The three-dimensional arrangement of chromosomes is critical for genome regulation and is the topic of intense research. Chromosome organization can be studied using Chromosome Conformation Capture (3C) - based assays1,2. 5C (“3C-Carbon Copy”) is an adaptation of 3C for high-throughput analysis of interaction networks and three-dimensional folding of chromosomes3.
5C combines 3C with multiplexed ligation-mediated amplification with pools of primers to detect millions of chromatin interactions in parallel (Fig. 1). The design of large numbers of 5C primers and the handling of large chromatin interaction maps can be daunting. To enable the community to adopt 5C we developed “my5C”, a publicly available set of webtools for all aspects of 5C studies. My5C is hosted at http://my5C.umassmed.edu. Here we highlight the main features of my5C (see also Supplemental Data 1–4).
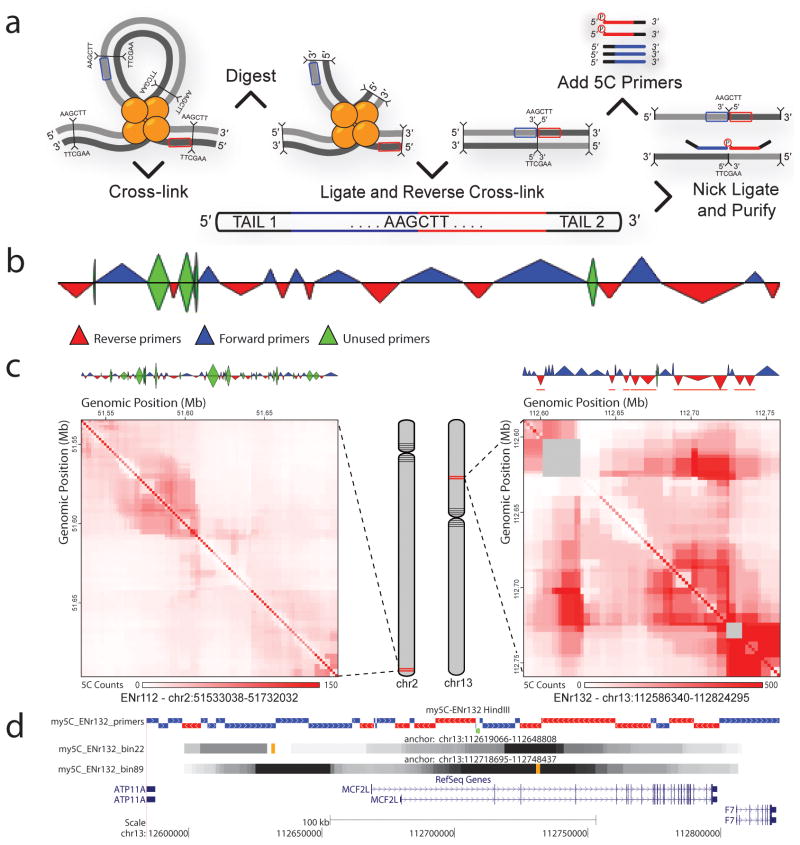
Figure 1.
Overview of my5C. (a) Top: 5C technology and locations of forward and reverse primers. Bottom: 5C ligation product. (b) 5C primer design output of my5C.primers showing an alternating design scheme. The triangles indicate restriction fragments. The height of the triangle scales with repetitiveness of the primer; width scales with size of the restriction fragment. (c) Heatmap representations of 5C data (bin size 30 Kb, step size 1.5 Kb) and primer designs for ENCODE regions ENr112 and ENr132 (Supplemental Data 1, 8, 9, 12, 13). The diagonal that is observed represents frequent interaction between loci located near each other in the genome. Prominent long-range looping interaction is visible in ENr132 (right panel) by the strong 5C signals away from the diagonal5. (d) Interaction data displayed in the UCSC genome browser (Supplemental Data 13). Orange position indicates the locus for which interactions (in grey scale) are displayed.
My5C is composed of two modules. The “my5C.primers” module is used to design 5C primers for restriction fragments throughout user-defined genomic regions (Supplemental Data 5). For analysis of overall three-dimensional conformation alternating primer design schemes3 (Fig. 1b) can be selected (Supplemental Data 6). For studies of networks of interactions between specific genomic elements, e.g. promoters and enhancers3, users can upload Datas containing genomic coordinates of these elements and my5C.primers will design forward and reverse primers for the two sets (Supplemental Data 7). Primer designs can be downloaded along with a custom microarray probe set for detection of all interactions that the primers interrogate.
In the second module, “my5C.heatmap”, datasets are visualized as two-dimensional heatmaps (Supplemental Data 8–13). Each datapoint corresponds to an interaction frequency between two loci (Fig. 1c). To facilitate exploration of large interaction maps the heatmaps are interactive: when moving the cursor over the heatmap detailed information is provided regarding the interaction at the cursor position. Clicking an interaction will display interaction proDatas across the dataset for each of the interacting loci.
