SUMMARY
The gastrointestinal tract harbors a diverse microbiota that has co-evolved with mammals for eons. Though most associations are symbiotic or commensal, some resident bacteria (termed pathobionts) have the potential to cause disease. Type VI secretion systems (T6SSs) are a novel mechanism for forging host-microbial interactions. We reveal a unique protective role for the T6SS of Helicobacter hepaticus, a Gram-negative bacterium of the intestinal microbiota. T6SS mutants display increased intracellular numbers within intestinal epithelial cells (IECs) and during intestinal colonization. Remarkably, T6SS directs an anti-inflammatory gene expression profile in IECs, and CD4+ T cells from T6SS mutant-colonized animals produce increased interleukin-17 (IL-17) in response to IECs presenting H. hepaticus antigens. These data demonstrate that H. hepaticus interacts with IECs and employs the T6SS to establish a balanced host relationship by limiting colonization and intestinal inflammation. We propose that altering host-bacterial equilibriums contribute to human disorders such as inflammatory bowel disease and colon cancer.
INTRODUCTION
The mammalian gastrointestinal (GI) tract represents an ecological niche of extraordinary microbial complexity (Hooper and Gordon, 2001). Humans and other mammals offer residence to ~1,000 microbial species which form an inter-kingdom bionetwork known to provide nutritional, metabolic and immunologic benefits to the host (Ley et al., 2006). Although the magnitude of this evolutionarily dynamic interaction is undeniably vast, surprisingly few molecules that mediate host-microbiota associations have been described. Microbial secretion systems are universal mechanisms dedicated to molecular interactions between bacterial and host cells, and can range from simple transporters to large, membrane-spanning structures. Type III secretion systems (T3SSs), T4SSs and T6SSs function as multi-subunit complexes that translocate effector substrates across the double membrane of Gram-negative bacteria, and act as a ‘needle and syringe’ to inject molecules directly into eukaryotic cells (Raskin et al., 2006). The temporal and spatial profile of secreted molecules can have profound effects on host biology. Therefore, these secretion systems represent well-evolved microbial strategies capable of mediating molecular interactions between the microbiota and mammals.
T6SS genes are present in over 25% of all sequenced bacterial genomes, and have been largely associated with bacterial pathogenesis. In recent seminal studies, Mekalanos and colleagues reported that Vibrio cholerae requires T6SS genes to cause cytotoxicity in macrophages (Pukatzki et al., 2006). A set of molecules were identified as the first T6SS substrates: Hcp (hemolysin co-regulated protein) and 3 VgrG (valineglycine-repeat) proteins. Further evidence supporting a role for T6SSs in bacterial virulence has been demonstrated in other Gram-negative pathogens including Pseudomonas aeruginosa (Mougous et al., 2006), enteroaggregative E. coli (Dudley et al., 2006), and Aeromonas hydrophila (Suarez et al., 2008). However, the function of bacterial T6SSs is not limited to eliciting disease. A growing body of research has shown that T6SSs may modulate virulence (Bladergroen et al., 2003; Das et al., 2002; Parsons and Heffron, 2005). Therefore in some contexts, T6SSs may modulate bacterial virulence in order to ‘balance’ the relationship between microbe and host.
T6SSs are almost exclusively found in α-, β-, and γ-proteobacteria (Bingle et al., 2008). Helicobacter hepaticus is unique in that it appears to contain the only bacterial genome in the epsilon (ε) subgroup of proteobacteria which encodes for a T6SS. H. hepaticus, a spiral microaerophilic bacterium, promotes inflammation in animal models of colon cancer and experimental colitis (inflammation of the colon). Interestingly, H. hepaticus only causes disease in immunocompromised animals that lack immune regulation and mount inflammatory responses toward intestinal bacteria (Erdman et al., 2009; Kullberg et al., 2001). Pathologies elicited by H. hepaticus remarkably resemble that of human disease based on molecular, cellular and histological parameters. Furthermore, colonization with H. hepaticus causes a chronic inflammatory response similar to human inflammatory bowel disease (IBD), in contrast to infections with Citrobacter rodentium or Salmonella typhimurium which induce acute and self-limiting infections. Therefore, the H. hepaticus-Mus musculus relationship represents a unique and invaluable tool in the study of intestinal pathologies highly similar to human disorders such as IBD and colon cancer.
Interestingly, multiple studies have shown that H. hepaticus sustains long-term colonization of the lower GI tract of wild-type mice without causing intestinal disease (reviewed in (Solnick and Schauer, 2001). In fact, H. hepaticus colonization is highly endemic in most laboratory animal facilities but is largely unnoticed due to the absence of clinical disease. We have previously proposed that H. hepaticus acts as an intestinal pathobiont— i.e., a symbiont that is able to promote pathology only when specific genetic or environmental conditions are altered in the host (Mazmanian et al., 2008; Round and Mazmanian, 2009). The concept of pathobionts is supported by clinical data which reveal that in IBD patients with underlying genetic mutations, inflammation is targeted to specific members of the microbiota and not to infectious pathogens (Packey and Sartor, 2009). T cell responses and antibody reactivity during IBD target certain subsets of symbiotic microbes such as Escherichia, Clostridium and Enterococcus species that are found in all people. Since H. hepaticus displays a potentially pathogenic association with its murine host similar to specific microbes found in humans, and the T6SS appears to mediate both symbiotic and pathogenic outcomes, we investigated a role for the H. hepaticus T6SS during intestinal inflammation.
Herein we report that the T6SS of H. hepaticus mediates critical protective functions during association with its mammalian host. In cell cultures, infection of intestinal epithelial cells (IECs) with H. hepaticus T6SS mutants results in increased bacterial association compared to wild-type bacteria. In animals, T6SS mutants colonize the lower GI tract to a higher degree. Most importantly, H. hepaticus defective in type VI secretion is unable to restrain potent innate and adaptive immune responses in an animal model of experimental colitis. Co-culture experiments with IECs presenting H. hepaticus antigens result in higher levels of pro-inflammatory cytokines when incubated with T cells from T6SS mutant-colonized animals compared to wild-type-colonized animals. Thus, our findings reveal that H. hepaticus has evolved a T6SS as a mechanism to actively maintain a non-pathogenic, symbiotic relationship in the GI tract by regulating bacterial colonization and host inflammation. Disturbances in the dynamic molecular interaction between gut bacteria and the intestinal immune system therefore lead to exacerbated host inflammation. As intestinal bacteria profoundly influence host biology, our findings support an emerging hypothesis that alteration in the composition of the microbiota, known as dysbiosis, is a critical factor in various human inflammatory disorders such as IBD and colon cancer.
RESULTS
H. hepaticus Possesses a Canonical and Functional T6SS
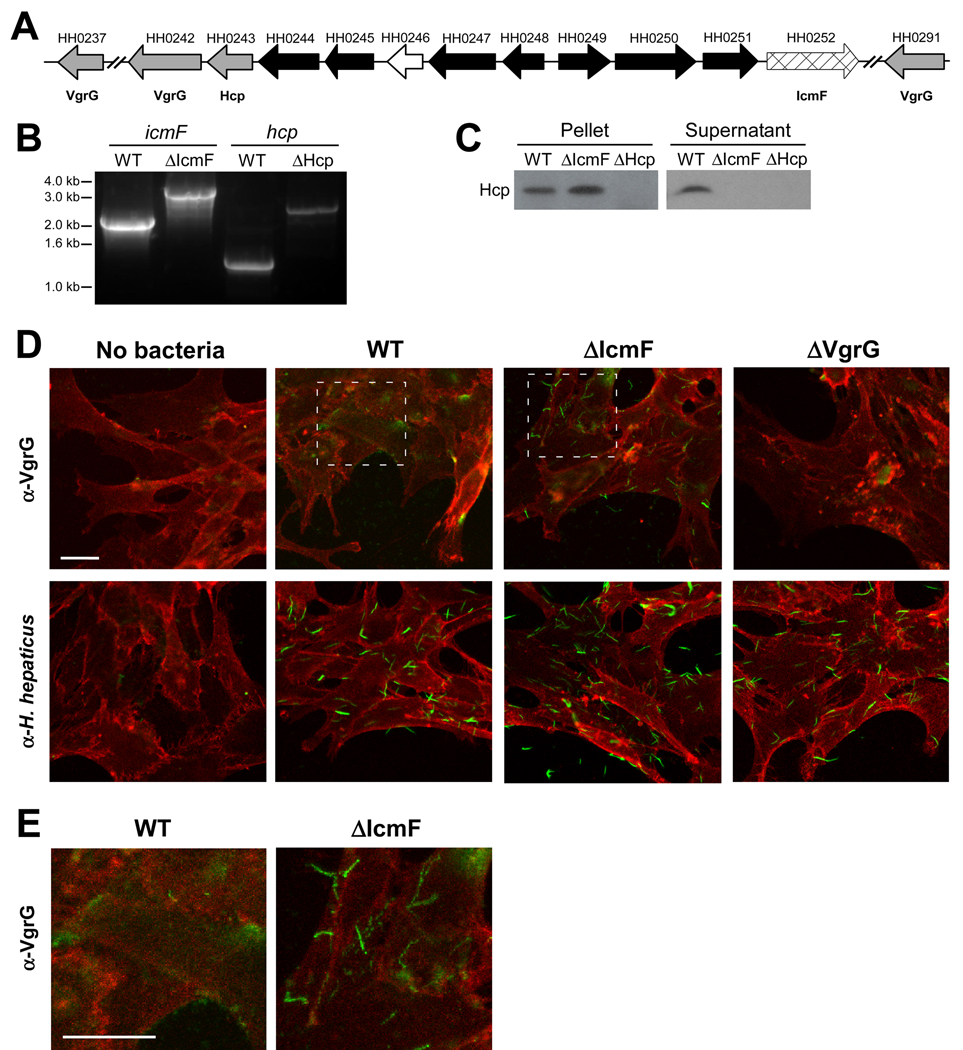
The genome of Helicobacter hepaticus contains a 71-kilobase genomic island (termed HHGI1 for Helicobacter hepaticus genomic island 1) which includes genes homologous to virulence factors (Suerbaum et al., 2003). Isolates missing the pathogenicity island are highly attenuated for inducing disease and deletion of a 19 gene segment of HHGI1 reduces virulence (Ge et al., 2008). However, the functions of the gene products encoded within HHGI1 remain entirely unknown. In recent years, studies have identified a novel protein secretion system referred to as a type VI secretion system (T6SS) (Raskin et al., 2006). We report herein that HHGI1 encodes for a set of T6SS genes that are homologous to those found in V. cholerae, P. aeruginosa, and other Gram-negative microorganisms (Table S1). H. hepaticus contains 12 homologous T6SS genes that are clustered and arranged in a genomic organization similar to other T6SSs (Figure 1A). A homolog of the icmF gene, thought to encode for a structural transmembrane protein of virtually all identified T6SSs, lies within this genomic locus (H. hepaticus gene HH0252). Furthermore, HHGI1 encodes for homologs of Hcp (HH0243) and 3 VgrG proteins (HH0237, HH0242, HH0291), highly conserved translocated substrates that play key roles in T6SSs.
