Abstract
Summary: A pangenome is the total of genes present in strains of the same species. Pangenome microarrays allow determining the genomic content of bacterial strains more accurately than conventional comparative genome hybridization microarrays. PanCGHweb is the first tool that effectively calls genotype based on pangenome microarray data.
Availability: PanCGHweb, the web tool is accessible from: http://bamics2.cmbi.ru.nl/websoftware/pancgh/
Contact: sacha.vanhijum@nizo.nl
1 INTRODUCTION
Pangenome microarrays contain probes that target all known genes within related strains of the same species (Tettelin et al., 2005).When compared to conventional comparative genome hybridization (CGH) microarrays that target the gene content of a single species, they allow to more accurately determine the genotype of a given bacterial strain (Bayjanov et al., 2009; Castellanos et al., 2009; Willenbrock et al., 2007). In pangenomes, orthologous genes can be defined as homologous genes derived by a strain divergence event from a single ancestral sequence. These orthologous genes (strain orthologs) share different levels of nucleotide sequence identity with paralogous genes (homologous genes derived by a duplication event from a single sequence) (Fitch, 1970). Effective genotyping can be achieved by grouping genes into ortholog groups (OGs) and subsequently genotyping at the level of OGs. Recently, we published an algorithm (PanCGH) that effectively deals with assigning OG presence/absence to each strain analyzed by pangenome microarrays (Bayjanov et al., 2009). Here, we describe a web tool—PanCGHweb—that uses this algorithm to effectively genotype strains based on pangenome microarray data.
2 METHODS
2.1 Implementation
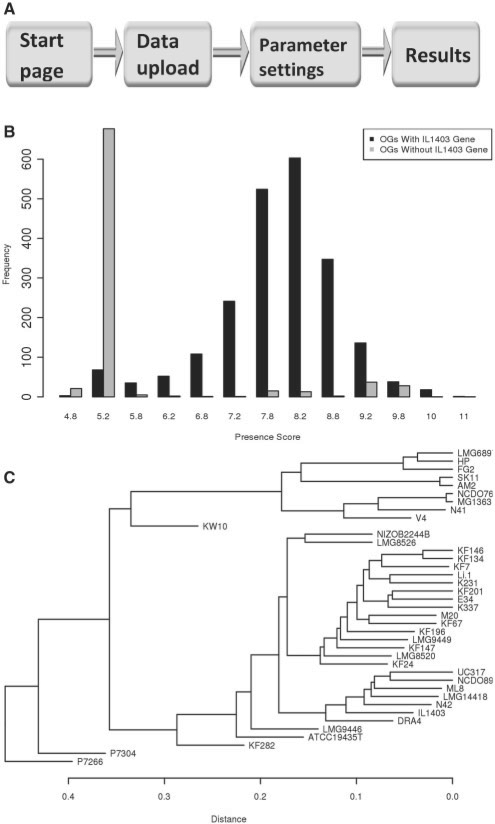
PanCGHweb is implemented in Python and R, and its wizard-like web-interface is generated by the FG-web framework (S.A.F.T.van Hijum et al., unpublished data). There are three major sections in the web-interface: (i) data upload; (ii) parameter settings; and (iii) displaying the results (Fig. 1A). The web tool works with major web browsers such as Internet Explorer, Firefox, Safari and Opera.
Fig. 1.
The PanCGHweb web tool. (A) Process flow in PanCGHweb. (B) Histogram of presence/absence of OGs for a reference strain (in this example Lactococcus lactis IL1403). Horizontal axis: presence score of OGs. Vertical axis: number of OGs with a corresponding presence score. Black bars: frequency of presence score of OGs that contain at least one gene from the reference strain. Gray bars: frequency of presence score of OGs that do not contain gene from the reference strain. (C) Phylogenetic tree of strains based on presence/absence of OGs in 39 L. lactis strains.
2.2 Input data
Open reading frame sequences for each reference bacterial strain and/or plasmid, on which probes were designed, should be provided by (i) selecting from the available daily updated Genbank sequences and (ii) optionally, uploading FASTA-formatted DNA sequences that are absent in the Genbank list. Normalized microarray hybridization data, where replicated measurements are represented by a single value (e.g. by averaging), should also be provided as tab-delimited file(s). Probe sequences should be provided in FASTA format.
