Abstract
Escherichia coli (E. coli) is a versatile organism for making nucleotides labeled with stable isotopes (13C, 15N, and/or 2H) for structural and molecular dynamics characterizations. Growth of a mutant E. coli strain deficient in the pentose phosphate pathway enzyme glucose-6-phosphate dehydrogenase (K10-1516) on 2-13C-glycerol and 15N-ammonium sulfate in Studier minimal medium enables labeling at sites useful for NMR spectroscopy. However, 13C-sodium formate combined with 13C-2-glycerol in the growth media adds labels to new positions. In the absence of labeled formate, both C5 and C6 positions of the pyrimidine rings are labeled with minimal multiplet splitting due to 1JC5C6 scalar coupling. However, the C2/C8 sites within purine rings and the C1′/C3′/C5′ positions within the ribose rings have reduced labeling. Addition of 13C-labeled formate leads to increased labeling at the base C2/C8 and the ribose C1′/C3′/C5′ positions; these new specific labels result in two- to three-fold increase in the number of resolved resonances. This use of formate and 15N-ammonium sulfate promises to extend further the utility of these alternate site specific labels to make labeled RNA for downstream biophysical applications such as structural, dynamics and functional studies of interesting biologically relevant RNAs.
Keywords: Alternate-site specific labeling, Formate enhanced isotope enrichment, Ribose and nucleobase, RNA, Structure and dynamics
Introduction
Nucleic acids and proteins can be labeled with stable isotopes for structural and dynamics studies (Dayie 2008) using E. coli as a common bacterial host (Ponchon and Dardel 2007; Ponchon et al. 2009), using enzymes from the pentose phosphate or de novo purine biosynthetic pathways (Gross et al. 1983; Parkin et al. 1984; Tolbert and Williamson 1996, 1997; Scott et al. 2000; Schultheisz et al. 2008), or using chemical synthesis (Milecki 2002).
Of these three methods, use of different E. coli bacteria grown on minimal media is attractive for a number of reasons. E. coli grown on chemically defined minimal media supplemented with 15N-labeled nitrogen and 13C-labeled carbon sources (Batey et al. 1995; Hoffman and Holland 1995; Nikonowicz 2001; Latham et al. 2005; Dayie 2008) produces isotopically labeled total cellular RNA that can be enzymatically digested to nucleoside monophosphates (NMPs). The NMPs can be phosphorylated to the corresponding nucleoside triphosphates (NTPs) and these labeled NTPs become the building blocks for making labeled RNA using T7 RNA polymerase. Of the different E. coli strains that offer potential for uniform and alternate site specific 13C isotopic labeling, the E. coli strain with a knockout of the zwf gene is particularly attractive. Deleting the zwf gene means this E. coli strain K10-1516 (hereafter referred to as K10) cannot encode the glucose-6-phosphate dehydrogenase (G6PDH) enzyme for use in the pentose phosphate pathway(PPP; Fraenkel 1968). Growth of this strain on 2-13C-glycerol without formate provides labeling mostly at the C2′ and C4′ ribose positions and at both C5 and C6 base positions of the cytidine monophosphate (CMP) with minimal multiplet splitting due to 1JC5C6 scalar coupling (Johnson et al. 2006; Hoogstraten and Johnson 2008).
These previous labeling methods offered the advantage of enriching only a few specific sites such as C8 in purine (Pur, adenine or Ade and guanine or Gua) and ribose C2′ and C4′. However, there are at least two disadvantages with such schemes. First, many useful sites are only fractionally enriched. For example, the ribose C1′ and C5′ atoms and the purine C2 and C8 positions are labeled at a very low level when using K10. Yet the C1′ carbon atoms have the most sugar chemical shift dispersion and C5′ carbon atoms along with their attached protons can provide valuable γ-torsion angle information (Wijmenga and van Buuren 1998). In addition, the use of 13C-formate and 12C-glucose as the only carbon sources limits the incorporation of an isotopic label to the C8 position of adenine and guanine bases and precludes their use for other NMR studies. Second, it is more time and cost efficient if the same sample can be used for numerous applications such as the analyses of the function, structure and dynamics of RNAs. Limiting the labels to a few selected ribose and base atoms greatly lessens the general usefulness of the resulting nucleotides. Here we sought to extend the utility of these labels while preserving the advantage of isolating most of the spin systems of interest and boosting the isotopic enrichment level of the other useful NMR sites.
We reasoned that by combining 13C-glycerol with 13C-formate, all the protonated carbons of high interest for RNA NMR spectroscopy will be labeled in both purine and pyrimidine (Pyr, that is cytosine or Cyt, uracil or Ura, and thymine or Thy) bases. In addition, the isotopic enrichment at sites within the ribose ring will increase without introducing multiplet splitting at key atomic sites such as C1′ and C5′. We can augment the level of isotopic enrichment at all the sites useful for proton-detected heteronuclear NMR experiments by adding 13C-formate to the 13C-glycerol growth media. Plus, the use of 15(NH4)2SO4 as the sole source of nitrogen leads to uniform and complete 15N labeling of all nitrogen positions in all nucleotides and it provides an alternative method for estimating the level of isotopic enrichment. This labeling method appears to be general and flexible, and affords a wide variety of purine and pyrimidine isotope labeling patterns useful for structural, functional and dynamics studies.
Materials and methods
Bacterial strains
The mutant strain K10-15-16 (CGSC # 4858 Hfr fhuA22, zwf-2, relA1, T2R, pfk-10) used in this work was obtained from the Yale Coli Genetic Stock Center.
Isotopes
All labeled compounds were bought from Cambridge Isotope Laboratory (Andover, MA) and Isotec-Sigma–Aldrich (Miamisburg, OH) with the following isotopic enrichments: [13C] sodium formate (99%), [2-13C] glycerol (99%) and [15N]-(NH4)2SO4 (99%).
Stock solutions
The stock solutions required for bacterial growth were prepared using distilled and deionized heat sterilized water. The Studier medium (SPG) contained the following (Studier 2005): 25 mM·(NH4)2SO4, 50 mM·KH2PO4, 50 mM·Na2HPO4, 2 mM·MgSO4 and trace metal solution; SPG was then supplemented with labeled glycerol or labeled formate and glycerol as needed.
Media for bacterial growth
Luria–Bertani (LB) and SPG minimal media were prepared as described (Sambrook and Russell 2001; Studier 2005). Each media was supplemented with the amount of the carbon source (sodium formate and glycerol) and nitrogen source (ammonium sulfate) that gives optimal growth conditions.
Protocol for growth optimization
The growth of each bacterial strain was optimized for the highest production of biomass per input gram of carbon source. Briefly, the first evening (Day 1) a glycerol stock of the K10 strain was plated on fresh LB agar plates with no antibiotic and incubated overnight at 37°C. The following evening (Day 2), a 5 ml starter culture in SPG medium supplemented with unlabeled carbon sources was inoculated with a single colony and incubated overnight at 37°C (i.e., 12–16 h). At the end of this incubation period (Day 3), the solution was pelleted at 3000 rpm for 5 min; the pellet was washed two times in 1x PBS and centrifuged at 3000 rpm for 5 min. This cell pellet was resuspended in 5 ml of fresh SPG medium without any carbon source; 1 ml of this resuspension was added to a 50 ml culture in SPG medium with unlabeled carbon source and incubated at 37°C for 12–14 h with shaking at 270 rpm, making sure the OD600 did not saturate. At the end of this incubation, the solution was centrifuged, the resultant pellet washed twice in 1x PBS and the pellet was resuspended in 50 ml fresh SPG medium without any carbon source. Then 5 ml was added to two 500 ml SPG medium containing labeled carbon source. About 10 ml of labeled media was saved for resuspension of the 500 ml culture pellet. The 500 ml labeled culture was incubated at 37°C for 12 h.
