Abstract
We have transcriptionally profiled the genes differentially expressed in E. coli prey cells when predatorily attacked by Bdellovibrio bacteriovorus just prior to prey cell killing. This is a brief, approximately 20–25 min period when the prey cell is still alive but contains a Bdellovibrio cell in its periplasm or attached to and penetrating its outer membrane. Total RNA was harvested and labelled 15 min after initiating a semi-synchronous infection with an excess of Bdellovibrio preying upon E. coli and hybridised to a macroarray spotted with all predicted ORFs of E. coli. SAM analysis and t-tests were performed on the resulting data and 126 E. coli genes were found to be significantly differentially regulated by the prey upon attack by Bdellovibrio. The results were confirmed by QRT-PCR. Amongst the prey genes upregulated were a variety of general stress response genes, potentially “selfish” genes within or near prophages and transposable elements, and genes responding to damage in the periplasm and osmotic stress. Essentially, the presence of the invading Bdellovibrio and the resulting damage to the prey cell elicited a small “transcriptional scream”, but seemingly no specific defensive mechanism with which to counter the Bdellovibrio attack. This supports other studies which do not find Bdellovibrio resistance responses in prey, and bodes well for its use as a “living antibiotic”.
Introduction
Bdellovibrio are small, highly motile, Gram-negative bacteria that act as “living antibiotics” in that they prey upon and kill a wide variety of Gram-negative bacteria; including human and animal pathogens. Despite years of studies in laboratories, there is only one report of resistance to Bdellovibrio predation [39] and resistance by most populations is thought to be a plastic (reversible) phenotype by a small minority of a susceptible strain [34] or by a sub-population that is less susceptible, but not completely resistant to predation [8]. In predation Bdellovibrio attach to the outer membrane of prey bacteria and pass through it into the periplasm before they attach to the prey cell cytoplasmic membrane and the prey cell dies after 20–25 min [30, 40]. Here, we investigate the transcriptional response of E. coli prey to 15 min of periplasmic invasion by Bdellovibrio predators. We find that there is a transcriptional stress response, to the osmotic changes associated with Bdellovibrio breaching the outer membrane and the damage caused by having another cell resident in the periplasm, rather than a response specifically to resist Bdellovibrio attack. We do not find upregulation of many genes reported to be upregulated in responses to phage attack or ppGpp alarmone systems [17, 37]. That E. coli transcriptionally “screams” slightly, but does not resist Bdellovibrio predation, bodes well for the potential use of Bdellovibrio as a therapeutic agent [35].
Materials and Methods: Culture and Predatory Infection Conditions
E. coli S17-1 and Bdellovibrio bacteriovorus 109J were grown as described previously [22]. Semi-synchronous infections were set up by using centrifugation concentrated Bdellovibrio. Control mock-infections of Bdellovibrio only and E. coli only were carried out concurrently using filtered respective supernatant samples in place of the missing cells. At 15 min post-infection, 4 ml samples of experimental and controls were taken for RNA extraction as described elsewhere [22].
Arrays, cDNA and Hybridisation, Data Acquisition and Analysis
Panorama E. coli gene arrays (Sigma Genosys) were used according to the manufacturer’s instructions and as described elsewhere [29]. Hybridisation and stringent washes were carried out according to the manufacturer’s instructions. Five independent experiments were performed using a total of 4 arrays with array swapping and use of control E. coli.
The ArrayVison software was used to analyse the phosphor-images with the template provided by Sigma Genosys for the Panorama arrays. The normalised data were analysed using SAM [36] in a paired test with 1000 permutations and a delta value of 0.86. In order to exclude the possibility that any of the significant genes, called as differentially regulated, were a result of the presence of cross-hybridising Bdellovibrio RNA, the pair sample of E. coli only control versus Bdellovibrio only control were analysed at the lowest possible stringency, i.e. with a delta of 0. This resulted in a false discovery rate of 4.7%. Any genes called in the Bdellovibrio control datasets, as well as the experimental infection datasets, were therefore excluded from the analysis. Finally, for further confidence, the data for each significant gene was subjected to a paired t-test. Only for 1 gene was the P value >0.05 (minE P = 0.051) showing the robustness of the significant gene lists to different statistical tests. The data discussed are deposited in NCBIs Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) via GEO Series accession number GSE9495.
Quantitative PCR
Quantitative PCR (QPCR) was used for sample genes, to verify the array results using the Statagene Full Velocity SYBR Green QRT-PCR kit as described previously [7] except in two-step reactions, with reverse transcription as described elsewhere [10]. Serial dilutions of the cDNA were used for QPCR with primers designed to specifically anneal to 3 genes that were upregulated in E. coli in response to Bdellovibrio attack (treA, cstA and creB) and 3 genes that were not significantly upregulated (hns, dnaK and arcA). Absolute expression was calculated against a standard curve of a template of extracted PCR product and the ratios of expression in the two samples were compared. At least two independent experiments were carried out and included relevant controls such as no template, and also with B. bacteriovorus 109J RNA as a putative “mock” template. A t-test was carried out on each set of data to ascertain significance. The PCR products were sequenced to check for the accuracy and specificity of the reactions. Standard semi-quantitative RT-PCR to check for co-transcribed operons was carried out as described previously [22].
Results and Discussion
In order to investigate the response of E. coli to Bdellovibrio attack, we used the most studied prey and predator strain combination of E. coli S17-1 and Bdellovibrio bacteriovorus 109J, and macroarrays with all open reading frames of E. coli K12 MG1655 spotted. We checked the similarity of the S17-1 and K12 sequences with a Nimblegen tiling array, and discovered that there is a very high level of sequence identity between the strains to allow correct hybridisation between the cDNA from the S17-1 strain tested and the array (data not shown).
