SUMMARY
The Drosophila STAT transcription factor Stat92E regulates diverse functions, including organ development and stem cell self-renewal. However, the Stat92E functional effectors that mediate these processes are largely unknown. Here we show that chinmo is a cell-autonomous, downstream mediator of Stat92E that shares numerous functions with this protein. Loss of either gene results in malformed eyes and head capsules due to defects in eye progenitor cells. Hyperactivation of Stat92E or misexpression of Chinmo results in blood cell tumors. Both proteins are expressed in germline (GSCs) and cyst stem cells (CySCs) in the testis. While Stat92E is required for the self-renewal of both populations, chinmo is only required in CySCs, indicating that Stat92E regulates self-renewal in different stem cells through independent effectors. Like hyperactivated Stat92E, Chinmo misexpression in CySCs is sufficient to maintain GSCs non-autonomously. Chinmo is therefore a key effector of JAK/STAT signaling in a variety of developmental and pathological contexts.
INTRODUCTION
The evolutionarily conserved JAK/STAT pathway plays critical roles in numerous developmental processes in mammals and Drosophila including embryonic development, hematopoeisis and stem cell self-renewal. In mammals Leukemic Inhibitory Factor activates Stat3 to maintain long-term murine embryonic stem cells (mESCs) (Matsuda et al., 1999). Consistent with this result, deletion of the Stat3 gene causes embryonic lethality, indicating its crucial role during fetal development (Takeda et al., 1997). Humans with loss-of-function mutations in Stat1, Stat3, Tyk2 or Jak3 present with immunodeficiency and Hyper-IgE syndrome due to the requirement of this pathway in developing blood cells (Dupuis et al., 2003; Minegishi et al., 2007; Paulson et al., 2008; Pesu et al., 2005). Laron-type human dwarfism is associated with mutations in the Growth Hormone receptor, which activates Stat5a/b, and is a condition mimicked by Stat5a/b deficiency in mice (Laron, 2002; Teglund et al., 1998). Fibroblasts expressing a constitutively-active Stat3 protein cause tumors in nude mice (Bromberg et al., 1999). Consistent with the latter result, persistent activation of Stat3 is associated with a dozen types of human cancers, including all classes of carcinoma (Darnell, 2005). Furthermore, germline activating mutations in Jak2 cause human blood cell cancers like polycythemia vera (Levine, 2009).
In Drosophila, the JAK/STAT pathway plays important roles in growth and patterning of the eye, in hematopoiesis and in stem cell self-renewal. The eye-antennal disc, derived from ~50 progenitor cells, gives rise to the adult eye, antenna and head capsule. These progenitors undergo exponential rates of growth during the first two of three larval stages or instars (reviewed in (Dominguez and Casares, 2005)). In wildtype eye discs, Notch signaling leads to activation of the JAK/STAT pathway and this promotes proliferation and maintenance of eye progenitors, as well as formation of the eye field (Bach et al., 2003; Chao et al., 2004; Ekas et al., 2006; Reynolds-Kenneally and Mlodzik, 2005; Tsai and Sun, 2004). Hematopoiesis in Drosophila occurs in the larval lymph gland (reviewed in (Evans et al., 2003)). At the third larval instar, the primary lobe of this organ is divided into three compartments: the posterior signaling center (PSC, the niche), the medullary zone (MZ) where multipotent progenitors called prohemocytes reside, and the cortical zone (CZ) where differentiating blood cells called hemocytes are found (Fig. 2G and (Jung et al., 2005)). Maintenance of MZ blood cell progenitors requires JAK/STAT signaling (Krzemien et al., 2007). This pathway also acts intrinsically to maintain bona fide stem cells in Drosophila (Gregory et al., 2008). These include GSCs and CySCs (also called cyst progenitor cells (CPCs) and/or somatic stem cells (SSCs) (Davies and Fuller, 2008)) in the testis, escort stem cells in the ovary, neuro-epithelial cells in the optic lobe of the brain and intestinal stem cells in the midgut (Buchon et al., 2009; Cronin et al., 2009; Decotto and Spradling, 2005; Jiang et al., 2009; Kiger et al., 2001; Leatherman and Dinardo, 2008; Tulina and Matunis, 2001; Yasugi et al., 2008). In the Drosophila testis, as well as in the other stem cells that depend upon JAK/STAT signaling for their maintenance, no effector genes activated by Stat92E that promote self-renewal have as yet been reported, with the sole exception of zfh1, which was found to be a Stat92E-regulated gene required for CySC self-renewal (Leatherman and Dinardo, 2008; Terry et al., 2006). Therefore, the identification of additional Stat92E effector genes that mediate self-renewal is an important area of stem cell research.
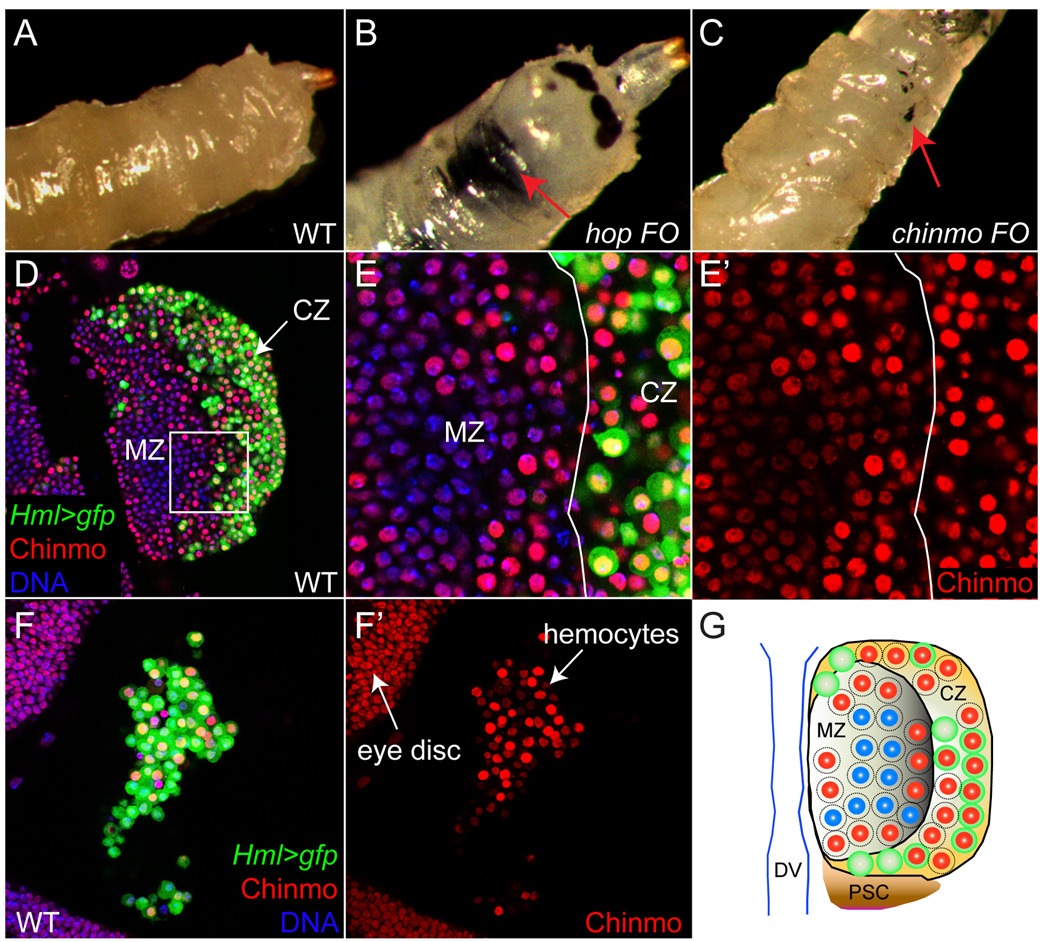
Figure 2. Mis-expression of chinmo causes melanotic tumors.
(A–C) Melanotic tumors are present in third instar larvae harboring hop (B, arrow) or chinmo (C, arrow) mis-expressing clones but not in WT (A). (D) In the lymph gland, Chinmo (red) is present in some MZ prohemocytes and some CZ hemocytes. E,E’ are close-ups of D. (F) Chinmo is present in mature hemocytes in the eye disc. (G) Schematic diagram of Chinmo expression in the lymph gland. In the MZ, some cells with blue nuclei (DNA) lack Chinmo while those with red nuclei express Chinmo. In the CZ, some Hml+ (green) cells lack Chinmo, but most express both Hml and Chinmo. In the CZ, some cells lack Hml but express Chinmo. Hml>gfp (green); DNA (blue) in D–F.
Drosophila serves as an excellent model for identifying and characterizing the conserved genes involved in these processes because it has a simple yet complete JAK/STAT pathway (Arbouzova and Zeidler, 2006). In flies, three related interleukin-6-like cytokines Unpaired (Upd) (also called Outstretched), Upd2 and Upd3 activate one dimerized gp-130-like cytokine receptor called Domeless (Dome). This leads to the phosphorylation of the sole JAK, called Hopscotch (Hop), which in turn activates the single STAT transcription factor, called Stat92E. Activated Stat92E dimers translocate to the nucleus and regulate gene transcription.
We recently identified chronologically inappropriate morphogenesis (chinmo) as a potential Stat92E downstream effector (Flaherty et al., 2009). Here, we show that chinmo is positively and cell-autonomously regulated at the transcriptional level by JAK/STAT pathway activity. Loss- and gain-of-function in chinmo or Stat92E in developing eye discs and in hemocytes results in similar phenotypes, including aberrations of the eye, antenna and head capsule and the formation of melanotic tumors. We also show that Chinmo and Stat92E regulate the expression of a common gene (Serrate (Ser)), suggesting that Chinmo can repress gene expression directly or indirectly. Stat92E is intrinsically required for the self-renewal of both CySCs and GSCs. While Chinmo is expressed in both of these stem cell populations, it is required only for the maintenance of CySCs. Mis-expression of chinmo in somatic cells in the testis results in expansion of both GSCs and CySCs outside of the niche, the same phenotype as hyperactivation of the JAK/STAT pathway or misexpression of zfh1. Furthermore, epistasis experiments revealed that chinmo does not act through zfh1 to promote stem cell expansion outside of the niche. Thus, Chinmo is an important downstream effector of JAK/STAT signaling in a variety of developmental and pathological processes.
