Abstract
The major fungal pathogen Candida albicans has the metabolic flexibility to assimilate a wide range of nutrients in its human host. Previous studies have suggested that C. albicans can encounter glucose-poor microenvironments during infection and that the ability to use alternative non-fermentable carbon sources contributes to its virulence. JEN1 encodes a monocarboxylate transporter in C. albicans and we show that its paralogue, JEN2, encodes a novel dicarboxylate plasma membrane transporter, subjected to glucose repression. A strain deleted in both genes lost the ability to transport lactic, malic and succinic acids by a mediated mechanism and it displayed a growth defect on these substrates. Although no significant morphogenetic or virulence defects were found in the double mutant strain, both JEN1 and JEN2 were strongly induced during infection. Jen1-GFP (green fluorescent protein) and Jen2-GFP were upregulated following the phagocytosis of C. albicans cells by neutrophils and macrophages, displaying similar behaviour to an Icl1-GFP fusion. In the murine model of systemic candidiasis approximately 20–25% of C. albicans cells infecting the kidney expressed Jen1-GFP and Jen2-GFP. Our data suggest that Jen1 and Jen2 are expressed in glucose-poor niches within the host, and that these short-chain carboxylic acid transporters may be important in the early stages of infection.
Introduction
The opportunistic commensal organism, Candida albicans, is part of the normal gastrointestinal microflora in healthy individuals. C. albicans is a common cause of mucosal infections in immunodefective individuals, can be responsible for life-threatening systemic infections, in severely immunocompromised patients, and remains a common cause of nosocomial bloodstream infections in humans (Odds, 1988; Calderone, 2002; Perlroth et al., 2007). As well as defective immune defences, a number of additional risk factors have been associated with invasive C. albicans infections. These include the use of broad spectrum antibiotics, the application of catheters or prosthetic devices, and the disruption of normal skin barriers (for a review see Perlroth et al., 2007).
The pathogenicity of C. albicans is promoted by virulence factors such as adhesins, secretion of hydrolytic enzymes, morphogenesis and phenotypic switching (Odds, 1988; Brown and Gow, 1999; Calderone and Fonzi, 2001; Calderone, 2002). In addition, fitness attributes, such as its robust stress responses and metabolic flexibility, promote the pathogenicity of C. albicans (Wysong et al., 1998; Alonso-Monge et al., 1999; Hwang et al., 2002). This fungus exhibits a notable capacity to sense and adjust to environmental changes, such as alterations in nutrient availability and extracellular pH, allowing it to survive within diverse niches such as the skin, mucous membranes, blood and internal organs of its human host (Odds, 1988; Calderone, 2002). Analyses of genome-wide expression profiles of C. albicans cells undergoing phagocytosis by mammalian macrophages showed a reprogramming of basal metabolism, which occurs in two successive steps (Prigneau et al., 2003; Lorenz et al., 2004). The first step involved the upregulation of genes encoding gluconeogenic, glyoxylate cycle and fatty acid β-oxidation enzymes, and the concomitant downregulation of glycolytic genes. The upregulation of genes encoding key glyoxylate cycle enzymes was also observed when C. albicans cells were exposed to human blood and neutrophils (Rubin-Bejerano et al., 2003; Fradin et al., 2005). The second step occurred as C. albicans cells escaped from macrophages and was coincident with the morphogenetic switch from the budding yeast form to the hyphal form. This late response was characterized by a resumption of glycolysis and a derepression of the translation machinery. The early response appears to be a major feature of the adaptation to non-fermentable carbon sources and was presumed to reflect glucose deprivation. C. albicans mutants that lack isocitrate lyase (icl1) display attenuated virulence in a mouse model of systemic candidiasis (Lorenz and Fink, 2001; Barelle et al., 2006). This suggests a requirement for the synthesis of C4 compounds from acetyl-CoA, at some stage in the infection process (Lorenz and Fink, 2001; Lorenz and Fink, 2002). Because the early response also included the upregulation of genes involved in lipid degradation, it was postulated that lipids could be the precursors of acetyl-CoA, and thus that peroxisomal fatty acid β-oxidation is important for fungal virulence (Lorenz and Fink, 2002).
The use of green fluorescent protein (GFP) fusions to monitor gene activity has reinforced these findings. Using this approach it was shown that glyoxylate cycle and gluconeogenic genes are induced in individual C. albicans cells following phagocytosis by macrophages and neutrophils, but not during cell-to-cell contact (Barelle et al., 2006). However, it has now been shown that fatty acids are not the only sources of acetyl-CoA available to C. albicans in vivo. A pex5 mutant, which is impaired in peroxisomal protein import and hence in fatty acid β-oxidation, is still virulent in the mouse model of systemic candidiasis (Piekarska et al., 2006). Therefore it was postulated that this fungus uses non-fermentable carbon sources, such as acetate or lactate, to survive within the glucose-poor environment of the phagolysosome (Piekarska et al., 2006). This idea was also supported by Ramirez and Lorenz (2007) who reported that the ability to assimilate alternative non-fermentable carbon sources contributes to the virulence of C. albicans. They showed that C. albicans requires carnitine acetyltransferases of the carnitine shuttle for growth on acetate as sole carbon source (Prigneau et al., 2003; Strijbis et al., 2008; Zhou and Lorenz, 2008).
The transport of carboxylic acids across the plasma membrane presumably is essential for the assimilation of these alternative carbon sources. These substrates are weak organic acids that partially dissociate in aqueous solution, according to their pKa(s) and to the pH value of the medium. The uptake of the undissociated form of these compounds can occur through the plasma membrane by a simple diffusion. However at pH of above 5, the carboxylic acids are predominantly present in their anionic form and their assimilation depends on transporter-mediated uptake (for a review see Casal et al., 2008). In Saccharomyces cerevisiae it has been reported that the Fps1 channel promotes the facilitated diffusion of the undissociated form of acetic acid at low pH (4.5) (Mollapour and Piper, 2007), and the ScJen1 transporter is responsible for the active transport of lactate and pyruvate (Casal et al., 1999; Akita et al., 2000). C. albicans Jen1 was identified by sequence homology with ScJen1 (Casal et al., 1999) and it was subsequently characterized as a lactate, pyruvate, propionate/proton symporter in this pathogen (Soares-Silva et al., 2004). JEN1 is upregulated about fivefold when C. albicans cells are phagocytosed by macrophages (Piekarska et al., 2006). Similarly, CYB2, which encodes an l-lactate dehydrogenase, is upregulated following macrophage attack (Piekarska et al., 2006), reinforcing the idea that carboxylate assimilation is an integral part of the C. albicans response to phagocytosis.
A recent study based on synteny analysis, sequence similarity and motif analysis revealed the existence of at least 35 fungal homologues of the S. cerevisiae JEN1 gene in 13 different Hemiascomycetes and four Euascomycetes (Lodi et al., 2007). A phylogenetic tree of ScJen1p homologues (Lodi et al., 2007) showed the existence of two main clusters. The first cluster represents a Jen1 group of proteins that have been functionally characterized as monocarboxylate transporters. The second cluster comprises Jen2-like proteins. This cluster includes a new Jen1-like protein from C. albicans, Jen2, which has not been functionally characterized. In addition, the cluster contains KlJen2, which is a succinate and D,L-malate plasma membrane transporter in Kluyveromyces lactis (Lodi et al., 2004; Queiros et al., 2007). Schizosaccharomyces pombe also expresses a dicarboxylate transporter, but the Mae1 protein has no significant homology to the Jen protein family. Mae1 transporter is constitutively expressed and not subjected to glucose repression (Grobler et al., 1995).