Semi-synchronous predatory infections were set up, such that at the 15-min time point, at which the RNA samples for expression studies were taken, greater than 95% of all E. coli cells had at least one Bdellovibrio cell attached or fully periplasmically invaded, but before prey killing and RNA degradation was extensive. Phase contrast microscopy revealed that 90% of E. coli cells had at least one Bdellovibrio cell attached to its surface at 15 min. Initial attachment was rapid, within 5 min more than 75% of cells had a Bdellovibrio attached. Burnham et al. [4] used a similar semi-synchronous infection process and showed that at 15 min post-mixing, the Bdellovibrio cells could not be removed from prey by mechanical agitation and so had started the penetration process; which they monitored by electron microscopy (EM), revealing damage to the prey outer membrane. We have also observed similar invasion by EM at this time point. Thus, very many of the “attached” Bdellovibrio would indeed be breaching the outer membrane at the 15-min time point we sampled. This fraction cannot be enumerated by live dead staining as the prey cells are still alive and the extent of the Bdellovibrio cell body length that had entered the periplasm of the prey cannot be measured by light microscopy. However, as 5% of the E. coli cells at the 15-min time point were rounded, infected bdelloplasts indicating that they had reached the stage of prey cell death, unsuitable for recovery of mRNA it was clear that significant prey penetration had occurred at 15 min and so maximal invasion of prey, with minimal prey cell death was achieved.
Pre-incubation of bacteria in Ca/HEPES buffer for 3 h prior to infection was to stabilise expression (viability was unchanged during this, as assayed by plaque and colony counts) and a portion of the supernatant from each was 0.22 μm filtered for use in the controls. Experiments were carried out 5 times with duplicate spots on the arrays and the data analysed by t-test and SAM. Genes found, by both tests, to be differentially regulated in the infected E. coli relative to uninfected E. coli control from these analyses were considered, excluding any which flagged as significant in the control of E. coli compared to Bdellovibrio; giving a final dataset of 126 differentially regulated E. coli genes in response to predation.
Upregulated Genes
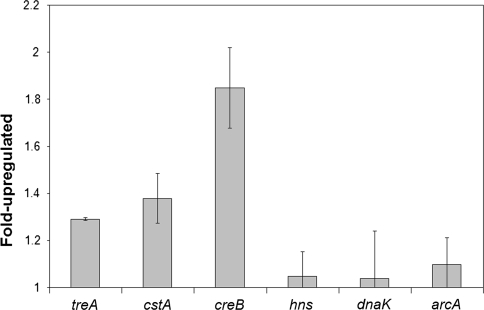
92 genes were found to be significantly upregulated by E. coli in response to periplasmic attack by Bdellovibrio. It is very important to realise, in comparison to more usual array experiments for bacteria, that all the small upregulated changes we have tabulated (we have not used a twofold cut-off) are significant and that they are small because a predatory Bdellovibrio is acting on the periplasm of the E. coli, as the prey cell’s transcription is measured—i.e. the E. coli is not in an optimum physiological state. Hughes et al. [16] demonstrated how transcriptional profiles with levels of upregulation less than 1.5-fold are very meaningful, even in bacteria that are not subject to insults such as predation. Statistical analyses on genes found to be differentially expressed, shown in the results Tables 1 and 2, confirm that the small changes in expression were significant. Quantitative RT-PCR analysis was carried out on three representative upregulated prey genes (with array fold-change values of 2.22, 1.32 and 1.16) of different functional classes; (treA, cstA and creB) and on three prey genes that were not significantly upregulated (hns, dnaK and arcA) (Fig. 1) showing the three upregulated genes to be all significantly upregulated when the three control genes were not. Tables 1 and 2 show up- or downregulated genes, their predicted functions and cellular locations and we discuss them by function below.
Table 1.
Significantly up-regulated E. coli genes at 15 minutes of Bdellovibrio predation
| Gene | Putative function | Predicted location | Fold-upregulated | P value from t-test |
|---|---|---|---|---|
| Stress response | ||||
| cstA | Carbon starvation induced protein | Inner membrane | 2.22 | 0.019816 |
| pqiB | Paraquat inducible protein | Unknown | 2.01 | 0.001174 |
| tehA | Multiresistance efflux pump | Inner membrane | 1.46 | 0.009733 |
| yadF | Putative carbonic anhydrase—possible pH regulation | Cytoplasmic | 1.43 | 0.042138 |
| treA | Trehalase, periplasmic—osmoregulation | Periplasmic | 1.32 | 0.000179 |
| yafO | Toxin of the yafO-yafN toxin–antitoxin system | Cytoplasmic | 1.31 | 0.000402 |
| sufI | Inhibits ftsI and hence septation | Periplasmic | 1.28 | 0.029411 |
| mazG | Regulator involved in amino acid starvation response | Cytoplasmic | 1.22 | 0.038161 |
