) | 20, 21 | ||||
1H and 13C NMR date were measured at 600.17 and 150.9, respectively.
Recorded in DMSO-d6 solution.
HMBC correlations are from the proton(s) stated to the indicated carbons.
Recorded in MeOH-d4 solution.
Assignments made by interpretation of the HSQC and HMBC spectra as well as through comparison of the observed 13C NMR spectrum with the 13C spectrum calculated using ACDLab v 11 (See Supporting Information for calculated values).
Gymnochrome F (2)
dark-brown oil; UV (MeOH) λmax (log ε): 300 (4.45), 333 (4.36), 498 (4.11), 554 (4.23) and 598 (4.46) nm; CD (MeOH, c = 10 µg/mL) reported in figure 2; IR (Film) 3388, 2928, 1705, 1609, 1451, 1245, 1128 and 1004 cm−1; 1H NMR (DMSO-d6) see table 1; 13C NMR (MeOH-d4) see table 1; ESI-MS/MS (m/z): 875 and 803 [M - H]−; HRESIMS: m/z 960.8463 [M + H]+ (calcd for C38H29Br4O10, 960.8489).
7-bromoemodic acid (5)
yellow oil; IR (Film) 3402, 2921, 1684, 1629, 1210 and 1135 cm−1; UV (MeOH) λmax (log ε): 271 (4.08) and 437 (3.88) nm; 1H NMR (MeOH-d4): δ 8.25 (H-1, s, 1H), 7.78 (H-3, s, 1H) and 7.27 (H-8, s, 1H); ESI-MS/MS (m/z): 334, 297, 268 and 225; HRESIMS: m/z 376.9273 [M - H]− (calcd for C15H6 BrO7, 376.9296).
Cytotoxicity Assays
1 and 2 were evaluated for their effects on proliferation of the PANC-1 human pancreatic carcinoma (ATCC No. CRL-1469); DLD-1 human colorectal adenocarcinoma (ATCC No. CCL-221); and NCI-ADR-RES (formerly MCF-7/ADR) human ovarian carcinoma cell lines. The PANC-1 and DLD-1 cell lines were obtained from the American Type Culture Collection (Rockville, MD). The NCI/ADR-RES cell line was obtained from the NCI-Frederick Cancer DCTD Tumor/Cell Line Repository (Bethesda, MD). Assays were run using protocols described previously.11 All samples were assayed a minimum of three times to derive the final IC50 value.
HDAC Assays
This assay was run as per the protocol described in Sambucetti et al.12
MCL-1 Assays
This assay was run as per the protocol described in Calcul et al.5
Antimicrobial Assays
The antimicrobial activities of 1-5 against Staphylococcus aureus (ATCC 29213), MRSA (methicillin-resistant Staphylococcus aureus, ATCC 700787), Pseudomonas aeruginosa (ATCC 27853), and Candida albicans (ATCC 44506) were determined as described in Park et al.13
Supplementary Material
Acknowledgment
The work described in this paper was funded by NIH grant 2U19-CA529955. We thank Prof. Dr. C. Hertweck and Dr. K. Ishida from the Leibniz Institute for Natural Product Research and Infection Biology (Germany) for the CD measurement. We thank G. Samples for sample preparation and D. Harmody and P. Linley for bioassays. We thank the governments of the Netherlands Antilles and Curacao for allowing us to operate in their territorial waters and collect the specimens used in this research. We thank A. Debrot, and W. L. Bakhuis of the CARMABI Foundation (Caribbean Marine Biological Institute), Curacao; and L. Pors and K. van Dongen of the Curacao Marine Park, who provided exceptional assistance in planning and collaboration during the expedition. This is Harbor Branch Oceanographic Institute at Florida Atlantic University Contribution Number 1799.
Footnotes
Supporting Information Available: LRMS, 1H and 13C NMR and 2D NMR spectra for 1 and 2 as well as the 1H NMR spectra for 4 and 5. This material is available free of charge via the Internet at http://pubs.acs.org.
Reference and Notes
- 1.Simmons TL, Andrianasolo E, McPhail K, Flatt P, Gerwick WH. Mol. Cancer Ther. 2005;4:333–342. [PubMed] [Google Scholar]
- 2.Pettit GR, Yoshiaki Kamano Y, Herald CL, Tuinman AA, Boettner FE, Kizu H, Schmidt JM, Baczynskyj L, Tomer KB, Bontems RJ. J. Am. Chem. Soc. 1987;109:6883–6885. [Google Scholar]
- 3.Gunasekera SP, Gunasekera M, Longley RE, Schulte GK. J. Org. Chem. 1990;55:4912–4945. [Google Scholar]
- 4.Bai R, Paull KD, Heraldy CL, Malspeis L, Pettit GR, Hamel E. J. Biol. Chem. 1991;266:15882–15889. [PubMed] [Google Scholar]
- 5.Calcul L, Chow R, Oliver AG, Tenney K, White KN, Wood AW, Fiorilla C, Crews P. J Nat Prod. 2009;72:443–449. doi: 10.1021/np800737z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Riccardis F, Iorizzi M, Minale L, Riccio R, De Forges BR, Debitus C. J. Org. Chem. 1991;56:6781–6787. [Google Scholar]
- 7.Alvi KA, Nair B, Gallo C, Baker D. J. Antibiot. 1997;50:264–266. [PubMed] [Google Scholar]
- 8.Skalkos D, Tatsis E, Gerothanassis IP, Troganis A. Tetrahedron. 2002;58:4925–4626. [Google Scholar]
- 9.Pace N, Mackinney G. J. Am. Chem. Soc. 1941;63:2570–2574. [Google Scholar]
- 10.Falk H. Angew. Chem. Int. Ed. 1999;38:3116–3136. [PubMed] [Google Scholar]
- 11.Gunasekera SP, Zuleta IA, Longley RE, Wright AE, Pomponi SA. J. Nat. Prod. 2003;66:1615–1617. doi: 10.1021/np030292s. [DOI] [PubMed] [Google Scholar]
- 12.(a) Sambucetti LC, Fischer DD, Zabludoff S, Kwon PO, Chamberlin H, Trogani N, Xu H, Cohen D. J. Biol. Chem. 1999;274:34940–34947. doi: 10.1074/jbc.274.49.34940. [DOI] [PubMed] [Google Scholar]; (b) Remiszewski SW, Sambucetti LC, Atadja P, Bair KW, Cornell WD, Green MA, Howell KL, Jung M, Kwon P, Trogani N, Walker H. J. Med Chem. 2002;45:753–757. doi: 10.1021/jm015568c. [DOI] [PubMed] [Google Scholar]
- 13.Park YC, Gunasekera SP, Lopez JV, McCarthy PJ, Wright AE. J. Nat. Prod. 2006;69:580–584. doi: 10.1021/np058113p. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.