Candida albicans JEN2 is closely related to JEN1 (Casal et al., 2008). Furthermore, JEN2 is strongly upregulated during phagocytosis by macrophages (160-fold, compared with fivefold for JEN1) (Piekarska et al., 2006), and is strongly repressed by glucose (18-fold). Therefore, we reasoned that JEN2 might play a significant role in the assimilation of short-chain carboxylic acids by C. albicans. We have confirmed this, showing that Jen2 executes a distinct function from Jen1 in C. albicans. Jen2 is a plasma membrane transporter, rapidly degraded following exposure to glucose, responsible for the saturable kinetics observed for malate and succinate uptake. Given the importance of non-fermentable carbon sources to the growth of C. albicans in some host niches (Prigneau et al., 2003; Strijbis et al., 2008; Zhou and Lorenz, 2008) and the potential importance of short-chain carboxylates as a carbon source in vivo, we have also compared the expression of Jen1 and Jen2 in disease models.
Results
JEN2 encodes a dicarboxylate transporter in C. albicans
The C. albicans JEN2 (orf19.5307) locus was annotated as a putative carboxylic acid plasma membrane transporter on the basis of its sequence similarity to ScJEN1 (http://www.candidagenome.org/). However, Jen2 function has not been tested experimentally. There is only one JEN locus in S. cerevisiae, even though this species has undergone whole genome duplication (Scannell et al., 2007). Yet there are two JEN loci in C. albicans, although this pathogen has not undergone whole genome duplication. We reasoned therefore, that the functions of C. albicans JEN1 and JEN2 might differ in C. albicans. Consequently, to determine the physiological role of JEN2 in C. albicans, both chromosomal copies of this gene were disrupted in the RM1000 strain, resulting in strain CNV3 (jen2Δ/jen2Δ: Table 1). Additionally, a double jen1jen2 mutant was constructed, resulting in C. albicans strain CNV4 (jen1Δ/jen1Δ, jen2Δ/jen2Δ: Table 1).
Table 1.
Saccharomyces cerevisiae and Candida albicans strains used in this study.
| Strain | Genotype | Reference |
|---|---|---|
| S. cerevisiae | ||
| W303-1A | MATa ade2 leu2 his3 trp1 ura3 | Thomas and Rothstein, 1989 |
| BLC 491-U2 | MATa ura3-52 JEN1: :GFP Kanr | Paiva et al., 2002 |
| jen1Δ | W303-1A jen1::KanMx4 | Paiva et al., 2004 |
| jen1Δ– p416GPD | jen1Δtransformed with p416GPD | Soares-Silva et al., 2004 |
| jen1Δ–p416GPDCaJEN2 | jen1Δtransformed with p416GPDCaJEN2 | This work |
| C. albicans | ||
| RM1000 | ura3::imm434/ura3::imm434, his1::hisG/his1::hisG | Negredo et al., 1997 |
| CPK2 | ura3::imm434/ura3::imm434 his1::hisG/his1::G jen1::HIS1/jen1::URA3 | Soares-Silva et al., 2004 |
| CNV3 | ura3::imm434/ura3::imm434 his1::hisG/his1::G jen2::HIS1/jen2::URA3 | This work |
| CNV4 | ura3::imm434/ura3::imm434 his1::hisG/his1::G jen1::HIS1/jen1::ura3-, jen2::ura3-/jen2::URA3 | This work |
| CNV2-1 | CPK2 with RPS1-CIp20 | This work |
| CNV2-2 | CPK2 with RPS1-CIp20-JEN1 | This work |
| CNV3-1 | CNV3 with RPS1-CIp20 | This work |
| CNV4-1 | CNV4 with RPS1-CIp20 | This work |
| CNV4-1 | CNV4 with RPS1-CIp20-JEN1 | This work |
| CPK20-5 | RM1000 with JEN1/JEN1-GFP-URA3 | Soares-Silva et al., 2004 |
| CNV30-5 | RM1000 with JEN2/JEN2-GFP-URA3 | This work |
| CJB | ura3::λimm434/ura3::λ imm434, pACT1-GFP | Barelle et al., 2004 |
| CJB-1 | ura3::λimm434/ura3::λ imm434, pGFP | Barelle et al., 2004 |
| CJB-3 | ura3::λimm434/ura3::λ imm434, pICL1GFP | Barelle et al., 2004 |
The ability of these new jen2 and jen1jen2 mutants to take up succinic acid and l-malic acid was compared with wild-type and jen1 cells. We examined the uptake of succinic acid and malic acid because JEN2 is found in the same phylogenetic cluster as KlJEN2, which is a succinate and malate transporter in Kluyveromyces lactis (Lodi et al., 2004; Queiros et al., 2007). Cells were grown in succinic acid and malic acid containing medium, at pH 5.0, under derepressing conditions (i.e. in the absence of glucose), and then the initial uptake rates of labelled succinic acid and l-malic acid were measured. Computer-assisted non-linear regression analyses to the experimental data suggested the presence of a mediated transport system for succinic acid in wild-type C. albicans cells (RM1000: Table 1) with the following kinetic parameters: Km 0.49 ± 0.27 mM and Vmax 0.25 ± 0.040 nmol s−1 mg dry wt.−1 (for concentrations between 0.1 mM and 4 mM, pH 5.0) (Fig. 1A). Malic acid uptake by wild-type cells also displayed saturable kinetics with a Km of 0.12 ± 0.019 mM, and a Vmax of 0.18 ± 0.0070 nmol s−1 mg dry wt.−1 (for concentrations between 0.04 mM and 2 mM, pH 5.0) (Fig. 1B).
Fig. 1.
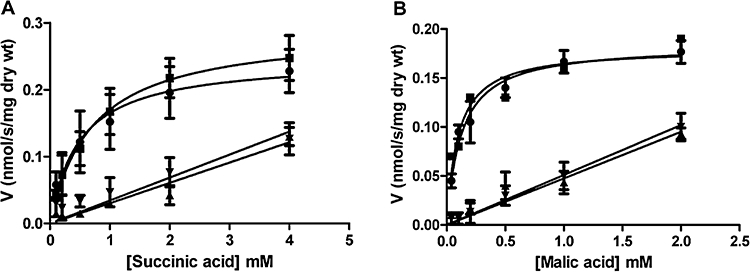
Transport of carboxylic acids in Candida albicans. A.Initial uptake rates of [2,3-14C] succinic acid, at pH 5.0, as a function of succinic acid concentration, after growth in medium containing succinic acid: RM1000 (wild-type), •; CPK2 (jen1),  ; CNV3 (jen2),
; CNV3 (jen2),  ; CNV4 (jen1jen2),
; CNV4 (jen1jen2),  . B.Initial uptake rates of L-[1,4(2,3)-14C] malic acid, at pH 5.0, as a function of malic acid concentration after growth in medium containing malic acid: symbols as for (A).