Figure 1. H. hepaticus Encodes for a Functional T6SS.
(A) Schematic diagram of the genetic organization of H. hepaticus T6SS genes. Gray arrows represent Hcp and VgrG genes, cross-hatched arrow indicates IcmF homologue, black arrows represent T6SS homologues of unknown function, and white arrow indicates a gene non-homologous to other T6SS genes. See also Table S1.
(B) Genomic DNA collected from mid-log cultures of WT, ΔIcmF, or ΔHcp H. hepaticus was amplified by PCR using icmF or hcp specific primers. Insertion of the eryR gene was detected by a 1.1kb-increase in the resulting PCR band.
(C) Hcp is undetected in supernatants from ΔIcmF and ΔHcp T6SS mutants. Equal amounts of mid-log bacterial cultures of WT, ΔIcmF, or ΔHcp were centrifuged to separate bacterial pellets and supernatant. Supernatants were subsequently filtered to ensure removal of all bacteria. Bacterial pellets and supernatants were analyzed by Western blot. Membranes were blotted with anti-Hcp antibody.
(D, E) Confocal images of bacteria incubated with MODE-K cells. WT, ΔIcmF, or ΔVgrG (ΔHH0242) H. hepaticus were incubated with MODE-K cells for 5hr. MODE-K cells were subsequently rinsed with PBS, fixed in 4% PFA, and stained for the eukaryotic cell membrane marker wheat germ agglutinin (red) and either H. hepaticus or VgrG (green). Outlined regions for WT and ΔIcmF in (D) are shown at higher magnification (E). Scale bar represents 20µm.
We constructed insertional mutants in the icmF or hcp genes, as previous studies in several pathogens have shown that deletion of these genes leads to T6SS defects. An antibiotic selection marker (erythromycin acetyl transferase, eryR) (Mehta et al., 2007) was inserted within each open reading frame by homologous recombination. PCR amplification verified integration of the eryR gene to create mutants ΔIcmF and ΔHcp (Figure 1B). To validate a functional defect in the T6SS, in vitro cultured bacteria were assayed by immunoblot for Hcp. In wild-type bacteria, Hcp was detected in the culture supernatant and cell pellet fraction as expected (Figure 1C). However, in the ΔIcmF mutant, Hcp was absent from supernatant fractions and accumulated to a higher degree in the cell pellet, indicating Hcp is still produced but not secreted in the absence of a functional T6SS. Therefore, deletion of the icmF homolog results in a secretion defect for Hcp, demonstrating functional inactivation of the T6SS.
Bacteria possessing T6SSs bind host cells into which substrates can be injected. Since H. hepaticus colonizes the murine gut and is in close association with intestinal crypts (see data below), we investigated the association of H. hepaticus with murine intestinal epithelial cells (IECs). H. hepaticus was co-cultured with the mouse intestinal epithelial cell line MODE-K (Vidal et al., 1993), and examined for translocalization of VgrG by confocal immunofluorescence microscopy. Labeling of MODE-K cells with antisera specific for the T6SS substrate VgrG showed diffuse staining upon incubation with wild-type bacteria (Figures 1D and 1E). Conversely, VgrG labeling of MODE-K cells incubated with ΔIcmF mutant showed punctate staining patterns suggesting VgrG produced by T6SS mutants is bacterially-associated. As whole bacterial antisera raised against H. hepaticus reveals a punctate staining pattern as well (Figure 1D), the diffuse staining of VgrG in wild-type bacteria suggests substrate translocation in the presence of MODE-K cells (Figure 1E). Taken together, our results demonstrate that deletion of T6SS genes results in mutants unable to secrete effector substrates during bacterial growth and co-culture with IECs.
H. hepaticus is Internalized into Murine IECs
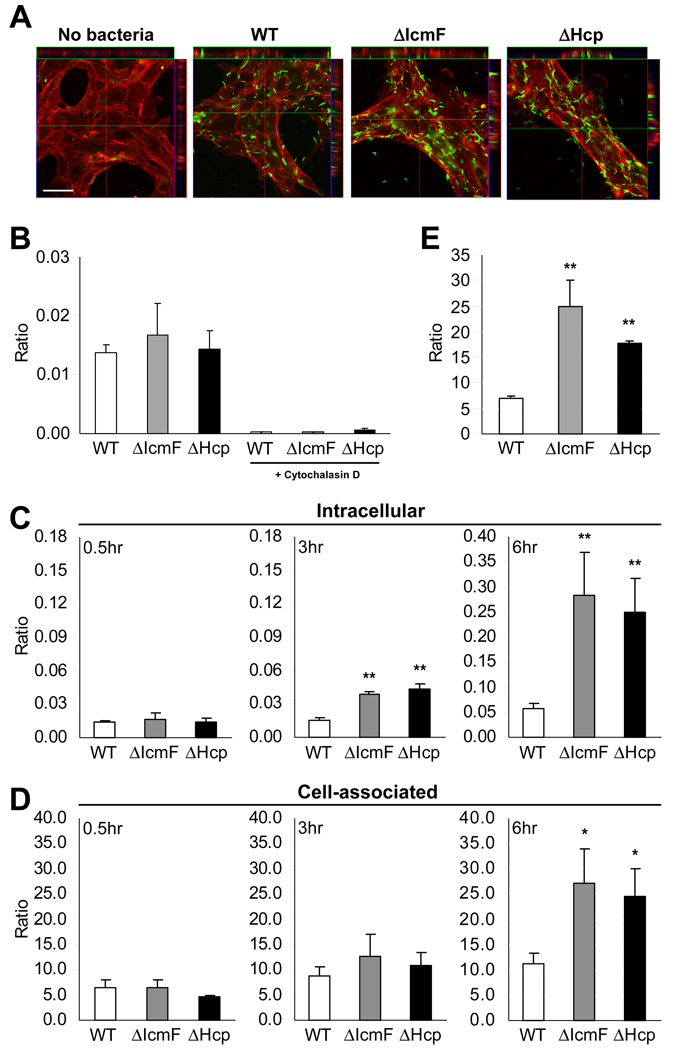
Many enteric pathogens such as Listeria monocytogenes and Shigella flexneri have been shown to enter IECs during infection. In several cases, T3SSs either facilitate entry or are required for internalization (Galan and Wolf-Watz, 2006). To determine if H. hepaticus enters IECs, we infected bacteria with MODE-K cells and observed that both wild-type and T6SS mutants were capable of entering host cells (Figure 2A). Confocal microscopy demonstrated that all strains were found associated with the cell surface as well. Z-stacks reconstructed from confocal images further revealed significant numbers of intact bacteria within the intracellular compartment of IECs (Movie S1). To determine if uptake was an active process, we incubated wild-type and T6SS mutants with MODE-K cells for 30 min in the presence or absence of cytochalasin D, an inhibitor of actin polymerization. Cells were subsequently treated with gentamicin to kill extracellular bacteria and plated for colony forming units (CFUs). Figure 2B reveals that while all strains could be recovered from the intracellular compartment of MODE-K cells, cytochalasin D inhibited bacterial uptake. Therefore, H. hepaticus appears to be actively internalized into cultured IECs through a requirement for actin rearrangement.
Figure 2. T6SS Mutants Display Higher Intracellular and Cell-associated Accumulation in MODE-K cells.
(A) Confocal image of bacteria inside MODE-K cells. WT, ΔIcmF, or ΔHcp was incubated with MODE-K for 6hr. MODE-K cells were rinsed with PBS, fixed in 4% PFA, and stained for H. hepaticus (green) and the eukaryotic cell membrane marker wheat germ agglutinin (red). Scale bar represents 30µm. See also Movie S1.
(B) Cytochalasin D inhibits uptake of H. hepaticus. Prior to incubation with bacteria, MODE-K cells were treated with 10µM cytochalasin D for 1hr. Bacteria were added at an MOI of 100. After 0.5hr incubation at 37°C under mi croaerophilic conditions, cells were treated with 100µg/ml gentamicin, and intracellular bacteria plated for enumeration. Results are expressed as colony-forming units (CFUs) of intracellular bacteria divided by number of MODE-K cells. Error bars indicate SEM from 3 experiments.
(C, D) Gentamicin protection assay in which MODE-K cells were incubated with bacteria at an MOI of 100. After 0.5, 3, or 6hr incubation, media was replaced with 100µg/ml gentamicin for enumeration of intracellular bacteria (C) or without gentamicin for cell-associated bacteria (D). Cells were washed and bacteria plated for quantification. Ratios are expressed as CFUs of bacteria divided by number of MODE-K cells. Error bars indicate SEM from 3–5 experiments. *p<0.05, **p<0.01 vs WT.
