Abstract
Background
Interleukin-6 (IL6) is a pleiotropic pro-inflammatory and immunomodulatory cytokine which likely plays an important role in the pathogenesis of COPD. There is a functional single nucleotide polymorphism (SNP), −174G/C, in the promoter region of IL6. We hypothesized that IL6 SNPs influence susceptibility for impaired lung function and COPD in smokers.
Methods
Seven and 5 SNPs in IL6 were genotyped in two nested case-control samples derived from the Lung Health Study (LHS) based on phenotypes of rate of decline of forced expiratory volume in one second (FEV1) over 5 years and baseline FEV1 at the beginning of the LHS. Serum IL6 concentrations were measured for all subjects. A partially overlapping panel of 9 IL6 SNPs was genotyped in 389 COPD cases from the National Emphysema Treatment Trial (NETT) and 420 controls from the Normative Aging Study (NAS).
Results
In the LHS, three IL6 SNPs were associated with FEV1 decline (0.023 ≤ P ≤ 0.041 in additive models). Among them the IL6_−174C allele was associated with rapid decline of lung function. The association was more significant in a genotype-based analysis (P = 0.006). In the NETT-NAS study, IL6_−174G/C and four other IL6 SNPs, all of which are in linkage disequilibrium with IL6_−174G/C, were associated with susceptibility to COPD (0.01 ≤ P ≤ 0.04 in additive genetic models).
Conclusion
Our results suggest that the IL6_−174G/C SNP is associated with rapid decline of FEV1 and susceptibility to COPD in smokers.
Keywords: genetic polymorphism, IL6, forced expiratory volume in one second (FEV1), lung function, chronic obstructive pulmonary disease (COPD)
INTRODUCTION
IL6 is a pleiotropic pro-inflammatory and immunomodulatory cytokine secreted by airway epithelial cells, alveolar macrophages, adipocytes and myocytes as well as other tissues and cells.1,2 The potential importance of IL6 in the pathogenesis of COPD is suggested by studies showing that high levels of serum or sputum IL6 are associated with impaired lung function or a faster decline in lung function.1,2 IL6 has been related to skeletal muscle weakness in COPD3 as well as to exacerbations4 and pulmonary infections5 in COPD patients. In addition, IL6 overexpression in the murine lung resulted in airway inflammation and emphysema-like airspace enlargement.6 Furthermore, IL6 is an important mediator of the acute phase response and can upregulate C-reactive protein (CRP) at the transcriptional level.7 CRP has been associated with lung function levels in healthy individuals and/or lung function decline in smoking-induced COPD.8,9 Taken together, these data support IL6 as an appealing candidate gene for smoking-induced lung function impairment and COPD.
The IL6 gene is located on chromosome 7p21. Previous studies have identified a functional SNP, −174G/C, in the promoter region of IL6.10 Before initiation of the current study, a small study reported there was no association of an IL6 SNP with COPD.11 Recently, another group showed that the IL6_−572C allele was associated with COPD.12 Large well-designed studies with carefully-defined COPD phenotypes are required to unravel the exact role of IL6 genetic variants in the pathogenesis of COPD.
We investigated smokers with mild to moderate airflow obstruction who were participants in the Lung Health Study (LHS) cohort and hypothesized that there would be significant associations between SNPs and haplotypes in IL6 with the rate of decline and/or the level of lung function and these association would be mediated through influencing IL6 serum concentrations. The LHS cohort provides an excellent opportunity to explore associations between gene polymorphisms and haplotypes with FEV1% predicted13,14 as well as the rate of decline in FEV1.15,16 To validate novel associations between IL6 SNPs with lung function phenotypes, replication of results was sought in COPD cases from the National Emphysema Treatment Trial (NETT) with participants from the Normative Aging Study (NAS) serving as controls.17,18
METHODS
Study participants
LHS participants: A total of 1488 subjects were selected from the ~ 4,800 LHS subjects for whom DNA and serum were available. The selection generated two nested case-control studies based on the extremes of rate of decline in lung function and baseline lung function. In the decline of lung function study, we selected the 266 and 293 non-Hispanic whites with the fastest and slowest rate of decline of lung function, respectively during the 5 year follow up (arbitrary cut-off points of ≥ 3.0% predicted decrease /year and ≥ 0.4% predicted increase /year in FEV1 were used for rapid decliners and non-decliners, respectively). The rationale to select nested case-control studies with the indicated sample sizes is that 1) this approach has the advantage of reducing cost while keeping satisfactory statistical efficiency when compared with the full cohort approach;19,20 2) the Common Disease/Common Variants hypothesis (CD/CV) was suggested one decade ago which states that disease susceptibility alleles of common diseases will be present at high frequencies,21 and 3) this sample size has relatively adequate power to detect common genetic risk variants as shown in our previous power analyses.22 The baseline lung function study consisted of the 532 and 527 participants who had the highest and lowest baseline FEV1% predicted, respectively (arbitrary cut-off points of FEV1 % predicted ≥ 88.9% and ≤ 67.0% were used for the high and low baseline groups, respectively). There were 130 participants that overlapped between the two sets of nested cases and controls due to the fact that subjects in the rate of decline study group had baseline lung function within one of the categories for baseline lung function.