. B.Initial uptake rates of L-[1,4(2,3)-14C] malic acid, at pH 5.0, as a function of malic acid concentration after growth in medium containing malic acid: symbols as for (A).
Similar observations were made for C. albicans jen1 mutant (CPK2: Table 1). For succinic acid transport, this mutant displayed a Km of 0.71 ± 0.27 and a Vmax of 0.29 ± 0.038 nmol s−1 mg dry wt.−1 (Fig. 1), and these jen1 cells transported malic acid with a Km of 0.096 ± 0.033 mM and a Vmax of 0.18 ± 0.015 nmol s−1 mg dry wt.−1 (Fig. 1). In contrast, the uptake of succinic and malic acid by jen2 and jen1jen2 cells fitted to a first order kinetics (Fig. 1). These results show that JEN2 codes for a saturable (second order kinetics) transport system of succinic and malic acid across the plasma membrane in C. albicans. In contrast, the kinetics of lactic acid transport was not affected by deletion of JEN2, but fitted to a first order kinetics in the strains lacking JEN1 (not shown). Therefore, the Jen1 and Jen2 transporters have different specificities in C. albicans: Jen1 transports short-chain monocarboxylic acids such as lactate, whereas Jen1 transports short-chain dicarboxylic acids such as succinate and malate.
jen1jen2 mutant is affected in the growth on mono- and dicarboxylic acids
Given that C. albicans jen mutants have defects in carboxylic acid transport, it was conceivable that these mutants might also display growth defects on the corresponding carbon sources. Therefore, the growth of wild-type and jen mutants was evaluated on solid media containing different carbon and energy sources, such as glucose, lactic, pyruvic, malic and succinic acids. Growth was evaluated at pH 5.0 and pH 7.0, and the cells were incubated across a range of temperatures: 18°C, 30°C and 37°C. When incubated at 37°C, for 96 h, jen1 and jen1jen2 mutants displayed a growth defect on media containing lactic or pyruvic acids, as sole carbon source (Fig. 2). The jen2 and jen1jen2 mutants exhibited a growth defect on succinic and malic acids (Fig. 2). The residual growth observed for the mutant strains could be attributed to simple diffusion; however, because it also persisted in the media with pH 7.0, it is also likely due to the presence of other transport systems, not identified yet.
Fig. 2.
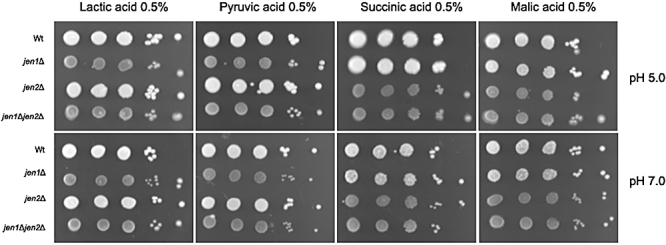
Growth phenotypes, at 37°C, of Candida albicans RM1000 (wild-type), CPK2 (jen1), CNV3 (jen2), CNV4 (jen1jen2), incubated for 96 h, in the following solid media: SC-lactic acid (0.5%, w/v, pH 5.0 or 7.0); SC-pyruvic acid (0.5%, w/v, pH 5.0 or 7.0); SC-succinic acid (0.5%, w/v, pH 5.0 or 7.0); SC-malic acid (0.5%, w/v, pH 5.0 or 7.0). Cells were serially diluted; 3 µl of drops of each dilution were spotted onto the plates.
Heterologous expression of C. albicans JEN2 in S. cerevisiae
No transporters for dicarboxylic acids have been so far assigned in S. cerevisiae, being assumed that these substrates cross the plasma membrane by a simple diffusion mechanism of the undissociated molecules (Salmon, 1987; Queirós et al., 2003). S. cerevisiae jen1 cells that express KlJEN2 in the plasmid p416-GPD (glyceraldehyde 3-phosphate dehydrogenase) acquire the ability to transport succinic and malic acids by a mediated mechanism (Queiros et al., 2007). Therefore C. albicans JEN2 was cloned into the same S. cerevisiae expression vector, p416-GPD (Mumberg et al., 1995), resulting in the plasmid pNV3 (Table 2). S. cerevisiae jen1 cells were transformed with this plasmid (pNV3) and with the control empty vector (p416-GPD: Table 2). We then determined the kinetic parameters for succinate uptake at pH 5.0 by these strains. S. cerevisiae jen1 cells expressing C. albicans JEN2 acquired the ability to transport succinate by a mediated mechanism with a Vmax of 0.17 ± 0.020 nmol s−1 mg dry wt.−1 and a Km of 0.50 ± 0.16 mM (Fig. 3). In contrast, control cells containing p416-GPD behaved as the wild-type. The kinetic parameters obtained were equivalent to the ones found in C. albicans wild-type cells. These data confirm the function of C. albicans Jen2 as a dicarboxylate transporter.
Table 2.
Plasmids used in this study.
| Plasmids | Source or reference |
|---|---|
| CIp20 | Murad et al., 2000 |
| pNV1 (CaJEN1 in Cip20) | This work |
| pGFP-URA3 | Gerami-Nejad et al., 2001 |
| pLUL | Dennison et al., 2005 |
| pLHL | Dennison et al., 2005 |
| pDDB57 | Wilson et al., 2000 |
| p416GPD | Mumberg et al., 1995 |
| pNV3 (CaJEN2 in p416GPD) | This work |
Fig. 3.
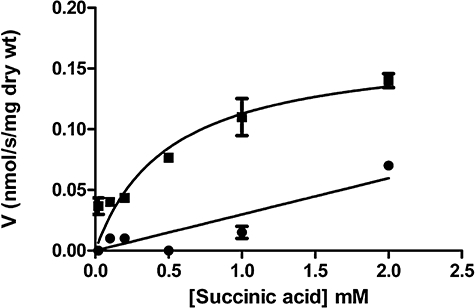
Heterologous expression of Candida albicans JEN2 in Saccharomyces cerevisiae. Initial uptake rates of [2,3-14C] succinic acid at pH 5.0, as a function of succinic acid concentration, by YNB-glucose-grown cells of S. cerevisiae W303-1A jen1Δ strain transformed with p416GPD (•) or pNV3 ( ).
).