| ycfQ | Predicted tetR-family response regulator | Cytoplasmic | 1.21 | 0.03176 |
| galR | lacI-type transcriptional regulator | Cytoplasmic | 1.19 | 0.033147 |
| betA | Choline dehydrogenase—osmoregulation | Cytoplasmic | 1.18 | 0.019939 |
| glnG | Nitrogen response regulator | Cytoplasmic | 1.18 | 0.030032 |
| iscR | DNA-binding transcriptional repressor | Unknown | 1.17 | 0.019957 |
| creB | Global response regulator | Cytoplasmic | 1.16 | 0.043154 |
| csiE | Global response regulator | Cytoplasmic | 1.16 | 0.005442 |
| Putative wall or membrane repair | ||||
| ybaL | Na/H exchanger | Inner membrane | 4.41 | 0.00837 |
| gltJ | Glutamate and aspartate transporter subunit | Inner membrane | 1.82 | 0.042358 |
| phnD | Phosphonate transporter, periplasmic domain | Periplasmic | 1.74 | 0.044892 |
| ydcV | Predicted spermidine/putrescine transporter subunit | Inner membrane | 1.70 | 0.039148 |
| frdA | Fumarate reductase catalytic and NAD/flavoprotein subunit | Periplasmic | 1.62 | 0.02433 |
| yqaA | Putative inner membrane protein | Inner membrane | 1.55 | 0.045115 |
| mdlA | ABC transporter | Inner membrane | 1.48 | 0.004624 |
| kefF | Potassium efflux | Cytoplasm | 1.45 | 0.027868 |
| yjjP | Predicted inner membrane protein | Inner membrane | 1.41 | 0.004595 |
| ydcT | Predicted spermidine/putrescine transporter subunit | Unknown | 1.41 | 0.030283 |
| yihO | Predicted transporter | Inner membrane | 1.40 | 0.032868 |
| flgN | Export chaperone for FlgK and FlgL | Flagellar basal body | 1.36 | 0.049212 |
| yaiW | Putative lipoprotein | Non-cytoplasmic | 1.30 | 0.013018 |
| fliG | Flagellar basal body protein | Flagellar basal body | 1.29 | 0.032728 |
| livJ | Leucine/isoleucine/valine transporter subunit | Periplasmic | 1.26 | 0.021585 |
| plsC | Phospholipid biosynthesis | Unknown | 1.24 | 0.042348 |
| tsr | Methyl-accepting chemotaxis protein | Inner membrane | 1.23 | 0.033582 |
| tnaB | Tryptophan/tyrosine permease family | Inner membrane | 1.23 | 0.01368 |
| cdsA | Cytidylyltransferase | Inner membrane | 1.21 | 0.001263 |
| ygfQ | Permease | Inner membrane | 1.19 | 0.020053 |
| hyfB | Hydrogenase 4 | Inner membrane | 1.19 | 0.024532 |
| nrfE | Cytochrome C assembly protein | Inner membrane | 1.19 | 0.002099 |
| hofQ | Predicted fimbrial transporter | Outer membrane | 1.18 | 0.024062 |
| hyfF | Hydrogenase 4 | Inner membrane | 1.17 | 0.010739 |
| yaaH | Predicted inner membrane protein | Inner membrane | 1.16 | 0.035842 |
| ybjL | Permease | Inner membrane | 1.15 | 0.02009 |
| ampG | Muropeptide transporter | Inner membrane | 1.15 | 0.042636 |
| fhiA | Flagellar basal body protein | Flagellar basal body | 1.12 | 0.043355 |
| Putative periplasmic content repair | ||||
| ykgF | Putative electron transport protein | Cytoplasmic | 2.28 | 0.02889 |
| hypA | Hydrogenase nickel insertion protein | Cytoplasmic | 2.09 | 0.044557 |
| hybA | Hydrogenase 2 4Fe-4S ferredoxin-type component | Periplasmic | 1.85 | 0.001521 |
| hycA | Formate hydrogenlyase regulatory protein | Unknown | 1.62 | 0.025271 |
| hypF | Hydrogenase maturation protein | Unknown | 1.58 | 0.048483 |
| hypB | Hydrogenase isoenzyme nickel incorporation protein | Cytoplasmic | 1.54 | 0.011291 |
| hycH | Hydrogenase maturation protein | Cytoplasmic | 1.42 | 0.030986 |
| ycjS | Putative oxidoreductase | Unknown | 1.33 | 0.003842 |
| glmU | Peptidoglycan biosynthesis; lipopolysaccharide biosynthesis | Cytoplasmic | 1.28 | 0.021109 |
| hycI | Hydrogenase maturation protease | Cytoplasmic | 1.23 | 0.002523 |
| fhlA | Formate hydrogenlyase transcriptional activator | Cytoplasmic | 1.22 | 0.001627 |
| Putative phage or transposon genes | ||||
| ykfC | Reverse transcriptase | Unknown | 1.40 | 0.000281 |
| insL | IS186/IS421 transposase | Cytoplasmic | 1.31 | 0.02964 |
| insD | IS2 insertion element transposase InsAB′ | Cytoplasmic | 1.20 | 0.005684 |
| Biosynthesis/metabolism | ||||
| ansA | Asparaginase | Cytoplasmic | 1.82 | 0.010869 |
| metJ | Methionine repressor | Cytoplasmic | 1.82 | 0.015532 |
| malQ | 4-alpha-glucanotransferase | Cytoplasmic | 1.68 | 0.00488 |
| aidB | Acetyl CoA dehydrogenase | Unknown | 1.60 | 0.004193 |
| hemC | Hydroxymethylbilane synthase | Unknown | 1.42 | 0.040394 |
| paaA | Catabolism of phenylacetic acid | Cytoplasmic | 1.41 | 0.005881 |
| paaH | 3-Hydroxybutyryl Co-A dehydrogenase | Inner membrane | 1.38 | 0.001737 |
| yeiT | Pyridine nucleotide-disulphide oxidoreductase | Cytoplasmic | 1.37 | 0.000155 |
| tktA | Transketolase | Cytoplasmic | 1.31 | 0.019749 |
| dfp | Flavoprotein | Unknown | 1.28 | 0.001176 |
| ilvY | DNA-binding transcriptional dual regulator | Cytoplasmic | 1.24 | 0.016897 |