(E) Increased adherence of T6SS mutants is not dependent on bacterial internalization. Prior to co-culture, MODE-K cells were treated with 10µM cytochalasin D. Bacteria were added at an MOI of 100 for 6hr at 37°C under microa erophilic conditions. Bacteria were plated for enumeration. Results are expressed as CFUs of bacteria divided by number of MODE-K cells. Error bars indicate SD from 2 experiments. **p<0.01 vs WT. See also Figure S1.
T6SS Mutants Display Increased Cell Association within IECs
We sought to determine if the T6SS affects bacterial internalization. MODE-K cells were incubated with H. hepaticus, treated with (or without) gentamicin, and plated for bacterial enumeration. At early time points (30 min), no differences were observed in the proportions of intracellular and cell-associated bacteria between wild-type and T6SS mutants (Figures 2C and 2D). Remarkably, by 3 hr and 6 hr, ΔIcmF and ΔHcp had significantly higher levels of intracellular bacteria (Figure 2C). In addition, T6SS mutants displayed greater numbers of MODE-K-associated bacteria after 6 hr (Figures 2D). Total numbers of bacteria recovered from co-cultures (which includes all non-cell associated bacteria) as well as cultures grown in the same media without MODE-K cells were comparable (Figure S1A and data not shown), demonstrating that the mutations did not affect bacterial growth. These results reveal that the T6SS of H. hepaticus limits intracellular bacterial numbers within IECs.
To investigate if the increased cell association by T6SS mutants was due to increased adherence, we blocked internalization of H. hepaticus into MODE-K cells. Cytochalasin D was added for the duration of the co-culture (6 hr). In the absence of bacterial internalization, T6SS mutants still exhibited an increase in adherence to the extracellular surface of MODE-K cells (Figure 2E). An increase in cell adherence in the absence of internalization was also observed for T6SS mutants when MODE-K cells were fixed with 4% paraformaldehyde prior to co-incubation (data not shown). Despite the increase in MODE-K adherence, the T6SS mutants did not affect the viability of MODE-K cells compared to wild-type bacteria. Staining for MODE-K cell apoptosis with Annexin V and propidium iodide showed that neither wild-type nor T6SS mutants had a significant effect on IEC viability (Figure S1B). Our studies are consistent with previous reports (for other bacteria) suggesting that T6SSs limit entry of additional microorganisms once bacteria have already gained access to the host cytoplasm (Ma et al., 2009).
T6SS Mutants Display Increased Colonization of the Murine Intestine
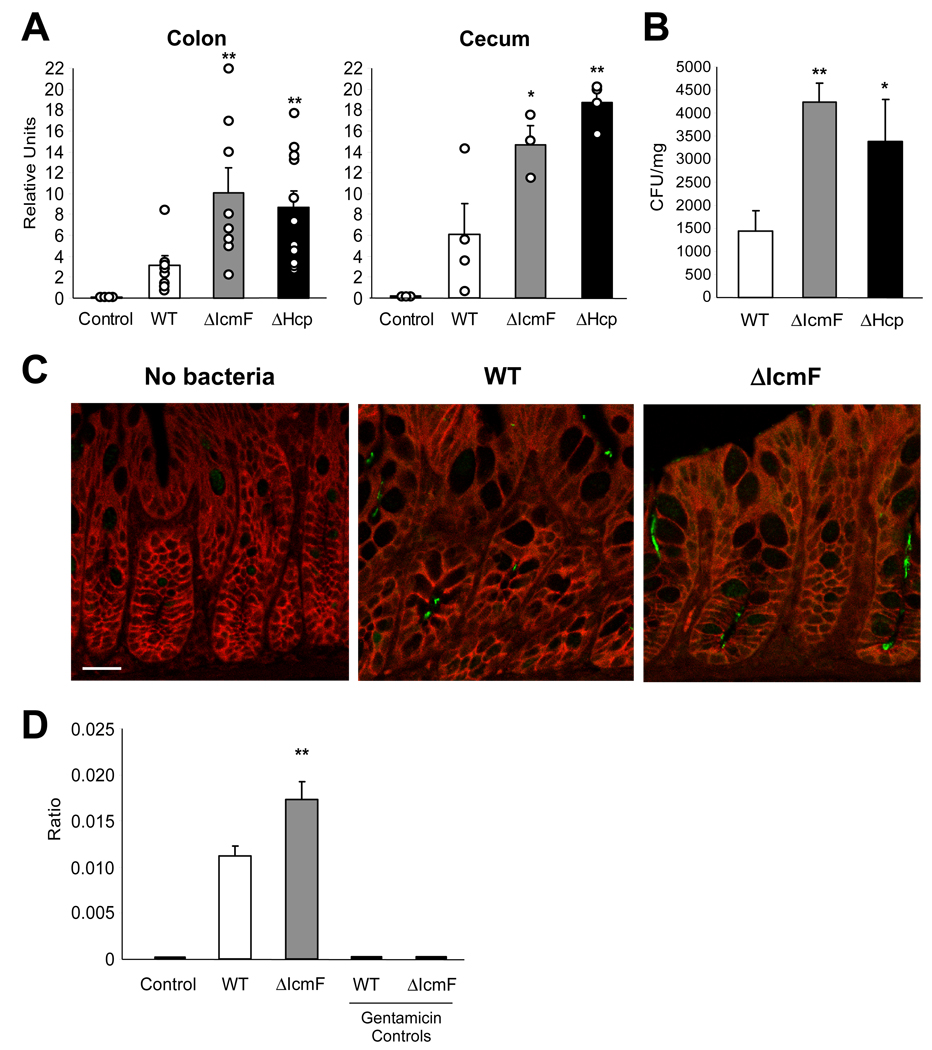
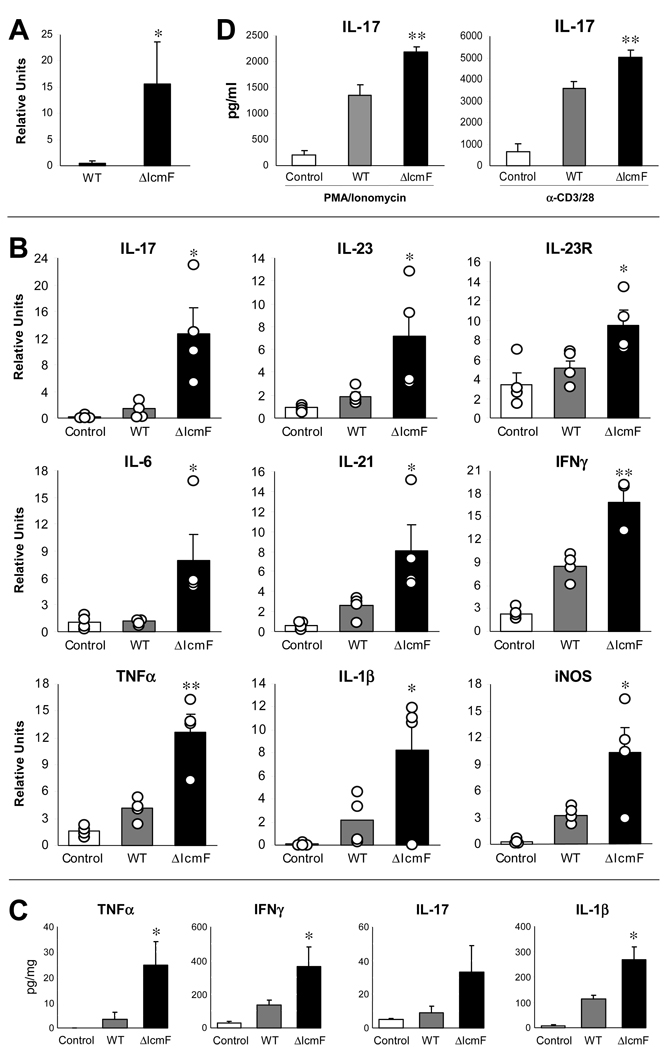
Germ-free animals (devoid of all microorganisms from birth) were employed to measure in vivo intestinal colonization. Mice were mono-colonized with wild-type, ΔIcmF, or ΔHcp H. hepaticus (ranging from 2–13 weeks of colonization), at which time RNA was isolated from colonic tissue. Colonization levels were determined by quantitative RT-PCR (qRT-PCR) for H. hepaticus-specific 16S ribosomal RNA, as previously described (Nanda Kumar et al., 2008). Interestingly, RNA extracted from the ceca and colons of mono-colonized animals revealed that H. hepaticus colonization by ΔIcmF and ΔHcp mutants was increased compared to wild-type (Figure 3A). In vitro analysis of RNA (and DNA) levels demonstrated that mutating the T6SS does not affect 16S rRNA expression (Figure S2A). Accordingly, plating for viable bacterial CFUs from homogenized colonic tissue confirmed significantly elevated levels of colonization for T6SS mutants (Figure 3B). We observed a similar increase for T6SS mutants following colonization for up to 3 months in two strains of germ-free mice, outbred Swiss Webster and inbred C57Bl/6 mice (data not shown). Images of colon sections from mono-colonized animals showed that both wild-type and ΔIcmF were found intimately associated with host cells (Figure 3C). Consistent with previous reports for wild-type H. hepaticus (Kullberg et al., 2002), we did not observe any cases of intestinal disease in colonized animals. Examination of colonic tissues (e.g. epithelial hyperplasia, infiltration of leukocytes, crypt abscesses) revealed no signs of histopathology (Figure S2B and data not shown). qRT-PCR analysis of colonic tissues for pro-inflammatory cytokines showed comparable levels of tumor necrosis factor-α (TNFα) and interferon-γ (IFNγ) between germ-free and mono-colonized animals (Figure S2C). In addition, the percentage of CD4+TCRβ+ cells in the mesenteric lymph nodes (MLNs) did not differ between groups (Figure S2D).