Expression of JEN1/JEN2 and subcellular localization of Jen1-GPF and Jen2-GFP in living C. albicans cells in response to different carbon sources
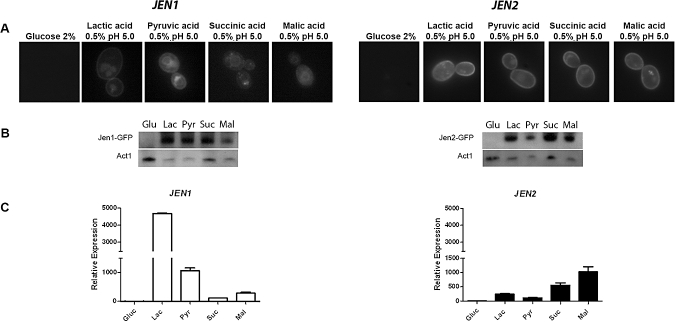
Candida albicans Jen2 was tagged with the reporter gene GFP at its carboxy-terminus (Table 1) to study the expression and sub-cellular localization of this transporter. The behaviour of this Jen2-GFP fusion in C. albicans was compared with a previously constructed Jen1-GFP fusion (Table 1). Cells expressing Jen1-GFP and Jen2-GFP were grown in minimal medium supplemented with 2% glucose to mid-exponential phase. Cells were then washed and transferred to fresh minimal media containing different carbon sources. After 4 h of incubation, at 30°C, samples were collected to examine cells by epifluorescence microscopy, to prepare extracts for further analyses by western immunoblotting with anti-GFP antibody and to prepare mRNA for quantitative real-time polymerase chain reaction (qRT-PCR) studies. The results show that the Jen2-GFP fusion was expressed and mainly localized to the plasma membrane, after derepression in lactic, succinic, pyruvic and malic acids (Fig. 4A), although some intracellular fluorescence was also observed. The levels of Jen2 protein expression were measured by Western blot analyses with an anti-GFP antibody, and compared with the internal control, Act1. The Jen2-GFP signal was detected in all of the conditions tested, except in glucose grown cells (Fig. 4B), which was consistent with the fluorescence data. We also quantified JEN2 mRNA levels by qRT-PCR (Fig. 4C). The data show lower JEN2 mRNA levels during growth on lactic or pyruvic acid compared with succinic or malic acid grown cells. This result correlates reasonably well with the levels of the corresponding Jen2-GFP fusions (Fig. 4B and C), although some post-translational regulation of Jen2-GFP levels cannot be excluded.
Fig. 4.
Expression of JEN1/JEN2 and localization of Jen1-GFP and Jen2-GFP in living Candida albicans cells grown on different carbon sources. Mid-exponential C. albicans cells grown in minimal medium containing 2% w/v glucose were washed twice with deionized water and then transferred, for 4 h at 30°C, to fresh minimal media containing different carbon sources: glucose 2%, w/v (glu); lactic acid 0.5%, v/v (lac); pyruvic acid 0.5%, v/v (pyr); succinic acid 0.5%, v/v (suc); malic acid 0.5%, v/v (mal). Samples were collected after induction, to examine cells by epifluorescence microscopy, to pepare mRNA and protein extracts. A.Subcellular localization of Jen1-GFP and Jen2-GFP in living cells. B.Protein extracts were separated by SDS-PAGE and analysed for Jen1-GFP and Jen2-GFP by western immunoblotting with an anti-GFP antibody. The blots were reprobed with an anti-ACT1 to provide loading controls. C.Expression analysis of JEN1 and JEN2 was followed by qRT-PCR. JEN1 and JEN2 mRNA expression levels were normalized to ACT1. The results are presented as the mean ± SD of two independent experiments with duplicates for each experiment.
With respect to Jen1-GFP subcellular expression, the images show a faint plasma membrane localization in lactic and pyruvic acid derepressed cells, although some intracellular fluorescence was also detected (Fig. 4A). Very low Jen1-GFP fluorescence levels were observed in malic and succinic acid derepressed cells. This was consistent with the Jen1-GFP signals on Western blots, which were lower for malic and succinic acid-derepressed cells relative to the Act1 loading control (Fig. 4B). Furthermore JEN1 transcript levels were lower in these cells compared with those in cells derepressed on lactic or pyruvic acids (Fig. 4C). However, in lactic and succinic acids derepressed cells there appeared to be a lack of correlation between the levels of JEN1 mRNA and Jen1-GFP protein (Fig. 4B and C) in that the relatively high levels of JEN1 mRNA in lactic acid were not reflected in equivalent Jen1-GFP levels; also the signal obtained for Jen1-GFP by Western blot in succinic acid was not consistent with the very low levels of JEN1 mRNA and the weak fluorescence found (Fig. 4A–C). Similar observations were reproduced in three independent experiments. We have, previously, confirmed that Jen1 fused with GFP is a functional lactate transporter with identical Km and Vmax as the wild-type. Therefore, the GFP fusion does not seem to affect the folding and/or localization of Jen1. This apparent inconsistency between JEN1 mRNA and proteins levels in lactic and succinic acid-grown cells could be explained by post-translational control of Jen1 expression levels, but this remains to be tested. Nevertheless, we have already shown that post-translational control mechanisms exist at the level of Jen1/Jen2 protein turnover in response to glucose (Andrade and Casal, 2001; Paiva et al., 2002; Queirós et al., 2003).
Taken together these results indicate that monocarboxylic acids induce JEN1 expression to a greater degree than JEN2, whereas JEN2 is induced more strongly than JEN1 during growth on dicarboxylic acids. These data, which suggest that the Jen transporters play distinct physiological roles in C. albicans cell, are consistent with the results obtained from the transport and growth assays. It still remains to be explained why JEN genes and proteins are also expressed under conditions where they do not appear to be functional: Jen1 is expressed in the presence of dicarboxylic acids and Jen2 in monocarboxylic acids.
Inactivation of Jen1-GFP and Jen2-GFP by glucose in C. albicans
Jen proteins are generally subjected to tight glucose repression (Paiva et al., 2002; Lodi et al., 2004; Queiros et al., 2007; Soares-Silva et al., 2007). In S. cerevisiae the addition of glucose to lactic acid-grown cells triggers the loss of ScJen1p activity and the repression of ScJEN1 expression (Andrade and Casal, 2001; Paiva et al., 2002). The loss of ScJen1-GFP is the result of its End3-dependent internalization and its subsequent targeting to the vacuole for degradation (Paiva et al., 2002). This glucose-regulated endocytosis of ScJen1 in S. cerevisiae has been characterized in detail and it is one of the few examples for which ubiquitin-K63 linked chain(s) have been shown to be required for correct trafficking at two stages of endocytosis: endocytic internalization and sorting at Multi Vesicular Bodies (MVBs) (Paiva et al., 2009). To compare the impact of glucose upon C. albicans Jen proteins with ScJen1, C. albicans cells carrying the JEN1-GFP or JEN2-GFP fusions were compared with S. cerevisiae cells expressing ScJEN1-GFP. These strains were grown in minimal medium containing 2% glucose and then derepressed for 4 h in minimal medium containing 0.5% lactic acid at pH 5.0, a condition where all transporters were shown to be expressed and localized to the plasma membrane (Fig. 4A). These cells were then treated with glucose, at final concentrations ranging from 0.01% to 1%. These conditions were selected to cover the range of glucose concentrations found in human blood, which generally varies between 3 and 5 mM (equivalent to about 0.06–0.1% glucose) (Barelle et al., 2006). Control cells were treated with sorbitol (final concentration 110 mM) as a control for the osmotic changes imposed by the glucose addition. After these treatments the cells were examined by epifluorescence microscopy (Fig. 5).
Fig. 5.
