Abstract
Infection with high-risk human papillomaviruses (HPV) and in particular the expression of the viral proteins E6 and E7 have been associated with the etiology of a subset of head and neck squamous cell cancer (HNSCC). However, the individual consequences of E6 and E7 expression in an in vivo model have not been examined in these tissues. We have used transgenes that direct expression of the HPV16 E6 and E7 proteins to the head and neck tissues of mice to dissect the contribution of these proteins to head and neck carcinogenesis. We report here that E7 is the major transforming oncogene in HPV-associated HNSCC, whereas E6 is more likely to play a secondary role in contributing to later stages of carcinogenesis. Furthermore, a conditional deletion of Rb, a prominent target for E7, in the same tissues did not recapitulate all E7-mediated phenotypes. Although our results do not preclude an important role for the E7-pRbinteraction, they highlight the importance of pRb-independent functions of E7 in head and neck carcinogenesis.
Introduction
High-risk human papillomaviruses (HPV), now well accepted to be causative agents for the majority of cervical cancers, more recently have been associated with a subset of head and neck cancers. The oncogenic properties of high-risk human papillomaviruses have been attributed largely to the two early proteins E6 and E7. Expression of E6 and E7 has been detected from both integrated and extrachromosomal genomes in HPV-positive head and neck squamous cell cancer (HNSCC) consistent with the hypothesis that these oncoproteins contribute to these cancers (1–3). E6 and E7 are known to bind and inactivate the tumor suppressors p53 and pRb, respectively (4–9), and these respective activities have been associated with their oncogenic properties (10, 11). Consistent with the contribution of E6 and E7 to HPV-positive HNSCC, their expression correlates with a paucity of mutations in p53 (12), decreased levels of pRb, and increased levels of p16 (13, 14). Conversely, in HPV-negative HNSCC, p53 is often mutated, levels of pRb are normal, and levels of p16 are decreased. The distinct nature of the alterations in the p16/pRb and p53 pathways not only distinguishes the HPV-positive cancers from the HPV-negative ones but also implies roles for both the HPV oncogenes. However, despite the overwhelming evidence from the study of human tumor samples, the individual contributions of these oncogenes has not been studied in vivo in the context of head and neck carcinogenesis.
Previous work, with strategies that focused on the use of transgene-directed expression of E6 and E7, has shed light on the in vivo roles of these proteins not only in tissues that are considered the natural host for HPV infection, such as the epidermis and the stratified epithelium of the cervix, but also tissues such as the retina, lens, and the central nervous system (15–20). In these mice, the HPV oncogenes are directed in their expression from the keratin 14 (K14) promoter, which targets expression of the viral oncogenes to the same basal compartment of stratified epithelia as observed in natural HPV infections. Furthermore, extensive analyses of these mice have shown that the HPV16 E6 and E7 oncoproteins display the same capacity to inactivate relevant cellular targets, including but not limited to p53 and pRb, respectively, as has been documented to occur in HPV-infected human epithelia (10, 21–25). From our studies, we have learned that these oncogenes not only possess properties that distinguish one from the other, but also that those properties may vary in a tissue-dependent manner. Furthermore, different activities of the oncogenes have been shown to be important for short-term acute phenotypes and/or long-term oncogenic phenotypes (21, 22, 26).
In this study, we have dissected the roles of E6 and E7 in an in vivo model for HPV-associated HNSCC. As previously described in a model for cervical cancer (20, 27), E7 was found in this study to be the major transforming oncogene in the head and neck with some evidence for a role of E6 in later stages of carcinogenesis. The HPV E7 oncoprotein is best known for its ability to target the tumor suppressor pRb for degradation. However, we describe here that the conditional deletion of Rb in the head and neck epithelia does not fully recapitulate the effects of E7, thus targets of E7 other than the tumor suppressor pRb must be important in its ability to mediate carcinogenesis.
Materials and Methods
Mice
The generation of K14E6 (originally called K14E6:E7TTL) and K14E7 (originally called K14E7:E6TTL) transgenic mice has been previously described (18, 19). Mice used were derived from founder lines, which were extensively characterized and chosen from a larger set of founder lines. These lines were chosen for their physiologic level of expression of the HPV oncogenes (27).1 All experiments were performed in the hemizygous state to minimize the possibility of effects that might stem from transgene insertion. To obtain bitransgenic K14E6K14E7 mice, female K14E6H mice were crossed with male K14E7 mice. In all the studies described, heterozygous singly transgenic mice were used. Mice carrying the K14Cre transgene were obtained from Dr. Anton Berns at the Netherlands Cancer Institute (Amsterdam, the Netherlands; ref. 28). Mice carrying a conditional Rb allele were generated by and obtained from Drs. Julien Sage (Department of Pediatrics and Genetics, Stanford University, Stanford, CA) and Tyler Jacks (Center for Cancer Research, Massachusetts Institute of Technology, Cambridge, MA; ref. 29). Tail clippings were collected from all the offspring at the time of weaning and DNA was isolated for genotyping by means of PCR.
For studies described in Fig. 1 and Fig. 2, all mice were maintained on the FVB/N inbred genetic background. For the studies described in Fig. 3 and Fig. 4, the studies were performed in a mixed FVB/N and C57BL/6 genetic background. For all the experiments that were performed at a mixed background, the mice were maintained at backcross 6 to FVB/N such that all the mice contained a similar degree of genetic heterogeneity. Mice were housed in the Association for Assessment of Laboratory Animal Care–approved McArdle Laboratory Animal Care Unit at the University of Wisconsin Medical School. All protocols for animal work were approved by the University of Wisconsin Medical School Institutional Animal Care and Use Committee.
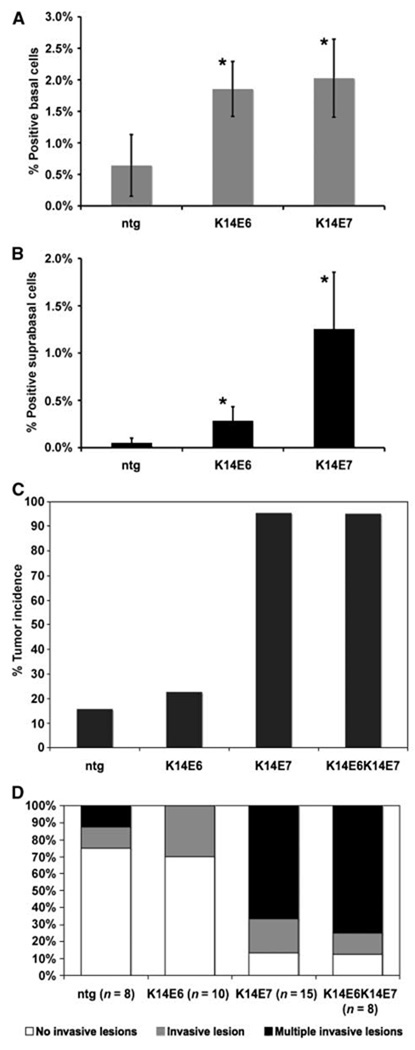
Figure 1.