Figure 3. T6SS Mutants Have Increased Colonization Levels within the Colon.
(A) ΔIcmF- and ΔHcp-mono-colonized animals have greater amounts of H. hepaticus 16S rRNA in the colon and cecum compared to WT-mono-colonized animals. RNA was collected from colon and cecum. Levels of 16S were quantified by qRT-PCR using H. hepaticus-specific 16S primers. Error bars show SEM, n=4–11 animals per group. *p<0.05, **p<0.01 vs WT. See also Figure S2A.
(B) ΔIcmF- and ΔHcp-mono-colonized animals have higher levels of viable H. hepaticus in the colon compared to WT-mono-colonized animals. Colon tissues were homogenized in BHI and plated for quantification. Bacterial numbers were normalized to colon tissue weights. Units are expressed as CFUs per mg of tissue. Error bars show SEM from n=4 animals per group. *p<0.05, **p<0.01 vs WT.
(C) H. hepaticus is found in colonic intestinal crypts. Colons from WT- and ΔIcmF-mono-colonized mice were paraffin-embedded and sectioned. Colon sections were stained for H. hepaticus (green) and E-cadherin (red). Animals were colonized for 8 weeks. Scale bar represents 20µm. See also Figures S2B–D.
(D) ΔIcmF-colonized animals have increased levels of intracellular H. hepaticus compared to WT-colonized animals. Purified IECs were treated with gentamicin either prior to or following lysis with saponin (which selectively permeablizes eukaryotic membranes). Bacteria were enumerated by plating on TVP (trimethoprim, vancomycin, polymyxin B) plates which are known to select for H. hepaticus, as confirmed by control mice which received no H. hepaticus. In ‘Gentamicin Controls’ IECs were lysed and then treated with gentamicin. Ratio is expressed as CFUs of bacteria divided by number of IECs. Error bars indicate SEM from n=12 animals per group. **p<0.01 vs WT. See also Figure S2E.
To investigate the intracellular association of H. hepaticus within IECs in vivo, we colonized SPF (specific pathogen free) C57Bl/6 mice with wild-type, ΔIcmF or no H. hepaticus. Colonic IECs were purified and treated with gentamicin either prior to or following IEC lysis. Internalized bacteria were enumerated as the gentamicin resistant population. Interestingly, ΔIcmF showed increased intracellular numbers in IECs compared to wild-type bacteria (Figure 3D). This increase in bacterial internalization by ΔIcmF was also observed in IECs recovered from the colons and ceca of C57Bl/6 Rag1−/− and IL-10−/− mice (Figure S2E). IECs lysed with saponin prior to incubation with gentamicin showed no viable H. hepaticus bacteria, demonstrating the complete killing of bacteria (Figure 3D). Our findings reveal that contrary to most secretion systems of enteric bacteria which promote infections, the H. hepaticus T6SS limits bacterial numbers during colonization of the mouse intestine.
The T6SS Directs an Anti-inflammatory Gene Expression Profile in IECs
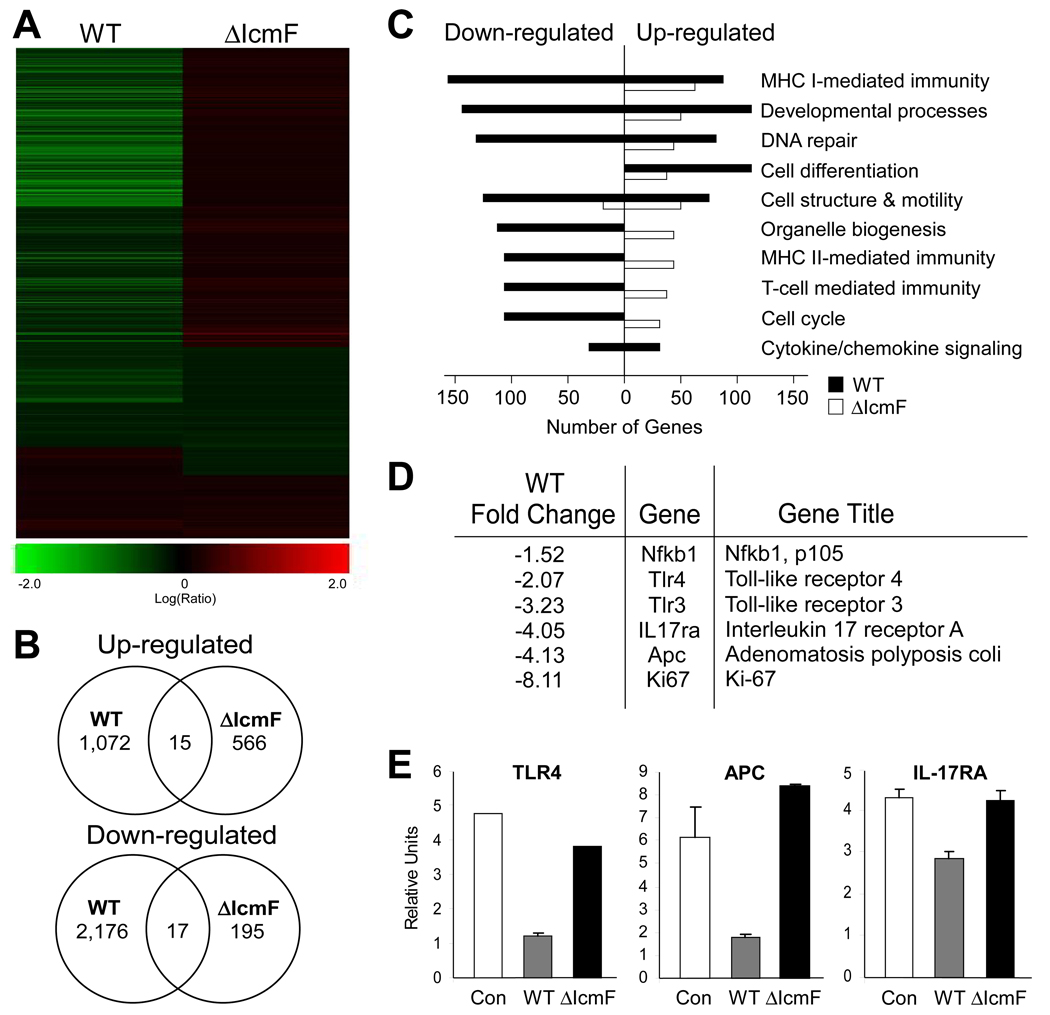
Based on the evidence from these studies of T6SS mutants, it appears that H. hepaticus intimately interacts with IECs. To appreciate the magnitude of this interaction, we examined the gene expression profile of MODE-K cells incubated with either wild-type or ΔIcmF bacteria. GeneChip analysis showed clear down-regulation of numerous transcripts in MODE-K cells co-cultured with wild-type bacteria possessing a functional T6SS (Figures 4A and 4B). Examination of various gene families exhibited an unmistakable pattern of transcriptional repression by wild-type H. hepaticus, including many immune-related genes (Figure 4C). Previous studies have shown H. hepaticus reduces activation of innate immune molecules such as Toll-like receptor 4 (TLR4) in IECs (Sterzenbach et al., 2007). We reveal by both microarray and qRT-PCR analysis of MODE-K RNA that suppression of TLR4 expression by H. hepaticus is T6SS-mediated (Figures 4D and 4E). Moreover, wild-type bacteria reduced the transcript levels for several additional mediators of innate (NF-κB, TLR3) and adaptive immunity (interleukin-17 receptor; IL-17RA). H. hepaticus promotes colon cancer in animal models; the molecular mechanisms underlying this outcome are unknown. Intriguingly, our data show that T6SS may serve a protective activity in the development of colorectal cancer, as a critical oncogene in hereditary colon cancers (Adenomatosis polyposis coli; APC) and a marker for cell proliferation during epithelial hyperplasia (Ki-67) are significantly reduced by wild-type bacteria (Figures 4D and 4E). The extent of the contribution by T6SS to H. hepaticus-mediated cancer remains to be determined. Collectively, these studies strongly support a model whereby the T6SS of H. hepaticus functions to reduce immune responses elicited by IECs during bacterial colonization.
Figure 4. Wild-type H. hepaticus Induces a Wide Repertoire of Responses in MODE-K cells.
RNA was harvested from MODE-K cells incubated for 6hr with either wild-type H. hepaticus, ΔIcmF mutant, or no bacteria and analyzed by an Agilent Whole Mouse Genome Microarray. Gene expression of MODE-K cells incubated with bacteria was compared to transcript levels from RNA of untreated MODE-K cells. Only genes with a p-value <0.5 and fold change >1.5 were used for subsequent analysis.
(A) Heat-map analysis of MODE-K gene expression in the presence of WT or ΔIcmF shows massive down-regulation by wild-type H. hepaticus.
(B) Venn diagram showing up- and down- regulation of gene expression in the presence of WT or ΔIcmF.
(C) Gene ontogeny analysis of changes in MODE-K transcript levels for various functional groups. Wild-type bacteria down-regulate numerous cellular pathways.
(D) Fold change of select innate and adaptive immune genes in the presence of WT H. hepaticus.
(E) qRT-PCR analysis of RNA from MODE-K cells for genes associated withinflammation and colon cancer. Error bars show SEM.
H. hepaticus T6SS Mutant Elicits Elevated Intestinal Inflammatory Responses
During homeostasis, a defined balance exists among commensal, symbiotic, and pathobiotic bacteria in the intestinal microbiota. We and others have hypothesized that disturbances in this equilibrium may lead to intestinal disorders in humans, reviewed in (Packey and Sartor, 2009; Round and Mazmanian, 2009). Dysbiosis can result from an overgrowth of pathobionts or a loss of beneficial commensal bacteria, leading to elevated host inflammation toward the microbiota. We therefore tested this hypothesis through association of animals with T6SS mutants to induce dysbiosis by experimentally increasing colonization of a defined pathobiont.