NETT-NAS participants: We selected 389 non-Hispanic white subjects who were enrolled in the NETT Genetics Ancillary Study. The control group was composed of 420 participants with normal spirometry from the NAS, a longitudinal study over the past four decades of healthy adult males that was initiated by the Boston Veterans Administration.
More information on the participants is included in the online supplement.
TagSNP selection and genotyping methods
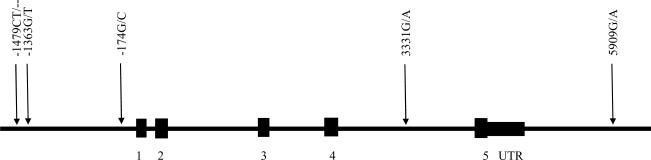
In the LHS, 5 tagSNPs were chosen from the SeattleSNPs database using the LDSelect program based on a relatively stringent LD threshold of r2 ≥ 0.8 and minor allele frequency cutoff of 10%. An additional 2 SNPs selected for the NETT-NAS study were subsequently chosen for genotyping in the decline of lung function study, in order to make the two studies more comparable. The nomenclature for the polymorphisms utilized in the study is summarized in Table E1 in the online supplement. SNP genotyping was performed using the TaqMan method (Applied Biosystems, Foster City, CA) for 5 tagSNPs and the Illumina Bead Array System for additional 2 SNPs. The positions of the selected and successfully genotyped 5 tagSNPs are shown in Figure 1.
Figure 1.
The IL6 gene structure and position of single nucleotide polymorphisms genotyped in the Lung Health Study subjects. Numbered regions represent exons. A: adenine; C: cytokine; G: guanine; T: thymidine; UTR: untranslated region.
In the NETT-NAS, the same criteria were used to select six LD-tagging IL6 SNPs and three additional IL6 SNPs were also selected for genotyping. The SNPs were genotyped on an Illumina BeadStation 500G System utilizing the GoldenGate assay technology (Illumina Golden Gate Assay, San Diego, CA).
SNP selection criteria are shown in more detail in the online supplement.
Measurements of serum IL6 concentration in the LHS participants
After collection, the blood samples were separated into their various components and shipped to the LHS data co-coordinating center on dry ice and were kept in −70°C freezers until use. The serum samples were thawed once for IL6 measurements. The concentrations of IL6 were measured using a highly sensitive chemiluminescent multiplexed sandwich immunoassay (SearchLight Proteome Array System®, Rockford, IL).
Statistical Analysis
In the LHS, Hardy-Weinberg equilibrium tests and linkage disequilibrium estimation were calculated using the genetics package for R (www.r-project.org). Multiple logistic regressions for rate of decline and baseline lung function were performed to test for the association with IL6 SNPs and with IL6 serum levels. Confounding factors included body mass index (BMI), age, gender, pack years of smoking, and smoking status. Multiple linear regression was performed for the complete data set to test for association of IL6 SNPs with log IL6 serum levels. Haplotype analysis was done using the R hapassoc package.
In the NETT-NAS, similar analyses were performed with SAS Genetics (Cary, NC).
Statistical Analysis is described in more detail in the online supplement.
RESULTS
Characteristics of the study participants
In the total of 1488 participants from the LHS, genotyping success rates were 96.4% to 98.6% for the 5 studied IL6 tagSNPs in all subjects and 97.9% for additional 2 SNPs in the rate of decline study. The demographic characteristics are shown in Table 1.
Table 1.
The distribution of demographic characteristics for all subjects and those in the two nested case control study groups in the LHS.
| All participants (N = 1488) | Rate of decline study | Baseline lung function study | |||||
|---|---|---|---|---|---|---|---|
| Fast Decliners (n = 266) | Non Decliners (n = 293) | p value | High Function (n = 532) | Low Function (n = 527) | p value | ||
| Men/Women | 948/540 | 158/108 | 197/96 | N/A | 352/180 | 325/202 | N/A |
| Age (years) | 48.41 (0.177) | 49.47 (0.397) | 47.48 (0.399) | <0.001 | 46.21 (0.772) | 50.76 (0.262) | <0.001 |
| Smoking history (pack-yrs)* | 40.41 (0.483) | 43.23 (1.178) | 38.35 (1.064) | 0.002 | 35.33 (0.125) | 45.24 (0.809) | <0.001 |
| Smoking status during 5 years follow-up† | |||||||
| Continuing smokers | 979 | 266 | 293 | N/A | 264 | 286 | N/A |
| Intermittent quitters | 315 | N/A | N/A | N/A | 157 | 158 | N/A |
| Sustained quitters | 194 | N/A | N/A | N/A | 111 | 83 | N/A |
| Body Mass Index (kg/m2) | 25.49 (0.099) | 25.22 (0.248) | 25.77 (0.210) | 0.091 | 25.39 (0.150) | 25.57 (0.183) | 0.455 |
| ΔFEV1/yr (% predicted pre)‡ | −0.98 (0.053) | −4.13 (0.066) | 1.087 (0.042) | <0.001 | −0.55 (0.067) | −1.22 (0.078) | <0.001 |
| ΔFEV1/yr (% predicted post)§ | −0.85 (0.04) | −3.44 (0.078) | 0.695 (0.054) | <0.001 | −0.75 (0.064) | −0.74 (0.078) | 0.949 |
| Baseline FEV1 (% predicted pre)∥ | 74.15 (0.302) | 72.68 (0.542) | 75.51 (0.472) | <0.001 | 86.49 (0.125) | 61.07 (0.180) | <0.001 |
| Baseline FEV1 (% predicted post)** | 77.57 (0.336) | 75.00 (0.561) | 79.80 (0.467) | <0.001 | 91.81 (0.099) | 62.57 (0.144) | <0.001 |
| IL6 concentration (pg/ml) | 2.60 (1.80–4.20) | 2.75 (2.00, 4.03) | 2.70 (1.70, 4.50) | 0.1840 | 2.50 (1.70, 3.90) | 2.70 (1.85, 4.55) | 0.12 |
Values are means (SEM) for continuous data.