Nucleic acid digestion and cis diol affinity column purification
The isotopically enriched ribonucleotides were isolated from E. coli as described earlier (Batey et al. 1995). The cell pellet was resuspended in 20 ml STE buffer (0.1 M NaCl, 10 mM Tris, 1 mM EDTA, pH 8) for two grams of frozen cells and lysed in STE with 0.5% SDS. The cellular proteins were removed with 25:24:1 Phenol:Chloroform:Isoamyl mix. The upper aqueous layer, pooled from multiple extractions of the organic and inclusion layers, was back-extracted with an equal volume of 24:1 Chloroform:Isoamyl alcohol. The residual nucleic acids were precipitated overnight with ethanol and acetate buffer (pH 5.2) at −20°C. The cellular nucleic acids were digested with nuclease P1 in 15 mM sodium acetate, 0.1 mM·ZnSO4, pH 5.2. The deoxyribonucleotides were separated from the ribonucleotides using a cis-diol boronate affinity chromatography in a 20 × 2.5 cm glass column. The digested nucleic acid solution was filtered and loaded onto a boronate affinity resin (10 g of Affigel 601 from Biorad) pre-equilibrated with 1 M TEABC at pH 9.5 at 4°C. The deoxyribonucleotides, salts, and other impurities were washed from the boronate column using five column volumes of 1 M TEABC. The ribonucleotides were eluted with water acidified with CO2 and the purity of the rNMPs was verified by NMR.
NMR experiments
NMR experiments were run on a four channel Bruker Avance 600 MHz spectrometer equipped with actively shielded z-axis gradient triple resonance probe. NMR data sets were processed and the peak positions and intensities were analyzed with Bruker’s TOPSPIN 2.1. The rNMP fractions were analyzed by 1H and 13C NMR experiments. One dimensional (1D) 13C spectra were collected with a 45° and 90° tip angles, and GARP (Shaka et al. 1985) 1H decoupling was applied only during acquisition. 1D 1H spectra were also collected without 13C decoupling during acquisition. All spectra, unless indicated, were collected at 25°C. Two-dimensional non-constant-time (1H, 13C) HSQC (Mueller 1979; Bodenhausen and Ruben 1980; Bax et al. 1990) were recorded to resolve ambiguities arising from overlap in the 1D spectra. Spectra requiring quantitative analysis were acquired with a long recycle delay (5 s) to ensure sufficient recovery of magnetization.
For the base region, the two-dimensional (2D) experiments were acquired with sweep widths of 8013 Hz in the 1H acquisition dimension and 3322 Hz in the 13C dimension. For each data set, 8 scans and 1024 complex points were collected in t 2 and 256 complex points were collected in t 1 using the Echo-Anti echo method (Palmer et al. 1991; Kay et al. 1992) for quadrature detection. 13C-GARP (Shaka et al. 1985) decoupling was applied during the acquisition period or omitted to obtain residual labeling information. Proton and carbon carrier was placed at 4.7 ppm and 142.5 ppm respectively. An INEPT delay of 2.5 ms (optimized for 1JHC = 200 Hz for the base region and corresponding to 1/(2*JHC)) was used for magnetization transfer. For the ribose region 2D experiments were acquired with sweep widths of 8013 Hz in the 1H dimension and 5058 Hz in the 13C dimension. For each data set, 8 scans and 1024 complex points were collected in t 2 and 256 complex points were collected in t 1 using the Echo-Anti echo method (Palmer et al. 1991; Kay et al. 1992) for quadrature detection. 13C-GARP decoupling was applied during the acquisition period. Proton and carbon carrier was placed at 4.7 ppm and 76.5 ppm respectively. An INEPT delay of 3.29 ms (optimized for 1JHC = 152 Hz for the ribose region and corresponding to 1/(2*JHC)) was used for magnetization transfer. The time domain data was zero filled in t 1 and t 2 before Fourier transformation to give a final real matrix size of 2048 × 1024 points.
Two- and three-bond 2D (1H, 15N) HSQC experiments were acquired as follows. For the 1H acquisition period (t 2), sweep widths of 2761 Hz (4.6 ppm) were used, and for the 15N evolution period (t 1), sweep widths of 4563.8 or 6815.2 Hz (75 or 112 ppm) were used. For each data set, 16 or 32 scans and 1024 complex points were collected in t2 and 128 complex points were collected in t1 using the Echo-Anti echo method for quadrature detection. Proton, carbon and nitrogen carrier was placed respectively at 4.7 ppm, 120.0 ppm and 202.5 (or 192) ppm. 15N-GARP decoupling (Shaka et al. 1985) was applied during the acquisition period, and 13C-GARP decoupling (Shaka et al. 1985) was also applied or omitted during the acquisition period. An INEPT delay of 15.6 or 25 ms (optimized for 2JHN = 16 or 20 or 25 Hz for the two bond coupling to the purine nitrogen and corresponding to 1/(2*JHN)) was used for magnetization transfer. These delays did not affect the % level labeling prediction (see below).
Relative peak intensities were determined by integrating peaks observed with proton decoupling during acquisition only and a long recycle delay (5 s) to allow sufficient magnetization recovery for the direct carbon experiments. For the 2D experiments, all data were plotted to the same base level, level of increment and number of contour levels before peak picking and peak integration.
To simulate the effect of a medium-sized RNA, we ran non-constant time 2D 13C HSQC and 13C TROSY (Meissner et al. 1998; Pervushin et al. 1998; Czisch and Boelens 1998; Weigelt 1998; Rance et al. 1999; Zhu et al. 1999; Schulte-Herbrüggen and Sorensen 2000) on a mixture of all four nucleotides dissolved in 95% w/w perdeuterated glycerol (Cambridge Isotope Labs, Andover, MA) at 30°C.
Results and discussion
The ability to transcribe RNA (or DNA) labeled with various isotopes, such as 13C, 15N and 2H, has enabled the application of heteronuclear multi-dimensional NMR techniques to characterize the structure and dynamics of interesting biological RNA molecules (D’Souza et al. 2004; Gumbs et al. 2006; Lu et al. 2010). Unsurprisingly a number of research groups have developed and continue to develop techniques for the biosynthetic production of isotopically labeled nucleotides. Some produce uniformly labeled nucleotides from E. coli (Nikonowicz et al. 1992; Michnicka et al. 1993), M. methylotrophus (Batey et al. 1992), or Methylophilus extorquens (Hines et al. 1994). Others produce site specifically labeled nucleotides from E.coli (Latham et al. 2005; Johnson et al. 2006), and still others produce these labels using the pentose phosphate and de novo purine synthetic pathways (Schultheisz et al. 2008). Similar methods for the production of uniform isotopically labeled deoxynucleosides have been proposed (Zimmer and Crothers 1995; Louis et al. 1998; Masse et al. 1998; Werner et al. 2001; Nelissen et al. 2009). Previously, 13C-formate added to an unlabeled glucose minimal medium enabled selective labeling of purine C8 positions (Latham et al. 2005), and use of E. coli deficient in the G6PDH gene enabled the site-labeling of pyrimidine C5 or C6 positions as well as various ribose carbon positions (Johnson et al. 2006). To our knowledge, no published reports have shown the combined advantages of both: Grow E. coli strain K10 on 13C-labeled glycerol with 13C-formate to overcome the limitations of each separate method. Using 2 g of 13C-2-glycerol and 0.7 g of NH4(SO4)2 as carbon and nitrogen sources, we obtained 4 g of wet cell pellet for a liter of E. coli K10 cell culture. Addition of 0.2 g of 13C formate to this medium was sufficient to label sites not otherwise labeled. Yields of up to 66 mg of rNMPs and 13 mg of dNMPs obtained are comparable to previous reports of labeling (Batey et al. 1992). To obtain optimal yields it is important to use minimal media other than M9. As reported by others, M9 is inferior to other buffers (Paliy and Gunasekera 2007; Dayie unpublished).