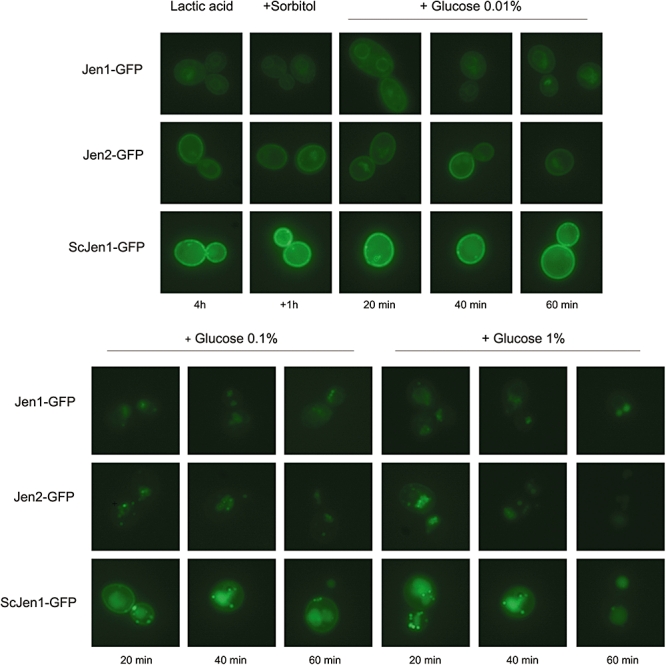
Timecourse of Jen1-GFP, Jen2-GFP and ScJen1-GFP inactivation at different glucose concentrations. Induced cells were treated with 0.01% (0.55 mM), 0.1% (5.5 mM) and 1% (55 mM) glucose or with sorbitol at the same concentrations (as a control for osmotic shock) and examined by fluorescence microscopy, after continued incubation: CPK20-5 (Candida albicans JEN1-GFP); CNV30-5 (C. albicans JEN2-GFP); BLC 491-U2 (Saccharomyces cerevisiae JEN1-GFP).
The first observation of note in these experiments was that fluorescence levels were generally higher in S. cerevisiae than C. albicans (Fig. 5). This was consistent with published comparisons of GFP levels in these two yeasts (Cormack et al., 1997). The next observation was that even 1 h after the addition of 0.01% glucose to the C. albicans and S. cerevisiae cells, Jen1-GFP, Jen2-GFP and ScJen1-GFP levels persisted in their plasma membranes (Fig. 5). However, within 20 min of incubation with 0.1 or 1% glucose, all the Jen-GFP fluorescence in C. albicans cells had disappeared from the plasma membrane (Fig. 5). In the S. cerevisiae cells incubated with 0.1% glucose, not all of the ScJen1-GFP signal was removed from the plasma membrane, but punctuate structures were observed in the cytoplasm (Fig. 5). When S. cerevisiae cells were treated with 1% glucose the ScJen1-GFP fluorescence signal finally disappeared from the plasma membrane after 60 min of incubation (Fig. 5). This indicates that the Jen1 and Jen2 transporters are internalized more quickly in response to glucose in C. albicans than ScJen1 in S. cerevisiae. We set to determine the threshold glucose concentration which triggers C. albicans Jen degradation. Because the results showed that Jen1 and Jen2 respond similarly to glucose in C. albicans, we carried out a Western blot analysis to follow Jen2-GFP expression, over time, after a pulse of 0.01% and 0.05% glucose, to lactic acid induced cells. In the presence of 0.05% (close to the minimal blood glucose concentration) the level of Jen2-GFP remained stable, even after 60 min of the pulse of glucose (data not shown). However, the addition of 0.1% glucose triggered Jen2-GFP degradation after 20 min of incubation. This indicates that Jen proteins may still be expressed in niches where only low amounts of glucose are present (≤ 0.05%). This is consistent with the view that C. albicans cells may be able to utilize some alternative carbon sources in such microenvironments. This view is strengthened by the observation that gluconeogenic and glyoxylate cycle genes are expressed by some C. albicans cells infecting the kidney (Barelle et al., 2006).
The inactivation of JEN1 or JEN2 does not affect C. albicans morphogenesis
The ability of C. albicans to switch between hypha and yeast growth forms is considered to be a virulence attribute of this fungus (Berman and Sudbery, 2002). The hyphal form facilitates the adhesion and invasion of human tissues as well as the evasion of phagocytic cells. In contrast, the yeast form is better adapted to dissemination (Gow et al., 2002). Hence the ability to switch between morphological forms is important for C. albicans virulence (Berman and Sudbery, 2002; Gow et al., 2002). Environmental pH is a trigger for C. albicans morphological differentiation (Buffo et al., 1984) and influences nutrient uptake via the functionality of plasma membrane transporters. The responses of C. albicans to changes in extracellular pH have been analysed by global expression analysis, revealing links between extracellular pH and iron acquisition (Bensen et al., 2004), another virulence determinant. Furthermore, the activities of plasma membrane proton transport systems are important in controlling internal pH and these are also associated with the regulation of dimorphism (Stewart et al., 1988; 1989; Kaur and Mishra, 1994). The yeast form is favoured at low ambient pHs, for example around pH 4.0 when undissociated forms of carboxylic acids predominate and the simple diffusion of these acids into the cell is favoured. On the other hand, hyphal growth is stimulated at neutral ambient pH, when the mediated transport systems are more active (Buffo et al., 1984; El Barkani et al., 2000). Therefore, environmental pH, morphogenesis and carboxylic acid uptake are linked, a reason why we tested whether the inactivation of JEN1 or JEN2 affects morphogenesis in C. albicans.
Morphogenesis can be triggered by several different treatments such as exposure of C. albicans cells to temperatures higher than 37°C, ambient pH above 6.5, serum and low concentrations of dissolved O2. Serum has been shown to be the strongest inducer of hyphal development, when combined with temperatures of 37°C. For that reason, C. albicans RM1000 (wild-type), CPK2 (jen1), CNV3 (jen2) and CNV4 (jen1jen2) cells were grown under conditions that normally induce JEN1 and JEN2 expression (minimal medium containing 0.5% lactic acid at pH 5.0). These cells were then transferred to hypha inducing conditions by supplementing the medium with 10% (v/v) fetal calf serum and incubating at 37°C. The morphology of cells in these cultures was then monitored after 1, 2 and 3 h. Filamentation was also assessed in solid media supplemented with 10% (v/v) fetal bovine serum (FBS). Both wild-type and mutant strains formed hyphae at similar rates (data not shown). To strengthen these findings C. albicans cells were also subjected to pH (Buffo et al., 1984) and GlcNAc switch (Mattia et al., 1982). Again, both at higher pHs and in the presence of poor carbon and nitrogen sources, at temperatures above 37°C, yeast to hypha transition was induced in a similar way in all the strains tested. Therefore, the Jen1 and Jen2 carboxylate transporters were not required for hyphal development under the conditions tested.
Regulation of JEN1 and JEN2 following contact with the host
Human blood is a complex and hostile environment for microorganisms. C. albicans cells adapt rapidly to this hostile environment, displaying dramatic changes in its transcript profile (Fradin et al., 2003). The first line of defence against C. albicans is provided by phagocytic cells such as macrophages and neutrophils (Lorenz and Fink, 2002). Therefore, we examined the impact of phagocytosis upon the expression of the C. albicans carboxylate transporters.
First we examined the behaviour of control C. albicans strains carrying pGFP, ACT1-GFP or ICL1-GFP fusions. The strains were incubated with murine macrophages or human neutrophils for 2.5–3 h at 37°C, and GFP expression was detected by fluorescence microscopy of the phagocytosed and non-phagocytosed C. albicans cells. All of the control strains behaved as expected (Fig. 6). No GFP fluorescence was detected in phagocytosed or non-phagocytosed cells in the negative control (pGFP). In contrast, the positive control (ACT1-GFP) was expressed under both conditions. Furthermore, ICL1-GFP was induced following phagocytosis, as reported previously (Lorenz et al., 2004; Fradin et al., 2005).