| aroA | 5-Enolpyruvylshikimate-3-phosphate synthetase | Unknown | 1.23 | 0.019383 |
| fadA | Fatty acid oxidation | Cytoplasmic | 1.23 | 0.02383 |
| dxr | 1-Deoxy-d-xylulose 5-phosphate reductoisomerase | Unknown | 1.23 | 7E-05 |
| hemB | Porphobilinogen synthase | Cytoplasmic | 1.21 | 0.017421 |
| guaA | GMP synthetase | Cytoplasm | 1.20 | 0.036066 |
| abgA | Aminobenzoyl-glutamate utilisation protein | Unknown | 1.18 | 0.028809 |
| rfaG | Glucosyltransferase I | Cytoplasmic | 1.16 | 0.021738 |
| deoD | Purine-nucleoside phosphorylase | Cytoplasmic | 1.15 | 0.021485 |
| ugd | UDP-glucose 6-dehydrogenase | Cytoplasm | 1.13 | 0.031243 |
| purT | Phosphoribosylglycinamide formyltransferase 2 | Cytoplasmic | 1.13 | 0.025227 |
| Others | ||||
| minE | Cell division topological specificity factor | Cytoplasm | 2.38 | 0.05137 |
| b2973 | Unknown function | Unknown | 1.16 | 0.035317 |
| rph | RNase PH exoribonuclease | Unknown | 1.61 | 0.003009 |
| ynjH | Unknown function | Unknown | 1.53 | 0.036073 |
| rtn | Resistance to phage N—this is an EAL domain protein | Inner membrane | 1.44 | 0.03824 |
| yhfV | Phosphotriesterase | Cytoplasmic | 1.26 | 0.014469 |
| yejH | Type III restriction endonuclease and helicase | Cytoplasm | 1.21 | 0.046185 |
| b2345 | Unknown function | Unknown | 1.18 | 0.039688 |
| yahG | Conserved protein of unknown function | Inner membrane | 1.18 | 0.033265 |
| yoaF | Unknown function | Unknown | 1.18 | 0.037459 |
| ybiU | Conserved protein of unknown function | Cytoplasm | 1.17 | 0.024414 |
| dnaA | Chromosome replication initiation | Cytoplasmic | 1.17 | 0.004566 |
| clpX | Protease | Cytoplasm | 1.15 | 0.011204 |
| holA | DNA polymerase III, delta subunit | Unknown | 1.13 | 0.00926 |
Table 2.
Significantly down-regulated E. coli genes at 15 minutes of Bdellovibrio predation
| Gene | Putative function | Predicted location | Fold-downregulated | P value from t-test |
|---|---|---|---|---|
| rdgB | dITP/XTP pyrophosphatase | Unknown | 0.89 | 0.001542 |
| msrA | Methionine sulfoxide reductase A | Unknown | 0.88 | 0.005384 |
| lsrB | AI2 transporter | Periplasmic | 0.87 | 0.004696 |
| dusC | tRNA-dihydrouridine synthase C | Cytoplasmic | 0.86 | 0.002917 |
| ydfH | Predicted DNA-binding transcriptional regulator | Cytoplasmic | 0.86 | 0.001975 |
| sdaC | Predicted serine transporter | Inner membrane | 0.86 | 0.003618 |
| yieK | Predicted 6-phosphogluconolactonase | Unknown | 0.85 | 0.003358 |
| yfiD | Pyruvate formate lyase subunit | Cytoplasmic | 0.85 | 0.005335 |
| pepE | Aspartyl-dipeptidase | Cytoplasmic | 0.85 | 4.38E-06 |
| malY | Bifunctional beta-cystathionase, PLP-dependent/regulator of maltose regulon | Unknown | 0.84 | 0.007903 |
| hpf | Hibernation promoting factor | Cytoplasmic | 0.84 | 0.000864 |
| insG | KpLE2 phage-like element; IS4 predicted transposase | Unknown | 0.84 | 0.004375 |
| yhdZ | Predicted amino acid transporter subunit | Cytoplasmic/inner membrane | 0.84 | 0.004034 |
| puub | Gamma-Glu-putrescine oxidase, FAD/NAD(P)-binding | Cytoplasmic | 0.82 | 0.005585 |
| yjdA | Conserved protein with nucleoside triphosphate hydrolase domain | Cytoplasmic | 0.81 | 0.002824 |
| cmoB | Predicted S-adenosyl-l-methionine-dependent methyltransferase | Cytoplasmic | 0.80 | 5.69E-05 |
| ilvH | Acetolactate synthase III, thiamine-dependent, small subunit | Cytoplasmic | 0.80 | 0.002524 |
| katE | Hydroperoxidase HPII(III) | Cytoplasmic | 0.79 | 0.003291 |
| sacY | Preprotein translocase membrane subunit | Inner membrane | 0.78 | 0.007958 |
| talB | Transaldolase B | Unknown | 0.78 | 0.003563 |
| yfcQ | Predicted fimbrial-like adhesin protein | Unknown | 0.77 | 0.004262 |
| glpG | Predicted intramembrane serine protease | Inner membrane | 0.76 | 0.003553 |
| ygcE | Predicted kinase | Unknown | 0.74 | 0.000554 |
| yebS | Conserved inner membrane protein | Inner membrane | 0.74 | 0.001472 |
| pspB | Phage shock protein B | Cytoplasmic/inner membrane | 0.74 | 0.000354 |
| pfkA | 6-Phosphofructokinase I | Cytoplasmic | 0.74 | 0.000155 |
| surA | Peptidyl-prolyl cis–trans isomerase | Periplasmic | 0.71 | 0.00257 |
| secG | Preprotein translocase membrane subunit | Inner membrane | 0.70 | 0.011531 |
| yfhL | Predicted 4Fe-4S cluster-containing protein | Cytoplasmic | 0.69 | 0.01222 |
| nrdR | Transcriptional repressor of ribonucleotide reductase genes | Cytoplasmic | 0.69 | 0.010921 |
| rutA | Predicted monooxygenase | Unknown | 0.66 | 0.003513 |
| srmB | ATP-dependent RNA helicase | Cytoplasmic | 0.64 | 0.00445 |
| tolB | Periplasmic protein | Periplasmic | 0.63 | 0.004886 |
| purM | Phosphoribosylaminoimidazole synthetase | Unknown | 0.51 | 0.010088 |
Fig. 1.
