Abstract
cAMP-dependent protein kinases (PKAs) are central mediators of cAMP signaling in eukaryotic cells. Previously we identified a cDNA which encodes for a PKA catalytic subunit (PKA-C) in Schistosoma mansoni (SmPKA-C) that is required for adult schistosome viability in vitro. As such, SmPKA-C could potentially represent a novel schistosome chemotherapeutic target. Here we sought to identify PKA-C subunit orthologues in the other medically important schistosome species, S. haematobium and S. japonicum, to determine the degree to which this potential target is conserved and could therefore be exploited for the treatment of all forms of schistosomiasis. We report the identification of PKA-C subunit orthologues in S. haematobium and S. japonicum (ShPKA-C and SjPKA-C respectively) and show that PKA-C orthologues are highly conserved in the Schistosoma, with over 99 % amino acid sequence identity shared among the three human pathogens we examined. Furthermore, we show that the recently published S. mansoni and S. japonicum genomes contain sequences encoding for several putative PKA substrates with homology to those found in Homo sapiens, Caenorhabditis elegans, and Saccharomyces cerevisiae.
Keywords: Schistosoma haematobium, Schistosoma japonicum, cAMP-dependent protein kinase
1. Introduction
Schistosomiasis, a disease caused by parasitic blood flukes of the genus Schistosoma, afflicts over 200 million people in tropical and sub-tropical regions worldwide and accounts for approximately 280,000 deaths annually in sub-Saharan Africa alone (Gryseels, et al., 2006). S. mansoni and S. haematobium are endemic in sub-Saharan Africa while S. japonicum is endemic in Asia, most notably in China. Currently, the anthelminthic praziquantel (PZQ) is the only drug that is used for the treatment of schistosomiasis, due to its ability to kill the adult worms of all the medically important Schistosoma species (S. mansoni, S. haematobium, and S. japonicum) (Cioli and Pica-Mattoccia, 2003). However, reliance on PZQ alone for both treatment and transmission control efforts may not be sustainable in the long term. PZQ tolerant strains have been selected for in the laboratory (Fallon and Doenhoff, 1994) and there is indirect evidence of decreased PZQ sensitivity in the field (Doenhoff, et al., 2008, Melman, et al., 2009). The potential for PZQ resistance is real and current research is focused on the identification of novel anti-schistosome chemotherapeutic targets (Caffrey, 2007).
Protein kinases have recently been explored as potential anti-schistosome targets (Dissous, et al., 2007). Through the reversible phosphorylation of specific tyrosine (Tyr) and serine/threonine (Ser/Thr) amino acid residues, protein kinases are important mediators in signal transduction pathways (Hanks, et al., 1988). cAMP-dependent protein kinase (PKA) is the major transducer of cAMP signaling and is involved in a wide variety of cellular processes in eukaryotic cells (Das, et al., 2007). In its inactive state, PKA is a tetrameric holoenzyme consisting of two identical regulatory subunits (PKA-R) which are bound to two identical catalytic subunits (PKA-C) (Taylor, et al., 1993). Binding of cAMP to the PKA-R subunits releases and activates the PKA-C subunits. Recently, we provided biochemical and molecular evidence that adult S. mansoni have a functional PKA (Swierczewski and Davies, 2009). Furthermore, we identified a cDNA that encodes for a PKA-C subunit in S. mansoni (SmPKA-C; GenBank Accession No. GQ168377) and showed that SmPKA-C is required for parasite viability in vitro through chemical inhibition and RNA interference studies (Swierczewski and Davies, 2009). As the SmPKA-C protein could potentially represent a new drug target in S. mansoni, we sought to identify PKA-C subunit orthologues in S. japonicum and S. haematobium to determine the extent to which this potential new target is conserved in the other two medically important schistosome species. Here we report the identification of PKA-C subunit orthologues in S. japonicum and S. haematobium (SjPKA-C and ShPKA-C respectively) which exhibit high levels of nucleotide and protein sequence identity to SmPKA-C.