Fig. 6.
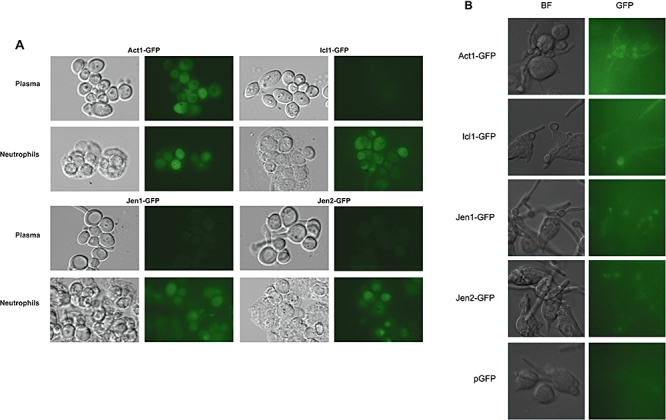
Differential regulation of Jen1-GFP and Jen2-GFP in Candida albicans following phagocytosis by immune cells. A.Interaction of C. albicans with neutrophils. C. albicans cells containing ACT1-GFP, ICL1-GFP, JEN1-GFP, JEN2-GFP or the control pGFP plasmid were mixed in a 1:1 ratio with human neutrophils, and examined microscopically after 2.5 h at 37°C. Corresponding light and fluorescence images are shown. B.Interaction of C. albicans with macrophages. Cultured murine macrophages (RAW.264.7) were mixed in a 1:3 ratio with the C. albicans strains, incubated for 3 h and analysed by light and fluorescence microscopy.
The fluorescence of C. albicans CPK20-5 (Jen1-GFP) and CNV30-5 (Jen2-GFP) cells was examined under equivalent conditions. Both Jen1-GFP and Jen2-GFP were expressed, although weakly, following phagocytosis by macrophages or neutrophils. In contrast, the non-phagocytosed cells in human plasma (a glucose-containing medium) displayed no significant Jen1-GFP and Jen2-GFP expression. As a further control, CPK20-5 and CNV30-5 cells were examined after 3 h of incubation at 37°C, 5% CO2, in glucose-containing cell culture medium [Dulbecco's modified Eagle's medium (DMEM)]. No fluorescence was observed (not shown). Therefore Jen1-GFP and Jen2-GFP expression mirrored that of the Icl1-GFP fusion. Expression of Jen1 and Jen2 carboxylate transporters was only observed following phagocytosis, presumably as a result of the transfer of C. albicans cells into the glucose poor environment of the phagolysosome (Fig. 6A and B).
If Jen1 and Jen2 are expressed following phagocytosis, do these carboxylate transporters promote the survival of phagocytosed C. albicans cells? We tested this by comparing the survival of jen1 and jen2 mutants with wild-type cells following exposure to human blood. RM1000 (wild-type), CPK2 (jen1), CNV3 (jen2) and CNV4 (jen1jen2) cells were grown to mid-exponential phase in minimal media at pH 5.0 containing either 0.5% lactic acid or 1% succinic acid. These mid-exponential cells were incubated with human blood for 30 min at 37°C, and then viable cell counts determined. As a negative control cells were plated after incubation in sterile water. In independent replicate experiments using blood from several different donors, we observed no significant difference in the blood killing between the wild-type cells and the jen mutant cells (60–65% survival). We conclude that the survival of phagocytosed C. albicans cells is not dependent upon Jen1 or Jen2.
Single cell profiling of C. albicans carboxylate transporter genes during systemic infections and virulence of C. albicans null jen mutants
After assessing the induction of the Jen1 and Jen2 carboxylate transporters in ex vivo infection models, we examined their expression in vivo in the mouse model of systemic candidiasis. Mice were infected with C. albicans strains containing JEN1-GFP, JEN2-GFP, ACT1-GFP or pGFP and humanely terminated after the animals lost approximately 20% of their body weight and/or showed signs of illness (3 or 4 days). The kidneys were removed aseptically and kidney sections prepared. Fungal cells in these sections were identified by Calcofluor white staining, and then GFP fluorescence in these cells detected by fluorescence microscopy (Gow and Gooday, 1982). As expected no GFP flucorescence was observed for the negative control (pGFP) (Fig. 7A). In contrast, over 80% of ACT1-GFP containing cells displayed fluorescence in the kidney sections. Jen1-GFP and Jen2-GFP were expressed in approximately 20–25% of the cells in infected kidneys (Fig. 7A and B). This is consistent with a previous report which suggested that about one-third of C. albicans cells infecting the mouse kidney assimilate carbon via gluconeogenesis (Barelle et al., 2006). Interestingly, the intensity of Jen1-GFP and Jen2-GFP fluorescence was comparable in vivo and in vitro grown fungal cells, suggesting that in those C. albicans cells that activate JEN1 and JEN2 in the kidney, the level of JEN expression is similar to that observed in gluconeogenic cells in vitro.
Fig. 7.
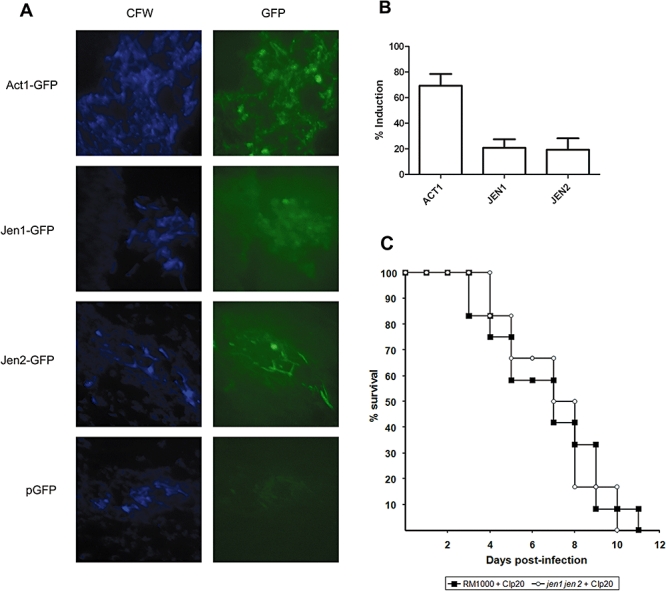
Expression of Jen1-GFP and Jen2-GFP in Candida albicans cells infecting the kidney. A.C. albicans cells infecting the kidneys of mice were visualized by staining with Calcofluor white (CFW), and then those that expressed their GFP fusions were imaged by fluorescence microscopy (GFP): Act1-GFP, Jen1-GFP and Jen2-GFP. B.The proportion of C. albicans cells infecting the kidney that display GFP fluorescence above background levels. C.Comparison of the virulence of wild-type and jen1jen2 cells in the mouse model of systemic candidiasis.