Examination of acute effects of E6 and E7 on DNA synthesis. To determine the DNA synthesis occurring at the basal (A) and suprabasal (B) compartments, mice were injected with BrdUrd before sacrifice and their tissues were stained for BrdUrd. At least three mice of each genotype, nontransgenic, K14E6, and K14E7h, were used and ∼8 to 10 frames of cells from tongue epithelium were quantified for each mouse. The amount of positive nuclei over the number of total cells was plotted in each case (columns); bars, SD. *, both in terms of basal and suprabasal DNA synthesis, the increase seen in K14E6 and K14E7 animals was increased compared with the amount of DNA synthesis seen in the nontransgenics. The differences were statistically significant according to a two-sided Wilcoxon rank sum test (K16E6 versus nontransgenic basal P = 0.01, suprabasal P = 0.04; K14E7 versus nontransgenic basal P = 0.009, suprabasal P = 0.009; K14E6 versus K14E7 basal P = 0.6, suprabasal P = 0.006). ntg, nontransgenic control animals. C, overt tumor incidence in carcinogen-treated mice. Animals represented in this group were treated with 4-NQO for 16 wk and held for a further 8 wk or until they were moribund. At the end point, necropsy was performed and the overt, visible tumors on the mouse tongue and esophagus were counted. The differences in tumor incidence between nontransgenic and K14E7 groups (P = 1.4 × 10−7), and K14E6K14E7 groups (P = 3.0 × 10−7), are statistically significant. Also, the differences between K14E6 and K14E7 (P = 1.1 × 10−6) and K14E6K14E7 groups (P = 1.1 × 10−6) are statistically significant. The differences seen between nontransgenic and K14E6 (P = 0.7) and K14E7 and K14E6K14E7 (P = 1) groups are not statistically significant. All statistical comparisons were performed using a two-sided Fisher’s exact test. D, multiplicity of cancers. Data from subset of mice randomly chosen for full histopathologic analysis are represented here in terms of cancer multiplicity. Animals with no cancers or benign tumors are assigned to the first group and the other two groups contain animals with one or more than one invasive cancers. The differences between the nontransgenic and K14E7 (P = 0.004) and K14E6K14E7 (P = 0.01) groups are statistically significant. The differences between the K14E6 and K14E7 and K14E6K14E7 groups are also statistically significant. The nontransgenic and K14E6 (P = 0.95) and the K14E7 and K14E6K14E7 (P = 0.72) comparisons are not statistically significant. All comparisons were performed using the Wilcoxon rank sum test (two-sided). The tumor incidences of the negative (nontransgenic) and positive (K14E6K14E7) control groups, which were further characterized in this figure, were previously reported in ref. 30.
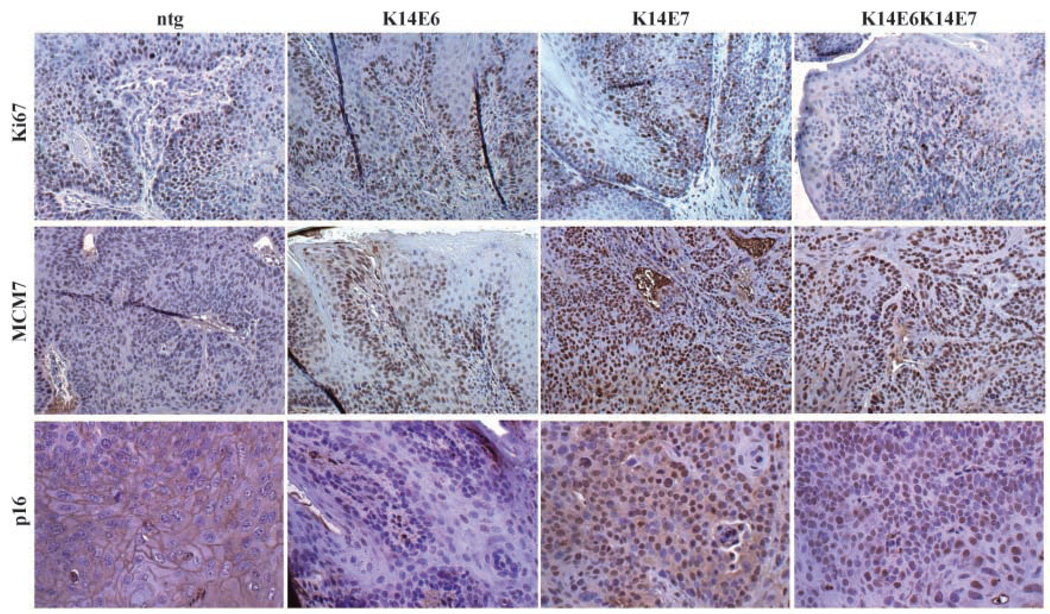
Figure 2.
Molecular characterization of cancers arising in treated K14E6 and K14E7 mice. To characterize molecular differences between the cancers generated in nontransgenic, K14E6, K14E7, and K14E6K14E7 animals, sets of cancers from these animals were stained for Ki67, MCM7, and p16. Representative images.
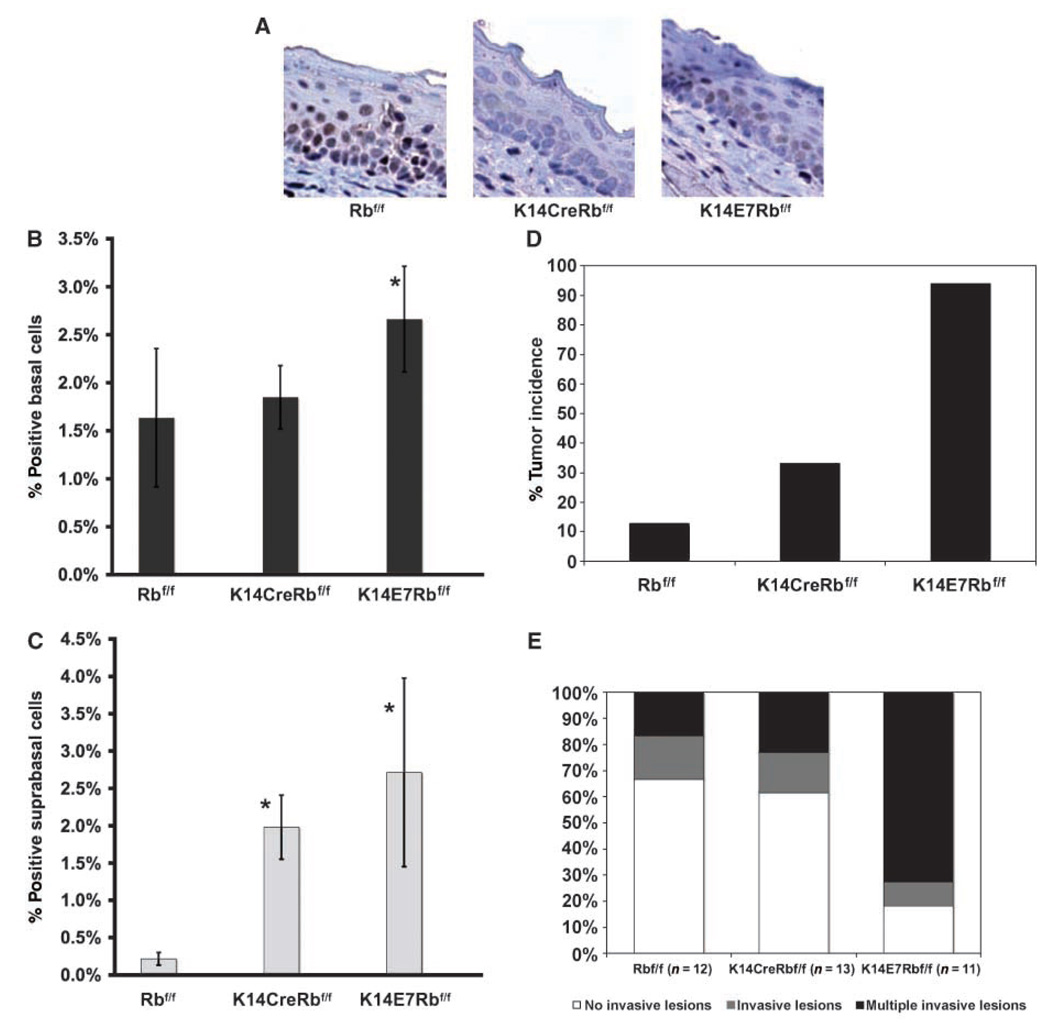
Figure 3.