RESULTS
chinmo is autonomously regulated by Stat92E in the eye-antennal imaginal disc
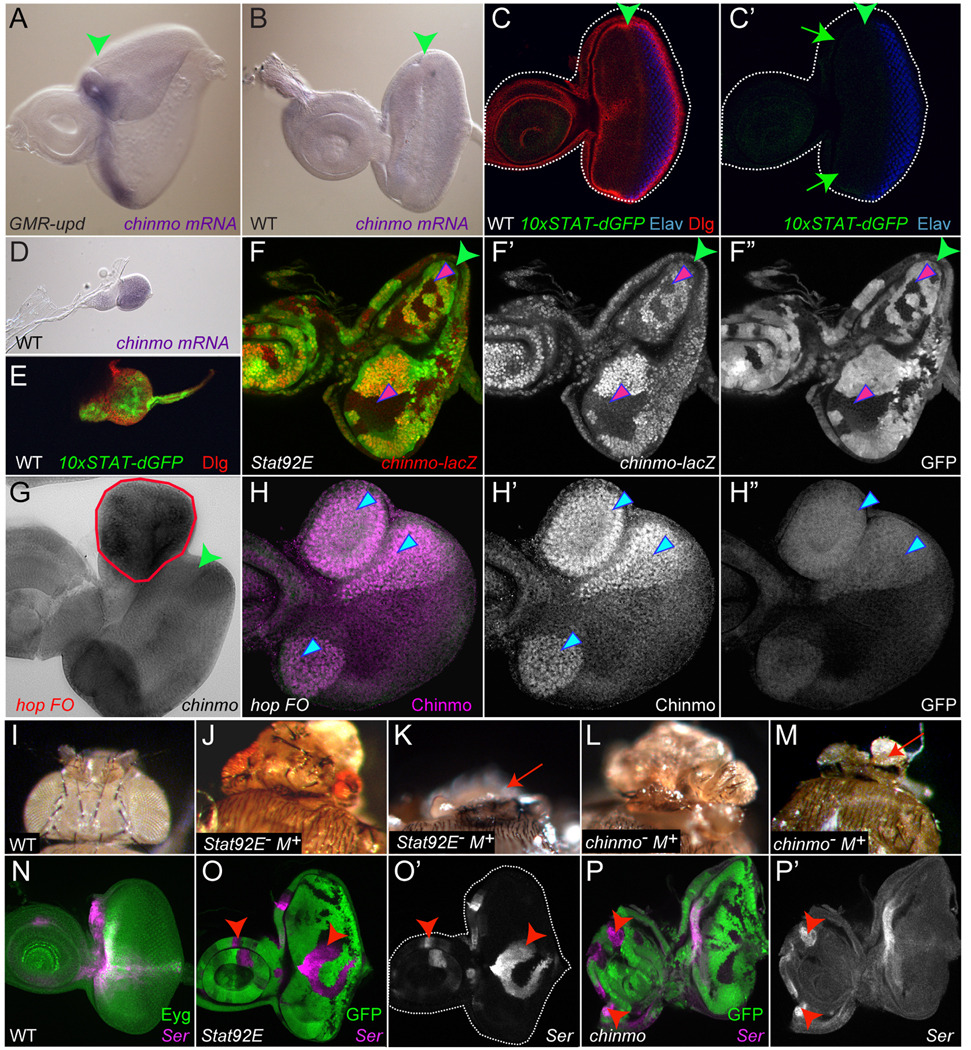
Like well established Stat92E target genes socs36E and dome, chinmo mRNA is upregulated only in cells located anterior to the furrow in GMR-upd eye discs (Fig. 1A and (Flaherty et al., 2009)). In addition, like a reporter of Stat92E transcriptional activity (10xSTAT-dGFP), chinmo mRNA is not expressed in wildtype third instar eye discs (Fig. 1B,C, C’ and (Bach et al., 2007; Flaherty et al., 2009)). upd and 10xSTAT-dGFP are expressed in first and second instar wildtype eye discs (Fig. 1E and (Ekas et al., 2006)). Similarly, chinmo mRNA is also expressed in this pattern (Fig. 1D and data not shown). To assess the requirement of Stat92E in regulation of chinmo expression, we generated Stat92E clones in the eye-antennal disc using the FLP/FRT technique and eyeless (ey)-FLP, which is active from formation of the eye disc primordium (Newsome et al., 2000; Xu and Rubin, 1993). chinmo expression, monitored by a chinmo-lacZ enhancer trap, is lost in a cell-autonomous manner in Stat92E mosaic clones located anterior to the furrow (Fig. 1F). Note that the perdurance of the β-gal protein transcribed from the chinmo enhancer trap allows us to monitor prior chinmo expression during third instar, despite the fact that chinmo mRNA is not expressed at this stage. Furthermore, chinmo mRNA and protein are strongly upregulated within clones misexpressing hop, which autonomously activates Stat92E (Fig. 1G,H and (Ekas et al., 2006)). These data indicate that the chinmo gene is autonomously regulated by JAK/STAT pathway activity. chinmo may indeed be a direct target gene of Stat92E since, as we previously reported, there are several single and a cluster of Stat92E binding sites in non-coding genomic regions of the chinmo gene (Flaherty et al., 2009).
Figure 1. chinmo is autonomously regulated by JAK/STAT pathway activity.
(A) chinmo mRNA is upregulated in cells anterior to the furrow in a third instar GMR-upd eye disc. (B) chinmo mRNA is absent at this stage in a wildtype (WT) eye disc. (C) Activity of the JAK/STAT pathway, as assessed by the 10xSTAT-dGFP transcriptional reporter, is not detected in a mid-third instar WT eye disc (arrows). (D,E) chinmo mRNA is expressed in a second instar eye disc, like 10xSTAT-dGFP. (F–F”) Expression of chinmo-lacZ (red) is autonomously lost in Stat92E clones, which lack GFP (pink arrowheads). (G) chinmo mRNA (black) is autonomously increased in a hop flip-out clone (hop FO) (red outline). (H–H”) Chinmo protein (red) is increased in hop FO clones (arrowheads). The furrow is marked by a green arrowhead in A–C,F,G. Dorsal is up and anterior to the left. (I) A dorsal view of a WT adult Drosophila head. (J,K) Animals with Stat92E− M+ clones in the eye-antennal disc exhibit small eyes, excess head cuticle, and are frequently headless. (L,M) Animals carrying chinmo− M+ clones also exhibit these phenotypes. Arrow points to an adult with only a rudimentary head (K,M). (N) Ser-LacZ expression (magenta) in a WT eye-antennal disc. Eyegone (Eyg) is green. (O,P) Ser-lacZ is ectopically expressed in the antennal disc in mosaic Stat92E (O’, arrowheads) or chinmo (P’, arrowheads) clones. Clones lack GFP in O,P.
chinmo was identified in a micro-array for Ey target genes, suggesting that Chinmo may function like Stat92E and play a critical role in growth and/or regional specification of the eye-antennal disc (Ekas et al., 2006; Ostrin et al., 2006). We previously reported that a moderately over-grown eye was observed in ey>hop flies (Bach et al., 2003). We attempted to address if mis-expression of Chinmo at the earliest stages of eye disc development would result in a similar phenotype. Unfortunately, frequent lethality induced by strong misexpression of chinmo, globally or locally, confounded a detailed analysis of gain-of-function phenotypes in the eye.
chinmo and Stat92E share similar loss-of-function phenotypes
We next addressed the issue of whether chinmo is a functionally-important effector of Stat92E in the eye-antennal disc. To test this prediction, we induced large Stat92E or chinmo clones in the eye disc using ey-FLP and Minute techniques (Morata and Ripoll, 1975). Minutes are mutations in ribosomal genes, which cause slow growth and recessive lethality in cells possessing the wildtype chromosome (Lambertsson, 1998; Morata and Ripoll, 1975). When used together, the ey-FLP and Minute techniques produce an eye composed almost entirely of homozygous mutant tissue (e.g., Stat92E or chinmo), which hereafter are referred to as Stat92E−M+ and chinmo−M+.
As we previously reported, 90% of Stat92E− M+ pupae with clones in the eye-antennal disc did not eclose from their pupal cases and displayed increased head cuticle, reduction of the eye field, partial or complete loss of antennal segments and often only a rudimentary head (Fig. 1J,K and (Ayala-Camargo et al., 2007; Ekas et al., 2006; Tsai et al., 2007)). 90% of pupae with chinmo− M+ clones in the eye-antennal disc also did not eclose, and they displayed Stat92E-like loss-of-function phenotypes (Fig. 1L,M). Furthermore, visual inspection of eye-antennal discs with chinmo− M+ clones revealed a similar morphology to those with Stat92E− M+ clones, suggesting that their common adult phenotypes arise from similar defects in larval eye-antennal disc progenitor cells (data not shown and (Ekas et al., 2006)). Lastly, our data suggest that Chinmo, like Stat92E, promotes proliferation of eye-antennal disc progenitor cells, since chinmo mosaic clones are always smaller than the twin spot (data not shown).
Chinmo and Stat92E both repress transcription of Ser
We recently published that Ser expression is repressed cell-autonomously by JAK/STAT signaling in the eye-antennal disc (Fig. 1O and (Flaherty et al., 2009)). Chinmo contains one Bric–a-brac, Tramtrack, Broad Complex (BTB) domain at the N-terminus and two C2H2 zinc finger (ZF) domains at the C-terminus (Fig. 6D) and was isolated based on its requirement for the temporal identity of mushroom body neurons (Zhu et al., 2006). BTB-domain proteins can act as transcriptional repressors or as adaptors for Cullin 3 (Cul-3) E3 ligases, which can promote protein degradation (Perez-Torrado et al., 2006). To determine if Chinmo, like activated Stat92E, could also affect the Ser gene, we examined the expression of a Ser-lacZ transcriptional reporter in chinmo1 mosaic clones in the eye-antennal disc (Bachmann and Knust, 1998). This Ser reporter was frequently ectopically expressed in a cell-autonomous manner (12 chinmo clones located in 7 antennal discs, Fig. 1P). The upregulation of Ser observed in chinmo1 or chinmoM33 positively-marked MARCM clones in antennal disc was invariably rescued (10/10 clones, data not shown) by overexpression of a wildtype chinmo transcript (UAS-5'UTR-chinmo-3'UTR; (Zhu et al., 2006)). Note that activated Stat92E and chinmo mRNA are not present in third instar eye discs (Fig. 1B,C). The ectopic Ser visible in Stat92E and chinmo clones at this stage is a consequence of de-repression of this gene at earlier larval stages. These results suggest that Chinmo functions either downstream of or in parallel to Stat92E in the antennal disc to regulate Ser expression.