Additionally, we determined the impact of jen mutations upon C. albicans virulence. Prototrophic wild-type cells and the jen1jen2 double mutant were tested in a murine model of systemic candidiasis (MacCallum and Odds, 2005). No statistically significant differences in the virulence of the wild-type and jen1jen2 cells were found. Furthermore similar fungal burdens were observed in the kidneys of infected animals. Therefore, the inactivation of Jen1 and Jen2 does not impair the virulence of C. albicans, at least in the mouse model of systemic candidiasis (Fig. 7C).
Discussion
Candida albicans has the ability to survive within different host niches in part due to its metabolic flexibility. This versatile microorganism is a Krebs-positive yeast (Rao et al., 1962) that can use different nutrients, including short-chain carboxylic acids, as sole carbon and energy sources. In this study we have shown that Jen2 is a dicarboxylate transporter that mediates the uptake of malate and succinate through the plasma membrane. This is in contrast to its C. albicans paralogue, Jen1, which transports lactate, pyruvate and propionatec (Soares-Silva et al., 2004). This conclusion was based on three complementary observations. First, Jen2 localized to the plasma membrane (Fig. 4A). Second, the inactivation of JEN2 (but not JEN1) was shown to abolish the saturable uptake of malic acid and succinic acid by C. albicans cells (Fig. 1) and to affect the growth in media containing those substrates, as sole carbon and energy sources (Fig. 2). Third, the heterologous expression of C. albicans JEN2 in S. cerevisiae cells conferred upon them the ability to take up succinic acid by a Michaelis-Menten kinetics (Fig. 3). These experiments confirmed that JEN1 and JEN2 execute distinct transport functions in C. albicans.
The Jen1 and Jen2 transporters displayed distinct regulatory profiles in response to carbon source, both at the protein and at the mRNA level. Both were expressed during growth on lactic, pyruvic acid, succinic and malic acids (Fig. 4B and C). However, the monocarboxylic acids tested induced the expression of JEN1 stronger than of JEN2 whereas JEN2 is more induced than JEN1 in the presence of dicarboxylic acids (Fig. 4C). This differential regulation reinforces the notion that these transporters play distinct roles in C. albicans.
Both Jen1 and Jen2 are sensitive to glucose (Fig. 5). Following the addition of glucose at concentrations ≥ 0.1% to C. albicans cells growing on lactic acid, both Jen1-GFP and Jen2-GFP were rapidly internalized. Interestingly, the rates of Jen1-GFP and Jen2-GFP internalization in C. albicans were significantly faster than the rate of ScJen1-GFP internalization in S. cerevisiae (Fig. 5). Therefore, mechanistic differences might exist between C. albicans and S. cerevisiae with regard to the glucose mediated downregulation of Jen transporters in these yeasts. These results corroborate suggestions that C. albicans and S. cerevisiae may sense sugars differently (Brown et al., 2006) despite both of these yeasts being sensitive to low concentrations of glucose (Rodaki et al., 2009).
Considerable advances have been made in our understanding of how C. albicans responds to the metabolic stimuli encountered in its human host, but large gaps in our knowledge remain. Genome-wide expression profiling of C. albicans cells in ex vivo infection models have highlighted the significance of central metabolic pathways for the adaptation of this organism to its host (Lorenz and Fink, 2001; Lorenz and Fink, 2002; Fradin et al., 2003), and it is now clear that these pathways are regulated in a niche-specific manner during infection (Barelle et al., 2006). It has been postulated that glucose levels are low in the phagolysosome, but this microenvironment is rich in fatty acids and their products (Lorenz and Fink, 2001). These compounds may be sources of acetyl-CoA that feed the glyoxylate cycle (Lorenz and Fink, 2001; Lorenz et al., 2004). However, acetate and lactate may be the main sources of acetyl-coA. This idea is consistent with microarray data, which reveal an upregulation of JEN1 and JEN2 in C. albicans cells phagocytosed by macrophages and neutrophils (Lorenz et al., 2004; Fradin et al., 2005; Piekarska et al., 2006). Moreover, S. cerevisiae and C. albicans acetyl-CoA synthases, which are responsible for the conversion of acetate to acetyl-CoA, are upregulated (8.7- and 6.1-fold, respectively) after phagocytosis by macrophages and by neutrophils (Fradin et al., 2005). It was suggested that acetate might be a product of lactate degradation that sustains C. albicans inside the phagolysosome (Piekarska et al., 2006). Our results revealed that both Jen1-GFP and Jen2-GFP are expressed inside macrophages and neutrophils, but not in the bloodstream, a glucose-rich environment (Fig. 6). Moreover, in the murine model of systemic candidiasis about 20–25% of C. albicans cells infecting the kidney express Jen1 and Jen2 (Fig. 7), a similar value to Icl1 (Barelle et al., 2006). These results support the idea that carboxylic acids such as lactate are present inside the phagolysosome, and suggest that both monocarboxylates and dicarboxylates, such as succinate and malate, may act as carbon sources that help to sustain C. albicans following phagocytosis. However, the inactivation of Jen1 and Jen2 does not attenuate the virulence of C. albicans (Fig. 7C). This result implies that maybe other, yet uncharacterized, carboxylate transporters play a role in the uptake of further carboxylic acids, such as acetic or citric acids.
The biochemical pathways that mediate monocarboxylate and dicarboxylate anabolism in C. albicans during the infection process remain to be confirmed. For example, it is not clear how malic acid is metabolized in this organism. According to the Candida Genome Database, orf19.1867, which displays homology to S. pombe MAE1 (Grobler et al., 1995), encodes a putative malate transporter that is induced during phagocytosis by macrophages (Prigneau et al., 2003). Also, orf19.3419 encodes a putative mitochondrial malic enzyme, with homology to S. cerevisiae MAE1 (Boles et al., 1998). This has resulted in a nomenclature conflict, because both orf19.1867 and orf19.3419 are referred to MAE1 in C. albicans. In S. pombe the malic enzyme is encoded by MAE2, and it is a cytoplasmatic protein that, unlike ScMae1, has a high affinity for its substrate (Viljoen et al., 1994). Our results suggest that those C. albicans genes currently presumed to be malate transporters are not involved in the uptake of dicarboxylic acids across the plasma membrane, because deletion of JEN2 abolished the mediated uptake of succinate and malate under the conditions tested (Fig. 1). Nevertheless, it is conceivable that another malate transporter exists in C. albicans, with a different pattern of regulation. Alternatively, C. albicans might express other malic acid transporters at alternative cellular locations, for example in the peroxisome (Tournu et al., 2005; Piekarska et al., 2008).
The same intriguing questions arise for a malic acid enzyme, whose activity has been reported in C. albicans in vitro (Rao et al., 1962). This enzyme converts malic acid into pyruvate. Depending upon yeast species and growth conditions, malic acid can be further converted into oxaloacetate and enter the Krebs cycle or can be metabolized to ethanol via acetaldehyde during maloalcoholic fermentation (Rodriguez and Thornton, 1990). S. cerevisiae can only use small amounts of malic acid under anaerobic conditions in the presence of glucose, by fermenting it to ethanol or to succinate via fumarate (Kuczynski and Radler, 1982). In S. cereviase it has been speculated that malic enzyme could be involved in the anaplerotic supply of pyruvate during growth on ethanol and acetate (Boles et al., 1998). These and other questions have arisen, particularly in the light of recent work that suggests that significant differences exist between the regulatory networks governing carbon metabolism in S. cerevisiae and C. albicans (Ramirez and Lorenz, 2007; 2009; Carman et al., 2008). A complete understanding of the precise function and regulation of C. albicans carboxylate transporters will require quantitative knowledge of how responsive C. albicans is to changes in the activity of these transporters, how responsive these transporters are to changes in the environment, and how they interact with other proteins. These answers will be essential to have a detailed view of the metabolic flexibility of this important pathogen.