2. Materials and Methods
Parasite materials
S. japonicum and S. haematobium adult worms, isolated from infected mice and hamsters respectively, were kindly provided by Dr. Fred Lewis (Biomedical Research Institute, Rockville, MD).
cDNA cloning and sequence analysis
Total RNA was extracted from adult S. japonicum and S. haematobium using the RNAzol B Method (IsoTex Diagnostics, Inc.). 1 μg ribonucleic acid (RNA) was used to synthesize complimentary deoxyribonucleic acid (cDNA) using the iScript Select cDNA Synthesis Kit and an oligo (dT)20 primer (Bio-Rad). For S. haematobium, a 1 kb fragment containing the entire open reading frame (ORF) of a PKA-C subunit was amplified by PCR from adult cDNA using the following primers designed from the SmPKA-C sequence: forward 5’-ATGGGTAATGCACAAGCTGC- 3’ and reverse 5’-AAATTCACTAAATTCTTTTGCACATTTCTCTGT- 3’. For S. japonicum, RNA ligase-mediated rapid amplification of 5’ and 3’ cDNA ends (RACE; 5’- 3’ RACE Kit (Invitrogen)) was used to isolate the 5’ end of the incomplete S. japonicum PKA-C sequence AY813860, using the gene-specific primers 5’-CGCCACCATTGACAAATTCCAGTACCAT-3’ and 5’-GGGTATGTTCAACTTGCTTTAATTTCACTA-3’. The complete ORF was then directly amplified from adult cDNA. 5’ RACE and PCR products were gel purified using the QIAquick Gel Extraction Kit (Qiagen), cloned into pCR4-TOPO vector (Invitrogen), and sequenced using the Big-DyeTerminator cycle sequencing kit (Applied Biosystems). Vector NTI software (Invitrogen) was used for nucleotide sequence editing and contig construction. Phylogenetic and molecular evolutionary analyses were conducted using MEGA version 4 (Tamura, et al., 2007). For nucleotide and amino acid sequences, sequences were aligned using the ClustalW algorithm (Eddy, 1995), phylogenetic trees were constructed using the Maximum Parsimony method (Eck and Dayhoff, 1966) and bootstrap consensus trees inferred from 500 replicates (Zharkikh and Li, 1995).
To identify sequences that may encode for putative PKA substrates in schistosomes, BLASTX searches of the S. mansoni and S. japonicum genome databases (http://www.genedb.org/genedb/smansoni/blast.jsp and http://lifecenter.sgst.cn/schistosoma/en/schistosomaCnIndexPage.do respectively) were performed using amino acid sequences of PKA substrates and PKA interacting proteins from Homo sapiens, Caenorhabditis elegans, and Saccharomyces cerevisiae (Gao, et al., 2008). PKA phosphorylation sites were identified using the ScanProsite database located on the ExPASy proteomics server (http://www.expasy.ch/tools/scanprosite/).
3. Results and Discussion
Identification of SjPKA-C and ShPKA-C subunit cDNAs
For S. japonicum, an incomplete cDNA (GenBank Accession No. AY813860) encoding for a putative PKA-C subunit was identified using the tblastn BLAST program, based on similarity with the SmPKA-C sequence. Approximately 150 nucleotides were missing from the 5’ end of this cDNA, resulting in truncation of the kinase domain and loss of the entire amino terminus from the predicted amino acid translation. The complete cDNA sequence of the putative S. japonicum PKA-C (SjPKA-C) was obtained using 5’ RACE and the complete ORF (GenBank Accession No. GU130553) was subsequently amplified from adult worm cDNA. For S. haematobium, the complete ORF of a PKA-C subunit (ShPKA-C, GenBank Accession No. GU116484) was amplified directly from adult worm cDNA using primers designed from the SmPKA-C sequence.