Number of packs of cigarettes smoked per day × number of years smoking.
Continuing smokers: participants who reported smoking at each annual visit. Sustained quitters: participants who were validated by salivary cotinine or exhaled CO as abstinent at every annual visit. Intermittent quitters: participants who were not sustained quitters or continuing smokers
Change in lung function over a 5-year period per year as % predicted FEV1 pre bronchodilator
Change in lung function over a 5-year period per year as % predicted FEV1 post bronchodilator
Lung function at the start of the Lung Health Study as measured by FEV1(%) predicted pre bronchodilator
Lung function at the start of the Lung Health Study as measured by FEV1(%) predicted post bronchodilator.
There were significant differences in several potential confounding factors, such as age, gender, pack years of smoking, and smoking status between study groups. Therefore, multiple regressions were performed to adjust for relevant confounding factors.
In the total of 809 participants in the NETT-NAS, the genotype call rate for IL6_615A/G (rs2069832) was 85%; for all other SNPs the call rates were ≥ 97%. The demographic characteristics for the study groups are shown in Table 2.
Table 2.
The distribution of demographic characteristics for NETT COPD cases and NAS controls.
| Demographic Characteristics* | NETT (n = 389) | NAS (n = 420) | p-value |
|---|---|---|---|
| Age (years) | 67 ± 6 | 68 ± 9 | 0.9 |
| Pack-years | 66 ± 30 | 39 ± 27 | <0.001 |
| FEV1 (% predicted, post BD)† | 28 ± 7 | 92 ± 11 | |
| Modified BODE score (median ± IQR)‡ | 5 ± 3 | NA | |
| Gender (% male) | 64% | 100% |
Values are ± standard deviation unless otherwise listed
FEV1 % predicted values for the NETT and NAS are based on the prediction equations of Crapo and Morris (47); however the NAS-1988 standards were used in the selection of the control group.
Modified BODE score incorporates the University of San Diego Shortness of Breath Questionnaire, IQR = interquartile range
The linkage disequilibrium pattern, Hardy-Weinberg disequilibrium and performance of tagSNPs
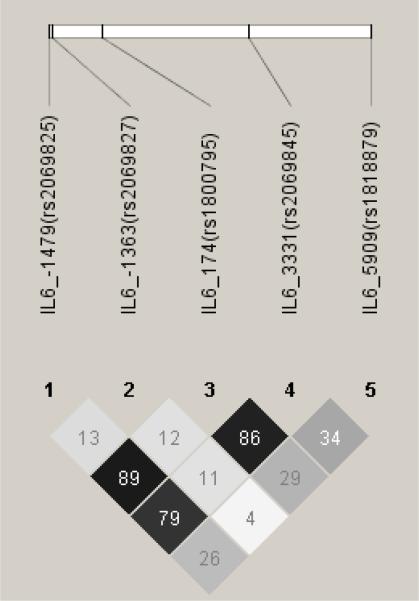
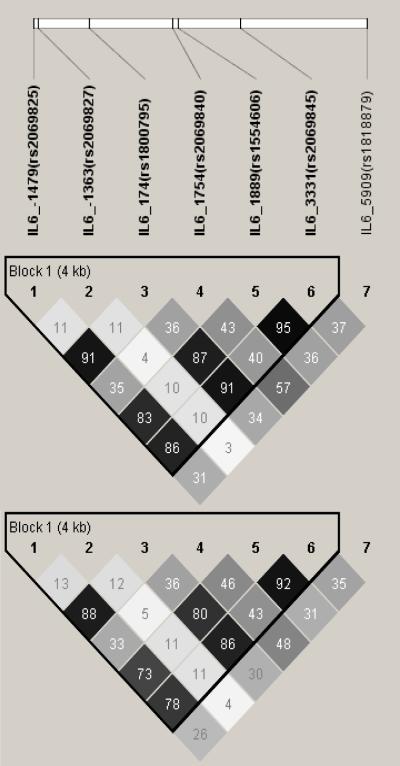
The LD pattern of the five IL6 tagSNPs in the full set of 1488 LHS study participants is shown in Figure 2A The r2 values were from 0.04 to 0.89. It is worth noting that the r2 values between IL6_−1479 (rs2069825) and IL6_−174 (rs1800795) as well as IL6_3331 (rs2069845) and IL6_−174 were greater than 0.86, which indicates that it is necessary to genotype only one of these 3 SNPs. The LD patterns of the low and high lung function subgroups were similar to that of all subjects (data not shown), as were those of the fast and non-decline subgroups, the LD pattern of all 7 SNPs genotyped in fast and non-decline subgroups are shown in Figure 2B. All the studied SNPs were in Hardy-Weinberg equilibrium.