Incorporation of 13C into ribose ring of nucleotides using the pentose phosphate pathway
To place our results within the context of the metabolic pathways in E. coli for nucleotide metabolism, we present a brief overview of some of the steps in glycolysis, gluconeogenesis and the tricarboxylic acid (TCA) cycle. Details can be found in standard textbooks (Voet et al. 2008; Nelson and Cox 2008).
Glycerol enters the metabolic cycle as dihydroxyacetone phosphate (DHAP) and equilibrates rapidly with glyceraldehyde-3-phosphate (GA3P) by the action of triosephosphate isomerase (TIM). From this point in the cycle, GA3P can be converted to ribose-5-phosphate (R5P) via gluconeogenesis using the oxidative pentose phosphate pathway (oPPP; Fig. 1). Alternatively GA3P and fructose-6-phosphate (F6P) can also be converted to R5P via the action of transketolase and transaldolase in the reverse of the non-oxidative PPP (noPPP; Fig. 1). In wild type E. coli both the oPPP and noPPP can be operative and the partitioning of the carbon flux through these two pathways leads to scrambling of labels in the ribose ring. However, for strains defective in the oPPP such as the K10 E. coli strain, most of the carbon flux is re-routed through the reverse noPPP via the action of transketolase and transaldolase (Edwards and Palsson 2000; Zhao et al. 2004; Nicolas et al. 2007). Assuming auxiliary biosynthetic pathways contribute negligibly to the central pathway, a 13C-label at the central C-2 carbon of glycerol would lead to isotopic enrichment for [2, 4-13C2]ribose and [4-13C]ribose in a 2:1 ratio (Fig. 1), and no label is expected at the 1, 3 or 5 ribose carbon positions (Johnson et al. 2006).
Fig. 1.
Major metabolic pathways involved in the production of nucleic acid nucleotides, including key steps in glycolysis, gluconeogenesis and one pass through the tricarboxylic (TCA) cycle. For E. coli carrying the zwf genotype (glucose 6-phosphate dehydrogenase (G6PDH) mutant), the oxidative branch of the pentose phosphate pathway is disabled (indicated by an X through the orange arrow) such that most of the carbon fluxes are shunted through the reverse non-oxidative pentose phosphate pathway (noPPP). Atom labels for the terminal (1, 3) carbons (magenta and thin circle) and central (2) carbon (cyan and thick circle) of glycerol are highlighted. Positions that are enriched due to the presence of 13CO2 (as bicarbonate) in the growth medium are shown with an encircled X, but this is lost through the first and subsequent pass through the TCA cycle. Pyrimidine bases derived from oxaloacetate (OAA) produced by carboxylation of phosphoenolpyruvate (PEP) is shown via the aspartate intermediate. This OAA is used as a substrate in the first and subsequent rounds of the TCA cycle to produce OAA with a pair of different labeling schemes as products due to the symmetric nature of the TCA cycle intermediate succinate. If [2-13C]glycerol is used Cα or Cβ or Cγ or Cβ and Cγ but not all three positions are labeled simultaneously. Similarly the labeling pattern of purines from glycine (Gly) derived from 3-phosphoglycerate (3PG) are shown such that if [2-13C]glycerol is used only the Cα position of Gly and therefore C5 position of the purine ring is labeled. The use of GA3P and F6P in the reverse of the non-oxidative PPP produces ribose labeled at the 2,4 and 4 positions if [2-13C]glycerol is used
Exogenous formate can enter the metabolic cycle by exchanging the carboxyl group of pyruvate by consuming acetyl-CoA (Thauer et al. 1972; Knappe et al. 1974) by the reversible action of pyruvate formate lyase (Kirkpatrick et al. 2001). This modified pyruvate may populate gluconeogenesis intermediates such as GA3P and F6P for use in the reverse of the noPPP. At the moment, the effect of exogenous formate on E. coli growth remains poorly characterized and poorly understood. Nonetheless, as we show later, addition of formate has an unexpected effect of increasing the level of enrichment at the ribose carbon positions predicted to have no label using the central metabolic pathway.
Incorporation of 13C into base ring of nucleotides via the tricarboxylic acid cycle, glycolysis, and gluconeogenesis
Various metabolic precursors make amino acids from which nucleotide bases are synthesized (Fig. 1). 3-phosphoglycerate (3PG) gives rise to serine (Ser) and glycine (Gly), and oxaloacetate (OAA) gives rise to aspartic acid (Asp). In turn, the six-membered Pyr ring is constructed from four atoms of Asp such that the NH amide group, the Cα-, Cβ- and Cγ-carbon positions of Asp becomes the N1, C6, C5 and C4 ring atoms respectively of Pyr (Fig. 1). The N3 and C2 positions are derived from glutamine amide and bicarbonate pools respectively. The bicarbonate single carbon pool is diluted by 12C carbons such that labeling at the Pyr C2 position is random at low levels unless this carbon pool is augmented with 13C-bicarbonate (Lundström et al. 2007). The larger Pur ring atoms C2 and C8 also derive from the formate pool. Thus, adding 13C-formate to the growth media is again expected to increase the level of 13C isotopic enrichment at the C2 and C8 sites. The amide group, the Cα- and carbonyl (CO)-carbon positions of glycine (Gly) become the N7, C5 and C4 ring atoms respectively (Fig. 1). We use Fig. 1 as a framework for interpreting some of our results with E. coli strain K10.
Label incorporation by E. coli strain K10 in the absence of 13C-labeled formate
E. coli strain K10 grown in 13C-2-glycerol media in the absence of labeled formate has varied labeling patterns in both ribose and base moieties (Fig. 2; Table 1). The ribose ring is labeled exclusively at the C2′ and C4′ positions (>80% label) as expected for the metabolic carbon flux going mostly through the transketolase/transaldolase branch of the noPPP. Little labeling is observed at C3′, and the negligible carbon–carbon splitting at C2′ and C4′ positions (Fig. 2a–c) further bears out the prediction from the analysis of the metabolic pathway. However, some residual labeling is observed at the C1′ (~1%) and C5′ (~1%) positions. The isotopic enrichment level at C1′ and C5′ increases in the presence of formate (as discussed further below). This residual labeling suggests gluconeogenesis might be significant in this mutant when grown on glycerol. Alternatively, a fraction (~7%) of serine molecules is predicted to be produced by a bypass of the disabled G6PDH in the zwf mutant (Fischer and Sauer 2003). Further studies such as metabolic flux analysis using gas chromatography–mass spectrometry (GC–MS) and NMR spectroscopy are needed to address the origin of these residual labels fully (Fischer and Sauer 2003).
Fig. 2.
Increased level of labeling in K10 without (red) and with (blue) 13C-formate in a 13C-2-glycerol background. The experiments were performed on mixtures of the four rNMPs isolated from the K10 bacterial culture. a Direct carbon detection 1D spectrum showing all the carbon positions for nucleotides labeled with glycerol and no formate (bottom, red) or glycerol with formate (top, blue). A long recycle delay of 5 s were used to allow for sufficient magnetization recovery and proton decoupling was limited to the acquisition period only. The level of enrichment at the adenine (Ade) and guanine (Gua) C8 positions increases by spiking with 13C-labeled formate. The C5′ region has an impurity that resonates in a distinct region in the 2D spectrum. b 2D non-constant time HSQC spectrum of a mixture all four labeled rNMPs showing the protonated base region. For ease of comparison the spectrum obtained without labeled formate (red contours) are displaced vertically relative to the formate labeled spectrum (blue contours). c 2D non-constant time HSQC spectrum of a mixture of all four labeled nucleotides showing the ribose region. The cytosine (Cyt) and Uracil (Ura) C5 resonances at 96.67 ppm and 102.69 ppm respectively are folded into the spectrum. The boxed resonances highlight the increased labeling level seen for C1′, C3′ and C5′ with spiking the growth medium with 13C-labeled formate
Table 1.