A, detection of pRb in control, Rb-deleted, and E7-expressing epithelial tongue sections from Rbf/f, K14CreRbf/f, and K14E7Rbf/f animals stained immunohistochemically for pRb. The results are similar to those seen on sections from esophagus. Although staining is abundantly detectable in Rbf/f controls, it is largely undetectable in K14CreRbf/f mice and greatly diminished in K14E7Rbf/f mice. B and C, acute effects of Rb deletion and E7 expression in epithelia: Rbf/f, K14CreRbf/f, and K14E7Rbf/f animals were injected with BrdUrd before sacrifice and BrdUrd-specific immunohistochemistry was performed on collected tissues. The number of positive basal or suprabasal over the total number of cells is shown in graphs B and C, respectively. At least three animals per group were quantified and 8 to 10 frames from tongue epithelium per animal were counted. Bars, SD. In terms of basal DNA synthesis, the increase was statistically significant only between K14E7Rbf/f and Rbf/f mice (P = 0.04, one-sided). In terms of suprabasal DNA synthesis both in K14CreRbf/f, and K14E7Rbf/f mice, a statistically significant increase over that seen in the Rbf/f control mice was observed (P = 0.03, P = 0.01, respectively, Wilcoxon rank sum test, two-sided). D, tumor incidence in carcinogen-treated animals: Following treatment with 4-NQO for 16 wk and a hold period for 8 wk, animals of the indicated genotypes were sacrificed and overt tumors in their tongue and esophagus were scored. The increase seen in K14E7Rbf/f compared with Rbf/f animals was statistically significant (P = 3.1 × 10−9), whereas that seen between K14CreRbf/f and Rbf/f animals was also statistically significant (P = 0.02). The difference between the K14E7Rbf/f and K14CreRbf/f groups was also statistically significant (P = 6.7 × 10−5). All comparisons were performed using a two-sided Fisher’s exact test. E, data from subset of mice randomly chosen for full histopathologic analysis are represented here in terms of cancer multiplicity. The differences between the K14E7Rbf/f and the Rbf/f (P = 0.02) and K14CreRbf/f (P = 0.009) are statistically significant. The difference between the Rbf/f and K14CreRbf/f is not statistically significant (P = 0.75). All comparisons were performed using a two-sided Wilcoxon rank sum test.
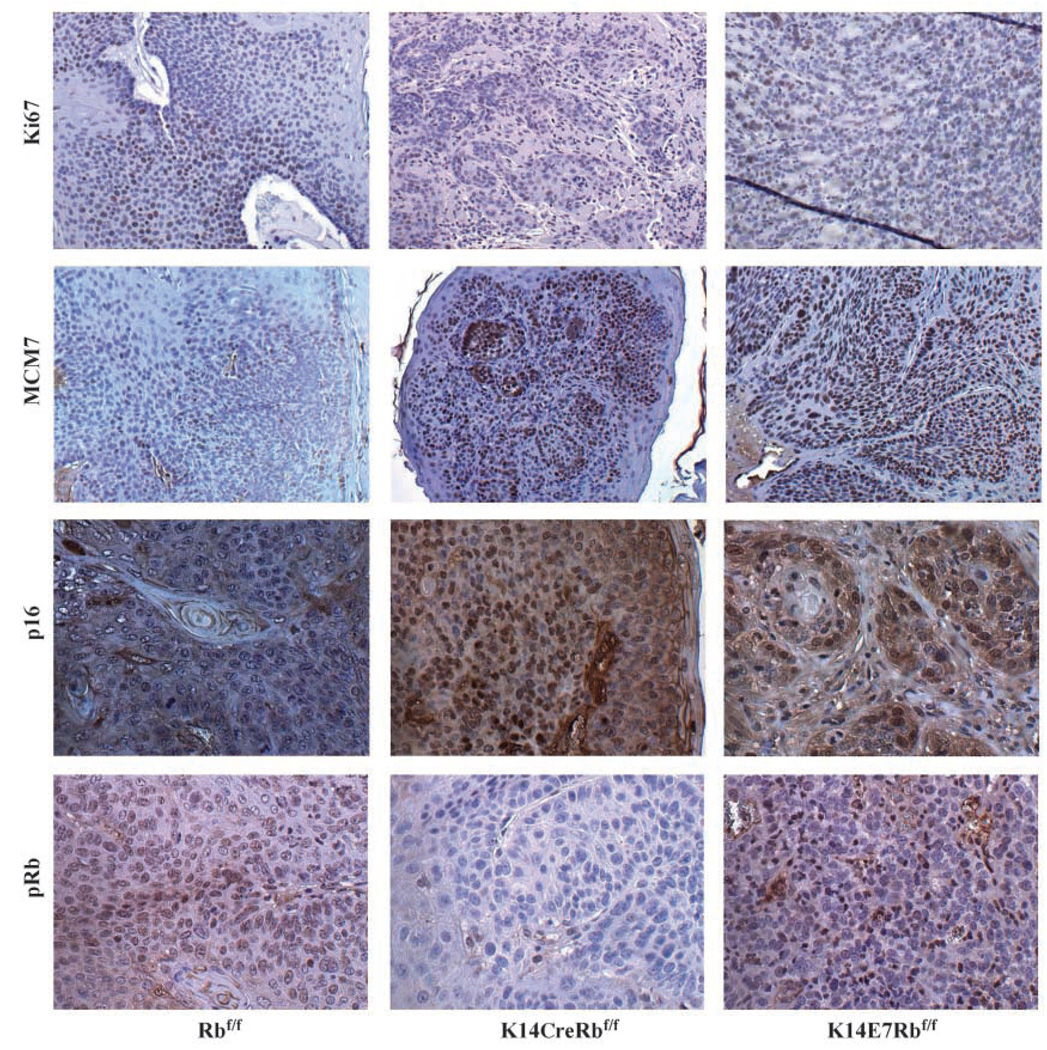
Figure 4.
Molecular characterization of cancers arising in Rbf/f, K14CreRbf/f, and K14E7Rbf/f mice: Cancers from Rbf/f, K14CreRbf/f, and K14E7Rbf/f animals were immunohistochemically stained and compared for the expression of these markers Ki67, MCM7, p16, and pRb. Representative panels for each genotype and marker.