Figure 2.
Linkage Disequilibrium of Single Nucleotide Polymorphisms (SNPs) of IL6 in the Lung Health Study subjects using HAPLOVIEW. The linkage disequilibrium (r2) between any two SNPs is listed in the cross cell. The darker the color indicates the higher the linkage disequilibrium between any two SNPs. Figure 2A: all subjects; Figure 2B: top: fast decline group; bottom: slow decline group.
More information on performance of tagSNPs is included in the online supplement.
Associations of SNPs and haplotypes in the IL6 gene with rate of decline and baseline of FEV1
Three of 7 IL6 SNPs were associated with FEV1 decline (0.023 ≤ P ≤ 0.041 in additive genetic models) (Table 3). The well known functional SNP IL6_−174G/C (rs1800795) was among them. The frequency of the IL6_−174C allele was significantly higher in the group with rapid decline of FEV1 than that in the non-decline group (45.2% versus 39.6%, OR 1.30, 95% CI = 1.01 – 1.66, P = 0.041). The association was more significant in the genotype-based analysis (P = 0.006) with 6 out of 7 SNPs reaching a significance level of P < 0.05 (Table 3).
Table 3.
Associations of SNPs in IL6 with rate of decline of FEV1 in the LHS and association with COPD in the NETT-NAS
| LD (r2 with rs1800795) | Rate of FEV1 decline study in the LHS | COPD case control study in the NETT-NAS | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bin | SNP ID | SNP in gene | LHS | NETT-NAS | Fast decline MAF % | Non-decline MAF % | p-value‡ genotype based | p-value‡ additive | NETT MAF % | NAS MAF % | p-value§ genotype based | p-value§ additive |
| 1 | rs1800797† | −598G/A | NA | 0.93 | 0.41 | 0.35 | 0.06 | 0.02 | ||||
| 1 | rs1800795*† | −174G/C | - | - | 0.45 | 0.40 | 0.006 | 0.041 | 0.42 | 0.36 | 0.03 | 0.01 |
| 1 | rs2069832† | 615A/G | NA | 0.98 | 0.42 | 0.37 | 0.1 | 0.09 | ||||
| 1 | rs1474348† | 1090G/C | NA | 0.97 | 0.41 | 0.36 | 0.1 | 0.04 | ||||
| 1 | rs1474347† | 1306G/T | NA | 0.98 | 0.42 | 0.36 | 0.06 | 0.03 | ||||
|
| ||||||||||||
| 2 | rs1554606*† | 1889G/T | 0.84 | 0.87 | 0.47 | 0.43 | 0.011 | 0.103 | 0.44 | 0.38 | 0.02 | 0.01 |
| 2 | rs2069845* | 3331G/A | 0.89 | NA | 0.47 | 0.42 | 0.007 | 0.078 | ||||
|
| ||||||||||||
| 3 | rs2069825* | −1479CT/− | 0.90 | NA | 0.44 | 0.37 | < 0.001 | 0.023 | ||||
|
| ||||||||||||
| 4 | rs2069840*† | 1754C/G | 0.36 | 0.33 | 0.32 | 0.37 | 0.005 | 0.064 | 0.35 | 0.36 | 0.3 | 0.7 |
|
| ||||||||||||
| 5 | rs1818879* | 5909G/A | 0.32 | NA | 0.29 | 0.34 | 0.012 | 0.035 | ||||
|
| ||||||||||||
| 6 | rs2069827*† | −1363G/T | 0.12 | 0.14 | 0.08 | 0.08 | 0.786∥ | 0.669 | 0.09 | 0.09 | 0.9 | 0.4 |
|
| ||||||||||||
| other | rs2069849† | 4338C/T | NA | 0.02 | 0.03 | 0.02 | 0.2 | 0.2 | ||||
Genotyped in the LHS,
Genotyped in the NETT-NAS
Adjustment for confounding factors such as age, gender, pack-years of smoking, and research center.
As the NAS controls were uniformly male smokers with normal lung function, the models were adjusted for age and pack-years.
The p values were from a dominant genetic model because the minor allele frequency of this SNP was very low.
It is worthwhile to note that the most significant association was found for the IL6_−1479CT in/del (rs2069825): the IL6_−1479CT deletion was associated with rapid decline of FEV1 (p < 0.001) (Table 3). Three other IL6 SNPs, were not significantly associated in the additive model, but were significant in the genotype-based analysis (Table 3). The IL6_−1479 CT in/del and another 2 associated IL6 SNPs were in high LD with IL6_−174G/C. Interestingly, the IL6_5909G/A and IL6_1754C/G, which were not in high LD with IL6_−174G/C and not in high LD each other (r2 = 0.52), were also significantly associated with decline of lung function (Table 3).
No association was found for IL6 haplotypes with rate of decline of FEV1; IL6 SNPs and haplotypes were not associated with the baseline level of FEV1 (data not shown).