13C enrichment levels at various carbon positions within ribonucleotides using [2-13C]-glycerol with and without 13C-labeled formate as carbon sources using E. coli strain K10
| Carbon position labeled | 13C-Carbon Source: 2-Glycerol only | 13C-Carbon Source: 2-Glycerol and Formate |
|---|---|---|
| Purinea | ||
| Ade C2 | 13.6 ± 2.7 | 26.4 ± 2.0 |
| Ade C8 | 10.0 ± 1.0 | 35.8 ± 1.8 (35.8)b |
| Gua C8 | 10.0 ± 1.0 | 37.8 ± 1.5 (39.8)b |
| Pyrimidinea | ||
| C5 | 44.7 ± 1.2 | 49.0 ± 2.9 |
| C6 | 45.7 ± 1.4 | 42.0 ± 5.5 |
| Ribosec | ||
| C1′ | 0.7 ± 0.3 | 3.0 (5.0 ± 1.9)d |
| C2′ | 90 ± 10 | 90 ± 10 |
| C3′ | <1 | 8.8 ± 1.5 |
| C4′ | 90 ± 10 | 90 ± 10 |
| C5′ | 0.7 ± 0.3 | 10.6 ± 0.9 |
aThe percentage label (Plabel) is calculated as the ratio of the sum of the intensities of satellite peaks to the sum of the intensities of the satellite and center peaks using the 2-bond 15N HSQC without 13C decoupling during acquisition; in Fig. 3c the satellite peaks are labeled I and II, the center peak is labeled III and PLabel = (I + II)/(I + II + III)
bThe numbers in parenthesis are calculated as the ratio of the sum of the intensities of satellite peaks to the sum of the intensities of the satellite and center peaks from the 1D 1H spectrum acquired without 13C decoupling
cFor the ribose region the degree of labeling is estimated using the percentage labeling relative to the C2′ and C4′ peak intensities, and each relative percentage labeling is scaled by 97%, assuming C2′ and C4′ positions are labeled at 97% level
dThe numbers in parenthesis are derived from the ratio of the sum of the intensities of satellite peaks to the sum of the intensities of the satellite and center peaks using the 2-bond 15N HSQC without 13C decoupling during acquisition
For the base atoms, both the protonated C5 and C6 carbon positions of Pyr are substantially labeled at ~45% close to the expected 50% level, whereas the protonated C2 and C8 carbon positions of Pur are labeled at a lower level (~10–14%; Fig. 2a–b). The C5 and C6 pyrimidine sites are constructed entirely from Asp which in turn is generated from OAA either by direct carboxylation of PEP or from the TCA cycle. Using [2-13C]-glycerol as the sole carbon source, Asp formed from carboxylation of PEP (using cellular bicarbonate breakdown to CO2 by pyruvate carboxylase) is expected to be 100% enriched exclusively at the Cα position or equivalently the C6 position of Pyr. A single pass through the TCA cycle leads, because the TCA cycle metabolite succinate is symmetric, to an equal probability of labeling either the Cα or the Cβ position. But both positions cannot be labeled simultaneously in the same molecule. Thus either the C5 or the C6 position of Pyr is labeled at 50% maximum enrichment with no undesired C5-C6 labeled pair. In the second pass through the TCA cycle, the C4 carbon is also labeled to a maximum value of 25%; subsequent passes through the cycle will reduce even further this level of labeling at C4. Those molecules labeled at C4 are predicted to have no label at either the C5 or the C6 position. Thus there should be no coupling between C4 and C5 or C4 and C6.
The Pur C2 and C8 positions arise from metabolic breakdown product of formate and the Pur C6 and Pyr C2 atomic positions arise from bicarbonate byproduct. As a result these sites are expected to be randomly labeled at very low levels in the absence of spiking the growth media with 13C-labeled formate or bicarbonate.
Label incorporation by E. coli strain K10 in the presence of 13C-labeled formate
Addition of 13C-formate leads to increased labeling in both ribose and base moieties (Fig. 2; Table 1). In the ribose ring, labeling increases for the C1′ (3–5%), C3′ (~9%) and C5′ (~11%) positions without introducing significant carbon–carbon coupling at these positions (C1′, C2′, C4′ and C5′; Fig. 2). These labeling efficiencies can be estimated from a comparative analysis of the 1D carbon spectra of uniformly labeled rNMPs and the site specific labeled rNMPs derived from the K10 bacteria culture. As discussed below a different method using two-bond 15N HSQC gives comparable results. Nonetheless it is unexpected that in the face of >80% labeling of C2′ and C4′, C4′–C5′ and C1′–C2′ splittings are not observed. Analysis of the reverse noPPP suggests oxaloacetate generated by several passes through the TCA cycle will populate a pyruvate intermediate that could ultimately label R5P at the C1′ and C5′ positions with exclusion of labels at C2′ and C4′ positions in the same molecule. This is in addition to the expected labels at C2′ and C4′ without adjacent labels at C1′ and C5′ in the same molecule. Alternately a bypass of the disabled G6PDH in the zwf mutant catalyzed by the perisplasmic glucose dehydrogenase (Fischer and Sauer 2003) could potentially produce a label at the C1′ and C5′ without any coupled adjacent labels. Further study using GC–MS and NMR are needed to resolve this empirical observation of label at the C1′ and C5′ positions.
A similar increase in the labeling level is observed in the base region on addition of labeled formate to the 13C-2-glycerol media. Significant isotopic enrichment of the C8 (~40%) and C2 (~26%) carbon positions of the Pur ring are observed, but those at the C6 and C5 Pyr positions remain unchanged (Fig. 2b; Table 1).