4-nitroquinoline-N-oxide–induced head and neck carcinogenesis study and histologic analysis
The treatment and guidelines for histologic analysis were previously described (30).Briefly, mice were treated with 4-nitroquinoline-N-oxide (4-NQO) in their drinking water for 16 weeks and then held off-treatment for 8 weeks. At the end point or when mice became moribund, they were euthanized, tumors in tongue and esophagus were scored, and tissues were collected for pathology.
Immunohistochemistry
The protocol for immunohistochemical analysis of tissues has been previously described (22). Briefly, primary incubation with anti-bromodeoxyuridine (BrdUrd) antibody (Calbiochem) was performed at a concentration of 1:50 in blocking solution at 4°C. For other antigens, the primary antibodies were used as follows: Ki67 (Dako, 1:25), Mcm7 (Neomarkers, 1:200), pRb (G3–245, PharMingen, 1:50), and p16 (M156, Santa Cruz Biotechnology, 1:50). Incubation with secondary antibodies was performed in a 1:100 dilution in PBS of biotinylated anti-rat immunoglobulin (PharMingen) for Ki67 or 1:100 Vectastain Universal Elite secondary antibody for all other antigens (Vector) for 30 min at room temperature. The signal was amplified using the ABC reagent from the Vectastain Universal Elite kit according to the manufacturer’s instructions. The signal was developed using 3,3′-diaminobenzidine substrate (Vector) for 2 to 9 min.
Results
Acute phenotypes of E6 or E7 in the head and neck epithelia
In K14E6/K14E7 bitransgenic mice, we observed an induction of aberrant DNA synthesis in the head and neck epithelia (30). To understand the individual roles of E6 and E7 in contributing to this phenotype, 7-week-old K14E6 and K14E7 transgenic mice, expressing either HPV16 E6 or HPV16 E7, respectively, were injected with the nucleotide analogue BrdUrd 1 h before sacrifice. Paraffin-embedded tissues were sectioned and stained with BrdUrd-specific antibodies. Both E6 and E7 led to an increase in the levels of basal DNA synthesis in the head and neck epithelia, suggesting that the expression of each viral oncoprotein in the undifferentiated cells of head and neck epithelia leads to an increase in the number of cells that are in the cell cycle at any given point (Fig. 1A). The expression of E6 and E7 each also led to an induction of DNA synthesis in the normally quiescent suprabasal compartment of the stratified epithelia of the head and neck region (Fig. 1B).
Contribution of E6 and E7 to tumorigenesis
Short-term phenotypes have in the past served as good indicators for E6 and E7 function in tissues; however, they have not necessarily correlated with the ability of E6 and E7 to contribute to tumorigenesis. We recently established a mouse model for head and neck cancers in which treatment with a low dose of 4-NQO led to the formation of more cancers and cancers of higher grade in bitransgenic K14E6K14E7 mice compared with nontransgenic mice (30). To evaluate the individual contributions of E6 and E7 to the tumors that formed in the bitransgenic mice, we treated K14E6 and K14E7 singly transgenic animals, with 4-NQO as previously described. At the end point or when they became moribund, mice were euthanized, tissues were excised, and the incidence of overt tumors on their tongue and esophagus was scored.
Out of a group of 22 4-NQO–treated K14E6 mice, only 5 (22%) had overt tumors at the study end point, not significantly higher than the frequency observed in the 4-NQO–treated nontransgenic mice (16%; see Fig. 1C). Esophagus and tongues harvested from a random subset of the mice of each genotype were sectioned throughout, and a thorough histopathologic analysis was carried out in which every 20th 5-µm section was stained with H&E and scored under the microscope for the presence of hyperplasia/ dysplasia, benign tumors, and grade of carcinoma. For each mouse, the worst phenotype found within the esophagus and tongue tissue was tabulated (Table 1). As seen in the nontransgenic animals, most of the E6 animals had benign disease such as hyperplasia or dysplasia in their tissues. The incidence of carcinoma in these two subgroups was not statistically different. The multiplicity of invasive disease likewise was not different between nontransgenic and K14E6 mice (Fig. 1D). Overall, we observed no significant differences in the nature of the head and neck disease between the nontransgenic and K14E6 mice based either on scoring the frequency of overt tumors or the histopathologic grade of disease.
Table 1.
Tissue pathology of 4-NQO–treated mice
| Hyperplasia/dysplasia (%) | Papilloma (%) | Grade 1 (%) | Grade 2 (%) | Grade 3 (%) | |
|---|---|---|---|---|---|
| Ntg (n = 8) | 50 | 25 | 0 | 25 | 0 |
| K14E6 (n = 10) | 70 | 0 | 30 | 0 | 0 |
| K14E7 (n = 15) | 7 | 7 | 27 | 13 | 47 |
| K14E6K14E7 (n = 8) | 0 | 13 | 0 | 13 | 75 |
NOTE: A randomly selected subgroup of mice from each genotype was selected for detailed histopathologic analysis of tongue and esophageal tissue for the phenotypes denoted above. Each mouse was assigned the worst phenotype found in at least two consecutive slides analyzed. Statistically significant differences were found only between the K14E7 versus nontransgenic groups (P = 0.015), and K14E6K14E7 and nontransgenic groups (P = 0.002). The other comparisons were not statistically different (nontransgenic versus K14E6, P = 0.417, two-sided; K14E7 versus K14E6K14E7, P = 0.062, one-sided, P = 0.125, two-sided). All statistical analyses were made using a Wilcoxon rank sum test.
Abbreviation: Ntg, nontransgenic control mice.
In contrast to the K14E6 mice, the K14E7 mice, when treated with 4-NQO, had an almost fully penetrant tumorigenic phenotype. Out of a group of 22 animals, 21 had overt tumors at the time of sacrifice (95%). This represents a statistically significant increase over the tumor incidence seen in the control group; however, it is not statistically different from that seen in the K14E6K14E7 group (95%). The subset of K14E7 mice that was randomly chosen for histopathologic analysis revealed that the majority of the mice had carcinomas (13 of 15) and, as seen in the K14E6K14E7 group, a large portion of the carcinomas in those mice were poorly differentiated, grade 3 lesions (7 of 15). The presence of high-grade carcinomas was observed in the K14E6K14E7 in a higher fraction (6 of 8) of mice analyzed, which suggests a contribution for E6 in late-stage carcinogenesis; however, that difference was just shy of the 95% confidence limit (P = 0.06; one-sided Wilcoxon rank sum test). The multiplicity of tumor load in these subgroups of mice histopathologically analyzed was also assessed. In the bitransgenic mice, 75% had multiple invasive lesions, whereas 67% of the K14E7 mice had multiple lesions (Fig. 1D). Again, this difference between the two groups, although it may be indicative of a function for E6 in the context of E7, is not statistically significant.
In this study, analyzing the individual contributions of HPV16 E6 and E7 in head and neck cancers, we found E7 to be the more potent oncogene. This finding correlates with that seen in our mouse model for cervical cancer, in which E7 also was found to be the dominant oncogene. Although a statistically significant contribution for E6 in the context of head and neck carcinogenesis was not observed under the experimental conditions used in this study, it is possible that under different experimental conditions, a contribution of E6 to carcinogenesis may be evident. A difference in the frequency of high-grade carcinomas between K14E7 and K14E6K14E7 mice, although not statistically significant, is consistent with a role for E6 in the later stages of tumor progression as observed in the skin and cervix.