Associations of IL6 SNPs and haplotypes with serum IL6 concentrations
The associations of IL6 SNPs with serum IL6 concentrations were analyzed in all LHS subjects for 5 tagSNPs and in rate of decline study subjects for 2 additional SNPs by linear regressions adjusted for BMI, age, gender, pack years of smoking, and smoking status (Table 4).
Table 4.
Association of serum concentrations of IL6 and IL6 genotypes (linear regression*)
| SNP | Genotype | IL6 level [ln IL6 (pg/ml)] |
||
|---|---|---|---|---|
| N | Coefficient (SE) | p value | ||
| II | 514 | |||
| IL6_−1479 | ID | 681 | −0.011 (0.046) | |
| (rs2069825) | DD | 229 | −0.080 (0.063) | 0.424† |
|
| ||||
| IL6_−1363 | GG | 1185 | ||
| (rs2069827) | GT+TT | 234 | 0.000 (0.057) | 0.997† |
|
| ||||
| GG | 483 | |||
| IL6_−174 | GC | 691 | 0.013 (0.047) | |
| (rs 1800795) | CC | 255 | −0.056 (0.061) | 0.486† |
|
| ||||
| CC | 234 | |||
|
| ||||
| IL6_1754 | CG | 234 | 0.000 (0.048) | |
|
| ||||
| (rs2069840) | GG | 72 | −0.030 (0.064) | 0.820‡ |
|
| ||||
| GG | 169 | |||
|
| ||||
| IL6_1889 | GT | 261 | 0.013 (0.044) | |
|
| ||||
| (rs 1554606) | TT | 110 | −0.057 (0.054) | 0.542‡ |
|
| ||||
| AA | 431 | |||
| IL6_3331 | AG | 689 | 0.015 (0.049) | |
| (rs2069845) | GG | 293 | −0.020 (0.060) | 0.819† |
|
| ||||
| AA | 685 | |||
| IL6_5909 | AG | 593 | 0.012 (0.044) | |
| (rs1818879) | GG | 137 | −0.025 (0.074) | 0.877† |
P values were from genotype-based analysis (co-dominant genetic models) except for IL6_−1363 where p values were from a dominant genetic model because the minor allele frequency for this SNP was very low.
Adjusted for BMI, age, gender, pack years of smoking, and smoking status;
Adjusted for BMI, age, gender, pack years of smoking.
No significant association was found for IL6 SNPs with IL6 concentrations. IL6 haplotypes were also not associated with IL6 concentrations (data not shown).
Associations of serum IL6 concentrations with rate of decline and baseline FEV1
As shown in Table 1, there were no significant differences in IL6 concentrations between the rapid decline and non-decline groups or between high and low FEV1 groups.
Replication of novel IL6 associations in the NETT-NAS participants
In the LHS, IL6 SNPs were significantly associated with rate of decline of FEV1 in mild COPD patients. Since rapid decline of lung function in smokers is the likely method of development of COPD we reasoned that the same SNPs would be associated with advanced COPD. To test this we used a case-control sample that has been very useful in revealing genes associated with COPD.17,18 In the NETT-NAS study cases had advanced COPD requiring lung volume reduction surgery and controls were derived from a population of smokers who have not developed COPD. The IL6_−174G/C and another four IL6 SNPs, which had high linkage disequilibrium with IL6_−174G/C, were associated with susceptibility to COPD (0.01 ≤ P ≤ 0.04 in additive genetic models). The IL6_−174C allele was associated with susceptibility to COPD (OR 1.3, 95% CI = 1.1 – 1.7, P = 0.01 in an additive genetic model). The frequency of the IL6_−174C allele was significantly higher in the NETT group than that in the NAS group (42.0% versus 36%). The association was also significant in genotype-based analysis (P = 0.03) (Table 3).
DISCUSSION
There are only three studies published on associations of IL6 SNPs with COPD. Seifart et al. reported that there was no association of IL6_−174 with COPD.11 Broekhuizen et al. did not find an association between IL6_−174 and a cachexia phenotype in COPD subjects.23 Recently, Córdoba-Lanús et al. reported that IL6_−572 but not IL6_−174 was associated with COPD.12 All three studies have relatively small sample sizes. The associations of IL6 SNPs with FEV1 decline in the current study are novel and are the most significant findings among all the studies we have published utilizing the LHS cohort.13–16,24–27 To strengthen our initial finding in the LHS, we incorporated an association study of IL6 SNPs with COPD in the NETT-NAS. All SNPs that were genotyped and in high LD with the IL6_−174G/C showed significant or borderline association with rapid decline of lung function in the LHS and with COPD in the NETT-NAS. We believe that the strength of the associations, the concordant results with several SNPs in high LD with the IL6_−174G/C SNP, the available previous functional data on IL6_−174G/C, the replication in a second population and the biologic plausibility for association provide strong evidence that this is a true association.
We examined the association of IL6 SNPs with IL6 serum levels as well as relationships between IL6 serum levels and lung function decline. We did not find any associations. We also found that adjusting the associations between IL6 SNPs and lung function for serum CRP levels in the LHS had no effect on the strength of the associations (data not shown). Therefore, we did not find evidence that the associations we report were mediated through an influence on production of IL6 or CRP.