My5C.heatmap provides a variety of tools to analyze interaction data. Users can display the difference, ratio or log ratio of two datasets. My5C.heatmap also enables users to identify elements that interact more frequently than expected, which may point to specific looping associations. To identify larger patterns, users can smooth data or perform sliding window analysis (Fig. 1c).
My5C.heatmap enables integrating chromosome conformation data with other genomic features. When a position on the heatmap is clicked, links to the UCSC genome browser appear that lead to the corresponding positions in the genome to explore other annotations. Further, lists of genomic annotations, e.g. promoters, can be uploaded and My5C.heatmap will highlight interaction data obtained for these loci in the heatmap.
Users can download any data displayed as a heatmap as tables or as lists of pairwise interactions that can be uploaded into Cytoscape for network visualization4. Data can be downloaded in UCSC BED format to display data as custom tracks in the UCSC genome browser (Fig. 1d, Supplemental Data 11). This allows users to integrate interaction data with publicly available genome annotations in the genome browser.
To ensure confidentiality all data are password protected and users can opt not to store data on the my5C server. My5C provides a critical resource to the emerging field of chromosome conformational studies.
Supplementary Material
Supplemental Data 1. my5C details, 5C methods and supplemental file descriptions.
Supplemental Data 2. A tutorial describing in detail how to use my5C.primers.
Supplemental Data 3. A tutorial describing in detail how to use my5C.uploads.
Supplemental Data 4. A tutorial describing in detail how to use my5C.heatmap.
Supplemental Data 5. An example file in the format of a FASTA file that contains the DNA sequence and genomic information of a genomic region that users can upload to My5C.primers.
Supplemental Data 6. An example file for defining variable step sizes of alternating design schemes (arbitrarily chosen for Enr122).
Supplemental Data 7. An example file with a list of genomic elements (arbitrarily chosen in Enr112) in the format required for upload in My5C.primers.
Supplemental Data 8. An example file for uploading interaction data to My5C.uploads linked to a primer pool.
Supplemental Data 9. An example file for uploading interaction data for ENr112 to My5C.uploads linked to a *CUSTOM* primer pool.
Supplemental Data 10. An example file for uploading interaction data for ENr132 to My5C.uploads linked to a *CUSTOM* primer pool.
Supplemental Data 11. An example of data display in the UCSC genome browser.
Supplemental Data 12. DNA sequences of 5C primers used for analysis of the conformation of ENr112. This is the standard output of My5C.primers.
Supplemental Data 13. DNA sequences of 5C primers used for analysis of the conformation of ENr132.
Acknowledgments
Labmembers, collaborators and Marian Walhout are acknowledged for discussions. This work was supported The National Institutes of Health (HG003143, HG004592) and the Keck Foundation.
Footnotes
COMPETING FINANCIAL INTERESTS
We hereby disclose that a provisional patent application has been filed that covers the set of web tools described in this paper. Bryan Lajoie and Job Dekker are listed as inventors on this patent application.
References
- 1.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing Chromosome Conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 2.Simonis M, Kooren J, de Laat W. An evaluation of 3C-based methods to capture DNA interactions. Nat Methods. 2007;4:895–901. doi: 10.1038/nmeth1114. [DOI] [PubMed] [Google Scholar]
- 3.Dostie J, et al. Chromosome Conformation Capture Carbon Copy (5C): A Massively Parallel Solution for Mapping Interactions between Genomic Elements. Genome Res. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dekker J. The 3 C’s of Chromosome Conformation Capture: Controls, Controls, Controls. Nat Methods. 2006;3:17–21. doi: 10.1038/nmeth823. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data 1. my5C details, 5C methods and supplemental file descriptions.
Supplemental Data 2. A tutorial describing in detail how to use my5C.primers.
Supplemental Data 3. A tutorial describing in detail how to use my5C.uploads.
Supplemental Data 4. A tutorial describing in detail how to use my5C.heatmap.
Supplemental Data 5. An example file in the format of a FASTA file that contains the DNA sequence and genomic information of a genomic region that users can upload to My5C.primers.
Supplemental Data 6. An example file for defining variable step sizes of alternating design schemes (arbitrarily chosen for Enr122).
Supplemental Data 7. An example file with a list of genomic elements (arbitrarily chosen in Enr112) in the format required for upload in My5C.primers.
Supplemental Data 8. An example file for uploading interaction data to My5C.uploads linked to a primer pool.
Supplemental Data 9. An example file for uploading interaction data for ENr112 to My5C.uploads linked to a *CUSTOM* primer pool.
Supplemental Data 10. An example file for uploading interaction data for ENr132 to My5C.uploads linked to a *CUSTOM* primer pool.
Supplemental Data 11. An example of data display in the UCSC genome browser.
Supplemental Data 12. DNA sequences of 5C primers used for analysis of the conformation of ENr112. This is the standard output of My5C.primers.
Supplemental Data 13. DNA sequences of 5C primers used for analysis of the conformation of ENr132.