The T cell transfer model of experimental colitis highlights the intricate interaction between mice and H. hepaticus. In this model, T and B cell deficient Rag1−/− mice are reconstituted with naïve, pathogenic CD4+CD45Rbhigh T cells to provide immune cells that can react to the microbiota. In the absence of H. hepaticus, SPF T cell recipient animals remain healthy. However, in the presence of H. hepaticus, mice develop a profound inflammatory response in the colon (Mazmanian et al., 2008). To test the effects of T6SS, we colonized T cell reconstituted Rag1−/− mice with wild-type or ΔIcmF strains of H. hepaticus. Initially, we examined colonization levels and again observed a significant increase by the T6SS mutant (Figure 5A). Most importantly, colonization with ΔIcmF led to higher levels of intestinal inflammation compared to wild-type (Figure 5B). qRT-PCR of RNA from colons revealed that animals colonized with the T6SS mutant displayed considerably increased cytokine transcript levels of numerous innate and adaptive immune responses. Many cytokines associated with the inflammatory T-helper 17 (TH17) arm of immunity were increased in response to ΔIcmF—IL-17, IL-21, IL-23, and IL-23R—suggesting the T6SS may help in controlling exaggerated TH17 responses toward bacteria. The pro-inflammatory cytokine TNF-α and inducible nitric oxide synthase (iNOS) were also significantly increased in response to the T6SS mutant. TNF-α and iNOS are highly expressed during H. hepaticus-induced colon inflammation and carcinogenesis, and are believed to be strong contributors to disease (Erdman et al., 2009). Interestingly, there was no marked difference in intestinal pathology by measures of cell proliferation, cellular infiltrates, and abscess formation between wild-type and ΔIcmF mutant-colonized animals (Figure S3A). Histopathology analysis by a blinded pathologist verified similar colitis scores between wild-type and mutant-colonized animals (Figure S3B). Although more subtle phenotypes cannot be excluded, the lack of increased disease by T6SS is not surprising given the fact that wild-type bacteria elicit very pronounced disease in the T cell transfer model. To measure inflammatory protein levels, organ cultures in which un-stimulated colon sections are cultured ex vivo, and supernatants assayed by ELISA showed an increase in the inflammatory molecules TNF-α, IL-1β and IL-17 in tissues harvested from ΔIcmF mutant-colonized animals compared to wild-type (Figure 5C). Furthermore, MLN cells from ΔIcmF-colonized animals that were re-stimulated in vitro with PMA/ionomycin or T cell stimuli (α-CD3/α-CD28) released increased IL-17 during in vitro cultures (Figure 5D). Analysis of the ceca also showed similar patterns of increased T6SS mutant colonization and elevated proinflammatory cytokine responses (Figures S3C–S3E). Collectively, these results reveal that experimentally induced dysbiosis results in increased inflammatory responses from both the innate and adaptive immune system, and the T6SS of H. hepaticus functions to reduce intestinal inflammation during colonization.
Figure 5. T6SS Mutant Leads to Greater Inflammation in the Colon in the Rag Adoptive Transfer Model of Colitis.
(A) ΔIcmF-colonized animals have greater amounts of H. hepaticus 16S rRNA in the colon compared to WT-colonized animals. RNA was collected from colons. Levels of 16S were quantified by qRT-PCR. Data are representative of 3 experiments, with n=4 per group. Error bars show SEM. *p<0.05 vs WT.
(B) RNA from the colon was analyzed by qRT-PCR for inflammatory cytokines. Experimental values were normalized against L32. Results are representative of 5 experiments, with n=4 per group. Each circle represents an individual animal. Error bars show SEM. *p<0.05, **p<0.01 vs WT. See also Figure S3.
(C) Colons were cultured ex vivo for 24hrs. Supernatants were assayed for cytokine by ELISA. Samples were normalized to total protein. Data are representative of 2 experiments, with n=4 per group. Each circle represents an individual animal. Error bars show SEM. *p<0.05 vs WT.
(D) Mesenteric lymph nodes pooled from each experimental group were restimulated with either PMA and ionomycin, or α-CD3 and α-CD28 for 24hrs. Supernatants were assayed by ELISA. Results are from 2 experiments, with n=4 mice per group. Error bars show SEM. **p<0.01 vs WT.
IECs are Capable of Stimulating CD4+ T cell Responses with H. hepaticus Antigen
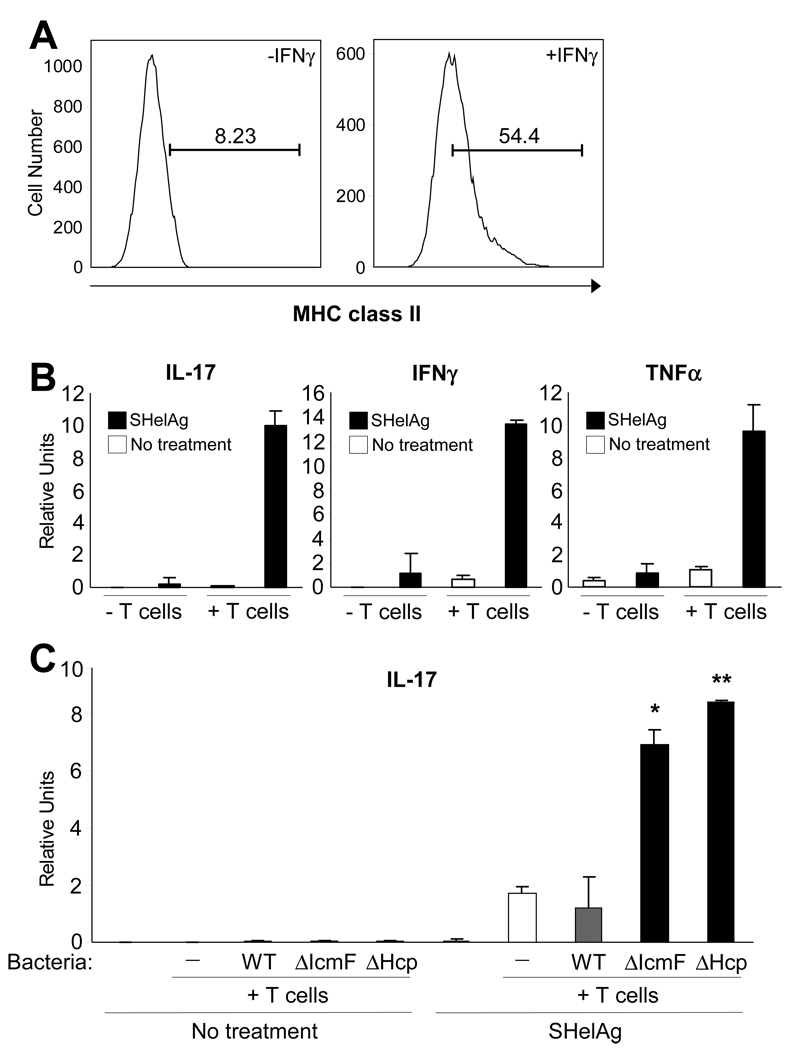
In the GI tract, IECs form a monolayer barrier that separates luminal contents from underlying host cells. Though IECs contribute to innate immunity by sensing through TLRs and secreting antimicrobial peptides, IECs are generally not believed to mediate adaptive immunity, unlike professional antigen-presenting cells (APCs) such as dendritic cells and macrophages. Interestingly, studies have shown that IECs are capable of expressing major histocompatibility complex (MHC) class II proteins (Bland and Warren, 1986; Mayer et al., 1991), and IEC presentation of antigens to CD4+ T cells via MHC class II molecules can result in lymphocyte proliferation (Westendorf et al., 2009; Vidal et al., 1993). MODE-K cells increase MHC class II expression in response to IFNγ treatment, when stained with antibodies directed against the I-Ak haplotype (Figure 6A) or to a non-polymorphic region of the I-A molecule (Figure S4). The T cell activating co-stimulatory molecules CD80 (B7.1) and CD86 (B7.2) were also increased (data not shown). To determine if IECs could present H. hepaticus antigens to T cells, MODE-K cells treated for 7 days with IFNγ were pulsed with soluble Helicobacter antigens (SHelAg). After 24hrs, MODE-K cells were washed and co-cultured with CD4+ T cells from MLNs of SPF Helicobacter-free C3H/HeJ animals. Total RNA was analyzed by qRT-PCR for cytokine levels. Co-cultures that had been treated with SHelAg showed an increase in numerous pro-inflammatory cytokine transcripts, such as IL-17, IFNγ, and TNFα (Figure 6B). These responses are T cell-specific, as no cytokine production was observed in the absence of T cells.
Figure 6. MODE-K Cells are Capable of Stimulating CD4+ T cell Responses with H. hepaticus Antigens.
(A) MODE-K cells were treated with 100U/ml of IFNγ for 7 days or left untreated. Cells were removed from the plate with trypsin, stained for MHC class II antigens, and analyzed by flow cytometry. See also Figure S4.
(B) MODE-K cells pre-treated with 100U/ml of IFNγ for 7 days were pulsed with either 20 µg/ml of soluble H. hepaticus antigen (SHelAg) from wild-type bacteria or no SHelAg for 24hrs. Media was changed prior to addition of CD4+ T cells harvested from syngeneic C3H/HeJ Helicobacter-free animals or no T cells. The ratio of MODE-K:T cells was 1:10. Cells were co-cultured for 3d. RNA was collected from total cells and analyzed by qRT-PCR. Error bars indicate SD from 2 independent experiments.
(C) MODE-K cells pre-treated with IFNγ were pulsed with either 20 µg/ml of SHelAg from wild-type bacteria or no SHelAg for 24hrs. Media was changed prior to addition of CD4+ T cells collected from animals colonized with wild-type, ΔIcmF, ΔHcp, or no H. hepaticus. The ratio of MODE-K:CD4+ T cells was 1:10. Cells were co-cultured for 3d. RNA was collected from total cells and analyzed by qRT-PCR. Error bars indicate SD from 2 independent experiments. *p<0.05, **p<0.01 vs WT treated with SHelAg.