Studies that have examined the effects of IL6 SNPs on IL6 mRNA and protein expression have led to conflicting results. The first reporter gene study demonstrated that a construct containing the −174G allele had higher reporter gene expression in HeLa cells, both under basal conditions and after LPS or IL1 stimulation10. However, a second reporter gene study showed that a construct containing −174C had higher IL1-induced expression in HeLa cells than that of the −174G construct, although the difference did not reach statistical significance.28 By comparison of the two different cell types, the authors concluded that there is a cell type-specific regulation of IL6 expression.28 Nine of the most recently published studies of IL6 SNPs with circulating IL6 concentrations are summarized in Table E2. A recent meta-analysis of 5659 subjects from seventeen studies concluded that the −174 IL6 SNP was not associated with circulating IL6 levels.29 There are several explanations for the lack of consistent associations. First, the IL6_−174G/C polymorphism might not be a strong determinant of serum IL6 levels. Second, the serum half-life of IL6 is short. Serum IL6 levels show marked diurnal variability.30 The blood samples for IL6 measurement in most studies, including our own, were not taken at a specific time of the day. A third explanation is that the SNPs studied may not be the actual functional SNPs. Recently, Samuel and colleagues have identified a novel IL6 transcriptional regulatory region (−5307 to −5202) much farther from the transcription initiation site than IL6_−17431 This report coupled with more recent identification of a novel functional SNP, IL6_−6331T/C (rs10499563), with the T allele preferentially binding to Oct-1 transcription factor and producing higher reporter gene expression, provides evidence that additional functional SNPs do exist in IL6.32 However, since IL6_−6331T/C is in low LD with IL6_−174, our finding is not likely to be explained by these new functional data.
If IL6 SNPs are not related to IL6 levels then what is the basis for their association with FEV1 decline and COPD? One possible explanation is that the association is truly driven via local pulmonary IL6 expression or that it is driven by serum IL6 levels but that the variability and lability of serum IL6 levels obscures this relationship; FEV1 may reflect the average IL6 levels and thus the degree of lung inflammation over the years of the study. In addition, the SNPs could influence IL6 levels and thus lung inflammation during exacerbations but not the constitutive levels during stable periods. IL6 is a pleiotropic cytokine which also modulates expression of many other genes.33 It may be that it is the effect of the IL6 variants on these genes that is the underlying mechanism for the associations we observed.
How can we explain the observation that IL6 SNPs were not associated with baseline FEV1 in the LHS but were associated with the presence of COPD in the NETT-NAS study? The mean age of the LHS participants was 48 years as opposed to a mean age of 68 years for the NETT-NAS participants. Baseline FEV1 at age 48 is influenced both by maximal attained FEV1 at ~ 25 years of age and by the rate of decline of lung function after age 25.34 However, the relative contribution of rate of decline in lung function will be much greater by age 68 than at age 48. Thus, FEV1 at age 68 in the NETT-NAS participants is likely to largely reflect the rate of decline of lung function during their long smoking history whereas there is likely a weaker relationship of FEV1 decline and baseline lung function at age 48.
Compared with previous studies, strengths of this study include larger sample size and good power. This sample size has adequate power to detect common genetic risk variants as shown in our previous power analyses, for example, it has 80% power to detect a relative risk of 2.0 when the frequency of the risk factor is 10% or above.22
There are several potential limitations of this study. Firstly, population stratification could have led to false-positive results. However, it has been reported that significant false-positive associations are unlikely to arise from population stratification in the non-Hispanic white population, especially in well-designed, moderately-sized, case-control studies such as ours.35 In addition, there was no significant evidence of population stratification in the NETT-NAS cases and controls.17 Second, false positive results might have arisen from multiple comparisons. However the consistent results in the NETT-NAS replication study make false positive results unlikely. Third, we have not identified the causal SNP for the associations. The identification of a novel functional SNP IL6_−6331T/C (rs10499563), which has low LD with IL6_−174G/C (rs1800795) with r2 of 0.169 in the CEU HapMap database, indicates that the control of IL6 transcription is likely to be complex.32 We cannot exclude the possibility that SNPs other than the IL6_−174G/C are also causal SNPs. Finally, serum IL6 levels were measured at year 5 of the LHS, therefore it may not be appropriate to link IL6 levels at year 5 with the baseline FEV1 as well as the rate of decline of FEV1 during 5 year follow-up.
In summary, we report associations of IL6 variants with rate of decline of lung function and with smoking-induced COPD.
ACKNOWLEDGEMENTS
This work was supported by grants from the Canadian Institutes of Health Research and National Institutes of Heath Grant 5R01HL064068-04. The Lung Health Study was supported by contract N01-HR-46002 from the Division of Lung Diseases of the National Heart, Lung, and Blood Institute. The NETT Genetics Ancillary Study was supported by National Institutes of Health grants HL075478 and HL71393. The Normative Aging Study is supported by the Cooperative Studies Program/ERIC of the U.S. Department of Veterans Affairs, and is a component of the Massachusetts Veterans Epidemiology Research and Information Center (MAVERIC). Dr. He is the recipient of Michael Smith Foundation for Health Research Fellowship and Izaak Walton Killam Memorial Scholarship Award. Dr. Foreman was supported by HL007427. Dr. DeMeo was supported by HL072918. Dr. Sandford is the recipient of a Canada Research Chair in genetics and a Michael Smith Foundation for Health Research Senior Scholar Award.