Proliferation and expression of biomarkers in carcinomas arising in K14E6 and K14E7 animals
In addition to the differences detected in tumor incidence between animals carrying the E6 versus E7 oncogenes, we wanted to determine whether there were other molecular differences that distinguished the tumors arising in these mice. Up-regulation of p16 has been reported in human cancers containing HPV DNA both in the cervix and the head and neck (13, 14, 31, 32). Our laboratory previously reported that MCM7 is another useful biomarker for carcinogenesis in the cervix of humans and mice (23). We previously described studies that showed that both p16 and MCM7 are induced in head and neck cancers arising in the K14E6K14E7 bitransgenic mice (30). We therefore carried out immunohistochemical studies on Ki67, p16, and MCM7 in the head and neck–treated tissues (tongue and esophagus) of our 4-NQO–treated bitransgenic, singly transgenic, and nontransgenic mice to determine whether characteristic marker expression was due to one or both oncogenes.
When comparing K14E6K14E7-positive cancers to nontransgenic cancers by immunohistochemistry, no marked differences in proliferation were noted using antibodies to Ki67, a marker for cell proliferation (ref. 30; see also Fig. 2). To assess whether tumors singly transgenic for E6 or E7 had any differences in the proliferation index, we stained those cancers with the same antibody (Fig. 2). Not surprisingly, there were no gross differences seen in the proliferation of the cancers from any of the genotypes; in fact, proliferation correlated with the state of disease rather than the genotype of the mice.
Tissues were also subjected to immunohistochemistry using an antibody specific for MCM7, previously seen to be up-regulated in cancers from K14E6K14E7 bitransgenic mice (ref. 30; see also Fig. 2). MCM7 is an E2F-responsive gene and therefore is predicted to be up-regulated owing to the inactivation of pRb by E7. In the K14E6 animals, MCM7 was slightly increased in cancers as opposed to normal or dysplastic epithelium; however, it was not increased to the same degree observed in bitransgenic cancers (Fig. 2). Not surprisingly, tissues from K14E7 mice showed a marked up-regulation in MCM7 not only in dysplastic epithelium, papillomas, and carcinomas but also in epithelium that was histopathologically normal (Fig. 2 and data not shown). The difference seen between the K14E7 animals and the nontransgenic animals was quite striking and analogous to the difference seen between the K14E6K14E7 animals and the nontransgenic animals (Fig. 2). The high frequency of MCM7-positive cells in the tissues from the K14E7 animals was greater than the frequency of Ki67-positive cells in the same tissues regardless of the grade of disease (Fig. 2), suggesting that E7 dysregulation of pRb is pervasive and is not restricted to proliferating cells. In contrast, in nontransgenic and K14E6 animals, MCM7, which is moderately expressed in carcinomas and restricted to the basal and parabasal layers of histopathologically normal epithelium, is more indicative of the proliferation state of the cells as the pattern of expression correlates well with the pattern of Ki67 expression (Fig. 2). We conclude that the ubiquitous expression of MCM7 seen in bitransgenic cancers is largely due to the action of E7 and less so to that of E6. The robust transcriptional activation of the E2F-responsive MCM7 protein is likely a result of the collective action of E7 on all the Rb family proteins. Unfortunately, due to the lack of adequate antibodies for immunohistochemistry specific for p107 and p130, we cannot verify that their overall levels are likewise diminished.
HPV-positive human tumors frequently show high levels of staining for p16 (13, 14, 31, 32). We hypothesized that this result is largely due to the action of E7 on pRb, which is downstream of p16 in the same pathway. We therefore performed p16-specific immunohistochemistry on tumors from our mice (Fig. 2). Our results were consistent with this hypothesis as abundant staining for p16 was observed on both K14E7 and K14E6K14E7 tumors. Very little p16 was detectable in the nontransgenic or K14E6 tumors.
Loss of pRb can recapitulate some but not all of the acute effects of E7 in inducing aberrant DNA synthesis in head and neck epithelia
A challenge in evaluating the individual interaction of E7 with pRb is posed by the fact that E7 interacts with all the RB family proteins (including p107 and p130) through the same binding pocket, the LxCxE binding motif. Thus, the use of a mutant form of E7 that cannot bind pRb is of limited value because this mutation will affect E7 binding to not only pRb but also p107 and p130. To determine specifically the importance of pRb as a relevant target in E7-mediated tumorigenesis, we made use of Rbf/f mice homozygous for a conditional null allele of Rb (29). In Rbf/f mice, exon 3 of the Rb allele is flanked by loxP sites. Excision of exon 3 by Cre recombinase leads to a frameshift and the premature termination of translation of pRb. The gene product of this recombined allele was shown to lack any function ascribed to full-length pRb. To direct expression of Cre to the same compartment in which we express E7, we obtained K14Cre mice in which the expression of the recombinase is driven from the same cytokeratin 14 promoter used in our K14E7 mice (28).
As evaluated by pRb-specific immunohistochemistry (Fig. 3A), pRb is expressed abundantly throughout the thickness of the head and neck epithelia of adult Rbf/f mice that are homozygous for conditional-null Rb allele but express no Cre recombinase. In these same epithelia in age-matched K14CreRbf/f animals, pRb was undetectable, validating that Cre was efficient at targeting recombination of the floxed allele of Rb in these tissues. When E7 is expressed in these same tissues in age-matched K14E7Rbf/f mice, pRb protein was diminished in its abundance but still detectable, consistent with E7 being able only partially to degrade pRb.
We have previously shown that the expression of E7 in head and neck epithelia can lead to an increase in DNA synthesis in the normally proliferating basal compartment of the epithelium, and induce unscheduled DNA synthesis in the normally quiescent, suprabasal compartment (Fig. 1). These same phenotypes were observed in same tissues of adult K14E7Rbf//f mice (Fig. 3B and C). In K14CreRbf/f mice, we observed a similar, statistically significant increase in DNA synthesis in the suprabasal (Fig. 3C) compartment of the head and neck tissues, but, interestingly, no significant increase in the basal compartment (Fig. 3B). It is unclear why the loss of pRb positively induces DNA synthesis in the suprabasal compartment of the epithelium yet leaves it unaffected in the basal compartment of the same tissue. A possible explanation may involve the amounts of the other pocket proteins that are expressed in the two compartments. Whereas in the head and neck epithelia, loss of Rb recapitulated some but not all E7-mediated effects on DNA synthesis, loss of Rb fully recapitulated the effects of E7 on DNA synthesis in the epidermis (22). Interestingly, loss of Rb recapitulated none of the E7-mediated phenotypes in the epithelium of the female reproductive tract (26). These observations highlight the significance of a particular tissue in studying the role of pRb in proliferation and differentiation. The differences observed between the tissues may offer clues to the biology of these different tissues, and, perhaps, specifically, to the pool of available p107 and/or p130 in each tissue.