Antigen-specific CD4+ T cell Responses are Increased in Animals Colonized with H. hepaticus T6SS Mutants
We investigated the novel hypothesis that IEC presentation of H. hepaticus antigens elicits increased pro-inflammatory responses from CD4+ T cells from animals colonized with T6SS mutants. MODE-K cells were pulsed with SHelAg or left untreated, washed and incubated with purified CD4+ T cells from mice colonized with either wild-type, ΔIcmF, ΔHcp or no H. hepaticus. In the presence of SHelAg, T cells from uncolonized and wild-type H. hepaticus-colonized animals produced comparable levels of IL-17 (Figure 6C). This illustrates that previous exposure to H. hepaticus antigens does not augment immune responses relative to naïve animals, consistent with our findings that wild-type bacteria do not induce inflammation. In contrast, CD4+ T cells harvested from both T6SS mutant-colonized animals elicited significantly increased levels of IL-17 compared to cells from wild-type-colonized animals. This response was antigen-specific, as no cytokine was produced in the absence of SHelAg. Therefore, colonization of animals with T6SS mutants leads to the generation of an increased Th17 cell response in the gut, revealing that H. hepaticus evolved this molecular mechanism to restrain unwanted intestinal inflammation. As Th17 cell responses are important mediators of IBD and colon cancer in experimental animals (Hue et al., 2006), our data suggest that increased gut inflammation in reaction to dysbiosis of the microbiota may be crucial in the onset and/or progression of human intestinal diseases.
DISCUSSION
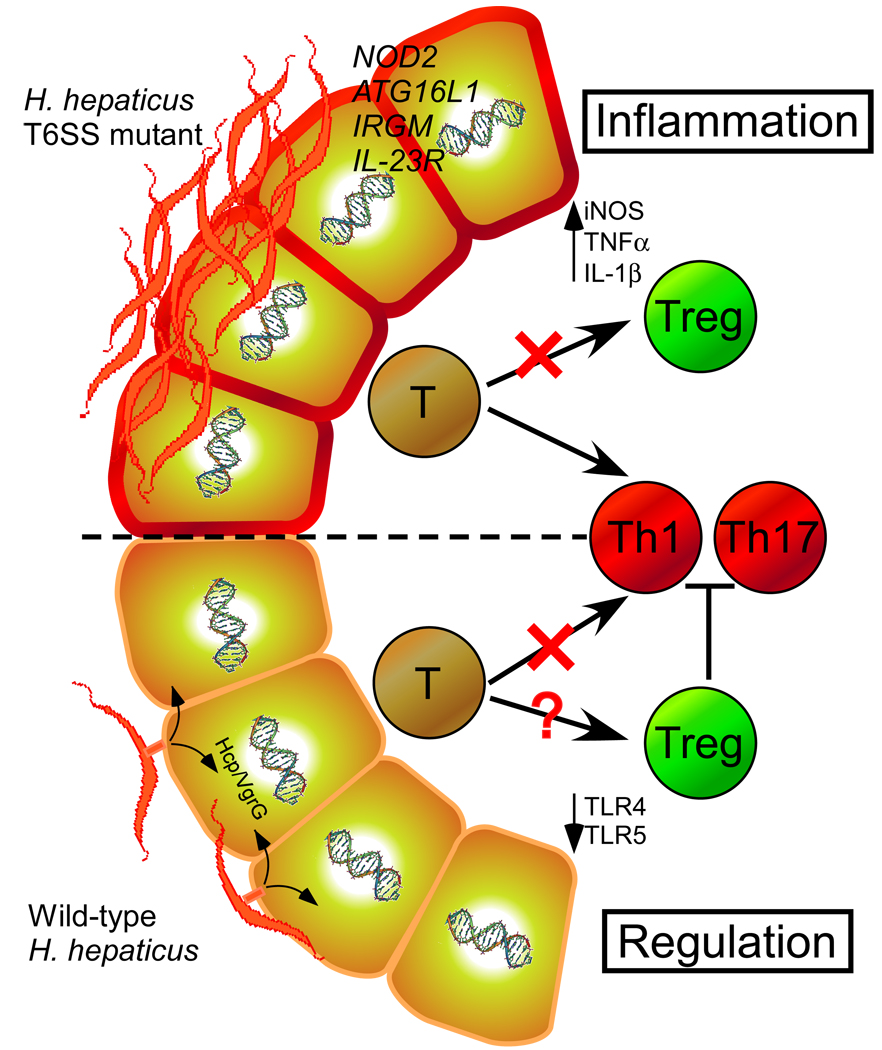
Host-bacterial interactions (whether beneficial or harmful) are defined by a dynamic exchange of molecules that mediate various biological outcomes. We characterize herein that the T6SS of H. hepaticus limits colonization of animals and actively suppresses innate & adaptive immune responses. Although T6SSs have been largely studied in the context of bacterial virulence, growing evidence supports the notion that T6SSs may have also evolved for non-pathogenic purposes in symbiotic bacteria. Previous studies have shown that H. hepaticus can reduce activation of TLR4 and TLR5 (Sterzenbach et al., 2007). Intriguingly, Kullberg et al. demonstrated that anti-inflammatory regulatory T cells (Tregs) from H. hepaticus-infected animals are able to prevent intestinal inflammation when transferred to naïve mice (Kullberg et al., 2002). As Treg cells are known to suppress Th1 and Th17 cell responses, this data correlates well with our findings. However, the molecular mechanism(s) underlying these observations have remained elusive. A large deletion of the HHGI1 has been reported to reduce inflammation caused by H. hepaticus resulting in the absence of typhocolitis in IL-10−/−mice (Ge et al., 2008). Though differences arising from animal models cannot be excluded, we speculate that additional virulence factors (not T6SS components) within HHGI1 may elicit pro-inflammatory responses. In support of this notion, several HHGI1 genes bear homology to known toxins (data not shown). Prospective studies will determine the precise function of gene products within HHGI1, and their effects on the induction of pathology in animal hosts. Collectively however, we reveal herein that type VI secretion functions to attenuate both innate and adaptive immunity to H. hepaticus. A summary of these findings are depicted in Figure 7, illustrating that the T6SS of H. hepaticus may shape an immunologically tolerant host immune system (i.e. reducing TLR, Th17 responses and promoting Tregs) through its interaction with IECs. Though numerous aspects of this host-bacterial interface remain to be determined, the T6SS appears to promote a symbiotic relationship between H. hepaticus and mammals.
Figure 7. Proposed Interactions between H. hepaticus T6SS and the Intestinal Immune Response during Colonization.
During prolonged intestinal colonization of animals, H. hepaticus intimately contacts the epithelium and uses its T6SS to create a tolerogenic immune environment (possibly through down-regulating TLR expression and/or promoting Treg development). Crosstalk between host and bacteria maintains a balanced symbiotic interaction. This balance can be disturbed by genetic mutations associated with IBD (NOD2, ATG16L1, IRGM, IL-23R) and/or dysbiosis caused by external disturbances (e.g. antibiotics, enteric infections, diet, etc.), which may result in elevated immune responses (increased Th17) in genetically susceptible hosts. Based on our and previous studies, it appears that the combination of host genotype and microbial status contributes to intestinal disease.
Understanding how H. hepaticus is able to maintain a long-term, non-pathogenic symbiosis with its murine host will be critical for insight into the biology of human intestinal Helicobacters. A handful of studies have implicated at least two species with tropism for humans, Helicobacter cinaedi and Helicobacter fennelliae, as being associated with enterocolitis, diarrhea, and bacteremia in a portion of infected patients (Flores et al., 1990; Fox et al., 2000). Moreover, studies have shown a higher association of Helicobacter species in the intestinal tract of Crohn’s disease and IBD patients compared to healthy patients, suggesting a potential role in pathogenicity (Bohr et al., 2004; Laharie et al., 2009). An important observation from these studies is that intestinal Helicobacter species are also detected in healthy patients. Similar to Helicobacter pylori whereby only 1% of colonized people develop gastric ulcers or gastric cancer, intestinal human Helicobacters do not appear to be pathogens as the majority of colonized people are asymptomatic. Unlike opportunistic pathogens that may not permanently colonize a host, understanding the dynamic molecular relationship between Helicobacters and mammals may provide paradigms for studies into human diseases caused by dysbiosis of pathobionts.