Footnotes
The Corresponding Author has the right to grant on behalf of all authors and does grant on behalf of all authors, an exclusive licence (or non exclusive for government employees) on a worldwide basis to the BMJ Publishing Group Ltd and its Licensees to permit this article to be published in Thorax editions and any other BMJPGL products to exploit all subsidiary rights, as set out in our licence http://thorax.bmjjournals.com/ifora/licence.pdf
REFERENCES
- 1.Donaldson GC, Seemungal TA, Patel IS, et al. Airway and systemic inflammation and decline in lung function in patients with COPD. Chest. 2005;128(4):1995–2004. doi: 10.1378/chest.128.4.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walston JD, Fallin MD, Cushman M, et al. IL-6 gene variation is associated with IL-6 and C-reactive protein levels but not cardiovascular outcomes in the Cardiovascular Health Study. Hum Genet. 2007;122(5):485–94. doi: 10.1007/s00439-007-0428-x. [DOI] [PubMed] [Google Scholar]
- 3.Yende S, Waterer GW, Tolley EA, et al. Inflammatory markers are associated with ventilatory limitation and muscle dysfunction in obstructive lung disease in well functioning elderly subjects. Thorax. 2006;61(1):10–16. doi: 10.1136/thx.2004.034181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wedzicha JA, Seemungal TA, MacCallum PK, et al. Acute exacerbations of chronic obstructive pulmonary disease are accompanied by elevations of plasma fibrinogen and serum IL-6 levels. Thromb Haemost. 2000;84(2):210–5. [PubMed] [Google Scholar]
- 5.Yende S, Tuomanen EI, Wunderink R, et al. Preinfection systemic inflammatory markers and risk of hospitalization due to pneumonia. Am J Respir Crit Care Med. 2005;172(11):1440–6. doi: 10.1164/rccm.200506-888OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuhn C, III, Homer RJ, Zhu Z, et al. Airway Hyperresponsiveness and Airway Obstruction in Transgenic Mice. Morphologic Correlates in Mice Overexpressing Interleukin (IL)-11 and IL-6 in the Lung. Am J Respir Cell Mol Biol. 2000;22(3):289–95. doi: 10.1165/ajrcmb.22.3.3690. [DOI] [PubMed] [Google Scholar]
- 7.Majello B, Arcone R, Toniatti C, et al. Constitutive and IL-6-induced nuclear factors that interact with the human C-reactive protein promoter. Embo J. 1990;9(2):457–65. doi: 10.1002/j.1460-2075.1990.tb08131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aronson D, Roterman I, Yigla M, et al. Inverse association between pulmonary function and C-reactive protein in apparently healthy subjects. Am J Respir Crit Care Med. 2006;174(6):626–32. doi: 10.1164/rccm.200602-243OC. [DOI] [PubMed] [Google Scholar]
- 9.Shaaban R, Kony S, Driss F, et al. Change in C-reactive protein levels and FEV(1) decline: A longitudinal population-based study. Respir Med. 2006;100(12):2112–20. doi: 10.1016/j.rmed.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 10.Fishman D, Faulds G, Jeffery R, et al. The effect of novel polymorphisms in the interleukin-6 (IL-6) gene on IL-6 transcription and plasma IL-6 levels, and an association with systemic-onset juvenile chronic arthritis. J Clin Invest. 1998;102(7):1369–76. doi: 10.1172/JCI2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seifart C, Dempfle A, Plagens A, et al. TNF-alpha-, TNF-beta-, IL-6-, and IL-10-promoter polymorphisms in patients with chronic obstructive pulmonary disease. Tissue Antigens. 2005;65(1):93–100. doi: 10.1111/j.1399-0039.2005.00343.x. [DOI] [PubMed] [Google Scholar]
- 12.Córdoba-Lanús E, de-Torres J-P, López-Aguilar C, et al. Association of IL-6 gene polymorphisms and COPD in a Spanish Population. Resp Med. 2008;102(12):1805–11. doi: 10.1016/j.rmed.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 13.He JQ, Burkett K, Connett JE, et al. Interferon gamma polymorphisms and their interaction with smoking are associated with lung function. Hum Genet. 2006;119(4):365–75. doi: 10.1007/s00439-006-0143-z. [DOI] [PubMed] [Google Scholar]
- 14.He JQ, Connett JE, Anthonisen NR, et al. Glutathione S-transferase variants and their interaction with smoking on lung function. Am J Respir Crit Care Med. 2004;170(4):388–94. doi: 10.1164/rccm.200312-1763OC. [DOI] [PubMed] [Google Scholar]
- 15.He JQ, Connett JE, Anthonisen NR, et al. Polymorphisms in the IL13, IL13RA1, and IL4RA genes and rate of decline in lung function in smokers. Am J Respir Cell Mol Biol. 2003;28(3):379–85. doi: 10.1165/rcmb.4885. [DOI] [PubMed] [Google Scholar]
- 16.He JQ, Ruan J, Connett J, et al. Antioxidant gene polymorphisms and susceptibility to a rapid decline in lung function in smokers. Am J Respir Crit Care Med. 2002;166(3):323–8. doi: 10.1164/rccm.2111059. [DOI] [PubMed] [Google Scholar]