The loss of pRb does not fully recapitulate the oncogenic phenotypes of E7
To compare the consequences of loss of Rb versus expression of E7 on the susceptibility of mice to HNSCC, we treated groups of Rbf/f, K14CreRbf/f, and K14E7Rbf/f animals using the same 4-NQO carcinogen treatment protocol. At the study end point, the animals from each group were evaluated for overt tumors and tissues from random subsets of mice were subjected to detailed histopathologic analyses (Fig. 3D and E; Table 2). Not surprisingly, the 4-NQO–treated K14E7Rbf/f animals had a high incidence of overt tumors (16 of 17 animals or 94%; Fig. 3D and E) similar to that seen in the K14E7 animals on the Rbwt/wtbackground (Fig. 1C). In the subset of K14E7Rbf/f mice for which we performed detailed histopathology, 9 of the 11 mice had invasive carcinomas, and the other two had noninvasive papillomas (Table 2). Furthermore, 8 of these 11 mice had multiple invasive lesions, and four of those had at least one high-grade carcinoma (grade 3; Table 2). The 4-NQO–treated Rbf/f animals had a low incidence (6 of 46 mice or 13%) of overt tumors (Table 2), similar to that observed in Rbwt/wt FVB/N animals. Of the 4-NQO–treated Rbf/f mice subjected to detailed histopathology, 8 of 12 (67%) of the animals had no invasive lesions (75% of FVB/N mice were free of invasive tumors and the difference between the two groups is not statistically significant). Furthermore, of the Rbf/fmice, none had high-grade lesions and only 2 of 12 had more than one invasive tumor. The 4-NQO–treated K14CreRbf/f animals had a statistically significant increase in the incidence of overt tumors (12 of 36 or 33%) over that observed in the in the 4-NQO–treated Rbf/f mice (Fig. 3D). However, this increase in tumor incidence seen in the K14CreRbf/f animals was significantly less than the incidence of tumors observed in the K14E7Rbf/f animals. Histopathologic analysis indicated that the tumor multiplicity (Fig. 3E) and grade of carcinoma (Table 2) was not statistically different to that observed in the 4-NQO–treated Rbf/f mice. The absence of a statistically significant difference in this analysis between the K14CreRbf/f mice and the Rbf/f group could be attributed to the small sample size of mice undergoing full histopathology. However, it is important to note that though the loss of Rb leads to a significant increase in tumor susceptibility, it is not nearly as dramatic as the susceptibility seen in the same tissues upon expression of E7, and that, interestingly, is despite the fact that E7 does not abolish detectable pRb as the conditional deletion does.
Table 2.
Histopathology of treated tissues
| Hyperplasia/dysplasia (%) | Papilloma (%) | Grade 1 (%) | Grade 2 (%) | Grade 3 (%) | |
|---|---|---|---|---|---|
| Rbf/f (n = 13) | 50 | 17 | 33 | 0 | 0 |
| K14CreRbf/f (n = 12) | 46 | 15 | 15 | 8 | 15 |
| K14E7Rbf/f (n = 11) | 0 | 18 | 27 | 18 | 36 |
NOTE: A randomly selected subset of the carcinogen-treated animals was examined in detail throughout the thickness of their tongue and esophageal epithelia for the phenotypes indicated above. Each mouse was assigned the worst phenotype found in two or more consecutive slides. The difference seen between the K14E7Rbf/f and Rbf/f groups was statistically significant (P = 0.001) as well as the difference between the K14E7Rbf/f and K14CreRbf/f groups (P = 0.01). The difference between the Rbf/f and K14CreRbf/f groups was not different (P = 0.56). All comparisons were performed using a two-sided Wilcoxon rank sum test.
Expression of molecular markers in cancers
We stained treated epithelia and cancer tissue from Rbf/f, K14E7Rbf/f, and K14CreRbf/f animals with an antibody specific to pRb (Fig. 4). In the control Rbf/f mice, pRb was detectable although the nuclear signal is quite modest. Positive staining for pRb in the Rbf/f mouse tissue can best be appreciated by comparing the signal with the complete absence of signal in the K14CreRbf/f mouse sample (Fig. 4). In K14E7Rbf/f tissues, detection of pRb either was diminished compared with Rbf/f mouse tissue (example shown in Fig. 4) or in some cases abolished (example not shown). Indeed, most cancers from Rbf/f mice and about half of the K14E7Rbf/f showed detectable pRb, although, in the case of the E7+ cancers, pRb still seemed diminished compared with that in the nontransgenic cancers. These findings are all consistent with prior findings in the mouse skin (22), mouse cervix,1 and in human keratinocytes (33) that all have shown that E7 only incompletely degrades pRb and further suggest that the complete loss of pRb is not necessary for the formation of cancers induced by E7. No K14CreRbf/f cancers had any detectable pRb as expected.
Specific to the cancers was the ubiquitous expression of p16 (Fig. 4) only in K14E7Rbf/f and K14CreRbf/f cancers. P16 silencing in human HPV-negative HNSCC is a frequent occurrence; other models using 4-NQO as an oral carcinogen have reported the inactivation of p16 in Rb wild-type animals (34). However, HPV-positive cancers are usually noted for their increased expression of p16 (13, 14). Given this information, we were not surprised to see the selective detection of p16 in E7-expressing cancers. The detection of p16 in Rb-deleted cancers was expected given the known role of pRb in the methylation of the INK4a locus (35). Thus, it makes sense that in both K14E7Rbf/f and K14CreRbf/f tissues in which pRb levels are at least diminished, methylation and silencing of the p16 locus does not occur. An interesting observation is that the loss of Rb in head and neck epithelia led to the expansion in the expression of MCM7 in the suprabasal compartment as was also observed in K14E7Rbf/f mice. Furthermore, MCM7 was abundantly expressed in tumors from K14CreRbf/f animals and not Rbf/fanimals (Fig. 4). Although there was a correlation in the activation of this E2F-responsive gene between tissues expressing E7 and lacking pRb, and both types of tissues had increased tumor incidence over that seen in the Rbf/f animals, the severity of the tumor phenotypes was not similar between the two types of tissues; it was much higher in E7-expressing tissue. This finding would suggest that E7 does not lead to tumorigenesis merely via the activation of E2F targets, or at least not only through those targets controlled primarily by pRb.