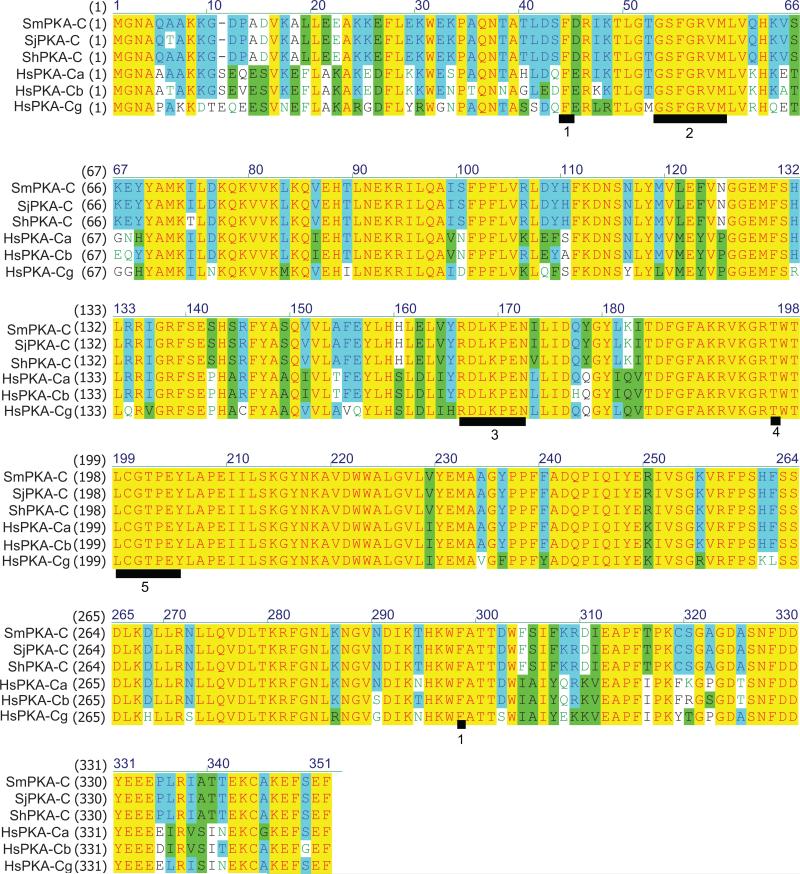
The complete ORFs for ShPKA-C and SjPKA-C are both 1053 bp and encode for proteins 350 amino acids in length, each with a predicted molecular mass of 40.4 kDa. These parameters are identical to those of the SmPKA-C ORF (Swierczewski and Davies, 2009). As with SmPKA-C and the human PKA-C subunit homologues, both proteins contained all conserved amino acid motifs of a PKA-C subunit, including an intact protein kinase domain (residues 43-297), the ATP-binding site GTGSFRGV (residues 50-57), the Ser/Thr active site RDLKPEN (residues 165 -171), a conserved autophosphorylation site, Thr 196, and a sequence motif which is required for regulatory subunit binding, LCGTPEY (198 - 204) (Hanks and Hunter, 1995, Kim, et al., 2005) (Fig. 1). At the amino acid level, the schistosome PKA-C subunit proteins share 99 % identity and only differ at three amino acid residues, all lying outside the regions that are critical for PKA-C function. There is a threonine residue at position 6 of SjPKA-C in lieu of an alanine in SmPKA-C and ShPKA-C. ShPKA-C protein differs from SmPKA-C and SjPKA-C at positions 73 (threonine in place of an isoleucine) and 172 (valine in place of an isoleucine) (Fig. 1).
Figure 1.
Amino acid alignment of PKA-C subunit sequences from Schistosoma. Amino acid sequences of PKA-C subunits from Schistosoma mansoni (SmPKA-C; GQ168377), S. haematobium (ShPKA-C; GU116484), S. japonicum (SjPKA-C; GU130533) and Homo sapiens PKA-Cα (HsPKA-Ca; P17612), H. sapiens PKA-Cβ (HsPKA-Cb; P22694), and H. sapiens PKA-Cγ (HsPKA-Cg; P22612) were aligned using the ClustalW algorithm. Underlines indicate conserved amino acid sequences as follows: 1, kinase domain (Phe43 – Phe297); 2, ATP-binding site (Gly50 - Val57); 3, Ser/Thr active site (Arg165 - Asn171); 4, conserved autophosphorylation site (Thr196); 5, conserved sequence motif for PKA regulatory subunit binding (Leu198 - Tyr204).