QRT-PCR results shown as fold increase in expression in the test infection sample compared to the E. coli-only control. treA, cstA and creB are all significantly upregulated whilst hns, dnaK and arcA are not significantly upregulated (or downregulated). These results agree with the array data and t-test on the QRT-PCR data showed that it was significant for the former, but not the latter genes. Error bars represent one standard deviation above and below the mean
Wall and Membrane Repair
Products of upregulated genes in this category include genes encoding enzymes potentially involved in synthesis and/or repair of the peptidoglycan, lipids and membrane bound proteins. The gene upregulation is likely to be an attempt by the prey to repair the damage caused by the physical entry of the Bdellovibrio via the outer membrane and peptidoglycan layer and the damage by Bdellovibrio degradative enzymes acting on both of these and the prey inner membrane. Interestingly, out of a possible 45+ prey flagellar genes only 3 (fliG, fhiA and flgN) were significantly upregulated. The prey flagellar driving rods pass from the cytoplasmic membrane to periplasm and then via washers in the cell wall and outer membrane, to the external flagellar filament propellers. The entry of the Bdellovibrio into the periplasm very likely prises apart the connection of many of the flagella across the periplasm. The 3 upregulated genes are amongst the first in the cascade to build new flagella and fliG encodes the essential rotor of the flagellar motor [25]. Thus, the E. coli are inducing genes to rebuild flagella and restore motility.
Interestingly there was very little overlap, (4 genes only, aroA, guaA, purM and pspB) between the genes that had altered expression due to Bdellovibrio attack and those (over 600) reported to be differentially regulated in E. coli in repones to chemical assaults on the cell membrane, by isobutanol in the work of Brynildsen and Liao [3]. We do not think the overlap to be significant.
Stress Response Genes
Amongst the prey genes upregulated in response to stress, were those specifically responding to disturbance to the periplasm by entry of the Bdellovibrio through the outer membrane, and its consequent osmolarity impacts on the cell, namely treA which encodes a periplasmic trehalase that breaks down trehalose in the periplasm to regulate osmolarity [2] and betA which encodes a protein of the choline–glycine betaine pathway involved in compatible osmolyte production [21]. At this point of the Bdellovibrio infection the E. coli prey cell is still alive and has an electrochemical proton gradient and is trying to restabilise its internal osmolarity after periplasmic invasion.
In general, it appears that prey stress responses to damaged proteins and disturbed periplasm and outer membrane are being elicited. As expected because the Bdellovibrio, at the 15-min infection stage, will not have yet secreted DNases into the prey [32]; no transcriptional response to DNA damage was detected as none of the classical prey SOS response genes were found to be upregulated although yafO, a toxin of a toxin–antitoxin system, may be indirectly involved in the SOS response [28]. No ppGpp alarmone type responses [37] were detected, probably as our sampling was early, but also it should be remembered that in Bdellovibrio predation, prey cell death is early, preceding the possibility of amino acid starvation. Many of the upregulated prey genes are predicted to encode parts of stress response systems including genes involved in responding to carbon starvation (cstA); [27], toxic stress caused by paraquat (pqiB); [19], potassium tellurite (tehA); [38] and pH (yadF); [12] as well as a number of genes involved in several different global responses to stress (creB; [14], sufI; [33] csiE; [26] yafO; [28] iscR; [44] glnG; [43] mazG; [23]). In some cases, such as for pqiB; [19], tehA; [38] and yadF; [12] the single gene products may have a function alone without other co-transcribed genes in their operon being expressed.
“Selfish DNA” Phage or Transposon Genes
A number of chromosomally integrated phage and transposon genes were transcribed by E. coli in response to Bdellovibrio challenge of prey. One, ykfC is predicted to encode a reverse transcriptase of the CP4-6 prophage and is also near IS elements [9], insD is an IS2 insertion element transposase [31] and insL is an IS186/IS421 transposase [6]. Induction of phage and transposable elements has been demonstrated in a number of studies of stressing bacteria [13, 20, 29, 41] and it is thought that it is a generic stress response whereby the transposable elements are in some way, detecting adverse conditions and are attempting to “jump ship”. Not all phage and transposon genes were upregulated; this has been seen before [13] and probably represents the fact that many prophage genes are no longer functional in the E. coli genome.
Biosynthesis and Housekeeping in the Periplasm
A significant number of genes encoding apparently housekeeping functions of prey were also upregulated upon attack by Bdellovibrio. 39 of these are predicted to encode periplasmic or membrane bound products, or those involved in production of periplasmic proteins (see Table 1) and so are likely induced to repair the damage caused to the prey by Bdellovibrio, by regenerating the membrane and periplasmic contents. The genes fhlA, hycA, hycH, hycI, hypA, hypB and hypF were all called as upregulated whilst the hypCDEFG and hycBCD genes within the same apparent operons were not called, but were just below the chosen significance threshold. It is likely therefore that both of these operons were synchronously upregulated in order that all of the gene products may act together, probably in an “attempt” to produce different hydrogenases in response to changing pH levels within the prey periplasm as a result of the invading Bdellovibrio [1].