- 17.Celedon JC, Lange C, Raby BA, et al. The transforming growth factor-beta1 (TGFB1) gene is associated with chronic obstructive pulmonary disease (COPD) Hum Mol Genet. 2004;13(15):1649–56. doi: 10.1093/hmg/ddh171. [DOI] [PubMed] [Google Scholar]
- 18.Hersh CP, Demeo DL, Lazarus R, et al. Genetic association analysis of functional impairment in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2006;173(9):977–84. doi: 10.1164/rccm.200509-1452OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allison DB, Heo M, Schork NJ, et al. Extreme selection strategies in gene mapping studies of oligogenic quantitative traits do not always increase power. Human Heredity. 1998;48(2):97–107. doi: 10.1159/000022788. [DOI] [PubMed] [Google Scholar]
- 20.Ernster VL. Nested case-control studies. Prev Med. 1994;23(5):587–90. doi: 10.1006/pmed.1994.1093. [DOI] [PubMed] [Google Scholar]
- 21.Lander ES. The new genomics: global views of biology. Science. 1996;274(5287):536–9. doi: 10.1126/science.274.5287.536. [DOI] [PubMed] [Google Scholar]
- 22.Tanaka G, Sandford AJ, Burkett K, et al. Tumour necrosis factor and lymphotoxin A polymorphisms and lung function in smokers. Eur Respir J. 2006;29(1):34–41. doi: 10.1183/09031936.00045206. [DOI] [PubMed] [Google Scholar]
- 23.Broekhuizen R, Grimble RF, Howell WM, et al. Pulmonary cachexia, systemic inflammatory profile, and the interleukin 1beta −511 single nucleotide polymorphism. Am J Clin Nutr. 2005;82(5):1059–64. doi: 10.1093/ajcn/82.5.1059. [DOI] [PubMed] [Google Scholar]
- 24.He JQ, Shumansky K, Connett JE, et al. Association of genetic variations in the CSF2 and CSF3 genes with lung function in smoking-induced COPD. Eur Respir J. 2008;32(1):25–34. doi: 10.1183/09031936.00040307. [DOI] [PubMed] [Google Scholar]
- 25.He JQ, Shumansky K, Zhang X, et al. Polymorphisms of interleukin-10 and its receptor and lung function in COPD. Eur Respir J. 2007;29(6):1120–6. doi: 10.1183/09031936.00002907. [DOI] [PubMed] [Google Scholar]
- 26.Joos L, McIntyre L, Ruan J, et al. Association of IL-1beta and IL-1 receptor antagonist haplotypes with rate of decline in lung function in smokers. Thorax. 2001;56(11):863–6. doi: 10.1136/thorax.56.11.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sandford AJ, Chagani T, Weir TD, et al. Susceptibility genes for rapid decline of lung function in the Lung Health Study. Am J Respir Crit Care Med. 2001;163(2):469–73. doi: 10.1164/ajrccm.163.2.2006158. [DOI] [PubMed] [Google Scholar]
- 28.Terry CF, Loukaci V, Green FR. Cooperative influence of genetic polymorphisms on interleukin 6 transcriptional regulation. J Biol Chem. 2000;275(24):18138–44. doi: 10.1074/jbc.M000379200. [DOI] [PubMed] [Google Scholar]
- 29.Huth C, Illig T, Herder C, et al. Joint analysis of individual participants' data from 17 studies on the association of the IL6 variant −174G >C with circulating glucose levels, interleukin-6 levels, and body mass index. Ann Med. 2008:1–21. doi: 10.1080/07853890802337037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sothern RB, Roitman-Johnson B, Kanabrocki EL, et al. Circadian characteristics of circulating interleukin-6 in men. J Allergy Clin Immunol. 1995;95(5 Pt 1):1029–35. doi: 10.1016/s0091-6749(95)70104-4. [DOI] [PubMed] [Google Scholar]
- 31.Samuel JM, Kelberman D, Smith AJ, et al. Identification of a novel regulatory region in the interleukin-6 gene promoter. Cytokine. 2008;42(2):256–64. doi: 10.1016/j.cyto.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 32.Smith AJ, D'Aiuto F, Palmen J, et al. Association of serum interleukin-6 concentration with a functional IL6 −6331T>C polymorphism. Clin Chem. 2008;54(5):841–50. doi: 10.1373/clinchem.2007.098608. [DOI] [PubMed] [Google Scholar]
- 33.Wegrzyn P, Jura J, Kupiec T, et al. A search for genes modulated by interleukin-6 alone or with interleukin-1beta in HepG2 cells using differential display analysis. Biochim Biophys Acta. 2006;1762(3):319–28. doi: 10.1016/j.bbadis.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 34.Tager IB, Segal MR, Speizer FE, et al. The natural history of forced expiratory volumes. Effect of cigarette smoking and respiratory symptoms. Am Rev Respir Dis. 1988;138(4):837–49. doi: 10.1164/ajrccm/138.4.837. [DOI] [PubMed] [Google Scholar]
- 35.Ardlie KG, Lunetta KL, Seielstad M. Testing for population subdivision and association in four case-control studies. Am J Hum Genet. 2002;71(2):304–11. doi: 10.1086/341719. [DOI] [PMC free article] [PubMed] [Google Scholar]