Phylogenetic analysis of schistosome PKA-C subunit cDNA sequences
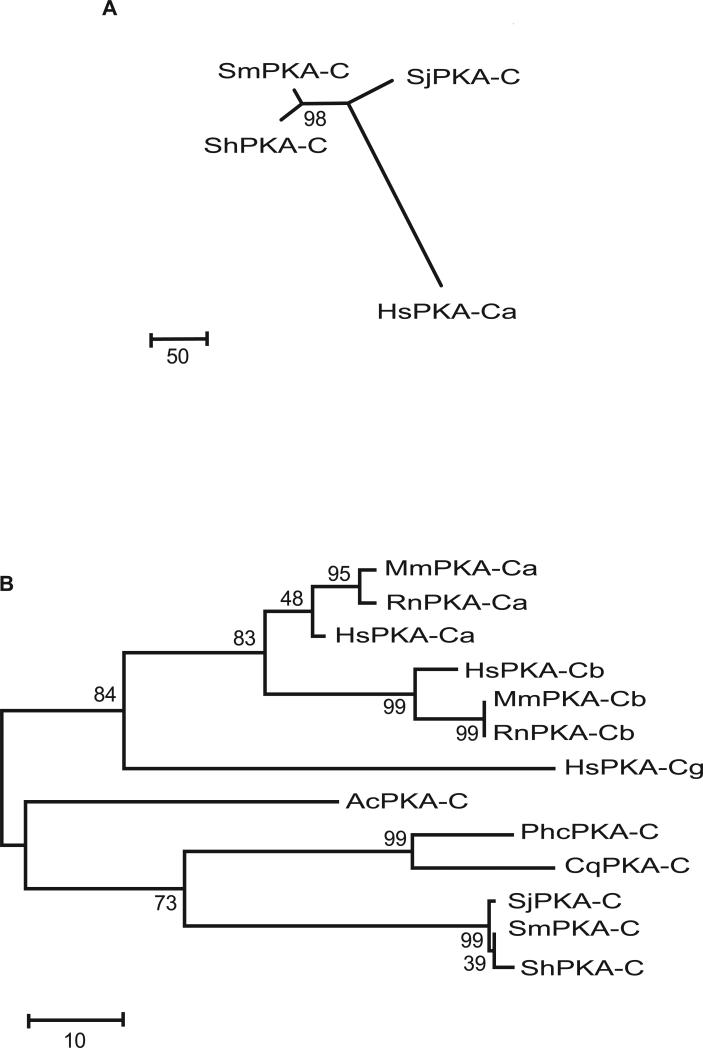
There is over 90% nucleotide identity among the three schistosome PKA-C cDNA sequences. Phylogenetic analysis of the schistosome nucleotide sequences showed that while all three schistosome PKA-C sequences are closely related, the sequences from the two African species (ShPKA-C and SmPKA-C) are most closely related, while the sequence from the Asian species (SjPKA-C) appears more divergent (Fig. 2A). This relationship is consistent with that determined from phylogenetic analysis of other nucleotide sequences from these three species – that the Asian schistosome species, including S. japonicum, form a group that is ancestral to the two more derived African species groups typified by S. mansoni and S. haematobium (Webster, et al., 2006).
Figure 2.
Phylogenetic analysis of PKA-C cDNA nucleotide and amino acid sequences from schistosomes and other species. A, Phylogenetic analysis of PKA-C cDNA nucleotide sequences of schistosomes. PKA-C ORF sequences were aligned using the ClustalW algorithm and the tree constructed using the Maximum Parsimony method. The bootstrap consensus tree inferred from 500 replicates is shown. The percentage of replicate trees in which the associated sequences clustered together in the bootstrap test is shown next to the branch. The tree is drawn to scale, with branch lengths calculated using the average pathway method (Tamura, et al., 2007) and are in the units of the number of changes over the whole sequence. There were a total of 1053 positions in the final alignment, out of which 52 were parsimony informative. Nucleotide sequences of the predicted open reading frames of the schistosome PKA-C subunits are labeled as follows: ShPKA-C, S. haematobium PKA-C (GU116484) SmPKA-C, S. mansoni PKA-C (GQ168377); SjPKA-C, S. japonicum PKA-C (GU130533). The H. sapiens PKA-Cα nucleotide sequence (BC108259), labeled HsPKA-Ca, is included for comparison. B, Phylogenetic analysis of PKA-C amino acid sequences from schistosomes and other species. Sequences were aligned using the ClustalW algorithm and the tree constructed using the Maximum Parsimony method. The bootstrap consensus tree inferred from 500 replicates is shown. The percentage of replicate trees in which the associated sequences clustered together in the bootstrap test is shown next to the branch. The tree is drawn to scale, with branch lengths calculated using the average pathway method (Tamura, et al., 2007) and are in the units of the number of changes over the whole sequence. There were a total of 350 positions in the final alignment, out of which 93 were parsimony informative. Amino acid sequences are labeled as follows: HsPKA-Ca, Homo sapiens PKA-Cα (P17612); HsPKA-Cb, H. sapiens PKA-Cβ (P22694); HsPKA-Cg, H. sapiens PKA-Cγ (P22612); MmPKA-Ca, Mus musculus PKA-Cα (P05132); MmPKA-Cb, M. musculus PKA-Cβ (P68181); RnPKA-Ca, Rattus norvegicus PKA-Cα (P27791); RnPKA-Cb, R. norvegicus PKA-Cγ (P68182); AcPKA-C, Ancylostoma caninum PKA-C (U15983); PhcPKA-C, Pediculus humanus corporis PKA-C (XM_0024273690); CqPKA-C, Culex quinquefasciatus PKA-C (XM_001868382).