Discussion
It is interesting to note the parallels and differences in terms of oncogene function in the various stratified epithelial tissues. In multistage carcinogenesis studies performed in the skin of E6 and E7 transgenic mice, E6 was shown to contribute weakly to the promotion stage to benign disease, whereas its primary role was in the progression to malignant lesions. In contrast, E7 played a major role in promotion, an early step in tumorigenesis (36). In the cervix of mice, E6 in addition to E7 led to an increase in the formation of large and more invasive cancers (20). In the studies described here, the combination of E6 and E7 seems to lead to a higher grade of disease and slightly increased multiplicity if tumors. The emerging pattern from studies in other tissues implicates E6 most prominently in the late stages of carcinogenesis, although the evidence is still weak for the head and neck model. The p53 tumor suppressor, an important target for E6, was also shown to be important at the late stages of carcinogenesis. When p53+/− mice were treated using multistage skin carcinogenesis protocols, the loss of p53 was shown to be important in the progression to more malignant disease (37). In addition, we have recently shown that inactivation of p53 in the cervical epithelia predisposes mice to cervical cancer.2 It is also informative that in human HPV-positive HNSCC, p53 mutations are notably absent, which supports an essential role for E6 in these cancers (12). Therefore, we predict that E6, at least through its inactivation of p53 if not its other properties, contributes to HNSCC. Supporting this view, our preliminary data indicate that by reducing the time of treatment with the cocarcinogen, 4NQO, a significant role of E6 in HPV-associated HNSCC becomes apparent.3
The E7 oncogene is more potent in inducing malignancies in both the head and neck and the cervix than the E6 oncogene in contrast to the skin. These results probably reflect not only the distinct roles of E6 and E7 at different stages of carcinogenesis but also the different behaviors of E6 and E7 depending on the tissue context. It should be noted that the different behaviors of the oncogenes cannot be merely attributed to differing levels of expression of the oncogenes. Both E6 and E7 expression in various epithelial tissues of these transgenic mice have been shown to be comparable with or lower than the levels expressed in HPV-positive cancer-derived cell lines (27).1
The results observed from the deletion of Rb in the head and neck epithelia lead to several conclusions about the action of E7 as an oncogene in these tissues. Because the induction of DNA synthesis in postmitotic cells seen in Rb-deleted head and neck epithelia does not correlate fully with oncogenic phenotypes, we conclude that the E7 oncoprotein cannot induce tumorigenesis purely through its ability to cause aberrant DNA synthesis in the differentiated compartment of these epithelia, although this is an activity that is thought to be key to the role of E7 in the viral life cycle (33). Furthermore, the activation of E2F-responsive genes that is seen in both pRb-deleted and E7-expressing tissue does not correlate quantitatively with oncogenesis in these tissues, indicating, at the very least, that activating E2F transcription is not sufficient to account for the role of E7 in carcinogenesis. The deletion of pRb leads to an activation of MCM7 as effectively as the expression of E7; however, it elevates the risk for cancer to a lesser degree than does expression of E7. It is still reasonable to posit that the dysregulation of at least some E2F-responsive genes are necessary for each outcome (aberrant DNA synthesis, tumorigenesis).
Evaluating the targets of the E7 oncoprotein that mediate its biological properties, pRb remains an important target because the loss of pRb contributes to ectopic proliferation as previously observed in stratified epithelia of the skin (Fig. 3), and in part also contributes to carcinogenesis (Fig. 3; Table 2). However, these studies show that E7 does not induce cancers merely by targeting pRb for degradation. This observation does not preclude the possibility that pRb is still an important target but clearly shows that its inactivation is not sufficient to recapitulate E7 activities. E7 can in fact interact with all of the pocket proteins, and it could be the combined effects of E7 on two or all of the pocket proteins that lead to its oncogenic effects. This would be in line with previously mentioned studies that implicate other pocket proteins with the combined loss of p107 and Rb, or p130 and Rb, resulting in tumorigenesis (29, 38, 39). However, the facts that there are still some acute phenotypes in the head and neck and increased tumor incidence upon the loss of Rb indicate that if there is indeed some functional redundancy and/or compensation, it cannot be complete; otherwise, no phenotypes would be observed.
Another explanation for the inability of loss of pRb to recapitulate the effects of E7 could involve a nonpocket protein target of E7 such as p21 or p27, either alone or in combination with other targets including pRb. The ability of E7 to bind p21 has been shown to be crucial for its in vitro transforming abilities (40).
In summary, we have performed studies to examine the individual contribution of the HPV16 oncogenes E6 and E7 in head and neck carcinogenesis. We have determined that E7 is the dominant oncogene in this model with the tumor incidence and biomarker expression seen in the bitransgenic K14E6K14E7 mice largely reflected in K14E7 mice. The effects of E7 in these tissues are not likely to be the singular outcome of the targeted degradation of the pRb tumor suppressor. Although the loss of pRb does contribute to HNSCC, it is likely that E7 mediates its cumulative effects through its action on one or more additional targets.
Acknowledgments
Grant support: NIH grants DE017315 and CA098428.
We thank the University of Wisconsin Comprehensive Cancer Center Histology Core Facility for its support.
Footnotes
S.J. Balsitis and P.F. Lambert, unpublished results.
A. Shai, H.C. P itot, and P.F. Lambert. The role of p53 in HPV-associated cancers, submitted for publication.
K. Strati and P.F. Lambert, unpublished studies.
References
- 1.Gillison ML, Koch WM, Capone RB, et al. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Inst. 2000;92:709–720. doi: 10.1093/jnci/92.9.709. [DOI] [PubMed] [Google Scholar]
- 2.Mellin H, Friesland S, Lewensohn R, Dalianis T, Munck-Wikland E. Human papillomavirus (HPV) DNA in tonsillar cancer: clinical correlates, risk of relapse, and survival. Int J Cancer. 2000;89:300–304. [PubMed] [Google Scholar]
- 3.Snijders PJ, Meijer CJ, van den Brule AJ, Schrijne-makers HF, Snow GB, Walboomers JM. Human papillomavirus (HPV) type 16 and 33 E6/E7 region transcripts in tonsillar carcinomas can originate from integrated and episomal HPV DNA. J Gen Virol. 1992;73:2059–2066. doi: 10.1099/0022-1317-73-8-2059. [DOI] [PubMed] [Google Scholar]
- 4.Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 5.Werness BA, Levine AJ, Howley PM. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 6.Huibregtse JM, Scheffner M, Howley PM. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J. 1991;10:4129–4135. doi: 10.1002/j.1460-2075.1991.tb04990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyer SN, Wazer DE, Band V. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 1996;56:4620–4624. [PubMed] [Google Scholar]
- 8.Dyson N, Howley PM, Munger K, Harlow E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 9.Jones DL, Thompson DA, Munger K. Destabilization of the RB tumor suppressor protein and stabilization of p53 contribute to HPV type 16 E7-induced apoptosis. Virology. 1997;239:97–107. doi: 10.1006/viro.1997.8851. [DOI] [PubMed] [Google Scholar]