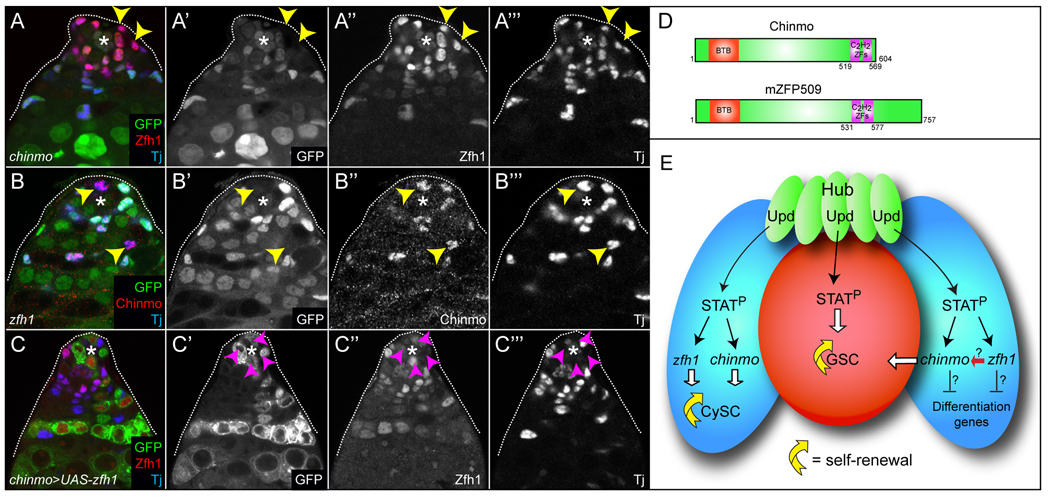
Figure 6. chinmo does not act through zfh1 to promote the self-renewal of CySCs.
(A-A’’’) Zfh1 (red) is not reduced in Tj+ CySCs/early cyst cells lacking chinmo (arrowheads). (B-B’’’) Chinmo (red) is not diminished in Tj+ CySCs/early cyst cells lacking zfh1 (arrowheads). Clones lack GFP expression in A,B. (C-C’’’) chinmo MARCM clones over-expressing zfh1 do not restore CySC characteristics to the clones. Only Zfh1−, Tj− GSCs mutant for chinmo were found close to the niche, pink arrowheads. Clones express GFP and Zfh1 in C. An * marks the Hub in A–C. (D) Schematic of the Chinmo protein and its putative mammalian homolog ZFP509. (E) A model of the autonomous requirement of activated Stat92E for self-renewal in GSCs and CySCs and autonomous requirement of Chinmo and Zfh1 in CySC self-renewal (left CySC (blue cell)). This is also a model of the non-autonomous expansion of CySCs and GSCs caused by hyperactivated JAK, misexpression of chinmo or of zfh1 in CySCs/early cyst cells (right CySC). STATP indicates activated Stat92E. See text for details.
Gain-of-function in Stat92E or chinmo causes melanotic tumors
We found that mis-expression of either hop or chinmo caused melanotic tumors (Fig. 2B,C), which are never observed in wildtype larvae (Fig. 2A). This phenotype is reminiscent of that seen in hopTum-l animals, which carry a dominant mutation in hop that activates Stat92E and causes extensive proliferation, precocious differentiation, and melanotic tumor formation among circulating blood cells (Hanratty and Dearolf, 1993; Luo et al., 1995). Subsequent antibody staining demonstrated that Chinmo is expressed in the larval lymph gland and in circulating hemocytes (Fig. 2D–F). In the lymph gland, Chinmo appears to be expressed throughout the organ, which includes both differentiating and progenitor cell types. Although variable, Chinmo is generally expressed at higher levels among differentiating CZ cells as compared to undifferentiated MZ cells (Fig. 2D,E and (Jung et al., 2005)). MZ progenitors, which also express dome, have been shown to require JAK/STAT signaling for their maintenance (Jung et al., 2005; Krzemien et al., 2007). Thus the expression of Chinmo in MZ cells may be indicative of a function in prohemocyte maintenance. We also find that Chinmo is expressed among circulating wildtype hemocytes that reside on the basal surface of the eye disc (Fig. 2F). In addition, over-expression of Chinmo in mature hemocytes using the Hemolectin (Hml) promoter results in the formation of melanotic tumors (data not shown), suggesting that Chinmo plays a role in the proliferation of developing hemocytes, as it does in eye disc progenitor cells. Taken together the shared loss- and gain-of-function phenotypes between Stat92E and chinmo support the conclusion that, at least in the eye-antennal disc and in hemocytes, chinmo is an important downstream mediator of the JAK/STAT pathway.
JAK/STAT pathway activity in the Drosophila testis
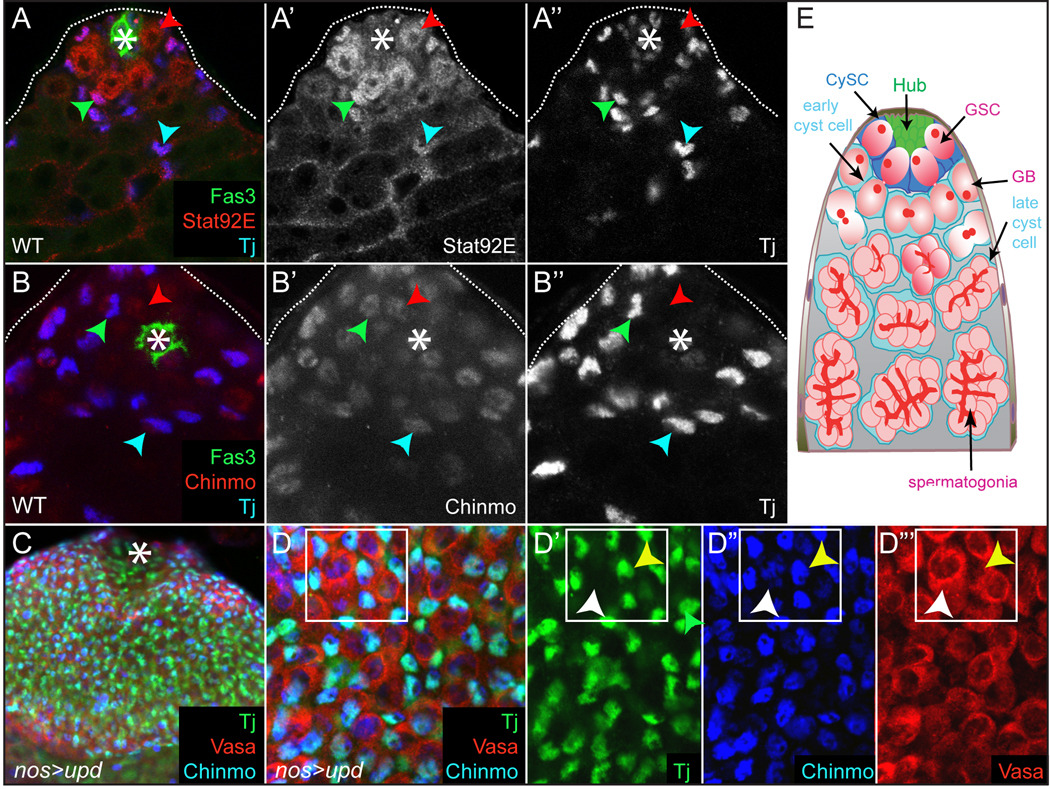
The niche of the Drosophila testis is composed of a group of somatic cells called the Hub, which maintain GSCs and CySCs (Fig. 3E and reviewed in (Davies and Fuller, 2008)). Both GSCs and CySCs maintain physical contact with the Hub, and CySCs envelope GSCs. This organization of niche and stem cells facilitates the spatially and temporally coordinated production of offspring from GSCs and CySCs, which then physically interact and co-differentiate. GSCs divide asymmetrically to produce one GSC, which remains in the niche and maintains the stem cell pool, and one gonialblast (GB), which is displaced from the Hub and begins differentiation. CySCs also divide asymmetrically to give rise to one CySC and one cyst cell. Two cyst cells envelop one GB, which undergoes transiently-amplifying synchronous mitotic divisions with incomplete cytokinesis. This results in a spermatogonium consisting of 16 germ cells that remain interconnected by cytoplasmic bridges, surrounded at all times by these same two cyst cells. Hub cells produce Upd, which activates Stat92E in adjacent GSCs and CySCs, as evidenced by the stabilization of Stat92E protein in these cells (Fig. 3A and (Boyle et al., 2007; Chen et al., 2002; Issigonis et al., 2009; Leatherman and Dinardo, 2008)). We also observe stabilized Stat92E in early cyst cells (Fig. 3A, blue arrowhead).
Figure 3. Stabilized Stat92E and Chinmo are present in GSCs and CySCs.
(A) In a WT testis Stat92E protein (red) is stabilized in GSCs, CySCs and early cyst cells (red, green, and blue arrowheads, respectively). CySCs and early cyst cells are Tj+ (blue). (B) Chinmo is expressed in a Tj− GSC, in a Tj+ CySC and in a Tj+ early cyst cell (red, green and blue arrowheads, respectively). (C) There is an increased number of GSCs (red) and CySCs (blue) in a nos>upd testis. (D-D’’’) Chinmo (blue) is upregulated in a nos>upd testis. In D’-D’’’ a yellow arrowhead indicates a Tj+ CySC that expresses Chinmo and a white one points to a Chinmo+ Vasa+ GSC. Fas3 is green in A,B and an * marks the Hub in A–C. Vasa is red and Tj is green in D. (E) Schematic of the Drosophila testis. See text for details.
Chinmo is expressed in GSCs and CySCs in the Drosophila testis
To assess if Chinmo expression overlapped with stabilized Stat92E in the testis, we co-stained wildtype adult testes with an antibody specific for Traffic Jam (Tj), a Maf-family transcription factor that is expressed highly in CySCs and early cyst cells (Li et al., 2003). We find that Chinmo, like stabilized Stat92E, is present at high levels in Tj+ CySCs and early cyst cells and at lower levels in GSCs (Fig. 3B’, green, blue and red arrowheads, respectively). Chinmo is also expressed in the Hub, in contrast to Stat92E, which is not stabilized in these cells (Fig. 3A’,B’). These observation raise the possibility (confirmed below) that Chinmo mediates a stem cell type-specific subset of STAT functions in the testis.