Phylogenetic analysis of schistosome PKA-C subunit amino acid sequences
Extensive conservation of PKA-C protein sequences among representatives of these three major groupings within the genus Schistosoma (Fig. 1) suggests that PKA-C is likely highly conserved in all Schistosoma species. BLAST comparison of the amino acid sequences with the non-redundant protein sequence database at NCBI showed that the putative schistosome PKA-C proteins shared approximately 70 % similarity with PKA-C subunits from other eukaryotic organisms. Comparison of the schistosome sequences with representative PKA-C amino acid sequences from other invertebrates (e.g. Pediculus humanus corporis, Culex quinquefasciatus, Ancylostoma caninum) and vertebrate host species (Mus musculus, Rattus norvegicus, Homo sapiens)(Fig. 2B) revealed that the schistosome PKA-C subunits are more similar to those of other invertebrates than to those of their hosts, suggesting there may be differences in the sensitivity of invertebrate and vertebrate PKA-C subunits to biochemical inhibition. Furthermore, mammalian PKA-C subunits segregate into distinct PKA-Cα, PKA-Cβ and PKA-Cγ subfamilies, while invertebrate PKA-C subunits do not. The absence of this diversification in schistosomes could potentially leave the parasite more vulnerable to PKA-C inhibition than host species, in which several different PKA-C isoforms are expressed. Differential susceptibility to PKA inhibition could potentially be exploited for therapeutic purposes.
Identification of potential PKA substrate homologues in S. mansoni and S. japonicum
In silico analysis of the S. mansoni and S. japonicum genomes revealed that schistosomes contain putative homologues of PKA-C substrates and PKA-C interacting proteins found in Homo sapiens, Caenorhabditis elegans, and Saccharomyces cerevisiae (Table 1)(Gao, et al., 2008). For example, nbc2 3 (XP_002572546) from S. mansoni and HDAC1/2 (CAX72896) from S. japonicum are predicted to encode for histone deacetylases that are homologues of human HDAC-8 (AAH50433), which has been shown to be negatively controlled by PKA phosphorylation (Lee, et al., 2004). Furthermore, the schistosome homologues contain putative PKA phosphorylation sites, conforming to the consensus sequence [RK](2) - x - [S or T] (Glass, et al., 1986), providing further support for the conclusion that these schistosome proteins are PKA substrates. Other putative PKA substrates we identified in the genomes of S. mansoni and S. japonicum are provided in Table 1. Further studies to identify schistosome PKA substrates are warranted as these downstream proteins could possibly represent additional novel drug targets in the parasite. Indeed, targeting of specific PKA substrates has led to the identification of possible new drug targets in the causative agent of Chagas’ disease, Trypanosoma cruzi (Bao, et al., 2008). Interestingly, potential homologues of other PKA substrates from H. sapiens (glial fibrillary acidic protein isoform 1 (GFAP; NP_002046) and another histone deacetylase (AAH50433)) could be identified in the S. mansoni genome (Smp_159900.2 (XP_002578164, identified as a putative lamin), and the SmHDAC3 histone deacetylase (ABN81194), respectively), but the predicted sequences of the schistosome proteins do not contain consensus PKA phosphorylation sites (data not shown), suggesting that the repertoire of PKA substrates in schistosomes may be different to those of host species. Such differences are suggestive of a divergence in the function of PKA between parasite and host that could be exploited therapeutically. Additional molecular and biochemical characterization of schistosome PKA substrates will be required to verify the existence of such a divergence in PKA function.
Table 1. Putative PKA substrates and interacting proteins in S. mansoni and S. japonicum.