- 10.Nguyen M, Song S, Liem A, Androphy E, Liu Y, Lambert PF. A mutant of human papillomavirus type 16 E6 deficient in binding α-helix partners displays reduced oncogenic potential in vivo. J Virol. 2002;76:13039–13048. doi: 10.1128/JVI.76.24.13039-13048.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heck DV, Yee CL, Howley PM, Munger K. Efficiency of binding the retinoblastoma protein correlates with the transforming capacity of the E7 oncoproteins of the human papillomaviruses. Proc Natl Acad Sci U S A. 1992;89:4442–4446. doi: 10.1073/pnas.89.10.4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balz V, Scheckenbach K, Gotte K, Bockmuhl U, Petersen I, Bier H. Is the p53 inactivation frequency in squamous cell carcinomas of the head and neck underestimated? Analysis of p53 exons 2–11 and human papillomavirus 16/18 E6 transcripts in 123 unselected tumor specimens. Cancer Res. 2003;63:1188–1191. [PubMed] [Google Scholar]
- 13.Hafkamp HC, Speel EJ, Haesevoets A, et al. A subset of head and neck squamous cell carcinomas exhibits integration of HPV 16/18 DNA and overexpression of p16INK4A and p53 in the absence of mutations in p53 exons 5–8. Int J Cancer. 2003;107:394–400. doi: 10.1002/ijc.11389. [DOI] [PubMed] [Google Scholar]
- 14.Wiest T, Schwarz E, Enders C, Flechtenmacher C, Bosch FX. Involvement of intact HPV16 E6/E7 gene expression in head and neck cancers with unaltered p53 status and perturbed pRb cell cycle control. Oncogene. 2002;21:1510–1517. doi: 10.1038/sj.onc.1205214. [DOI] [PubMed] [Google Scholar]
- 15.Arbeit JM, Munger K, Howley PM, Hanahan D. Neuroepithelial carcinomas in mice transgenic with human papillomavirus type 16 E6/E7 ORFs. Am J Pathol. 1993;142:1187–1197. [PMC free article] [PubMed] [Google Scholar]
- 16.Arbeit JM, Munger K, Howley PM, Hanahan D. Progressive squamous epithelial neoplasia in K14-human papillomavirus type 16 transgenic mice. J Virol. 1994;68:4358–4368. doi: 10.1128/jvi.68.7.4358-4368.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Griep AE, Herber R, Jeon S, Lohse JK, Dubielzig RR, Lambert PF. Tumorigenicity by human papillomavirus type 16 E6 and E7 in transgenic mice correlates with alterations in epithelial cell growth and differentiation. J Virol. 1993;67:1373–1384. doi: 10.1128/jvi.67.3.1373-1384.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herber R, Liem A, Pitot H, Lambert PF. Squamous epithelial hyperplasia and carcinoma in mice transgenic for the human papillomavirus type 16 E7 oncogene. J Virol. 1996;70:1873–1881. doi: 10.1128/jvi.70.3.1873-1881.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song S, Pitot HC, Lambert PF. The human papillomavirus type 16 E6 gene alone is sufficient to induce carcinomas in transgenic animals. J Virol. 1999;73:5887–5893. doi: 10.1128/jvi.73.7.5887-5893.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Riley RR, Duensing S, Brake T, Munger K, Lambert PF, Arbeit JM. Dissection of human papillomavirus E6 and E7 function in transgenic mouse models of cervical carcinogenesis. Cancer Res. 2003;63:4862–4871. [PubMed] [Google Scholar]
- 21.Balsitis S, Dick F, Lee D, et al. Examination of the pRb-dependent and pRb-independent functions of E7 in vivo. J Virol. 2005;79:11392–11402. doi: 10.1128/JVI.79.17.11392-11402.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balsitis SJ, Sage J, Duensing S, Munger K, Jacks T, Lambert PF. Recapitulation of the effects of the human papillomavirus type 16 E7 oncogene on mouse epithelium by somatic Rb deletion and detection of pRb-independent effects of E7 in vivo. Mol Cell Biol. 2003;23:9094–9103. doi: 10.1128/MCB.23.24.9094-9103.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brake T, Connor JP, Petereit DG, Lambert PF. Comparative analysis of cervical cancer in women and in a human papillomavirus-transgenic mouse model: identification of minichromosome maintenance protein 7 as an informative biomarker for human cervical cancer. Cancer Res. 2003;63:8173–8180. [PubMed] [Google Scholar]
- 24.Nguyen ML, Nguyen MM, Lee D, Griep AE, Lambert PF. The PDZ ligand domain of the human papillomavirus type 16 E6 protein is required for E6’s induction of epithelial hyperplasia in vivo. J Virol. 2003;77:6957–6964. doi: 10.1128/JVI.77.12.6957-6964.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song S, Gulliver GA, Lambert PF. Human papillomavirus type 16 E6 and E7 oncogenes abrogate radiation-induced DNA damage responses in vivo through p53-dependent and p53-independent pathways. Proc Natl Acad Sci U S A. 1998;95:2290–2295. doi: 10.1073/pnas.95.5.2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balsitis S, Dick F, Dyson N, Lambert PF. Critical roles for non-pRb targets of human papillomavirus type 16 E7 in cervical carcinogenesis. Cancer Res. 2006;66:9393–9400. doi: 10.1158/0008-5472.CAN-06-0984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shai A, Brake T, Somoza C, Lambert PF. The human papillomavirus E6 oncogene dysregulates the cell cycle and contributes to cervical carcinogenesis through two independent activities. Cancer Res. 2007;67:1626–1635. doi: 10.1158/0008-5472.CAN-06-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29:418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 29.Sage J, Miller AL, Perez-Mancera PA, Wysocki JM, Jacks T. Acute mutation of retinoblastoma gene function is sufficient for cell cycle re-entry. Nature. 2003;424:223–228. doi: 10.1038/nature01764. [DOI] [PubMed] [Google Scholar]
- 30.Strati K, Pitot HC, Lambert PF. Identification of biomarkers that distinguish human papillomavirus (HPV)-positive versus HPV-negative head and neck cancers in a mouse model. Proc Natl Acad Sci U S A. 2006;103:14152–14157. doi: 10.1073/pnas.0606698103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klussmann JP, Gultekin E, Weissenborn SJ, et al. Expression of p16 protein identifies a distinct entity of tonsillar carcinomas associated with human papillomavirus. Am J Pathol. 2003;162:747–753. doi: 10.1016/S0002-9440(10)63871-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.von Knebel Doeberitz M. New markers for cervical dysplasia to visualise the genomic chaos created by aberrant oncogenic papillomavirus infections. Eur J Cancer. 2002;38:2229–2242. doi: 10.1016/s0959-8049(02)00462-8. [DOI] [PubMed] [Google Scholar]
- 33.Collins AS, Nakahara T, Do A, Lambert PF. Interactions with pocket proteins contribute to the role of human papillomavirus type 16 E7 in the papillomavirus life cycle. J Virol. 2005;79:14769–14780. doi: 10.1128/JVI.79.23.14769-14780.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang XH, Knudsen B, Bemis D, Tickoo S, Gudas LJ. Oral cavity and esophageal carcinogenesis modeled in carcinogen-treated mice. Clin Cancer Res. 2004;10:301–313. doi: 10.1158/1078-0432.ccr-0999-3. [DOI] [PubMed] [Google Scholar]
- 35.Kotake Y, Cao R, Viatour P, Sage J, Zhang Y, Xiong Y. pRB family proteins are required for H3K27 trimethy-lation and Polycomb repression complexes binding to and silencing p16INK4α tumor suppressor gene. Genes Dev. 2007;21:49–54. doi: 10.1101/gad.1499407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song S, Liem A, Miller JA, Lambert PF. Human papillomavirus types 16 E6 and E7 contribute differently to carcinogenesis. Virology. 2000;267:141–150. doi: 10.1006/viro.1999.0106. [DOI] [PubMed] [Google Scholar]
- 37.Kemp CJ, Donehower LA, Bradley A, Balmain A. Reduction of p53 gene dosage does not increase initiation or promotion but enhances malignant progression of chemically induced skin tumors. Cell. 1993;74:813–822. doi: 10.1016/0092-8674(93)90461-x. [DOI] [PubMed] [Google Scholar]
- 38.MacPherson D, Conkrite K, Tam M, Mukai S, Mu D, Jacks T. Murine bilateral retinoblastoma exhibiting rapid-onset, metastatic progression and N-myc gene amplification. EMBO J. 2007;26:784–794. doi: 10.1038/sj.emboj.7601515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.HRobanus-Maandag E, Dekker M, van der Valk M, et al. p107 is a suppressor of retinoblastoma development in pRb-deficient mice. Genes Dev. 1998;12:1599–1609. doi: 10.1101/gad.12.11.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Helt AM, Funk JO, Galloway DA. Inactivation of both the retinoblastoma tumor suppressor and p21 by the human papillomavirus type 16 E7 oncoprotein is necessary to inhibit cell cycle arrest in human epithelial cells. J Virol. 2002;76:10559–10568. doi: 10.1128/JVI.76.20.10559-10568.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]