It was previously reported that misexpression of Upd in germ cells using the nanos (nos) promoter caused significant expansion of GSCs and CySCs outside of the niche, a result that we also observe (Fig. 3C and (Kiger et al., 2001; Tulina and Matunis, 2001; Van Doren et al., 1998)). Furthermore, stabilized Stat92E is detected in the expanded populations of both GSCs and CySCs in nos>upd testes, indicating that Upd can activate Stat92E in both stem cell populations (Leatherman and Dinardo, 2008). Consistent with the hypothesis that Chinmo is a downstream mediator of Stat92E function in the testis, chinmo transcripts were also significantly increased (4.4 fold) in a whole-genome micro-array analysis of nos>upd testes (Terry et al., 2006). In these testes, we found that the Chinmo protein is expressed at high levels in CySCs and at lower levels in GSCs (Fig. 3D–D”’, yellow and white arrowheads, respectively). However, expression of the chinmo enhancer trap or Chinmo protein was only modestly decreased in negatively-marked Stat92E clones in the testis (data not shown). The lack of reduction of chinmo in the absence of Stat92E may be an issue of perdurance of β-gal, Chinmo and/or Stat92E proteins. CySCs lacking Stat92E or chinmo differentiate within 3 days post-clone induction (pci) (see below), precluding the analysis of chinmo expression in Stat92E clones beyond this time point. Alternatively, factors in addition to Stat92E may regulate Chinmo expression in the adult testis.
Chinmo is required for the self-renewal of CySCs
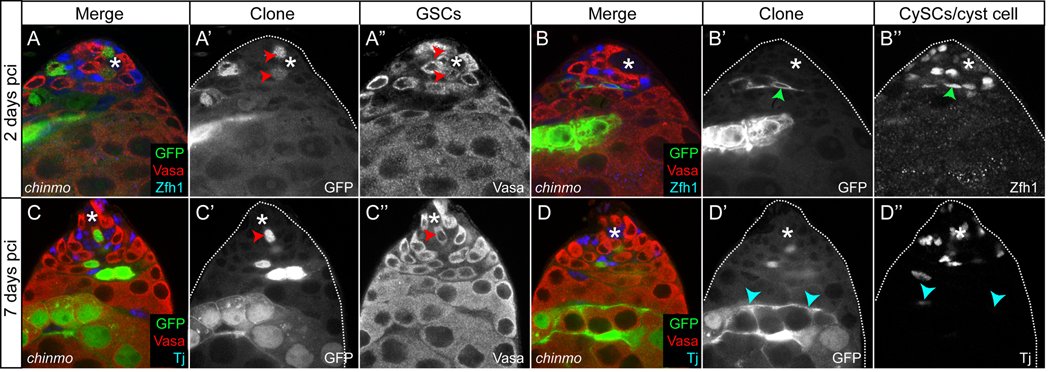
To assess if chinmo, like Stat92E, is required for the self-renewal of GSCs and CySCs, we used the MARCM technique to generate positively-marked FRT40 wildtype or FRT40 chinmo1 clones (Lee and Luo, 1999). We counted the number of testes with at least one mutant stem cell remaining in the niche at 2 and 7 days pci. As expected, in control testes containing FRT40 wildtype clones, we were able to find many positively-marked CySCs and GSCs in contact with the Hub at both time points (Table 1 and data not shown). At 2 days pci, we were also able to find chinmo mutant GSCs that were in contact with the Hub and that expressed the germ cell specific protein Vasa, and chinmo mutant CySCs that enveloped GSCs and expressed high levels of Zfh1 (Fig. 4A–A”,B–B”, red and green arrowheads, respectively, Table 1 and (Leatherman and Dinardo, 2008)). These data indicate that chinmo clones can be induced in these two stem cell populations. At 7 days pci, numerous GSCs mutant for chinmo could be identified in contact with the Hub, indicating that chinmo is not required for the self-renewal of GSCs (Fig. 4C–C”, red arrowheads, and Table 1). However, at 7 days pci, we were unable to find a single CySC mutant for chinmo, despite the analysis of 200 testes (Fig. 4D–D” and Table 1). These data indicate that CySCs lacking chinmo either differentiate or die. To distinguish between these possibilities, we looked for the differentiating progeny of CySCs mutant for chinmo at 7 days pci. At this time point, we found chinmo mutant somatic cells that resided outside the Hub in most of the testes we examined, indicating that CySCs lacking chinmo do indeed differentiate (Fig. 4D–D’’, blue arrowheads, n=36/48 testes examined (75%)). Furthermore, mis-expression of the pan-caspase inhibitor p35 in chinmo MARCM clones did not restore CySC characteristics to the clones (Fig. S1A–A”’). We also performed clonal analysis at 2 and 7 days pci using the FLP/FRT technique to induce negatively-marked clones, and we observed results similar to those seen with chinmo MARCM clones (Fig. S1B,C and Table 1). In the negatively-marked clone analysis, we monitored wildtype, chinmoM33 and chinmo1 clones and obtained similar results with either chinmo allele (Table 1). Taken together, these data indicate that chinmo, although expressed in both GSCs and CySCs, is only required in CySCs for their maintenance. Moreover, we also demonstrate that activated Stat92E regulates self-renewal through different effectors in these adjacent stem cells (Fig. 6E).
Table 1.
Quantification of chinmo clones
| Time pci and Genotype | Number of testes with positively-marked clones (MARCM) |
Number of testes with negatively-marked clones (FLP/FRT) |
||
|---|---|---|---|---|
| GSCs | CySCs | GSCs | CySCs | |
| 2 days FRT40A (control) | 20/30 (67%) | 15/30 (50%) | 11/13 (85%) | 8/13 (62%) |
| 7 days | 22/26 (85%) | 14/26 (54%) | 17/23 (74%) | 12/23 (52%) |
| 2 days FRT40A chinmo1 | 39/68 (57%) | 45/68 (66%) | 25/25 (100%) | 16/25 (64%) |
| 7 days | 38/48 (79%) | 0/48 (0%) | 50/50 (100%) | 0/50 (0%) |
| 2 days FRT40A chinmoM33 | n/d | n/d | 26/28 (93%) | 20/28 (71%) |
| 7 days | n/d | n/d | 48/58 (83%) | 0/58 (0%) |
pci = post clone induction; n/d = not done
Figure 4. chinmo is required autonomously for the self-renewal of CySCs.
(A,B) Vasa+ GSCs mutant for chinmo (A’,A’’, red arrowheads) and Zfh1+ CySCs mutant for chinmo (B’,B’’, green arrowhead) can be detected next to the Hub at 2 days pci. (C,D) At 7 days pci, chinmo− Vasa+ GSCs can be found next to the Hub (C’,C’’, red arrowhead). At this time point no chinmo− Tj+ CySCs can be found in the niche (C,D). Late cyst cells lacking chinmo are found far from the niche at 7 days pci (D’,D’’, blue arrowheads). In A–D chinmo clones are GFP+ and Vasa is red. Zfh1 is blue in A,B and Tj is blue in C,D. An * marks the Hub.
Sustained chinmo expression results in expansion of GSCs/GBs and CySCs/early cyst cells
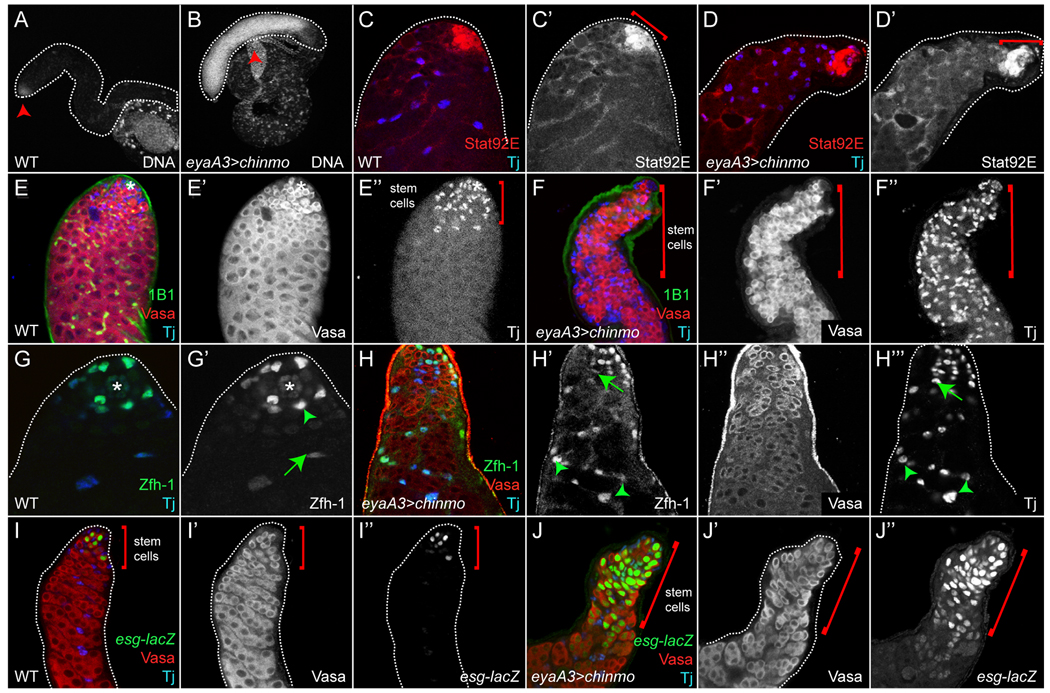
Autonomous hyperactivation of the JAK/STAT pathway by misexpression of hopTum-l only in CySCs (but not in GSCs) is sufficient to expand the number of CySCs and GSCs outside of the niche, a phenotype similar to that observed in nos>upd testes (Leatherman and Dinardo, 2008). To investigate whether chinmo misexpression mimics this phenotype, we employed the UAS/Gal4 technique to drive chinmo in the somatic lineage using eyaA3-Gal4, which is active at low levels in CySCs and at high levels in cyst cells (Brand and Perrimon, 1993; Leatherman and Dinardo, 2008). We analyzed eyaA3>chinmo testes for the presence of increased numbers of undifferentiated cells, which fluoresce brightly with DNA dyes (Leatherman and Dinardo, 2008). As predicted, eyaA3>chinmo testes were filled with brightly fluorescing cells, whereas in wildtype they were restricted to the niche (Fig. 5A,B, arrowheads). In eyaA3>chinmo testes, there were many individual or pairs of Vasa+ cells intermingled with Tj+ cells, presumably GSCs/GBs and CySCs/early cyst cells, respectively (Fig. 5F–F”). The excess of early germ cells was not a consequence of defective encystment, since DE-Cadherin+ extensions from somatic cells did, in fact, encyst individual or pairs of germ cells (Fig. S2F’, arrowheads, and (Leatherman and Dinardo, 2008; Sarkar et al., 2007; Schulz et al., 2002; Tazuke et al., 2002)). Moreover, we ruled out the possibility that the expansion of GSCs/GBs and CySCs/early cyst cells in eyaA3>chinmo testes was caused by the ectopic production of Upd or ectopic stabilization of Stat92E, as Stat92E is only stabilized in these testes in a pattern similar to wildtype (Fig. 5C,D). Importantly, misexpression of chinmo in male germ cells (i.e., nos>chinmo) did not generate any phenotypes, indicating that overexpression of chinmo in GSCs cannot promote expansion of GSCs and CySCs (Fig. S3A–A”). These data again support the model that only sustained Stat92E activity (and as a result misexpression of its effectors) in the somatic lineage can promote non-autonomous expansion of stem cells in the testis. These effects were dependent on the BTB and ZF domains of Chinmo (Fig. S3B–G).