Estimation of the degree of 13C isotope incorporation using two- and three-bond 15N HSQC
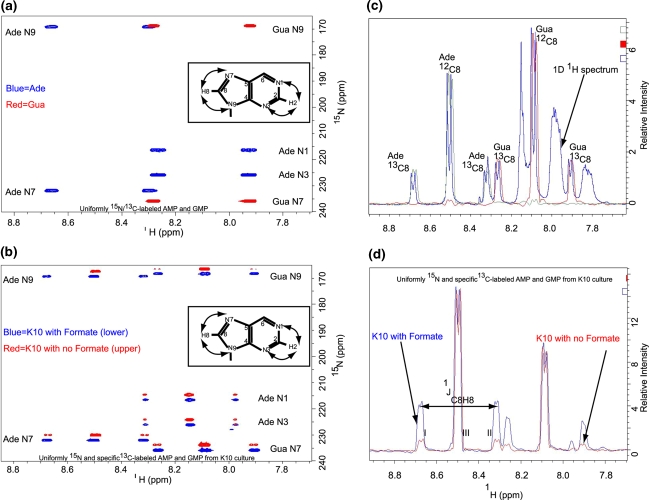
Finally, addition of labeled 15N-ammonium sulfate enables high level labeling of the aromatic nitrogens and estimation of the degree of 13C isotope incorporation. The level of 13C labeling efficiency is usually estimated using 1D 1H or natural abundance 13C carbon spectra. Lack of a central singlet peak and the presence of doublet satellite peaks indicate close to 100% labeling efficiency. Absence of the doublet satellite peaks and the presence of a dominant central peak are then taken as lack of 13C incorporation. Thus by comparing the intensity of each 13C satellite peak to the intensity of the center peak, the labeling efficiency is readily estimated. This 1D approach works well for single nucleotides that have no spectral overlap. For a mixture of the four rNMPs extracted from the K10 bacteria culture, there is significant overlap in both the base and ribose regions. For example Ade H1′ (6.02 ppm) overlaps completely with Cyt H5 (6.02 ppm) in the proton chemical shift region, and Ura H1′ (5.90 ppm) overlaps with Cyt H1′ (Fig. 2c). This overlap problem limits the usefulness of the 1D method. Long range (two- and three-bond) proton–nitrogen correlations in 15N-HSQC spectra make it possible to estimate the labeling efficiency of the C2 and C8 carbon sites within the Pur aromatic ring, the C5 and C6 carbon sites within the Pyr aromatic ring and the Pur C1′ carbon site (Fig. 3). Relaxation properties and transfer efficiencies are different for long range and one-bond magnetization transfers, and so it is important to validate the use of the long range 15N-HSQC method to estimate the level of 13C incorporation. The 1D slices from the 2D 2JHN HSQC spectra (Fig. 3d) overlay completely with the 1D 1H spectrum (Fig. 3c), suggesting the percentage label can be estimated using either the 2D or 1D experiment, but the 2D is preferable in case of overlap. With this experiment one can correlate the H2 proton resonances to the N1 and N3 nitrogen positions in the adenine (Ade) ring, and also the H8 proton resonances to the N7 and N9 nitrogen positions in the Pur ring (Fig. 3b). By omitting the carbon decoupling field during the proton acquisition period, the proton resonances are split by the directly attached 13C atom (C2 or C8) into a doublet (Fig. 3a–b). Using this method, the 1JCH coupling constants measured for uniformly labeled AMP, CMP, UMP, and GMP are in excellent agreement with previous reported measurements. For uniformly 13C/15N-labeled AMP and GMP, the 2D method, in excellent agreement with the 1D 1H method, gives 98.9% 13C isotopic enrichment at the C8 positions. As expected, each of the H2 and H8 proton resonance is split into a doublet with little central peak in the acquisition dimension (Fig. 3a). As the level of 13C isotopic enrichment decreases from 100 to 0%, each doublet gives rise to a central singlet. Analyses of the multiplet pattern of the four labeled nucleotides derived from the K10 bacteria cultures facilitated the estimation of the degree of isotopic incorporation. In the absence of formate, the level of enrichment was ~10% for the Pur C8 and ~14.0% for the Pur C2. In the presence of formate the level of enrichment increases to ~38% for Pur C8 and ~28% for Ade C2 (Table 1).
Fig. 3.
Estimation of C2 and C8-13C labeling efficiency using two- and three-bond 15N-HSQC experiment without carbon decoupling during acquisition. The 2D 1H-15N HSQC spectra depict H8-N7/N9 crosspeaks for Ade and Gua and H2-N1/N3 correlations for Ade. At each N1 and N3 nitrogen position a singlet is observed for the H2 proton at 8.14 ppm if the C2 carbon is unlabeled and a doublet if C2 carbon is 13C-labeled due to the large one bond 1H-13C coupling of ~202 Hz. Similarly at each N7 and N9 nitrogen position a singlet is observed for the H8 proton at 8.5 ppm (for Ade) and 8.08 ppm (for Gua) if C8 is unlabeled and a doublet if C8 is13C-labeled due to the large one bond 1H-13C coupling of ~215 Hz. Thus the ratio of each satellite peak to the central peak gives a good estimate of the degree of 13C- labeling. a The 2D 2JHN HSQC spectra for uniformly labeled NMPs (AMP, red; GMP, blue) are superimposed. The inset shows the observable long range 1H-15N correlations in the purine ring. b 2D 2JHN HSQC spectra for the mixture of four rNMPs obtained from the K10 bacterial culture are superimposed (the spectrum obtained without labeled formate, red contours and upper; formate labeled spectrum, blue contours and lower). The H2 protons and N1 and N3 nitrogen atoms and H8 protons and the N7 atoms in nucleotides labeled using K10 with formate in a 13C-2-glycerol background are depicted. The carbon decoupling field is turned off during acquisition. c The aromatic region of all 4 rNMPs extracted from K10 cultures. The 1H spectrum with no 13C-decoulpling during acquisition (blue) is superimposed on 1D slices of the rows corresponding to the nitrogen chemical shifts of Ade N7 (green) and Gua N7 (red; see Fig. 3b). The 1D slices from the 2D 2JHN HSQC spectra overlay completely with the proton spectrum, suggesting the percentage label can be estimated using either the 2D or 1D experiment, but the 2D is preferable in case of overlap. d 1D section of the Pur N7 position (see Fig. 3b) is depicted for labeled rNMPs without formate (red) and with formate (blue). The satellite peaks are labeled I and II, and the center peak is labeled III
Applications of selective labels for NMR study of nucleic acids
High levels of isotopic enrichment lead to considerable direct one-bond scalar couplings and residual dipolar couplings from adjacent carbons yielding complex spectra for macromolecules. These deleterious consequences can negate the benefits of uniform labeling for monitoring RNA-ligand interactions, assignment of resonances and structural characterizations, to name only a few. For example, spectral resolution is degraded and transfer of magnetization through multiple pathways can attenuate the resultant signal. Preparation of samples lacking 13C–13C one-bond coupled spin pairs is thus critical for reducing spectral complexity and improving spectral resolution for multidimensional NMR experiments for assignment and structure determination of RNAs. Figure 4 illustrates the negative effect of coupling in a uniformly labeled sample, even in the ideal case of four nucleotides with minimum overlap. For example, in the uniformly labeled nucleotides, the C2′ and C4′ positions form a doublet of a doublet arising from the splitting of C2′ by C1′ and C3′ and the splitting of C4′ by C3′ and C5′ (Fig. 4a–b). These couplings give rise to a triplet at both positions instead of the singlet obtained using the site specific labeling (Fig. 4). These C2′ and C4′ regions of the HSQC spectra demonstrate the nearly three-fold increase in the number of resolved resonances due to the site specific labeling. Similarly C1′ and C5′ positions form a doublet arising from the splitting of C1′ by C2′ and the splitting of C5′ by C4′ (Fig. 4c–d). The new site specific labels again result in a nearly two-fold increase in the number of resolved resonances in the C1′ and C5′ regions using non-constant time HQSC experiments.
Fig. 4.
2D non-constant time HSQC spectra of all four labeled nucleotides showing the increased level of labeling in K10 with formate in a 2-glycerol background without introducing significant multiplet splitting in the ribose ring carbons atoms which contrasts with the uniformly labeled nucleotides. The spectra of uniformly labeled nucleotides are shown to the right of the site specific labeled rNMPs. For the uniformly labeled nucleotide AMP = blue, GMP = red, CMP = blue and UMP = purple. Note how the uniformly labeled rNMPs suffer from multiplet splitting absent in the new labels. a Ribose C4′, b Ribose C2′, c Ribose C1′ and d Ribose C5′. The resonances from each of the four nucleotides are annotated for adenine (Ade), cytosine (Cyt), guanine (Gua), and uracil (Ura). Not shown is C3′ that has doublet splitting instead of triplet seen in the uniformly labeled NMP sample
Even though these unwanted splittings can be removed using constant time (Bax et al. 1979; Bax and Freeman 1981; Grzesiek and Bax 1992; van de Ven and Philippens 1992) or adiabatic band selective decoupling during the carbon evolution period (Kupce and Wagner 1996; Brutscher et al. 2001; Dayie 2005), both solutions to the splitting problem are unsatisfactory. Use of constant time evolution limits considerably the acquisition time that can be used to obtain adequate resolution, and the long constant-time delay needed for improved resolution typically leads to significant signal loss for medium-sized to large RNA molecules (Dayie 2005). Similarly, use of band selective decoupling means the sites decoupled are not available for analysis. For example, selectively decoupling C2′ during carbon evolution precludes its observation. The selective labeling presented here removes both of these complications. A very important problem in NMR of nucleic acids is monitoring how nucleic acids interact site specifically with their ligands. High quality uncluttered spectra is important for such studies and for efforts in monitoring RNA-drug interactions (e.g., Thomas and Hergenrother 2008).