Putative S. mansoni and S. japonicum homologues of known PKA substrates and interacting proteins were identified by BLAST analysis. GenPept names and accession numbers for all proteins are provided. Full GenPept descriptors of the proteins are as follows: SLC4A4, solute carrier family 4, sodium bicarbonate cotransporter, member 4 isoform 1; HDAC-8, histone deacetylase 8; PPP1R9B, protein phosphatase 1, regulatory subunit 9B; kin-2, type II PKA regulatory subunit; CNR_1, cannabinoid receptor 1 isoform a; Bub2p, protein containing a TBC (Tre-2/Bub2/Cdc16) domain; nbc2 3, sodium bicarbonate cotransporter; SmHDAC1, histone deacetylase; Sm06231, hypothetical protein; Smp_030400, type I-beta PKA regulatory subunit; Smp_043260, histamine-responsive G protein coupled-receptor; Smp_001420; Gh regulated tbc protein-1; HDAC1/2, histone deacetylase 1/2; SJCHGC09512, PDZ (post synaptic density) domain-containing protein; SJCHGC07612, hypothetical protein; SJCHGC08537, hypothetical protein.
| PKA substrate/interacting protein | Putative schistosome homologue | PKA phosphorylation sites in schistosome homologue |
|---|---|---|
| S. mansoni | ||
| Homo sapiens SLC4A4 (NP 001091954) | nbc2 3 (XP 002572546) | S253, T447, S1230 |
| H. sapiens HDAC-8 (AAH50433) | SmHDAC1 (XP 002571987) | S359 |
| H. sapiens PPP1R9B (NP 115984) | Sm06231 (XP 002575145) | S41, S766 |
| Caenorhabditis elegans kin-2 (NP 001076771) | Smp_030400 (XP 002574488) | S191 |
| H. sapiens CNR_1 (NP 057167) | Smp_043260 (XP 002575668) | T65 |
| Saccharomyces cerevisiae Bub2p (NP 013771) | Smp_001420 (XP 002571615) | S41 |
| S. japonicum | ||
| H. sapiens HDAC-8 (AAH50433) | HDAC1/2 CAX72896 | S407 |
| H. sapiens PPP1R9B (NP 115984) | SJCHGC09512 AAX26695 | S273 |
| H. sapiens CNR_1 (NP 057167) | SJCHGC07612 AAX28307 | T78 |
| Saccharomyces cerevisiae Bub2p (NP 013771) | SJCHGC08537 AAW24955 | S41 |
As PZQ is effective against all three medically important schistosome species, new anti-schistosome chemotherapeutics intended to replace or augment PZQ therapy should ideally have similar broad-spectrum activity. However, efforts to identify novel chemotherapeutic targets in schistosomes have focused primarily on S. mansoni, with the hope that targets identified in this species will be translatable to S. haematobium and S. japonicum (Caffrey, 2007). In this study, we show that S. haematobium and S. japonicum express orthologous PKA-C subunit proteins that are 99 % identical to the SmPKA-C protein. In our previous study, we showed that inhibition of SmPKA-C expression in adult S. mansoni by RNA interference produced lethality in treated adult worms, indicating that SmPKA-C is required for adult S. mansoni viability, at least in vitro (Swierczewski and Davies, 2009). Since the nucleotide and amino acid sequences of SjPKA-C and ShPKA-C are almost identical to that of SmPKA-C, we hypothesize that ShPKA-C and SjPKA-C are also essential gene products. RNA interference studies using specific dsRNA to ShPKA-C and SjPKA-C are underway to test this hypothesis. Together, our data suggest that a novel anti-schistosome drug that targets schistosome PKA-C subunits may be useful for the treatment of all forms of schistosomiasis.