Figure 5. Sustained chinmo expression results in expansion of GSCs and CySCs.
(A) In a WT testis, only stem cells at the apical tip fluoresce when stained with Hoechst 33342 (arrowhead). (B) In an eyaA3>chinmo testis DNA-bright cells accumulate throughout the testis (arrowhead). (C) In a WT testis, activated Stat92E (red) is found in GSCs and CySCs in contact with the Hub (bracket), as well as in early cyst cells. (D) In an eyaA3>chinmo testis, stabilized Stat92E is expressed in the same pattern as in WT (bracket). Tj is blue in C,D. (E) In a WT testis, Tj+ CySCs and early cyst cells are only found at the apical tip of the testis (E”, bracket). (F) In an eyaA3>chinmo testis, Tj+ somatic cells are found throughout the testis (F-F”, brackets). 1B1 (green), Vasa (red) and Tj (blue) in E,F. (G) In a WT testis, Zfh1 (green) is expressed highly in CySCs and at lower levels in early cyst cells. Both cell types express Tj (blue). (H) However, in an eyaA3>chinmo testis, Zfh1+ CySCs and early cyst cells are found throughout the testis. In G,H green arrowheads mark CySCs and green arrows mark early cyst cells. Vasa (red) and Tj (blue) in H. (I) In a wild type testis esg-lacZ (green) is restricted to GSCs and GBs (I-I”, brackets). (J) In an eyaA3>chinmo testis, esg-lacZ positive cells are expanded throughout the testis (J-J”, brackets). Vasa (red) and Tj (blue) in I,J. An * marks the Hub.
Expanded somatic and germ cells in testes with sustained chinmo expression have stem cell characteristics
To confirm that the expanded cells in eyaA3>chinmo testes have stem cell traits similar to those in eyaA3>hopTum-l testes, we analyzed the expression of different stem cell markers. Most expanded somatic cells were positive for Tj, a marker of CySCs and early cyst cells (Fig. 5F,F’’). Moreover, they had high levels of Zfh1, which accumulates in CySCs/early cyst cells, and had low or negligible levels of Eya (Fig. 5H,H”, Fig. S2J,J’, green arrowheads, and (Leatherman and Dinardo, 2008)). Taken together, these data strongly indicate that somatic cells in eyaA3>chinmo testes retained stem cell characteristics. Our results also support the conclusion that many of the expanded germ cells in eyaA3>chinmo testes are GSCs/GBs. First, germ cells residing far from the Hub expressed the escargot (esg) enhancer trap M5.4-lacZ, which is normally expressed only in GSCs/GBs (Fig. 5I–I”,J-J” and (Gonczy and DiNardo, 1996)). Second, individual or pairs of Vasa+ cells in eyaA3>chinmo testes contained dot fusomes, germ-cell organelles found only in GSCs/GBs in wildtype testes (Fig. 3E, Fig. S2G,G’,H,H’ and (Lin et al., 1994)). Third, excess germ cells and somatic cells often underwent mitosis as single cells, as evidenced by the expression of the M-phase marker phospho-histone 3 (Fig. S2A,A’,B,B’, red and green arrowheads, respectively). Fourth, groups of individual or pairs of germ cells located far from the niche did not express the differentiation factor Bag-of-marbles (Bam), which in wildtype testes is expressed in two- to four-cell spermatogonia but not in GSCs/GBs (Fig. S2C,C’,D,D’ and (Gonczy et al., 1997)). However, in some eyaA3>chinmo testes, we did identify groups of Vasa+ spermatogonia that were also Bam+, indicating that some excess germ cells in these testes could differentiate (Fig. S2D,D’, red arrowheads). Taken together, these data indicate that sustained expression of chinmo in the somatic lineage (1) autonomously induces the expansion of CySCs/early cyst cells and/or inhibits their differentiation, and (2) non-autonomously promotes expansion of GSCs/GBs.
chinmo does not act through zfh1 to maintain CySCs
In the Drosophila testis, no effectors activated by Stat92E that promote self-renewal have as yet been reported, with the sole exception of zfh1. CySCs lacking zfh1 differentiate within 1–2 days, in contrast to CySCs lacking chinmo, which differentiate in 2–3 days (Fig. 4 and (Leatherman and Dinardo, 2008)). This difference in timing suggests that zfh1 and chinmo regulate complementary downstream target genes in CySCs. To investigate the interaction between chinmo and zfh1, we first determined whether they regulate each other’s expression. We generated mosaic zfh1 clones using zfh165.34 and zfh175.26 hypomorphic alleles and analyzed Chinmo protein expression at 2 days pci (Broihier et al., 1998). In addition, we generated chinmo clones and examined Zfh1 protein expression at 3 days pci. Importantly, we found that Chinmo expression was not reduced in zfh1 clones of either allele and that Zfh1 expression was not altered in chinmo clones, indicating that these factors do not regulate each other’s expression (Fig. 6A,B, yellow arrowheads). In order to address if zfh1 and chinmo were epistatic one another, we assessed whether over-expression of Zfh1 could rescue the loss of CySCs lacking chinmo, and vice versa, using the MARCM technique. Over-expression of zfh1 in chinmo mutant somatic clones did not restore CySC characteristics to these clones (Fig. 6C, n=20 testes). Note that no somatic cells at the Hub are GFP+ in Fig. 6C. In fact, the only GFP+ cells residing in the niche are Tj− GSCs (Fig. 6C–C”’, pink arrowheads). These data indicate that chinmo does not act through zfh1. Unfortunately, due to technical limitations, we were unable to generate zfh1 mutant clones that overexpress chinmo and therefore cannot make conclusions about whether zfh1 acts through chinmo (Fig. 6E).
DISCUSSION
This study has revealed important information about a newly-identified JAK/STAT pathway effector gene, chinmo, and its role in Stat92E-dependent biological processes, including eye development, hematopoeisis and stem cell self-renewal. We identified chinmo as a cell-autonomously induced downstream mediator of JAK/STAT activity that shares loss- and gain-of-function phenotypes with Stat92E in several tissues. Although chinmo was originally identified in a screen for genes required for temporal identity of mushroom body neurons, no factors that regulate its expression had been identified (Zhu et al., 2006). The fact that chinmo and Stat92E exhibit a high degree of functional overlap suggests that chinmo performs multiple Stat92E-dependent functions, including growth of the eye disc, formation of melanotic tumors, proliferation of mature hemocytes, self-renewal of adult stem cells and repression of Ser. Furthermore, our results raise the interesting hypothesis that the JAK/STAT pathway is also required for the temporal identity of neurons in the mushroom body. We have also shown that Chinmo, like stabilized Stat92E, is expressed GSCs and in CySCs in the testis. However, unlike Stat92E, Chinmo is required intrinsically only for the self-renewal of CySCs and not of GSCs. These data clearly indicate that Stat92E acts through distinct effector genes in these stem cells to promote cell-autonomous self-renewal. Finally misexpression of chinmo in CySCs results in the expansion of GSCs and CySCs, a phenotype also observed with mis-expression of hopTum-l or of zfh1 in somatic cells (Leatherman and Dinardo, 2008). This provides additional evidence for the coordination of self-renewal and differentiation between adjacent GSCs and CySCs.
Chinmo as a negative regulator of gene expression
The BTB domain mediates protein–protein interactions, including dimerization, recruitment of transcriptional repressors to DNA, and protein degradation by acting as adaptors for Cul-3 E3 ubiquitin ligases (Perez-Torrado et al., 2006). In the antenna, we find that Ser is cell-autonomously repressed by both Stat92E and Chinmo (this study and (Flaherty et al., 2009)). These data suggest that Chinmo might act as a transcriptional repressor, at least in the antennal disc, and that Ser might be one of its transcriptional targets. It should be stressed that our results do not rule out the possibility that Chinmo can also act as an adaptor for Cul-3 and promote protein degradation. In fact, recent work has revealed that even BTB-ZF transcription factors like promyelocytic leukemia zinc finger (PLZF) can interact with Cul-3 (Furukawa et al., 2003). Whereas the role of the BTB domain in PLZF-dependent transcriptional repression has been well documented, the physiological role of the PLZF-Cul-3 interaction and the proteins it modifies are as yet unknown. Future experiments will be needed to determine if Chinmo can act both as a transcriptional repressor per se and as an adaptor for E3 ligases. In either scenario, factors modified by Chinmo, in addition to Ser, will need to be identified and characterized.
Chinmo and its role in CySC self-renewal
In this study, we show that Chinmo, like Zfh1, is essential for the self-renewal of CySCs in the Drosophila testis. Furthermore, we demonstrate that sustained expression of Chinmo in somatic cells, like that of Zfh1, is sufficient to induce the expansion of GSCs and CySCs. In addition, we prove that chinmo and zfh1 do not regulate each other’s expression. chinmo and zfh1 both appear to act downstream of Stat92E to maintain CySCs, which raises the possibility that these factors function either in an epistatic or parallel manner in the somatic lineage (this study and (Leatherman and Dinardo, 2008)). We show that chinmo does not act through zfh1 but were unable to determine if the reciprocal was true. However, we hypothesize that if zfh1 is upstream of chinmo then CySCs lacking either of these factors should differentiate at the same time point. In fact, CySCs lacking zfh1 differentiate faster than those lacking chinmo, suggesting that zfh1 may not reside upstream of chinmo (this study and (Leatherman and Dinardo, 2008)). Despite the unresolved genetic relationship between zfh1 and chinmo, our data are consistent with a model in which they function in a parallel pathway in the self-renewal of CySCs and the expansion of GSCs and CySCs (Fig. 6E).