Interestingly, the rtn gene, annotated for resistance to phage N [11], was upregulated in our study. That this gene renders E. coli resistant to phage in experiments exposing E. coli to phage [5] suggests that the prey cell may be detecting damage caused by infection and attempting to respond, however, the response which could be effective against phage (the details of which are as yet unknown in the literature) has no preventative effect on the invading Bdellovibrio. Interestingly, other genes that were reported to be upregulated upon phage infection were not upregulated in our study showing that there are different responses to phage and to Bdellovibrio infections [18]. Genes reported to be initially upregulated by phage infection are nagE and dhaK, involved in initial steps of cellular energy generation, with later genes such as rraA and rof involved in transcription processes and so these are likely useful for phage processing that is not necessary for Bdellovibrio. Particularly significant is the fact that the phage shock proteins are not upregulated by Bdellovibrio invasion [17, 24]. The phage shock protein (Psp) F regulon response in E. coli is thought to be induced by impaired inner membrane integrity and an associated decrease in proton motive force, although the mechanisms by which the Psp system detects the stress signal and responds have so far remained undetermined. PspA and PspG respond to a variety of inducing stimuli by switching the cell to anaerobic respiration and fermentation and by downregulating motility, thereby subtly adjusting and maintaining energy usage and proton motive force. It is surprising given the apparent response to periplasmic damage upon Bdellovibrio infection that the Psp system is not induced, but perhaps the damage induced by Bdellovibrio is too little at this point for a transcriptional response by the phage shock system, as after 15 min of infection there is little interference of Bdellovibrio with the prey inner cytoplasmic membrane (and thus the prey cells are still viable and transcriptionally active as intended and required by our experimental design). Intriguingly, pspB, an activator of the Psp regulon [42] appears to be downregulated (Table 2), giving rise to speculation that the Bdellovibrio may be actively repressing the Psp regulon by some unknown interference mechanism to prevent restoration of the prey proton-motive-force which is shortly deliberately disrupted by the Bdellovibrio as the infection proceeds [30].
Downregulated Genes
34 prey genes were significantly downregulated upon predation by Bdellovibrio and in most cases they do not seem to be of any obvious use to the prey cell. In some cases, the downregulated genes are repressors and so their downregulation could result in de-repression of operons. One such example is malY, which is a repressor of the mal operon and so its downregulation could result in the upregulation of the mal operon [45]. Another is phage shock regulator pspB, mentioned above; where it is interesting to speculate as to whether the invading predator has somehow repressed expression of a potentially repairing response of the prey to aid in quick killing.
In conclusion, periplasmic invasion of prey by Bdellovibrio elicits transcriptional responses by the E. coli prey cell which seem to be an attempt to repair the physical damage to the periplasm caused by the presence of a Bdellovibrio penetration and to stabilise the osmolarity. It seems unlikely that the products of the genes transcribed would help the cell to defend itself against the Bdellovibrio attack; rather that it is a transcriptional “scream” responding to damage caused by the Bdellovibrio. This transcriptional response is short-lived as the Bdellovibrio go onto kill the prey in the subsequent 10 min after our RNA sampling. That we have been able to profile the response to Bdellovibrio binding and invasion by E. coli, a susceptible prey bacterium, may allow future gene expression comparisons to non-susceptible prey, to confirm the long held hypothesis that prey range in Bdellovibrio is determined by the ability to bind productively to prey outer membrane surfaces rather than being a result of the presence or absence of prey defensive gene expression as is the case for defence against phage [15].
Acknowledgements
We thank Marilyn Whitworth, Michael Capeness and Rob Till for technical assistance, and Mark Gomelsky, Laura Hobley and Chien-Yi Chang for helpful discussions. This work was supported by grants AL067712 and AL077459 to RES for CL from the Wellcome Trust, and in part by Human Frontier Science Program Grant RGP57/2005.067712.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Bagramyan K, Mnatsakanyan N, Poladian A, Vassilian A, Trchounian A. The roles of hydrogenases 3 and 4, and the F0F1-ATPase, in H2 production by Escherichia coli at alkaline and acidic pH. FEBS Lett. 2002;516:172–178. doi: 10.1016/S0014-5793(02)02555-3. [DOI] [PubMed] [Google Scholar]
- 2.Boos W, Ehmann U, Bremer E, Middendorf A, Postma P. Trehalase of Escherichia coli. Mapping and cloning of its structural gene and identification of the enzyme as a periplasmic protein induced under high osmolarity growth conditions. J Biol Chem. 1987;262:13212–13218. [PubMed] [Google Scholar]