2.3 Algorithm
The PanCGH algorithm calls presence/absence of OGs based on pangenome microarray data. PanCGHweb performs the following steps: (i) orthology grouping; (ii) alignment of probes to genes; and (iii) genotype calling.
Step 1: Inparanoid (Remm et al., 2001) is used with its default settings (minimum bit score of 50 and confidence score of 0.25) for the orthology prediction among genes of the selected reference genomes (Genbank files; see above). The run time of Inparanoid is reduced by a few orders of magnitudes by adapting the software to use BLAT (Kent, 2002) for sequence alignments. Genes that are not part of the selected reference genomes can be grouped based on their homology, or each gene can form a separate group.
Step 2: the microarray probes are aligned by BLAT to the individual gene members of each OG. Probes that could not be aligned to any gene and genes with no matching probes are reported.
Step 3: using the PanCGH algorithm (Bayjanov et al., 2009) the fluorescence signal intensities of probes associated to each gene are summarized to a gene score (the most frequently occurring signal intensity). The maximum of gene scores of all gene members of an OG is used as the presence score for that OG. An OG is considered to be present if its presence score is above the threshold of 5.5 in log scale. The steps involved in determining the optimal threshold value are described on the web site of PanCGHweb.
2.4 Output of the algorithm
Results of PanCGHweb include: (i) projection plot, which overlays presence/absence of OGs on the selected genomes; (ii) histogram of presence score of OGs for any reference strain, which can be used to validate whether the default threshold of 5.5 is an optimal choice for presence/absence calling (Fig. 1B); (iii) receiver operating curves using all possible presence/absence calling thresholds for all reference strains; (iv) two different phylogenetic trees of strains, one based on presence/absence values and the other based on presence scores. Such trees enable estimating the genomic diversity among all strains (Fig. 1C); (v) hierarchical tree based on signal intensity values of all arrays; (vi) box and whisker plot that shows signal intensity distribution among all arrays; and (vii) orthology grouping information and presence/absence of genes in each strain. Additionally, the following tab-delimited files can be downloaded: OGs list, alignment of probes to genes, presence/absence of OGs and presence score of OGs.
3 CONCLUSIONS
For genotyping, pangenome microarrays offer a cost-effective alternative to DNA sequencing and allow to more accurately determine genomic content compared to standard CGH techniques. We have developed a web tool for pangenome microarray analysis based on our PanCGH algorithm. It enables researchers to analyze these complex hybridization data in a facile and transparent way to understand genomic diversity among related strains.
ACKNOWLEDGEMENTS
We thank Christof Francke and Bas Dutilh for useful discussions.
Funding: Besluit Subsidies Investeringen Kennisinfrastructuur (BSIK) grant [through the Netherlands Genomics Initiative (NGI)]; BioRange programme [as part of, the Netherlands Bioinformatics Centre (NBIC)]; and NGI (as part of the Kluyver Centre for Genomics of Industrial Fermentation).
Conflict of Interest: none declared.
REFERENCES
- Bayjanov JR, et al. PanCGH: a genotype-calling algorithm for pangenome CGH data. Bioinformatics. 2009;25:309–314. doi: 10.1093/bioinformatics/btn632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellanos E, et al. Discovery of stable and variable differences in the Mycobacterium avium subsp. paratuberculosis type I, II, and III genomes by pan-genome microarray analysis. Appl. Environ. Microbiol. 2009;75:676–686. doi: 10.1128/AEM.01683-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch WM. Distinguishing homologous from analogous proteins. Syst. Zool. 1970;19:99–113. [PubMed] [Google Scholar]
- Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remm M, et al. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol. 2001;314:1041–1052. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- Tettelin H, et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc. Natl Acad. Sci. USA. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willenbrock H, et al. Characterization of probiotic Escherichia coli isolates with a novel pan-genome microarray. Genome Biol. 2007;8:R267. doi: 10.1186/gb-2007-8-12-r267. [DOI] [PMC free article] [PubMed] [Google Scholar]