Zfh1 has been shown to act as a transcriptional repressor (Fortini et al., 1991; Leatherman and Dinardo, 2008). Our data suggest that Chinmo may inhibit transcription directly or by post-translational modification of factors that silence genes. Zfh1 is expressed highly in CySCs and at low levels in early cyst cells. It is not expressed in late cyst cells. In contrast, Chinmo is expressed at high and comparable levels in CySCs and early cyst cells, but not in late cyst cells. Taking into account all of these results, we propose two models to explain the function of Chinmo in the somatic lineage of the testis. In the first, we hypothesize that Zfh1 and Chinmo regulate distinct downstream effectors, all of which are required for the maintenance of CySCs. In this model, early cyst cells, which express high levels Chinmo but low levels of Zfh1, can only become late cyst cells when Chinmo expression is sufficiently decreased there, allowing for full cyst cell differentiation and the complete development of mature germ cells. In the second model, Chinmo and Zfh1 regulate different genes critical for CySC self-renewal, but in contrast to the first, Chinmo only has a function in CySCs and not in early cyst cells. In this second model, we invoke the existence of co-factors expressed only in CySCs that act in concert with Chinmo, thereby restricting Chinmo function only to CySCs.
Chinmo and its non-autonomous effects on GSCs
Activation of the JAK/STAT signaling pathway in CySCs is sufficient to promote self-renewal of both CySCs and GSCs, indicating that CySCs can influence GSC maintenance (Leatherman and Dinardo, 2008). Although the mechanisms by which CySCs regulate GSC self-renewal have not yet been elucidated at the molecular level, two models have been proposed. Activated Stat92E, through its functional effectors, could (1) block CySC differentiation intrinsically, thus also inhibiting GSC differentiation at the same time, or (2) send one or more non-cell-autonomous signals from CySCs to GSCs, thus promoting GSC self-renewal. Either model would result in an increase in the number of CySCs and GSCs (Fig. 6E and (Gilboa, 2008; Leatherman and Dinardo, 2008)). In the first model, sustained activation of JAK/STAT signaling in CySCs allows these cells to continue proliferating. Therefore, they accumulate outside of the niche. Previous studies have shown that the mitoses of GSCs and CySCs must be linked in order to have the appropriate number of GSCs and their progeny always encapsulated by two CySCs/cyst cells (Leatherman and Dinardo, 2008; Sarkar et al., 2007; Schulz et al., 2002; Tazuke et al., 2002). In the second model, the self-renewal of GSCs depends on two independent signals: one is Upd sent from the niche, which activates Stat92E in adjacent GSCs, and the other is an unknown factor presumed to come from the neighboring CySCs. A similar situation occurs in female flies where two signals are required for GSC maintenance. First, cap cells, which form the ovarian niche, produce Decapentaplegic (Dpp), which acts on GSCs by inhibiting the bam gene (Xie and Spradling, 1998). Second, an Upd cytokine, produced by ovarian somatic support cells adjacent to the niche, acts on cap cells to increase Dpp production, thus influencing GSC self-renewal in a non-autonomous manner (Lopez-Onieva et al., 2008; Wang et al., 2008). It is currently unknown whether the non-autonomous signal from CySCs to GSCs in the testis is a secreted factor.
Our results indicate that Chinmo has an important role in the self-renewal of CySCs, in the inhibition of CySC differentiation and in the transduction of the non-autonomous signal from CySCs to GSCs. Furthermore, we show that this expansion requires the BTB and ZF domains in Chinmo, suggesting that the molecular function of Chinmo, in this process may be transcriptional repression and/or protein degradation. The fact that somatic misexpression of Chinmo, like that of Zfh1, can promote GSC self-renewal/expansion indicates that at least three factors play important roles in regulating stem cell self-renewal in a non-autonomous manner: activated Stat92E, Zfh1 and Chinmo. These observations raise many questions, such as the importance of JAK/STAT pathway activity in the soma, whether chinmo can bypass the requirement for Stat92E in CySCs and the mechanism of non-autonomous self-renewal between adjacent stem cell populations. These issues need to be addressed at the molecular level in the future.
Chinmo and its potential mammalian ortholog
A protein BLAST search against the non-redundant mouse database identified mZFP509 as a potential Chinmo ortholog. mZFP509 is a 757 amino acid protein that has the same overall structure as Chinmo: an N-terminal BTB domain located between residues 20–120 separated from two C-terminal C2H2 Zinc fingers by a stretch of ~400 amino acids (Fig. 6D). mZFP509 is 27% identical to Chinmo (with 36% identity in the BTB domain and 31% identity the ZF region) and is 83.7% identical to hZFP509 (UniGene). Micro-array studies indicate that mzfp509 is enriched in normal and cancer stem cells. mzfp509 transcripts are present in mESCs but are substantially reduced during their differentiation (Gissel et al., 2005; Hailesellasse Sene et al., 2007). They are also significantly increased in PU.1 deficient pre-leukemic hematopoietic stem cells and normal mammary stem cells (Behbod et al., 2006; Steidl et al., 2006). These data suggest that ZFP509 and Chinmo are orthologs and that what is discovered about Chinmo in Drosophila may have a high probability of holding true for its mammalian counterpart. For example, blocking hZFP509 function may have therapeutic value in inhibiting cancer stem cells, thus offering better outcomes for human patients.
EXPERIMENTAL PROCEDURES
Fly stocks
These stocks are described in FlyBase: Stat92E85C9; Stat92E397; zfh165.34; zfh175.26; chinmok13009 (chinmo-lacZ); chinmo1, UAS-mCD8-GFP, FRT40A /CyO; chinmoM33, FRT40A/CyO; ey-Gal4; nos-Gal4 VPI6; Hml-Gal4; UAS-2XEGFP; UAS-5’UTR chinmo 3’UTR/CyO; UAS-hop; GMR-upd. We used 10xSTAT-dGFP (Bach et al., 2007); eyaA3-Gal4 (Leatherman and Dinardo, 2008), Ser-lacZ (Bachmann and Knust, 1998).
Clonal analysis
We used Stat92E85C9 and Stat92E397, strong hypomorhic alleles (Silver and Montell, 2001), and chinmo1 and chinmoM33, a null and hypomorphic allele, respectively (Zhu et al., 2006). Similar phenotypes were observed in either Stat92E or chinmo allele. Mosaic Stat92E clones in the eye-antennal disc were generated using ey-FLP; FRT82B ubi-GFP/TM6B or hs-FLP122; FRT82B ubi-GFP/TM6B. Large Stat92E− M+ clones were generated using ey-flp; FRT82B M(3)96C ubi-GFP/TM6B. Mosaic chinmo clones were generated using hs-FLP, FRT40A 2xubi-GFP. Large chinmo− M+ clones were generated using ey-FLP; FRT40A M(2)24F arm-lacZ/CyO. For the clonal analyses in the testes we generated both positively- and negatively-marked clones using the MARCM and FLP/FRT techniques, respectively. To generate MARCM clones, we heat-shocked 1-day old adult male flies twice for 1 hour at 37°C. The flies were allowed to rest for at least 2 hours between heat shocks. To generate negatively-marked clones, we heat-shocked 1-day old adult male flies for 30 minutes or 1 hour at 37°C. In both analyses heat-shocked flies were then maintained at 25°C and were given new food every two days. Control and mutant clones were analyzed at 2 and 7 days pci. To address if CySCs mutant for chinmo underwent apoptosis, chinmo MARCM clones over-expressing the pan-caspase inhibitor p35 (Hay et al., 1994) were generated in these males: hs-FLP, UAS-gfp, tub-Gal4/Y; tub-Gal80 FRT40A/chinmo1 FRT40A; UAS-p35/+.
Gain-of-function (flip-out) clones were induced by heat-shocking larvae from [AyGAL4]25 [UAS-GFP.S65T]T2; MKRS hs-flp 86E/TM6B x UAS-hop or x UAS- 5’UTR-chinmo-3’UTR for 1 hour at 39°C (Ito et al., 1997).
Immunostaining
We used these antibodies: Mouse (Ms) anti-Fascilin III (Fas3) (1:50); Ms anti-Eya (1:20); Rat anti-DE-Cadherin (1:20) (all from the Developmental Studies Hybridoma Bank (DSHB)); Ms anti-1B1 (1:20); Ms anti-Bam (1:10); Rabbit (Rb) anti-Vasa (1:1000); Rb anti-Chinmo (1:1000); Rb anti-Zfh1 (1:5000); Guinea pig anti-Tj (1:3000); Rb anti-phospho-histone H3 (1:500) (Upstate Biotech); Ms anti-β-galactosidase (β-gal) (1:1000) (Sigma); Rb anti-Stat92E (1:1000); Rb anti-cleaved Caspase-3 (1:200) (Cell Signaling); Goat anti-Vasa (1:400) (Santa Cruz, #SC-26877). Secondary antibodies conjugated to FITC, Cy3, or Cy5 (Molecular Probes and Jackson Immunologicals) were used at 1:200. Testes were stained for 5 minutes with Hoechst 33342 (Sigma) at 1.0 mg/ml. Testes were dissected in 1x phosphate buffered saline (PBS), fixed for 15 minutes in 4% formaldehyde in 1xPBS, washed for 1 hour at 25°C in 1xPBS with 0.5% Triton-X 100, blocked in PBTB (1xPBS 0.2% Triton-X 100 and 1% bovine serum albumin) for 1 hour at 25°C. Primary antibodies were incubated overnight at 4°C. They were washed 2 times for 30 minutes in PBTB and incubated 2 hours in secondary antibody in PBTB at 25°C and then washed 2 times for 30 minutes in 1xPBS with 0.2% Triton-X 100. They were mounted in Vectashield (Vector Laboratories). Eye discs were processed as described in (Ekas et al., 2006). Images were captures on an LSM510 Zeiss confocal microscope.
Lymph gland analysis
Hml-Gal4, UAS-2XEGFP and UAS-5’UTR-chinmo-3’UTR/CyO flies were reared under standard conditions except for the misexpression of Chinmo in Hml-Gal4, UAS-2XEGFP cells, where animals were reared at 29°C for maximal Gal4 activity.
Third instar larvae were dissected and tissues were fixed in 4% formaldehyde in 1xPBS, pH 7.4 for 30 minutes, washed three times in 1xPBST (1xPBS with 0.4% Triton-X 100) for 15 minutes, blocked with 10% normal goat serum in 1xPBST for 30 minutes. Rb anti-Chinmo antibody was used at 1:100 in block and incubated with tissues overnight at 4°C. Tissues were washed as above, reblocked, and incubated with anti-rabbit Cy3 (Jackson Laboratories) at 1:200 overnight at 4°C or for 3 hours at room temperature. Tissues were washed two times in 1xPBST with TOPRO-3 (Invitrogen) added to the second wash at 1:1000, followed by two washes with 1xPBS to remove detergent. Samples were mounted in VectaShield. Images were captured using a Biorad Radiance 2000 confocal scanning system attached to a Zeiss AxioScope microscope.