The complex consortium of microbes within our GI tract actively shapes mammalian immune responses (Mazmanian et al., 2005). The microbiota has been implicated in numerous human disorders such as IBD, colon cancer, allergies, asthma, and type 1 diabetes (Kinross et al., 2008; Mazmanian et al., 2008; Penders et al., 2007; Wen et al., 2008), highlighting the importance of understanding the individual species that make up a ‘healthy’ microbiota. Numerous investigations have shown a significant alteration in the microbiota of patients with IBD (Frank et al., 2007; Scanlan et al., 2006). A recent metagenomic (culture-independent analysis) case-control study comparing the microbiota of patients with IBD to that of non-IBD controls, revealed a statistically significant difference between the microbial compositions (Frank et al., 2007). Our understanding of how dysbiosis affects IBD is still preliminary. Further, genetic factors play an important role in the pathogenesis of IBD. Polymorphisms in bacterial sensing (NOD2/Card15) (Hampe et al., 2001), autophagy (ATG16L1) (Hampe et al., 2007) and T cell immunity (IL-23R) (Duerr et al., 2006) genes have highlighted the connection between microbes and inflammation in IBD (Figure 7). However, genetic variations appear to predispose, but not predict, disease development as concordance rates between monozygotic twins are only 30% for IBD, and many people with polymorphisms for IBD-related genes are healthy. Thus, environmental factors play a significant role in disease. Mounting evidence predicts that IBD, at least in part, results from dysbiosis of the normal microbiota (O'Hara and Shanahan, 2006). The convergent contributions of the host genetic landscape and epigenetic variables (i.e. the microbiota) should therefore be considered in evaluating the cause of complex immunologic diseases in humans. Our findings predict that discrete and identifiable bacterial species of the microbiota can drive intestinal inflammation if their ‘balance’ with the host is altered. If true, therapeutics which selectively target pathobionts may prove invaluable as a treatment for intestinal diseases such as IBD and colon cancer.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
Helicobacter hepaticus ATCC51449 (ATCC) was cultured on Brucella agar plates with 5% sheep’s blood (Teknova) or in BHI with 10% FBS. Cultures were grown at 37°C in 1% O2, 10% CO2, and 10% H2. For construction of mutants, ~2kb fragments of HH0252 (IcmF), HH0243 (Hcp), and HH0242 (VgrG) were PCR amplified and ligated into pGEMT (Promega). An erythromycin resistance gene digested from pSLB167 (Mehta et al., 2007) was inserted within the ORFs. Plasmid construction was carried out using E. coli JM109 with erythromycin and ampicillin used at 150µg/ml and 100µg/ml, respectively. Plasmid was introduced into H. hepaticus by electroporation. Mutants were selected on plates with 5µg/ml erythromycin.
MODE-K Cell Culture
MODE-K cells were cultured in DMEM supplemented with 10% FBS, 2mM L-glutamine, 50U/ml penicillin, 50µg/ml streptomycin, and 10mM hepes at 37°C in 5% CO 2 incubator. Cells were passaged using trypsin-EDTA. For bacterial incubations, media were changed to UltraDOMA-PF media (Lonza) supplemented with 10% FBS, 2mM L-glutamine, 10mM hepes, non-essential amino acids, 1mM sodium pyruvate, and 0.5mM β-mercaptoethanol. Cytochalasin D (Sigma) was used at 10µM and added 1hr prior. For gentamicin protection assays, bacteria were added at an MOI of 100. Incubations were carried at 37°C in 1% O 2. MODE-K cells were washed in PBS, and media added with or without 100µg/ml gentamicin for 2hr at 37°C. MODE-K cells were rinsed with PBS, lysed with 0.1% saponin for 15min at 25°C, and plated for bacterial quantification.
Generation of H. hepaticus Antibodies
~900bp fragments HH0243 (Hcp) and HH0242 (VgrG) were cloned into pQE30 6xHis-tagged expression vector (Qiagen) and transformed into E. coli JM109. E. coli were grown at 25°C with 0.5mM IPTG for 5hr. Peptides wer e purified using Ni-NTA columns (Qiagen) and injected into chickens for antibody production (QED Bioscience). Antibodies were collected from eggs using the EGGstract IgY kit (Promega).
Immunohistochemistry
MODE-K cells were fixed in 4% PFA. Paraffin-embedded tissues were deparaffinized in xylene and rehydrated in ethanol. Antigen retrieval in 10mM sodium citrate, pH 6.0 was carried out for 20min at 95°C. PBS with 5% FBS was used to block and dilute antibodies. Wheat germ agglutinin conjugated to tetramethylrhodamine was used at 1:1000 for 1hr at 4°C (Invitrogen). Anti- H. hepaticus and anti-VgrG were incubated at 20µg/ml overnight at 4°C. Rabbit anti-mouse E-cadherin antibody was d iluted at 1:250 (Santa Cruz Biotech). Samples were imaged using a Zeiss LSM 510 Upright confocal microscope.
Microarray Hybridization and Data Analysis
RNA was prepared using TRIzol. RNA was labeled and hybridized to Agilent microarrays (Whole Mouse Genome Microarray) following manufacturer's instructions. Microarrays were scanned using an Agilent DNA Microarray Scanner G2565CA, and data were acquired using Agilent's Feature Extraction Software version 10.1.1.1. Significant genes were selected based on p<0.05 and fold change >1.5. For enrichment analysis of biological process ontology, probe lists were analyzed in DAVID and selected based on p< 0.01.
Animal Housing
7- to 10-week-old animals were used for all experiments. SPF C57Bl/6 and C3H/HeJ mice were purchased from Taconic Farms and Jackson Laboratories, respectively. SPF C57Bl/6 Rag1−/− and C57Bl/6 IL-10−/− mice were bred and maintained in our facilities. Germ-free Swiss Webster and C57Bl/6 mice were kept in sterile isolators. Germ-free animals were screened weekly for bacterial, viral, and fungal contamination. Animals were cared for under established protocols and IACUC guidelines of California Institute of Technology.
IEC Isolation
Colons were cut longitudinally and 1cm fragments were incubated twice in HBSS (-Ca2+, Mg2+) with 5mM EDTA and 10mM Hepes for 20min at 37°C wi th gentle agitation. Cells were then treated with 5% FBS, 3U/ml Dispase, and 100ug/ml DNAse for 30min at 37°C. IECs were subsequently treated with 200ug/ml gentamicin for 2hr at 37°C, lysed with 0.5% saponin, and plated on TVP plates for selection of H. hepaticus.
Adoptive Cell Transfer
Single-cell suspensions of spleens from sex-matched mice were treated with red blood cell lysing buffer (Sigma). CD4+ T cells were isolated using a negative selection CD4+ isolation kit (Miltenyi Biotec). Cells were stained with 5µg/ml anti-CD4-FITC and 2µg/ml anti-CD45Rb-PE (eBioscience). CD4+CD45Rbhi cells were isolated by fluorescence activated cell sorting (FACS). 2×105 cells were injected intraperitoneally into Rag1−/−animals. 2 weeks later mice were orally gavaged with 1×108 wild-type, ΔIcmF, or ΔHcp H. hepaticus. Animals were sacrificed 2–4 weeks after. Colon tissues were fixed in Bouin’s fixative and sent out for paraffin-embedded sectioning and H&E staining (Pacific Pathology, San Diego)
Quantitative real-time PCR
RNA was extracted using TRIzol. RNA was treated with DNase (Sigma) prior to cDNA conversion using iScript cDNA synthesis kit (Bio-Rad). qRT-PCR was performed using iQ SYBR Green Supermix (Bio-Rad). Reactions were carried out on a Bio-Rad iCycler IQ5. For cytokine analysis, samples were normalized to the housekeeping gene L32.
Colon Organ Culture and MLN Re-stimulation
Colon tissues were washed in PBS and cultured in 48-well plates in serum-free complete RPMI for 24hr. Supernatants were collected and normalized to total protein concentration using Bradford reagent. Samples were analyzed by ELISA (eBioscience). For re-stimulation assays, MLNs were disrupted into single-cell suspensions and cultured in 48-well plates at 1×106 cells/ml in complete RPMI. Cell stimulants were added: PMA at 50ng/ml, ionomycin at 500ng/ml, anti-CD3 at 2µg/ml, and anti-CD28 at 2µg/ml. Supernatants were collected after 1d and analyzed by ELISA.
MODE-K Antigen Presentation
MODE-K cells were treated with 100U/ml of IFNγ for 7 days prior to experiments. MODE-K cells were pulsed with 20µg/ml of SHelAg for 24hrs. Cells were rinsed prior to the addition of CD4+ T cells collected from the MLNs of SPF C3H/HeJ mice colonized with wild-type, ΔIcmF, ΔHcp, or no H. hepaticus for 2 weeks. The ratio of MODE-K to T cells was 1:10. After 72 hrs, RNA was collected, and cytokine levels assayed by qRT-PCR. Preparation of SHelAg consisted of sonicating wild-type bacteria and centrifuging lysate to remove insoluble material. MODE-K cells were stained with anti-MHC class II antibodies obtained from eBioscience or ATCC (10.2.16) and analyzed by flow cytometry.
Statistical Analysis
Student’s t test was used for evaluating statistical significance. p<0.05 was considered significant.
Accession Numbers
Microarray data have been deposited in the GEO database with the accession number GSE20434.