While exceptional resolution is obtained with this new label, the fully enzymatic methods can yield >95% label at the C1′ position compared to the 3–5% obtained here. However, the fully enzymatic method is limited to piecemeal labeling of each ribose position using site specifically labeled glucose at increased cost. The enzymatic method also requires the coupling of the base moiety to the labeled sugar component. Unfortunately the selectively labeled bases required for coupling are not commercially available in useful forms and those available are quite pricey.
In addition to cost considerations, it is important to ascertain the usefulness of site specific labels under conditions of broadened resonances that accompany RNA of increased size. By dissolving the labeled nucleotides in 95% w/w per deuterated glycerol, we can take advantage of the increased viscosity of the glycerol as a function of temperature. At 30°C the viscosity of glycerol is about 240 times that of water and at this temperature most of the base resonances are reduced in intensity in the non-constant time 13C HSQC spectrum such that the resonances for Cyt C5 and Pur C8 are barely visible in the spectrum (Fig. 5a). The reduction in intensity is consistent with increased overall correlation time and rapid signal decay. Use of the non-constant time 13C TROSY experiment, as expected, rescues these signals (Fig. 5b). It is clear that these and other new experiments can be designed to probe RNA-ligand interactions at very high resolution.
Fig. 5.
Comparison of non-constant time sensitivity-enhanced a HSQC and b TROSY of selective 13C-enriched nucleotides dissolved in 95% w/v d8-glycerol at 30°C for all 4 rNMPs derived from K10 bacterial culture. Base correlations are depicted. The ribose C2′ resonances that normally resonate between 73.7 and 74.7 ppm and Cyt and Ura C5 resonances at 96.67 ppm and 102.69 ppm respectively (in a decoupled HSQC) are folded in. Identical acquisition and processing parameters were used. The time domain matrices were processed without apodization functions. As expected the TROSY peaks are right and down shifted from the decoupled HSQC peaks. Two resonances that are either very weak or absent in the HSQC spectrum are boxed
Additionally a number of important spin relaxation applications benefit significantly from the selective 13C labeling strategy. These include obtaining accurate relaxation parameters such as 13C-CPMG based relaxation dispersion rates for quantifying millisecond (ms) time-scale processes, as well as longitudinal relaxation rate (R 1) and proton–carbon heteronuclear Overhauser effect (NOE; Yamazaki et al. 1994; Dayie and Wagner 1997) important for quantifying ns-ps time-scale motions in RNA (Johnson et al. 2006; Johnson and Hoogstraten 2008).
Finally, conventional proton detected experiments are deemed more valuable because of higher sensitivity compared to carbon-detected ones. However, advances in cryoprobe technology and higher fields have made carbon-detected experiments entirely feasible and, in the case of extracting residual dipolar coupling, an excellent alternative to proton-detected methods that suffer from 1H–1H dipolar interactions (Fiala and Sklenár 2007). Thus selectively enriching the non-protonated carbon position is potentially valuable for obtaining additional structural information other than that associated with protonated sites.
Conclusion
We have taken advantage of the versatility of growing an E. coli strain deficient in the glucose-6-phosphate dehydrogenase enzyme (K10) of the pentose phosphate pathway in chemically defined minimal media to synthesize nucleotides labeled with stable isotopes of 13C and 15N for structural and molecular dynamics characterizations. By combining 13C-labeled glycerol with 13C-sodium formate, the enrichment of 13C label is increased for all the protonated carbon sites that are of considerable interest for RNA NMR spectroscopy without introducing significant multiplet splitting at C5/C6 in the Pyr ring and C1′/C2′/C4′/C5′ in the ribose ring. Introduction of 15N labeling provides another method for estimating the degree of 13C label incorporation using a long range 15N HSQC with the carbon decoupling field turned off during acquisition. This efficient and inexpensive method for preparing ribonucleotides with these distributions of 13C enrichment will likely minimize not just scalar couplings but also splittings from long-range dipolar couplings, thereby providing greater spectral quality than normally obtained with fully labeled nucleotides. Use of these nucleotides should therefore allow high resolution probing of RNA-ligand interactions, the measurement of structurally useful parameters such as chemical shift anisotropy-offsets, and the accurate extraction of relaxation parameters such as chemical exchange lifetimes from power dependence of R1ρ on the strength of the spinlock fields.
Acknowledgments
This work was supported in part by the Nano-Biotechnology Award and the National Institutes of Health grant GM077326 to T.K.D. The authors thank Profs. David Fushman and Vitali Tugarinov (University of Maryland) for useful discussions and Jacob Sama for assistance with the initial stages of the research.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- AMP
Adenosine 5′-monophosphate
- CMP
Cytidine 5′-monophosphate
- DHAP
Dihydroxyacetone phosphate
- FBP
Fructose-6-bisphosphate
- F6P
Fructose-6-phosphate
- G6PDH
Glucose-6-phosphate dehydrogenase
- GA3P
Glyceraldehyde-3-phosphate
- Gly
Glycine
- GMP
Guanosine 5′-monophosphate
- noPPP
Non-oxidative pentose phosphate pathway
- OAA
Oxaloacetate
- oPPP
Oxidative pentose phosphate pathway
- R5P
Ribose-5-phosphate
- rNMPs
Ribonucleoside monophosphates
- rNTPs
Ribonucleoside triphosphates
- Ser
Serine
- TIM
Triosephosphate isomerase
- UMP
Uridine 5′-monophosphate
References
- Batey RT, Inada M, Kujawinski E, Puglisi JD, Williamson JR. Preparation of isotopically labeled ribonucleotides for multidimensional NMR spectroscopy of RNA. Nucleic Acids Res. 1992;20:4515–4523. doi: 10.1093/nar/20.17.4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batey RT, Battiste JL, Williamson JR. Preparation of isotopically enriched RNAs for heteronuclear NMR. Methods Enzymol. 1995;261:300–322. doi: 10.1016/S0076-6879(95)61015-4. [DOI] [PubMed] [Google Scholar]
- Bax A, Freeman R. Investigation of complex networks of spin-spin coupling by two-dimensional NMR. J Magn Reson. 1981;44:542–561. [Google Scholar]
- Bax A, Mehlkopf AF, Smidt J. Homonuclear broad-band decoupled absorption spectra. J Magn Reson. 1979;35:167–169. [Google Scholar]
- Bax A, Ikura M, Kay LE, Torchia DA, Tschudin R. Comparison of different modes of two-dimensional reverse correlation NMR for the study of proteins. J Magn Reson. 1990;86:304–318. [Google Scholar]