Antibody generation
A peptide corresponding to the last 14 residues (GMADFDTITNFENF) of Stat92E (Chen et al., 2002) or one corresponding to the amino acids 142–155 (TGRRSVRNSLSGGS) of Chinmo (Zhu et al., 2006) was coupled to KLH and was injected into rabbits (by Proteintech Group, Inc or New England Peptide, respectively). Polyclonal antisera were affinity-purified and were found to be specific for Stat92E by ELISA, by Western blotting 3HA-Stat92E and by immuno-fluorescence of Stat92E397 clones, which lacks the epitope, and for Chinmo antibody by Western blotting endogenous and recombinant Chinmo (Fig. S3C and data not shown).
Supplementary Material
ACKNOWLEDGEMENTS
We thank A. Zamparini, R. Lehmann, J. Treisman, H.D. Ryoo, R. Dasgupta, S. DiNardo, E. Matunis, M. Fuller, L. Jones, D. McKearin, D. Godt, A. Bachmann, T. Lee and the Bloomington Stock Center for fly stocks and reagents. Several mAbs used in this study were obtained from the DSHB, The University of Iowa, Iowa City, IA 52242. We are indebted to M. Fuller, J. Morante-Oria and C. Desplan for sharing unpublished results. We thank A. Zamparini, J. Treisman, R. Dasgupta, S. Small, S. DiNardo, M. Fuller and D. Levy and members of the Bach, Dasgupta and Lehmann labs and two anonymous reviewers for helpful comments. We apologize to colleagues whose work was not cited to do space constraints. This project was supported in part by a Basil O’Connor Starter Scholar Research Award (Grant No.: 5-FY06) from the March of Dimes Foundation to EAB; by an NIH institutional training grant (T32 GM066704-03) to LAE; by a Research Scholar Grant (RSG-DDD-115829) from the American Cancer Society to EAB; and a National Institutes of Health grant from NIGMS (1R01GM085075) to EAB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors state no conflict of interests.
REFERENCES
- Arbouzova NI, Zeidler MP. JAK/STAT signalling in Drosophila: insights into conserved regulatory and cellular functions. Development. 2006;133:2605–2616. doi: 10.1242/dev.02411. [DOI] [PubMed] [Google Scholar]
- Ayala-Camargo A, Ekas LA, Flaherty MS, Baeg GH, Bach EA. The JAK/STAT pathway regulates proximo-distal patterning in Drosophila. Dev Dyn. 2007;236:2721–2730. doi: 10.1002/dvdy.21230. [DOI] [PubMed] [Google Scholar]
- Bach EA, Ekas LA, Ayala-Camargo A, Flaherty MS, Lee H, Perrimon N, Baeg GH. GFP reporters detect the activation of the Drosophila JAK/STAT pathway in vivo. Gene Expr Patterns. 2007;7:323–331. doi: 10.1016/j.modgep.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Bach EA, Vincent S, Zeidler MP, Perrimon N. A sensitized genetic screen to identify novel regulators and components of the Drosophila janus kinase/signal transducer and activator of transcription pathway. Genetics. 2003;165:1149–1166. doi: 10.1093/genetics/165.3.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann A, Knust E. Dissection of cis-regulatory elements of the Drosophila gene Serrate. Dev Genes Evol. 1998;208:346–351. doi: 10.1007/s004270050190. [DOI] [PubMed] [Google Scholar]
- Behbod F, Xian W, Shaw CA, Hilsenbeck SG, Tsimelzon A, Rosen JM. Transcriptional profiling of mammary gland side population cells. Stem Cells. 2006;24:1065–1074. doi: 10.1634/stemcells.2005-0375. [DOI] [PubMed] [Google Scholar]
- Boyle M, Wong C, Rocha M, Jones DL. Decline in self-renewal factors contributes to aging of the stem cell niche in the Drosophila testis. Cell Stem Cell. 2007;1:470–478. doi: 10.1016/j.stem.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Broihier HT, Moore LA, Van Doren M, Newman S, Lehmann R. zfh-1 is required for germ cell migration and gonadal mesoderm development in Drosophila. Development. 1998;125:655–666. doi: 10.1242/dev.125.4.655. [DOI] [PubMed] [Google Scholar]
- Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE., Jr Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- Buchon N, Broderick NA, Poidevin M, Pradervand S, Lemaitre B. Drosophila intestinal response to bacterial infection: activation of host defense and stem cell proliferation. Cell Host Microbe. 2009;5:200–211. doi: 10.1016/j.chom.2009.01.003. [DOI] [PubMed] [Google Scholar]
- Chao JL, Tsai YC, Chiu SJ, Sun YH. Localized Notch signal acts through eyg and upd to promote global growth in Drosophila eye. Development. 2004;131:3839–3847. doi: 10.1242/dev.01258. [DOI] [PubMed] [Google Scholar]
- Chen HW, Chen X, Oh SW, Marinissen MJ, Gutkind JS, Hou SX. mom identifies a receptor for the Drosophila JAK/STAT signal transduction pathway and encodes a protein distantly related to the mammalian cytokine receptor family. Genes Dev. 2002;16:388–398. doi: 10.1101/gad.955202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin SJ, Nehme NT, Limmer S, Liegeois S, Pospisilik JA, Schramek D, Leibbrandt A, Simoes Rde M, Gruber S, Puc U, et al. Genome-wide RNAi screen identifies genes involved in intestinal pathogenic bacterial infection. Science. 2009;325:340–343. doi: 10.1126/science.1173164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JE. Validating Stat3 in cancer therapy. Nat Med. 2005;11:595–596. doi: 10.1038/nm0605-595. [DOI] [PubMed] [Google Scholar]
- Davies EL, Fuller MT. Regulation of self-renewal and differentiation in adult stem cell lineages: lessons from the Drosophila male germ line. Cold Spring Harb Symp Quant Biol. 2008;73:137–145. doi: 10.1101/sqb.2008.73.063. [DOI] [PubMed] [Google Scholar]
- Decotto E, Spradling AC. The Drosophila ovarian and testis stem cell niches: similar somatic stem cells and signals. Dev Cell. 2005;9:501–510. doi: 10.1016/j.devcel.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Dominguez M, Casares F. Organ specification-growth control connection: new insights from the Drosophila eye-antennal disc. Dev Dyn. 2005;232:673–684. doi: 10.1002/dvdy.20311. [DOI] [PubMed] [Google Scholar]
- Dupuis S, Jouanguy E, Al-Hajjar S, Fieschi C, Al-Mohsen IZ, Al-Jumaah S, Yang K, Chapgier A, Eidenschenk C, Eid P, et al. Impaired response to interferon-alpha/beta and lethal viral disease in human STAT1 deficiency. Nat Genet. 2003;33:388–391. doi: 10.1038/ng1097. [DOI] [PubMed] [Google Scholar]
- Ekas LA, Baeg GH, Flaherty MS, Ayala-Camargo A, Bach EA. JAK/STAT signaling promotes regional specification by negatively regulating wingless expression in Drosophila. Development. 2006;133:4721–4729. doi: 10.1242/dev.02675. [DOI] [PubMed] [Google Scholar]
- Evans CJ, Hartenstein V, Banerjee U. Thicker than blood: conserved mechanisms in Drosophila and vertebrate hematopoiesis. Dev Cell. 2003;5:673–690. doi: 10.1016/s1534-5807(03)00335-6. [DOI] [PubMed] [Google Scholar]
- Flaherty MS, Zavadil J, Ekas LA, Bach EA. Genome-wide expression profiling in the Drosophila eye reveals unexpected repression of notch signaling by the JAK/STAT pathway. Dev Dyn. 2009;238:2235–2253. doi: 10.1002/dvdy.21989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortini ME, Lai ZC, Rubin GM. The Drosophila zfh-1 and zfh-2 genes encode novel proteins containing both zinc-finger and homeodomain motifs. Mech Dev. 1991;34:113–122. doi: 10.1016/0925-4773(91)90048-b. [DOI] [PubMed] [Google Scholar]
- Furukawa M, He YJ, Borchers C, Xiong Y. Targeting of protein ubiquitination by BTB-Cullin 3-Roc1 ubiquitin ligases. Nat Cell Biol. 2003;5:1001–1007. doi: 10.1038/ncb1056. [DOI] [PubMed] [Google Scholar]
- Gilboa L. A triumvirate at the testis tip: sharing the power of stem cell control. Dev Cell. 2008;15:5–6. doi: 10.1016/j.devcel.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Gissel C, Voolstra C, Doss MX, Koehler CI, Winkler J, Hescheler J, Sachinidis A. An optimized embryonic stem cell model for consistent gene expression and developmental studies: a fundamental study. Thromb Haemost. 2005;94:719–727. doi: 10.1160/TH05-05-0338. [DOI] [PubMed] [Google Scholar]
- Gonczy P, DiNardo S. The germ line regulates somatic cyst cell proliferation and fate during Drosophila spermatogenesis. Development. 1996;122:2437–2447. doi: 10.1242/dev.122.8.2437. [DOI] [PubMed] [Google Scholar]
- Gonczy P, Matunis E, DiNardo S. bag-of-marbles and benign gonial cell neoplasm act in the germline to restrict proliferation during Drosophila spermatogenesis. Development. 1997;124:4361–4371. doi: 10.1242/dev.124.21.4361. [DOI] [PubMed] [Google Scholar]
- Gregory L, Came PJ, Brown S. Stem cell regulation by JAK/STAT signaling in Drosophila. Semin Cell Dev Biol. 2008;19:407–413. doi: 10.1016/j.semcdb.2008.06.003. [DOI] [PubMed] [Google Scholar]
- Hailesellasse Sene K, Porter CJ, Palidwor G, Perez-Iratxeta C, Muro EM, Campbell PA, Rudnicki MA, Andrade-Navarro MA. Gene function in early mouse embryonic stem cell differentiation. BMC Genomics. 2007;8:85. doi: 10.1186/1471-2164-8-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanratty WP, Dearolf CR. The Drosophila Tumorous-lethal hematopoietic oncogene is a dominant mutation in the hopscotch locus. Mol Gen Genet. 1993;238:33–37. doi: 10.1007/BF00279527. [DOI] [PubMed] [Google Scholar]
- Hay BA, Wolff T, Rubin GM. Expression of baculovirus P35 prevents cell death in Drosophila. Development. 1994;120:2121–2129. doi: 10.1242/dev.120.8.2121. [DOI] [PubMed] [Google Scholar]