- 3.Brynildsen MP, Liao JC. An integrated network approach identifies the isobutanol response network of Escherichiacoli. Mol Syst Biol. 2009;5:277. doi: 10.1038/msb.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burnham JC, Hashimoto T, Conti SF. Electron microscopic observations on the penetration of Bdellovibriobacteriovorus into Gram-negative bacterial hosts. J Bacteriol. 1968;96:1366–1381. doi: 10.1128/jb.96.4.1366-1381.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chae K-S, Yoo OJ. Cloning of lambda resistant genes from Brevibacterium albidum and Proteus vulgaris into Escherichia coli. Biochem Biophys Res Commun. 1986;140:1101–1105. doi: 10.1016/0006-291X(86)90748-5. [DOI] [PubMed] [Google Scholar]
- 6.Chong P, Hui I, Loo T, Gillam S. Structural analysis of a new GC-specific insertion element IS186. FEBS Lett. 1985;192:47–52. doi: 10.1016/0014-5793(85)80040-5. [DOI] [PubMed] [Google Scholar]
- 7.Evans KJ, Lambert C, Sockett RE. Predation by Bdellovibriobacteriovorus HD100 requires type IV pili. J Bacteriol. 2007;189:4850–4859. doi: 10.1128/JB.01942-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gallet R, Alizon S, Comte PA, Gutierrez A, Depaulis F, van Baalen M, Michel E, Muller-Graf CD. Predation and disturbance interact to shape prey species diversity. Am Nat. 2007;170:143–154. doi: 10.1086/518567. [DOI] [PubMed] [Google Scholar]
- 9.Garcia-Vallve S, Palau J, Romeu A. Horizontal gene transfer in glycosyl hydrolases inferred from codon usage in Escherichia coli and Bacillussubtilis. Mol Biol Evol. 1999;16:1125–1134. doi: 10.1093/oxfordjournals.molbev.a026203. [DOI] [PubMed] [Google Scholar]
- 10.Gomelsky L, Sram J, Moskvin OV, Horne IM, Dodd HN, Pemberton JM, McEwan AG, Kaplan S, Gomelsky M. Identification and in vivo characterization of PpaA, a regulator of photosystem formation in Rhodobactersphaeroides. Microbiology. 2003;149:377–388. doi: 10.1099/mic.0.25972-0. [DOI] [PubMed] [Google Scholar]
- 11.Hall BG. The rtn gene of Proteusvulgaris is actually from Escherichia coli. J Bacteriol. 1997;179:2433–2434. doi: 10.1128/jb.179.7.2433-2434.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hashimoto M, Kato J. Indispensability of the Escherichia coli carbonic anhydrases YadF and CynT in cell proliferation at a low CO2 partial pressure. Biosci Biotechnol Biochem. 2003;67:919–922. doi: 10.1271/bbb.67.919. [DOI] [PubMed] [Google Scholar]
- 13.Herold S, Siebert J, Huber A, Schmidt H. Global expression of prophage genes in Escherichia coli O157:H7 strain EDL933 in response to norfloxacin. Antimicrob Agents Chemother. 2005;49:931–944. doi: 10.1128/AAC.49.3.931-944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirakawa H, Nishino K, Yamada J, Hirata T, Yamaguchi A. Beta-lactam resistance modulated by the overexpression of response regulators of two-component signal transduction systems in Escherichia coli. J Antimicrob Chemother. 2003;52:576–582. doi: 10.1093/jac/dkg406. [DOI] [PubMed] [Google Scholar]
- 15.Hoskisson PA, Smith MC. Hypervariation and phase variation in the bacteriophage ‘resistome’. Curr Opin Microbiol. 2007;10:396–400. doi: 10.1016/j.mib.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 16.Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, Bennett HA, Coffey E, Dai H, He YD, Kidd MJ, King AM, Meyer MR, Slade D, Lum PY, Stepaniants SB, Shoemaker DD, Gachotte D, Chakraburtty K, Simon J, Bard M, Friend SH. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/S0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 17.Jovanovic G, Lloyd LJ, Stumpf MP, Mayhew AJ, Buck M. Induction and function of the phage shock protein extracytoplasmic stress response in Escherichia coli. J Biol Chem. 2006;281:21147–21161. doi: 10.1074/jbc.M602323200. [DOI] [PubMed] [Google Scholar]
- 18.Karlsson F, Malmborg-Hager AC, Albrekt AS, Borrebaeck CA. Genome-wide comparison of phage M13-infected vs. uninfected Escherichia coli. Can J Microbiol. 2005;51:29–35. doi: 10.1139/w04-113. [DOI] [PubMed] [Google Scholar]
- 19.Koh YS, Roe JH. Isolation of a novel paraquat-inducible (pqi) gene regulated by the soxRS locus in Escherichia coli. J Bacteriol. 1995;177:2673–2678. doi: 10.1128/jb.177.10.2673-2678.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koide T, Vencio RZ, Gomes SL. Global gene expression analysis of the heat shock response in the phytopathogen Xylellafastidiosa. J Bacteriol. 2006;188:5821–5830. doi: 10.1128/JB.00182-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamark T, Kaasen I, Eshoo MW, Falkenberg P, McDougall J, Strom AR. DNA sequence and analysis of the bet genes encoding the osmoregulatory choline-glycine betaine pathway of Escherichia coli. Mol Microbiol. 1991;5:1049–1064. doi: 10.1111/j.1365-2958.1991.tb01877.x. [DOI] [PubMed] [Google Scholar]
- 22.Lambert C, Evans KJ, Till R, Hobley L, Capeness M, Rendulic S, Schuster SC, Aizawa S, Sockett RE. Characterizing the flagellar filament and the role of motility in bacterial prey-penetration by Bdellovibrio bacteriovorus. Mol Microbiol. 2006;60:274–286. doi: 10.1111/j.1365-2958.2006.05081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee S, Kim MH, Kang BS, Kim JS, Kim GH, Kim YG, Kim KJ. Crystal structure of Escherichia coli MazG, the regulator of nutritional stress response. J Biol Chem. 2008;283:15232–15240. doi: 10.1074/jbc.M800479200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lloyd LJ, Jones SE, Jovanovic G, Gyaneshwar P, Rolfe MD, Thompson A, Hinton JC, Buck M. Identification of a new member of the phage shock protein response in Escherichia coli, the phage shock protein G (PspG) J Biol Chem. 2004;279:55707–55714. doi: 10.1074/jbc.M408994200. [DOI] [PubMed] [Google Scholar]