- Bodenhausen G, Ruben DJ. Natural abundance nitrogen-15 NMR by enhanced heteronuclear spectroscopy. Chem Phys Lett. 1980;69:185–189. doi: 10.1016/0009-2614(80)80041-8. [DOI] [Google Scholar]
- Brutscher B, Boisbouvier J, Kupce E, Tisne C, Dardel F, Marion D, Simorre JP. Base-type-selective high-resolution 13C edited NOESY for sequential assignment of large RNAs. J Biomol NMR. 2001;19:141–151. doi: 10.1023/A:1008340210079. [DOI] [PubMed] [Google Scholar]
- Czisch M, Boelens R. Sensitivity enhancement in the TROSY experiment. J Magn Reson. 1998;134:158–160. doi: 10.1006/jmre.1998.1483. [DOI] [PubMed] [Google Scholar]
- D’Souza V, Dey A, Habib D, Summers MF. NMR structure of the 101-nucleotide core encapsidation signal of the Moloney murine leukemia virus. J Mol Biol. 2004;337:427–442. doi: 10.1016/j.jmb.2004.01.037. [DOI] [PubMed] [Google Scholar]
- Dayie KT. Resolution enhanced homonuclear carbon decoupled triple resonance experiments for unambiguous RNA structural characterization. J Biomol NMR. 2005;32:129–139. doi: 10.1007/s10858-005-5093-6. [DOI] [PubMed] [Google Scholar]
- Dayie KT. Key labeling technologies to tackle sizeable problems in RNA structural biology. Int J Mol Sci. 2008;9:1214–1240. doi: 10.3390/ijms9071214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayie KT, Wagner G. Carbonyl carbon probe of local mobility in 13C, 15N-enriched proteins using high resolution NMR. J Am Chem Soc. 1997;119:7797–7806. doi: 10.1021/ja9633880. [DOI] [Google Scholar]
- Edwards JS, Palsson BO. The Escherichia coli MG1655 in silico metabolic genotype: its definition, characteristics, and capabilities. Proc Natl Acad Sci. 2000;97:5528–5533. doi: 10.1073/pnas.97.10.5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala R, Sklenár V. 13C-detected NMR experiments for measuring chemical shifts and coupling constants in nucleic acid bases. J Biomol NMR. 2007;39:153–163. doi: 10.1007/s10858-007-9184-4. [DOI] [PubMed] [Google Scholar]
- Fischer E, Sauer U. Metabolic flux profiling of Escherichia coli mutants in central metabolism using GC-MS. Eur J Biochem. 2003;270:880–891. doi: 10.1046/j.1432-1033.2003.03448.x. [DOI] [PubMed] [Google Scholar]
- Fraenkel DG. Selection of Escherichia coli mutants lacking glucose-6-phosphate dehydrogenase or gluconate-6-phosphate dehydrogenase. J Bacteriol. 1968;95:1267–1271. doi: 10.1128/jb.95.4.1267-1271.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross A, Abril O, Lewis JM, Geresh S, Whitesides GM. Practical synthesis of 5-phospho-D-ribosyl. alpha.-1-pyrophosphate (PRPP): enzymatic routes from ribose 5-phosphate or ribose. J Am Chem Soc. 1983;105:7428–7435. doi: 10.1021/ja00363a037. [DOI] [Google Scholar]
- Grzesiek S, Bax A. Improved 3D triple resonance NMR techniques applied to a 31 kDa protein. J Magn Reson. 1992;96:432–440. [Google Scholar]
- Gumbs OH, Padgett RA, Dayie KT. Fluorescence and solution NMR study of the active site of a 160-kDa group II intron ribozyme. RNA. 2006;12:1693–1707. doi: 10.1261/rna.137006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines JV, Landry SM, Varani G, Tinoco I. Carbon-proton scalar couplings in RNA—3D heteronuclear and 2D isotope—edited NMR of A C-13 labeled extra-stable hairpin. J Am Chem Soc. 1994;116:5823–5831. doi: 10.1021/ja00092a037. [DOI] [Google Scholar]
- Hoffman DW, Holland JA. Preparation of C-13-labeled ribonucleotides using acetate as an isotope source. Nucleic Acids Res. 1995;23:3361–3362. doi: 10.1093/nar/23.16.3361-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogstraten CG, Johnson JE. Metabolic labeling: taking advantage of bacterial pathways to prepare spectroscopically useful isotope patterns in proteins and nucleic acids. Concepts Magn Reson Part A. 2008;32:34–55. [Google Scholar]
- Johnson JE, Jr, Hoogstraten CG. Extensive backbone dynamics in the GCAA RNA tetraloop analyzed using 13C NMR spin relaxation and specific isotope labeling. J Am Chem Soc. 2008;130:16757–16769. doi: 10.1021/ja805759z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JE, Julien KR, Hoogstraten CG. Alternate-site isotopic labeling of ribonucleotides for NMR studies of ribose conformational dynamics in RNA. J Biomol NMR. 2006;35:261–274. doi: 10.1007/s10858-006-9041-x. [DOI] [PubMed] [Google Scholar]
- Kay LE, Keifer P, Saarinen T. Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J Am Chem Soc. 1992;114:10663–10665. doi: 10.1021/ja00052a088. [DOI] [Google Scholar]
- Kirkpatrick C, Maurer LM, Oyelakin NE, Yoncheva YN, Maurer R, Slonczewski JL. Acetate and formate stress: opposite responses in the proteome of Escherichia coli. J Bacteriol. 2001;183:6466–6477. doi: 10.1128/JB.183.21.6466-6477.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knappe J, Blaschkowski HP, Gröbner P, Schmitt T. Pyruvate formate-lyase of Escherichia coli: the acetyl-enzyme intermediate. Eur J Biochem. 1974;50:253–263. doi: 10.1111/j.1432-1033.1974.tb03894.x. [DOI] [PubMed] [Google Scholar]
- Kupce E, Wagner G. Multisite band-selective decoupling in proteins. J Magn Reson B. 1996;110:309–312. doi: 10.1006/jmrb.1996.0048. [DOI] [PubMed] [Google Scholar]
- Latham MR, Brown DJ, McCallum SA, Pardi A. NMR methods for studying the structure and dynamics of RNA. Chembiochem. 2005;6:1492–1505. doi: 10.1002/cbic.200500123. [DOI] [PubMed] [Google Scholar]
- Louis JM, Martin RG, Clore GM, Gronenborn AM. Preparation of uniformly isotope-labeled DNA oligonucleotides for NMR spectroscopy. J Biol Chem. 1998;273:2374–2378. doi: 10.1074/jbc.273.4.2374. [DOI] [PubMed] [Google Scholar]
- Lu K, Miyazaki Y, Summers MF. Isotope labeling strategies for NMR studies of RNA. J Biomol NMR. 2010;46:113–125. doi: 10.1007/s10858-009-9375-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundström P, Teilum K, Carstensen T, Bezsonova I, Wiesner S, Hansen DF, Religa TL, Akke M, Kay LE. Fractional 13C enrichment of isolated carbons using [1–13C]- or [2–13C]-glucose facilitates the accurate measurement of dynamics at backbone Calpha and side-chain methyl positions in proteins. J Biomol NMR. 2007;38:199–212. doi: 10.1007/s10858-007-9158-6. [DOI] [PubMed] [Google Scholar]
- Masse JE, Bortmann P, Dieckmann T, Feigon J. Simple, efficient protocol for enzymatic synthesis of uniformly 13C, 15N-labeled DNA for heteronuclear NMR studies. Nucleic Acids Res. 1998;26:2618–2624. doi: 10.1093/nar/26.11.2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Schulte-Herbrueggen T, Briand J, Sorensen OW. Double spin-state-selective coherence transfer. Application for two-dimensional selection of multiplet components with long transverse relaxation times. Mol Phys. 1998;96:1137–1142. [Google Scholar]