- Issigonis M, Tulina N, de Cuevas M, Brawley C, Sandler L, Matunis E. JAK-STAT signal inhibition regulates competition in the Drosophila testis stem cell niche. Science. 2009;326:153–156. doi: 10.1126/science.1176817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development. 1997;124:761–771. doi: 10.1242/dev.124.4.761. [DOI] [PubMed] [Google Scholar]
- Jiang H, Patel PH, Kohlmaier A, Grenley MO, McEwen DG, Edgar BA. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell. 2009;137:1343–1355. doi: 10.1016/j.cell.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung SH, Evans CJ, Uemura C, Banerjee U. The Drosophila lymph gland as a developmental model of hematopoiesis. Development. 2005;132:2521–2533. doi: 10.1242/dev.01837. [DOI] [PubMed] [Google Scholar]
- Kiger AA, Jones DL, Schulz C, Rogers MB, Fuller MT. Stem cell self-renewal specified by JAK-STAT activation in response to a support cell cue. Science. 2001;294:2542–2545. doi: 10.1126/science.1066707. [DOI] [PubMed] [Google Scholar]
- Krzemien J, Dubois L, Makki R, Meister M, Vincent A, Crozatier M. Control of blood cell homeostasis in Drosophila larvae by the posterior signalling centre. Nature. 2007;446:325–328. doi: 10.1038/nature05650. [DOI] [PubMed] [Google Scholar]
- Lambertsson A. The minute genes in Drosophila and their molecular functions. Adv Genet. 1998;38:69–134. doi: 10.1016/s0065-2660(08)60142-x. [DOI] [PubMed] [Google Scholar]
- Laron Z. Growth hormone insensitivity (Laron syndrome) Rev Endocr Metab Disord. 2002;3:347–355. doi: 10.1023/a:1020905725012. [DOI] [PubMed] [Google Scholar]
- Leatherman JL, Dinardo S. Zfh-1 controls somatic stem cell self-renewal in the Drosophila testis and nonautonomously influences germline stem cell self-renewal. Cell Stem Cell. 2008;3:44–54. doi: 10.1016/j.stem.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Levine RL. Janus kinase mutations. Seminars in oncology. 2009;36:S6–S11. doi: 10.1053/j.seminoncol.2009.02.005. [DOI] [PubMed] [Google Scholar]
- Li MA, Alls JD, Avancini RM, Koo K, Godt D. The large Maf factor Traffic Jam controls gonad morphogenesis in Drosophila. Nat Cell Biol. 2003;5:994–1000. doi: 10.1038/ncb1058. [DOI] [PubMed] [Google Scholar]
- Lin H, Yue L, Spradling AC. The Drosophila fusome, a germline-specific organelle, contains membrane skeletal proteins and functions in cyst formation. Development. 1994;120:947–956. doi: 10.1242/dev.120.4.947. [DOI] [PubMed] [Google Scholar]
- Lopez-Onieva L, Fernandez-Minan A, Gonzalez-Reyes A. Jak/Stat signalling in niche support cells regulates dpp transcription to control germline stem cell maintenance in the Drosophila ovary. Development. 2008;135:533–540. doi: 10.1242/dev.016121. [DOI] [PubMed] [Google Scholar]
- Luo H, Hanratty WP, Dearolf CR. An amino acid substitution in the Drosophila hopTum-l Jak kinase causes leukemia-like hematopoietic defects. Embo J. 1995;14:1412–1420. doi: 10.1002/j.1460-2075.1995.tb07127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T, Nakamura T, Nakao K, Arai T, Katsuki M, Heike T, Yokota T. STAT3 activation is sufficient to maintain an undifferentiated state of mouse embryonic stem cells. Embo J. 1999;18:4261–4269. doi: 10.1093/emboj/18.15.4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minegishi Y, Saito M, Tsuchiya S, Tsuge I, Takada H, Hara T, Kawamura N, Ariga T, Pasic S, Stojkovic O, et al. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature. 2007;448:1058–1062. doi: 10.1038/nature06096. [DOI] [PubMed] [Google Scholar]
- Morata G, Ripoll P. Minutes: mutants of drosophila autonomously affecting cell division rate. Dev Biol. 1975;42:211–221. doi: 10.1016/0012-1606(75)90330-9. [DOI] [PubMed] [Google Scholar]
- Newsome TP, Asling B, Dickson BJ. Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development. 2000;127:851–860. doi: 10.1242/dev.127.4.851. [DOI] [PubMed] [Google Scholar]
- Ostrin EJ, Li Y, Hoffman K, Liu J, Wang K, Zhang L, Mardon G, Chen R. Genome-wide identification of direct targets of the Drosophila retinal determination protein Eyeless. Genome Res. 2006;16:466–476. doi: 10.1101/gr.4673006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson ML, Freeman AF, Holland SM. Hyper IgE syndrome: an update on clinical aspects and the role of signal transducer and activator of transcription 3. Curr Opin Allergy Clin Immunol. 2008;8:527–533. doi: 10.1097/ACI.0b013e3283184210. [DOI] [PubMed] [Google Scholar]
- Perez-Torrado R, Yamada D, Defossez PA. Born to bind: the BTB protein-protein interaction domain. Bioessays. 2006;28:1194–1202. doi: 10.1002/bies.20500. [DOI] [PubMed] [Google Scholar]
- Pesu M, Candotti F, Husa M, Hofmann SR, Notarangelo LD, O'Shea JJ. Jak3, severe combined immunodeficiency, and a new class of immunosuppressive drugs. Immunol Rev. 2005;203:127–142. doi: 10.1111/j.0105-2896.2005.00220.x. [DOI] [PubMed] [Google Scholar]
- Reynolds-Kenneally J, Mlodzik M. Notch signaling controls proliferation through cell-autonomous and non-autonomous mechanisms in the Drosophila eye. Dev Biol. 2005;285:38–48. doi: 10.1016/j.ydbio.2005.05.038. [DOI] [PubMed] [Google Scholar]
- Sarkar A, Parikh N, Hearn SA, Fuller MT, Tazuke SI, Schulz C. Antagonistic roles of Rac and Rho in organizing the germ cell microenvironment. Curr Biol. 2007;17:1253–1258. doi: 10.1016/j.cub.2007.06.048. [DOI] [PubMed] [Google Scholar]
- Schulz C, Wood CG, Jones DL, Tazuke SI, Fuller MT. Signaling from germ cells mediated by the rhomboid homolog stet organizes encapsulation by somatic support cells. Development. 2002;129:4523–4534. doi: 10.1242/dev.129.19.4523. [DOI] [PubMed] [Google Scholar]
- Silver DL, Montell DJ. Paracrine signaling through the JAK/STAT pathway activates invasive behavior of ovarian epithelial cells in Drosophila. Cell. 2001;107:831–841. doi: 10.1016/s0092-8674(01)00607-9. [DOI] [PubMed] [Google Scholar]
- Steidl U, Rosenbauer F, Verhaak RG, Gu X, Ebralidze A, Otu HH, Klippel S, Steidl C, Bruns I, Costa DB, et al. Essential role of Jun family transcription factors in PU.1 knockdown-induced leukemic stem cells. Nat Genet. 2006;38:1269–1277. doi: 10.1038/ng1898. [DOI] [PubMed] [Google Scholar]
- Takeda K, Noguchi K, Shi W, Tanaka T, Matsumoto M, Yoshida N, Kishimoto T, Akira S. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc Natl Acad Sci U S A. 1997;94:3801–3804. doi: 10.1073/pnas.94.8.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazuke SI, Schulz C, Gilboa L, Fogarty M, Mahowald AP, Guichet A, Ephrussi A, Wood CG, Lehmann R, Fuller MT. A germline-specific gap junction protein required for survival of differentiating early germ cells. Development. 2002;129:2529–2539. doi: 10.1242/dev.129.10.2529. [DOI] [PubMed] [Google Scholar]
- Teglund S, McKay C, Schuetz E, van Deursen JM, Stravopodis D, Wang D, Brown M, Bodner S, Grosveld G, Ihle JN. Stat5a and Stat5b proteins have essential and nonessential, or redundant, roles in cytokine responses. Cell. 1998;93:841–850. doi: 10.1016/s0092-8674(00)81444-0. [DOI] [PubMed] [Google Scholar]
- Terry NA, Tulina N, Matunis E, DiNardo S. Novel regulators revealed by profiling Drosophila testis stem cells within their niche. Dev Biol. 2006;294:246–257. doi: 10.1016/j.ydbio.2006.02.048. [DOI] [PubMed] [Google Scholar]
- Tsai YC, J.G. Y, P.H. C, Posakony JW, Barolo S, Kim J, Henry Sun Y. Upd/Jak/STAT signaling represses wg transcription to allow initiation of morphogenetic furrow in Drosophila eye development. Dev Biol. 2007;306:760–771. doi: 10.1016/j.ydbio.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Tsai YC, Sun YH. Long-range effect of upd, a ligand for Jak/STAT pathway, on cell cycle in Drosophila eye development. Genesis. 2004;39:141–153. doi: 10.1002/gene.20035. [DOI] [PubMed] [Google Scholar]
- Tulina N, Matunis E. Control of stem cell self-renewal in Drosophila spermatogenesis by JAK-STAT signaling. Science. 2001;294:2546–2549. doi: 10.1126/science.1066700. [DOI] [PubMed] [Google Scholar]
- Van Doren M, Williamson AL, Lehmann R. Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr Biol. 1998;8:243–246. doi: 10.1016/s0960-9822(98)70091-0. [DOI] [PubMed] [Google Scholar]
- Wang L, Li Z, Cai Y. The JAK/STAT pathway positively regulates DPP signaling in the Drosophila germline stem cell niche. J Cell Biol. 2008;180:721–728. doi: 10.1083/jcb.200711022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie T, Spradling AC. decapentaplegic is essential for the maintenance and division of germline stem cells in the Drosophila ovary. Cell. 1998;94:251–260. doi: 10.1016/s0092-8674(00)81424-5. [DOI] [PubMed] [Google Scholar]
- Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- Yasugi T, Umetsu D, Murakami S, Sato M, Tabata T. Drosophila optic lobe neuroblasts triggered by a wave of proneural gene expression that is negatively regulated by JAK/STAT. Development. 2008;135:1471–1480. doi: 10.1242/dev.019117. [DOI] [PubMed] [Google Scholar]
- Zhu S, Lin S, Kao CF, Awasaki T, Chiang AS, Lee T. Gradients of the Drosophila Chinmo BTB-zinc finger protein govern neuronal temporal identity. Cell. 2006;127:409–422. doi: 10.1016/j.cell.2006.08.045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.