Supplementary Material
ACKNOWLEDGMENTS
We thank Diana Perez and Rochelle Diamond for help with cell sorting, Vijaya Rao and Igor Antoshechkin of the Caltech Genomics Facility for the microarray studies, and the Beckman Imaging Center at Caltech for use of microscopes. We are grateful to Dr. Rob Maier (University of Georgia) and Stéphane Benoit (University of Georgia) for the kind gift of the pSLB167 plasmid, Dr. Dominique Kaiserlian (INSEM, France) for the generous gift of MODE-K cells, and Dr. Vincent T. Young (University of Michigan) for advice on generating mutants. Histopathology analysis was performed by Dr. Roderick T. Bronson (Harvard Medical School). We are grateful to members of the Mazmanian laboratory for their critical review of the manuscript. J. C. is supported by a pre-doctoral training grant (NIH GM007616). S.K.M. is a Searle Scholar. This work is supported by funding from the NIH/NIDDK (DK078938), Emerald Foundation, Damon Runyon Cancer Research Foundation and the Crohn’s and Colitis Foundation of America to S.K.M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bingle LE, Bailey CM, Pallen MJ. Type VI secretion: a beginner's guide. Curr Opin Microbiol. 2008;11:3–8. doi: 10.1016/j.mib.2008.01.006. [DOI] [PubMed] [Google Scholar]
- Bladergroen MR, Badelt K, Spaink HP. Infection-blocking genes of a symbiotic Rhizobium leguminosarum strain that are involved in temperature-dependent protein secretion. Mol Plant Microbe Interact. 2003;16:53–64. doi: 10.1094/MPMI.2003.16.1.53. [DOI] [PubMed] [Google Scholar]
- Bland PW, Warren LG. Antigen presentation by epithelial cells of the rat small intestine. I. Kinetics, antigen specificity and blocking by anti-Ia antisera. Immunology. 1986;58:1–7. [PMC free article] [PubMed] [Google Scholar]
- Bohr UR, Glasbrenner B, Primus A, Zagoura A, Wex T, Malfertheiner P. Identification of enterohepatic Helicobacter species in patients suffering from inflammatory bowel disease. J Clin Microbiol. 2004;42:2766–2768. doi: 10.1128/JCM.42.6.2766-2768.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Chakrabortty A, Banerjee R, Chaudhuri K. Involvement of in vivo induced icmF gene of Vibrio cholerae in motility, adherence to epithelial cells, and conjugation frequency. Biochem Biophys Res Commun. 2002;295:922–928. doi: 10.1016/s0006-291x(02)00782-9. [DOI] [PubMed] [Google Scholar]
- Dudley EG, Thomson NR, Parkhill J, Morin NP, Nataro JP. Proteomic and microarray characterization of the AggR regulon identifies a pheU pathogenicity island in enteroaggregative Escherichia coli. Mol Microbiol. 2006;61:1267–1282. doi: 10.1111/j.1365-2958.2006.05281.x. [DOI] [PubMed] [Google Scholar]
- Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, Abraham C, Regueiro M, Griffiths A, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdman SE, Rao VP, Poutahidis T, Rogers AB, Taylor CL, Jackson EA, Ge Z, Lee CW, Schauer DB, Wogan GN, et al. Nitric oxide and TNF-alpha trigger colonic inflammation and carcinogenesis in Helicobacter hepaticus-infected, Rag2-deficient mice. Proc Natl Acad Sci U S A. 2009;106:1027–1032. doi: 10.1073/pnas.0812347106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores BM, Fennell CL, Kuller L, Bronsdon MA, Morton WR, Stamm WE. Experimental infection of pig-tailed macaques (Macaca nemestrina) with Campylobacter cinaedi and Campylobacter fennelliae. Infect Immun. 1990;58:3947–3953. doi: 10.1128/iai.58.12.3947-3953.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox JG, Chien CC, Dewhirst FE, Paster BJ, Shen Z, Melito PL, Woodward DL, Rodgers FG. Helicobacter canadensis sp. nov. isolated from humans with diarrhea as an example of an emerging pathogen. J Clin Microbiol. 2000;38:2546–2549. doi: 10.1128/jcm.38.7.2546-2549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan JE, Wolf-Watz H. Protein delivery into eukaryotic cells by type III secretion machines. Nature. 2006;444:567–573. doi: 10.1038/nature05272. [DOI] [PubMed] [Google Scholar]
- Ge Z, Sterzenbach T, Whary MT, Rickman BH, Rogers AB, Shen Z, Taylor NS, Schauer DB, Josenhans C, Suerbaum S, et al. Helicobacter hepaticus HHGI1 is a pathogenicity island associated with typhlocolitis in B6.129-IL10 tm1Cgn mice. Microbes Infect. 2008;10:726–733. doi: 10.1016/j.micinf.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampe J, Cuthbert A, Croucher PJ, Mirza MM, Mascheretti S, Fisher S, Frenzel H, King K, Hasselmeyer A, MacPherson AJ, et al. Association between insertion mutation in NOD2 gene and Crohn's disease in German and British populations. Lancet. 2001;357:1925–1928. doi: 10.1016/S0140-6736(00)05063-7. [DOI] [PubMed] [Google Scholar]
- Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, Albrecht M, Mayr G, De La Vega FM, Briggs J, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science. 2001;292:1115–1118. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- Hue S, Ahern P, Buonocore S, Kullberg MC, Cua DJ, McKenzie BS, Powrie F, Maloy KJ. Interleukin-23 drives innate and T cell-mediated intestinal inflammation. J Exp Med. 2006;203:2473–2483. doi: 10.1084/jem.20061099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinross JM, von Roon AC, Holmes E, Darzi A, Nicholson JK. The human gut microbiome: implications for future health care. Curr Gastroenterol Rep. 2008;10:396–403. doi: 10.1007/s11894-008-0075-y. [DOI] [PubMed] [Google Scholar]
- Kullberg MC, Jankovic D, Gorelick PL, Caspar P, Letterio JJ, Cheever AW, Sher A. Bacteria-triggered CD4(+) T regulatory cells suppress Helicobacter hepaticus-induced colitis. J Exp Med. 2002;196:505–515. doi: 10.1084/jem.20020556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullberg MC, Rothfuchs AG, Jankovic D, Caspar P, Wynn TA, Gorelick PL, Cheever AW, Sher A. Helicobacter hepaticus-induced colitis in interleukin-10-deficient mice: cytokine requirements for the induction and maintenance of intestinal inflammation. Infect Immun. 2001;69:4232–4241. doi: 10.1128/IAI.69.7.4232-4241.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laharie D, Asencio C, Asselineau J, Bulois P, Bourreille A, Moreau J, Bonjean P, Lamarque D, Pariente A, Soule JC, et al. Association between entero-hepatic Helicobacter species and Crohn's disease: a prospective cross-sectional study. Aliment Pharmacol Ther. 2009;30:283–293. doi: 10.1111/j.1365-2036.2009.04034.x. [DOI] [PubMed] [Google Scholar]
- Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Ma AT, McAuley S, Pukatzki S, Mekalanos JJ. Translocation of a Vibrio cholerae type VI secretion effector requires bacterial endocytosis by host cells. Cell Host Microbe. 2009;5:234–243. doi: 10.1016/j.chom.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer L, Eisenhardt D, Salomon P, Bauer W, Plous R, Piccinini L. Expression of class II molecules on intestinal epithelial cells in humans. Differences between normal and inflammatory bowel disease. Gastroenterology. 1991;100:3–12. doi: 10.1016/0016-5085(91)90575-6. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- Mehta NS, Benoit SL, Mysore J, Maier RJ. In vitro and in vivo characterization of alkyl hydroperoxide reductase mutant strains of Helicobacter hepaticus. Biochim Biophys Acta. 2007;1770:257–265. doi: 10.1016/j.bbagen.2006.09.022. [DOI] [PubMed] [Google Scholar]
- Nanda Kumar NS, Balamurugan R, Jayakanthan K, Pulimood A, Pugazhendhi S, Ramakrishna BS. Probiotic administration alters the gut flora and attenuates colitis in mice administered dextran sodium sulfate. J Gastroenterol Hepatol. 2008;23:1834–1839. doi: 10.1111/j.1440-1746.2008.05723.x. [DOI] [PubMed] [Google Scholar]
- O'Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packey CD, Sartor RB. Commensal bacteria, traditional and opportunistic pathogens, dysbiosis and bacterial killing in inflammatory bowel diseases. Curr Opin Infect Dis. 2009;22:292–301. doi: 10.1097/QCO.0b013e32832a8a5d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DA, Heffron F. sciS, an icmF homolog in Salmonella enterica serovar Typhimurium, limits intracellular replication and decreases virulence. Infect Immun. 2005;73:4338–4345. doi: 10.1128/IAI.73.7.4338-4345.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penders J, Stobberingh EE, van den Brandt PA, Thijs C. The role of the intestinal microbiota in the development of atopic disorders. Allergy. 2007;62:1223–1236. doi: 10.1111/j.1398-9995.2007.01462.x. [DOI] [PubMed] [Google Scholar]
- Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, Heidelberg JF, Mekalanos JJ. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci U S A. 2006;103:1528–1533. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin DM, Seshadri R, Pukatzki SU, Mekalanos JJ. Bacterial genomics and pathogen evolution. Cell. 2006;124:703–714. doi: 10.1016/j.cell.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009 doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlan PD, Shanahan F, O'Mahony C, Marchesi JR. Culture-independent analyses of temporal variation of the dominant fecal microbiota and targeted bacterial subgroups in Crohn's disease. J Clin Microbiol. 2006;44:3980–3988. doi: 10.1128/JCM.00312-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solnick JV, Schauer DB. Emergence of diverse Helicobacter species in the pathogenesis of gastric and enterohepatic diseases. Clin Microbiol Rev. 2001;14:59–97. doi: 10.1128/CMR.14.1.59-97.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterzenbach T, Lee SK, Brenneke B, von Goetz F, Schauer DB, Fox JG, Suerbaum S, Josenhans C. Inhibitory effect of enterohepatic Helicobacter hepaticus on innate immune responses of mouse intestinal epithelial cells. Infect Immun. 2007;75:2717–2728. doi: 10.1128/IAI.01935-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez G, Sierra JC, Sha J, Wang S, Erova TE, Fadl AA, Foltz SM, Horneman AJ, Chopra AK. Molecular characterization of a functional type VI secretion system from a clinical isolate of Aeromonas hydrophila. Microb Pathog. 2008;44:344–361. doi: 10.1016/j.micpath.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suerbaum S, Josenhans C, Sterzenbach T, Drescher B, Brandt P, Bell M, Droge M, Fartmann B, Fischer HP, Ge Z, et al. The complete genome sequence of the carcinogenic bacterium Helicobacter hepaticus. Proc Natl Acad Sci U S A. 2003;100:7901–7906. doi: 10.1073/pnas.1332093100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal K, Grosjean I, evillard JP, Gespach C, Kaiserlian D. Immortalization of mouse intestinal epithelial cells by the SV40-large T gene. Phenotypic and immune characterization of the MODE-K cell line. J Immunol Methods. 1993;166:63–73. doi: 10.1016/0022-1759(93)90329-6. [DOI] [PubMed] [Google Scholar]
- Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, Hu C, Wong FS, Szot GL, Bluestone JA, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westendorf AM, Fleissner D, Groebe L, Jung S, Gruber AD, Hansen W, Buer J. CD4+Foxp3+ regulatory T cell expansion induced by antigen-driven interaction with intestinal epithelial cells independent of local dendritic cells. Gut. 2009;58:211–219. doi: 10.1136/gut.2008.151720. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.