- Michnicka MJ, Harper JW, King GC. Selective isotopic enrichment of synthetic RNA: application to the HIV-1 TAR element. Biochemistry. 1993;32:395–400. doi: 10.1021/bi00053a002. [DOI] [PubMed] [Google Scholar]
- Milecki J. Specific labelling of nucleosides and nucleotides with 13C and 15N. J Label Comp Radiopharm. 2002;45:307–337. doi: 10.1002/jlcr.553. [DOI] [Google Scholar]
- Mueller L. Sensitivity enhanced detection of weak nuclei using heteronuclear multiple quantum coherence. J Am Chem Soc. 1979;101:4481–4484. doi: 10.1021/ja00510a007. [DOI] [Google Scholar]
- Nelissen FH, Girard FC, Tessari M, Heus HA, Wijmenga SS. Preparation of selective and segmentally labeled single-stranded DNA for NMR by self-primed PCR and asymmetrical endonuclease double digestion. Nucleic Acids Res. 2009;37:e114. doi: 10.1093/nar/gkp540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DL, Cox MM. Lehninger principles of biochemistry. New York: W.H. Freeman; 2008. [Google Scholar]
- Nicolas C, Kiefer P, Letisse F, Krömer J, Massou S, Soucaille P, Wittmann C, Lindley ND, Portais JC. Response of the central metabolism of Escherichia coli to modified expression of the gene encoding the glucose-6-phosphate dehydrogenase. FEBS Lett. 2007;581:3771–3776. doi: 10.1016/j.febslet.2007.06.066. [DOI] [PubMed] [Google Scholar]
- Nikonowicz EP. Preparation and use of 2H-labeled RNA oligonucleotides in nuclear magnetic resonance studies. Methods Enzymol. 2001;338:320–341. doi: 10.1016/s0076-6879(02)38227-2. [DOI] [PubMed] [Google Scholar]
- Nikonowicz EP, Sirr A, Legault P, Jucker FM, Baer LM, Pardi A. Preparation of 13C and 15N labelled RNAs for heteronuclear multi-dimensional NMR studies. Nucleic Acids Res. 1992;20:4507–4513. doi: 10.1093/nar/20.17.4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paliy O, Gunasekera TS. Growth of E-coli BL21 in minimal media with different gluconeogenic carbon sources and salt contents. Appl Microbiol Biotechnol. 2007;73:1169–1172. doi: 10.1007/s00253-006-0554-8. [DOI] [PubMed] [Google Scholar]
- Palmer AG, Cavanagh J, Wright PE, Rance M. Sensitivity improvement in proton-detected two-dimensional heteronuclear correlation NMR spectroscopy. J Magn Reson. 1991;93:151–170. [Google Scholar]
- Parkin DW, Leung HB, Schramm VL. Synthesis of nucleotides with specific radiolabels in ribose. Primary 14C and secondary 3H kinetic isotope effects on acid-catalyzed glycosidic bond hydrolysis of AMP, dAMP, and inosine. J Biol Chem. 1984;259:9411–9417. [PubMed] [Google Scholar]
- Pervushin K, Wider G, Wuthrich K. Single transition-to-single transition polarization transfer (ST2-PT) in [15 N, 1H]-TROSY. J Biomol NMR. 1998;12:345–348. doi: 10.1023/A:1008268930690. [DOI] [PubMed] [Google Scholar]
- Ponchon L, Dardel F. Recombinant RNA technology: the tRNA scaffold. Nat Methods. 2007;4:571–576. doi: 10.1038/nmeth1058. [DOI] [PubMed] [Google Scholar]
- Ponchon L, Beauvais G, Nonin-Lecomte S, Dardel F. A generic protocol for the expression and purification of recombinant RNA in Escherichia coli using a tRNA scaffold. Nat Prot. 2009;4:947–959. doi: 10.1038/nprot.2009.67. [DOI] [PubMed] [Google Scholar]
- Rance M, Loria JP, Palmer AG., III Sensitivity improvement of transverse relaxation-optimized spectroscopy. J Magn Reson. 1999;136:92–101. doi: 10.1006/jmre.1998.1626. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
- Schulte-Herbrüggen T, Sorensen OW. Clean TROSY: compensation for relaxation-induced artifacts. J Magn Reson. 2000;144:123–128. doi: 10.1006/jmre.2000.2020. [DOI] [PubMed] [Google Scholar]
- Schultheisz HL, Szymczyna BR, Scott LG, Williamson JR. Pathway engineered enzymatic de novo purine nucleotide synthesis. ACS Chem Biol. 2008;3:499–511. doi: 10.1021/cb800066p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott LG, Tolbert TJ, Williamson JR. Preparation of specifically 2H- and 13C-labeled ribonucleotides. Methods Enzymol. 2000;317:18–38. doi: 10.1016/S0076-6879(00)17004-1. [DOI] [PubMed] [Google Scholar]
- Shaka AJ, Barker PB, Freeman R. Computer-optimized decoupling scheme for wideband applications and low-level operation. J Magn Reson. 1985;64:547–552. [Google Scholar]
- Studier FW. Protein production by auto-induction in high-density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Thauer RK, Kirchniawy FH, Jungermann KA. Properties and function of the pyruvate-formate-lyase reaction in clostridiae. Eur J Biochem. 1972;27:282–290. doi: 10.1111/j.1432-1033.1972.tb01837.x. [DOI] [PubMed] [Google Scholar]
- Thomas JR, Hergenrother PJ. Targeting RNA with small molecules. Chem Rev. 2008;108:1171–1224. doi: 10.1021/cr0681546. [DOI] [PubMed] [Google Scholar]
- Tolbert TJ, Williamson JR. Preparation of specifically deuterated RNA for NMR studies using a combination of chemical and enzymatic synthesis. J Am Chem Soc. 1996;118:7929–7940. doi: 10.1021/ja961274i. [DOI] [Google Scholar]
- Tolbert TJ, Williamson JR. Preparation of specifically deuterated and 13C-Labeled RNA for NMR studies using enzymatic synthesis. J Am Chem Soc. 1997;119:12100–12108. doi: 10.1021/ja9725054. [DOI] [Google Scholar]
- van de Ven FJM, Philippens MEP. Optimization of constant-time evolution in multidimensional experiments. J Magn Reson. 1992;97:637–644. [Google Scholar]
- Voet D, Voet JG, Pratt CW. Fundamentals of biochemistry. 3. New York: Wiley; 2008. [Google Scholar]
- Weigelt J. Single scan, sensitivity- and gradient-enhanced TROSY for multidimensional NMR experiments. J Am Chem Soc. 1998;120:10778–10779. doi: 10.1021/ja982649y. [DOI] [Google Scholar]
- Werner MH, Gupta V, Lambert LJ, Nagata T. Uniform 13C/15N-labeling of DNA by tandem repeat amplification. Methods Enzymol. 2001;338:283–304. doi: 10.1016/s0076-6879(02)38225-9. [DOI] [PubMed] [Google Scholar]
- Wijmenga SS, van Buuren BNM. The use of NMR methods for conformational studies of nucleic acids. Prog NMR Spectroscopy. 1998;32:287–387. doi: 10.1016/S0079-6565(97)00023-X. [DOI] [Google Scholar]
- Yamazaki T, Muhandiram R, Kay LE. NMR experiments for the measurement of carbon relaxation properties in highly enriched, uniformly 13C, 15 N-labeled proteins: application to 13Cα carbons. J Am Chem Soc. 1994;114:8266–8278. doi: 10.1021/ja00097a037. [DOI] [Google Scholar]
- Zhao J, Baba T, Mori H, Shimizu K. Effect of zwf gene knockout on the metabolism of Escherichia coli grown on glucose or acetate. Metab Eng. 2004;6:164–174. doi: 10.1016/j.ymben.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Zhu G, Xia Y, Kong XM, Sze KH. 2D and 3D TROSY-enhanced NOESY of 15N labeled proteins. J Biomol NMR. 1999;13:77–81. doi: 10.1023/A:1008398227519. [DOI] [PubMed] [Google Scholar]
- Zimmer DP, Crothers DM. NMR of enzymatically synthesized uniformly 13C, 15 N-labeled DNA oligonucleotides. Proc Natl Acad Sci. 1995;92:3091–3095. doi: 10.1073/pnas.92.8.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]