REFERENCES
- 1.Bao Y, Weiss LM, Braunstein VL, Huang H. Role of protein kinase A in Trypanosoma cruzi. Infection and Immunity. 2008;76:4757–4763. doi: 10.1128/IAI.00527-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caffrey CR. Chemotherapy of schistosomiasis: present and future. Current Opinion in Chemical Biology. 2007;11:433–439. doi: 10.1016/j.cbpa.2007.05.031. [DOI] [PubMed] [Google Scholar]
- 3.Cioli D, Pica-Mattoccia L. Praziquantel. Parasitology Research. 2003;90(Supp 1):S3–9. doi: 10.1007/s00436-002-0751-z. [DOI] [PubMed] [Google Scholar]
- 4.Das R, Esposito V, Abu-Abed M, Anand GS, Taylor SS, Melacini G. cAMP activation of PKA defines an ancient signaling mechanism. Proceedings of the National Academy of Sciences U S A. 2007;104:93–98. doi: 10.1073/pnas.0609033103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dissous C, Ahier A, Khayath N. Protein tyrosine kinases as new potential targets against human schistosomiasis. Bioessays. 2007;29:1281–1288. doi: 10.1002/bies.20662. [DOI] [PubMed] [Google Scholar]
- 6.Doenhoff MJ, Cioli D, Utzinger J. Praziquantel: mechanisms of action, resistance and new derivatives for schistosomiasis. Current Opinion in Infectious Diseases. 2008;21:659–667. doi: 10.1097/QCO.0b013e328318978f. [DOI] [PubMed] [Google Scholar]
- 7.Eck RV, Dayhoff MO. National Biomedical Research Foundation. Silver Spring; Maryland: 1966. Atlas of Protein Sequence and Structure. [Google Scholar]
- 8.Eddy SR. Multiple alignment using hidden Markov models. Proceedings of the International Conference on Intelligent Systems for Molecular Biology. 1995;3:114–120. [PubMed] [Google Scholar]
- 9.Fallon PG, Doenhoff MJ. Drug-resistant schistosomiasis: resistance to praziquantel and oxamniquine induced in Schistosoma mansoni in mice is drug specific. American Journal of Tropical Medicine and Hygiene. 1994;51:83–88. doi: 10.4269/ajtmh.1994.51.83. [DOI] [PubMed] [Google Scholar]
- 10.Gao X, Jin C, Ren J, Yao X, Xue Y. Proteome-wide prediction of PKA phosphorylation sites in eukaryotic kingdom. Genomics. 2008;92:457–463. doi: 10.1016/j.ygeno.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 11.Glass DB, el-Maghrabi MR, Pilkis SJ. Synthetic peptides corresponding to the site phosphorylated in 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase as substrates of cyclic nucleotide-dependent protein kinases. Journal of Biological Chemistry. 1986;261:2987–2993. [PubMed] [Google Scholar]
- 12.Gryseels B, Polman K, Clerinx J, Kestens L. Human schistosomiasis. Lancet. 2006;368:1106–1118. doi: 10.1016/S0140-6736(06)69440-3. [DOI] [PubMed] [Google Scholar]
- 13.Hanks SK, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB Journal. 1995;9:576–596. [PubMed] [Google Scholar]
- 14.Hanks SK, Quinn AM, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 15.Kim C, Xuong NH, Taylor SS. Crystal structure of a complex between the catalytic and regulatory (RIalpha) subunits of PKA. Science. 2005;307:690–696. doi: 10.1126/science.1104607. [DOI] [PubMed] [Google Scholar]
- 16.Lee H, Rezai-Zadeh N, Seto E. Negative regulation of histone deacetylase 8 activity by cyclic AMP-dependent protein kinase A. Molecular and Cellular Biology. 2004;24:765–773. doi: 10.1128/MCB.24.2.765-773.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Melman SD, Steinauer ML, Cunningham C, Kubatko LS, Mwangi IN, Wynn NB, Mutuku MW, Karanja DM, Colley DG, Black CL, Secor WE, Mkoji GM, Loker ES. Reduced Susceptibility to Praziquantel among Naturally Occurring Kenyan Isolates of Schistosoma mansoni. PLoS Neglected Tropical Diseases. 2009;3:e504. doi: 10.1371/journal.pntd.0000504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swierczewski BE, Davies SJ. A Schistosome cAMP-Dependent Protein Kinase Catalytic Subunit Is Essential for Parasite Viability. PLoS Neglected Tropical Diseases. 2009;3:e505. doi: 10.1371/journal.pntd.0000505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 20.Taylor SS, Zheng J, Radzio-Andzelm E, Knighton DR, Ten Eyck LF, Sowadski JM, Herberg FW, Yonemoto WM. cAMP-dependent protein kinase defines a family of enzymes. Philosophical Transactions of the Royal Society of London B: Biological Sciences. 1993;340:315–324. doi: 10.1098/rstb.1993.0073. [DOI] [PubMed] [Google Scholar]
- 21.Webster BL, Southgate VR, Littlewood DT. A revision of the interrelationships of Schistosoma including the recently described Schistosoma guineensis. International Journal for Parasitology. 2006;36:947–955. doi: 10.1016/j.ijpara.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 22.Zharkikh A, Li WH. Estimation of confidence in phylogeny: the complete-and-partial bootstrap technique. Molecular Phylogenetics and Evolution. 1995;4:44–63. doi: 10.1006/mpev.1995.1005. [DOI] [PubMed] [Google Scholar]