- 25.Macnab RM. How bacteria assemble flagella. Annu Rev Microbiol. 2003;57:77–100. doi: 10.1146/annurev.micro.57.030502.090832. [DOI] [PubMed] [Google Scholar]
- 26.Marschall C, Hengge-Aronis R. Regulatory characteristics and promoter analysis of csiE, a stationary phase-inducible gene under the control of sigma S and the cAMP-CRP complex in Escherichia coli. Mol Microbiol. 1995;18:175–184. doi: 10.1111/j.1365-2958.1995.mmi_18010175.x. [DOI] [PubMed] [Google Scholar]
- 27.Matin A. The molecular basis of carbon-starvation-induced general resistance in Escherichia coli. Mol Microbiol. 1991;5:3–10. doi: 10.1111/j.1365-2958.1991.tb01819.x. [DOI] [PubMed] [Google Scholar]
- 28.McKenzie GJ, Magner DB, Lee PL, Rosenberg SM. The dinB operon and spontaneous mutation in Escherichia coli. J Bacteriol. 2003;185:3972–3977. doi: 10.1128/JB.185.13.3972-3977.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richmond CS, Glasner JD, Mau R, Jin H, Blattner FR. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 1999;27:3821–3835. doi: 10.1093/nar/27.19.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rittenberg SC, Shilo M. Early host damage in the infection cycle of Bdellovibrio bacteriovorus. J Bacteriol. 1970;102:149–160. doi: 10.1128/jb.102.1.149-160.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ronecker HJ, Rak B. Genetic organization of insertion element IS2 based on a revised nucleotide sequence. Gene. 1987;59:291–296. doi: 10.1016/0378-1119(87)90337-4. [DOI] [PubMed] [Google Scholar]
- 32.Rosson RA, Rittenberg SC. Regulated breakdown of Escherichia coli deoxyribonucleic acid during intraperiplasmic growth of Bdellovibrio bacteriovorus 109J. J Bacteriol. 1979;140:620–633. doi: 10.1128/jb.140.2.620-633.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Samaluru H, SaiSree L, Reddy M. Role of SufI (FtsP) in cell division of Escherichia coli: evidence for its involvement in stabilizing the assembly of the divisome. J Bacteriol. 2007;189:8044–8052. doi: 10.1128/JB.00773-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shemesh Y, Jurkevitch E. Plastic phenotypic resistance to predation by Bdellovibrio and like organisms in bacterial prey. Environ Microbiol. 2004;6:12–18. doi: 10.1046/j.1462-2920.2003.00530.x. [DOI] [PubMed] [Google Scholar]
- 35.Sockett RE, Lambert C. Bdellovibrio as therapeutic agents: a predatory renaissance? Nat Rev Microbiol. 2004;2:669–675. doi: 10.1038/nrmicro959. [DOI] [PubMed] [Google Scholar]
- 36.Storey JD. A direct approach to false discovery rates. J R Stat Soc Series B (Stat Methodol) 2002;64:479–498. doi: 10.1111/1467-9868.00346. [DOI] [Google Scholar]
- 37.Traxler MF, Summers SM, Nguyen HT, Zacharia VM, Hightower GA, Smith JT, Conway T. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol Microbiol. 2008;68:1128–1148. doi: 10.1111/j.1365-2958.2008.06229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turner RJ, Taylor DE, Weiner JH. Expression of Escherichia coli TehA gives resistance to antiseptics and disinfectants similar to that conferred by multidrug resistance efflux pumps. Antimicrob Agents Chemother. 1997;41:440–444. doi: 10.1128/aac.41.2.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Varon M. Selection of predation-resistant bacteria in continuous culture. Nature. 1979;277:386–388. doi: 10.1038/277386a0. [DOI] [Google Scholar]
- 40.Varon M, Drucker I, Shilo M. Early effects of Bdellovibrio infection on the synthesis of protein and RNA of host bacteria. Biochem Biophys Res Commun. 1969;37:518–525. doi: 10.1016/0006-291X(69)90946-2. [DOI] [PubMed] [Google Scholar]
- 41.Walker D, Rolfe M, Thompson A, Moore GR, James R, Hinton JC, Kleanthous C. Transcriptional profiling of colicin-induced cell death of Escherichia coli MG1655 identifies potential mechanisms by which bacteriocins promote bacterial diversity. J Bacteriol. 2004;186:866–869. doi: 10.1128/JB.186.3.866-869.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weiner L, Brissette JL, Model P. Stress-induced expression of the Escherichia coli phage shock protein operon is dependent on sigma 54 and modulated by positive and negative feedback mechanisms. Genes Dev. 1991;5:1912–1923. doi: 10.1101/gad.5.10.1912. [DOI] [PubMed] [Google Scholar]
- 43.Weiss V, Claverie-Martin F, Magasanik B. Phosphorylation of nitrogen regulator I of Escherichia coli induces strong cooperative binding to DNA essential for activation of transcription. Proc Natl Acad Sci USA. 1992;89:5088–5092. doi: 10.1073/pnas.89.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yeo WS, Lee JH, Lee KC, Roe JH. IscR acts as an activator in response to oxidative stress for the suf operon encoding Fe-S assembly proteins. Mol Microbiol. 2006;61:206–218. doi: 10.1111/j.1365-2958.2006.05220.x. [DOI] [PubMed] [Google Scholar]
- 45.Zdych E, Peist R, Reidl J, Boos W. MalY of Escherichia coli is an enzyme with the activity of a beta C-S lyase (cystathionase) J Bacteriol. 1995;177:5035–5039. doi: 10.1128/jb.177.17.5035-5039.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]