Experimental procedures
Yeast strains, plasmids and growth conditions
The yeast strains and the plasmids used in this work are listed in Tables 1 and 2 respectively. Cultures were maintained on YPD (Sherman, 1991). Yeast cells were grown at 30°C in minimal medium (yeast nitrogen base 0.67% w/v) supplemented with the appropriate requirements for prototrophic growth (Sherman, 1991) and containing carbon sources at the following concentrations: glucose (2% w/v: SD), lactic acid (0.5% v/v, pH 5.0), pyruvic acid (0.5% w/v, pH 5.0), succinic acid (1% v/v, pH 5.0) or malic acid (1% v/v, pH 5.0). Solid media was prepared adding agar (2% w/v) to the respective liquid media, and the pH for media containg carboxylic acids was always set either to 5.0 or 7.0. Cultures were harvested during the exponential phase (OD640nm = 0.5). Yeast cells were grown under repression conditions in SD. For derepression conditions, glucose-grown cells were harvested, washed twice in ice-cold deionized water and inoculated into fresh minimal medium supplemented with the carbon source of choice.
C. albicans mutant construction
The C. albicans JEN2 gene was identified through homology to ScJEN1 using the blast program. The two JEN2 alleles in C. albicans strains RM1000 (JEN1/JEN1: Table 1) were inactivated sequentially with the loxP-URA3-loxP (LUL) and loxP-HIS1-loxP (LHL) markers (Dennison et al., 2005). jen2::LUL and jen2::LHL disruption cassettes designed to delete the complete JEN2 open reading frame were generated by PCR amplification with the primers CaJEN2-S1Fwd and CaJEN2-S2Rev (Table 3). The jen2::LUL cassette was transformed into C. albicans (Gietz and Woods, 1998), transformants selected on the basis of their uridine protrophy, and correct integration confirmed by diagnostic PCR with primers DCaURA3-1Rev and DCaJEN2lcFwd (Table 3) (Wilson et al., 1999). The resultant heterozygote (jen2::loxP-URA3-loxP/JEN2) was then transformed with the jen2::LHL cassette, and accurate disruption of the remaining JEN2 allele confirmed by PCR using primers DCaHIS1-1Rev and DCaJEN2lcFwd, and CaJEN2A1 and CaJEN2A2 (Table 3). This yielded the homozygous null mutant, CNV3 (jen2::LUL/jen2::LHL: Table 1). The genotypes of this mutant and all other mutants made in this study were confirmed by Southern blotting (not shown).
Table 3.
Oligonucleotides used in this study.
| Name | Sequence |
|---|---|
| CaJEN1Fwd | GCATGCTTAGCACTGGCACTGACGCGTATTAGTAAATAGACTTTAATTTAG CTTTTACCC |
| CaJEN1Rev | ATCGAATGCTTAACTGATCACGTCGACTTTTTAGTATTTGATTGAATT GAATTGGTTATAAGA |
| CaJEN2Fwd | GCATGCTTAGCACTGGCACTGACGCGTGAGCACTAACAATTAGTTGTACAGTTCAAAACTCCG |
| CaJEN2Rev | ATCGAATGCTTAACTGATCACGTCGACCCGTCTCATATTTCTAACCGATTGTGCCAGTGGCTC |
| DRPS10 | GTGGTTGGAGCTTTGATG |
| DCaJEN2Rev | AGCCATGAGAGCCATCTC |
| CaJEN2A1 | GGTGATACATATGGTAGA |
| CaJEN2A2 | GTGATCCACATTGGATGG |
| CaJEN2-S1Fwd | GGATGAATTGAAACAATATTCCTGGCACGAAGTTCTTAATCCGTTTGAACCATTAGTGGA CCTCTTCGCTATTACGCCAG |
| CaJEN2-S2Rev | CGGCACCTCTGTTTTCAGGACCAATAAAAACAACAAACATCAAGTAAGCCAAAACAGCAC GCAGATTACCCTGTTATCCCTA |
| DCaURA3-1Rev | CTGCTCTCTCACTATAGGTC |
| DCaHIS1-1Rev | CGGTCTGGTAAATGATTGAC |
| DCaJEN2lcFwd | CCCAATACATCACATTAC |
| CaJEN2-DB1Fwd | CTAACCATAGAAATATTATGACTGCTGCTGATACTCATTCTATCACTAGTGCTGATG TTC TTTTCCCAGTCACGACGTT |
| CaJEN2-DB2Rev | CCACCAACACTTACTCTTTATGTTCAACTTCTGGTTTTTCCAAAGTCTTTTGAG TAATATTAC TGTGGAATTGTGAGCGGATA |
| CaJEN2-DB3Fwd | CATAATAGACACATTATTCGTCCACCAAAATTCACTTGGCCCGCTATTCGAAAATATGCC TTTTCCCAGTCACGACGTT |
| CaJEN2-DB4Rev | CTTCAAGATCAGAATCACCTCTGTCTTCCCTATCACTATCGTACGCACTGTATTCGTCATC TGTGGAATTGTGAGCGGATA |
| DminiURA3Rev | TAGAAGGACCACCTTTGATTGT |
| CaJEN2GFPFwd | GAAGAAGGTAATATTACTCAAAAGACTTTGGAAAAACCAGAA GTTGAACATAAAGAG GGTGGTGGTTCTAAAGGTGAAGAATTATTC |
| CaJEN2GFPRev | CACACACATACTATTTTAACAAATCATAAACCCATTTATTATCAAAATAAACTATACTTG TCTAGAAGGACCACCTTTGATTG |
| DJEN2GFP_Fwd | CTTGGTCAGTGGTGCCAA |
| GFP Rev | AACATCACCATCTAATTCAAC |
| JEN2_416For | GGGATCCAATATTATGACTGCTGCTGATACTCATTCTATC |
| JEN2_416Rev | GAAGCTTTTAGTGATGGTGATGGTGATGCTCTTTATGTTCAACTTCTGGTTTTTC |
| JEN2416ForREQ | AAAACACCAAGAACTTAGTTTCGACGGATTCTAGAACTAGTGGATCCAATATTATGACTGCTGCTGATACTCATTCTATC |
| JEN2416RevREQ | CATGACTCGAGGTCGACGGTATCGATAAGCTTTTAGTGATGGTGATGGTGATGCTCTTTATGTTCAACTTCTGGTTTTTC |
| URA3-dpl200fwd | TAAAACGACGGCCAGTGAAT |
| URA3-dpl200rev | ACCATGATTACGCCAAGCTC |
| RPS10 | ACTAATTCTTCTCTTCAG |
| Ins CaJEN1 | AAGTCTATTTACTAATACG |
| TDH promoter | ACAAGGCAATTGACCCACGCATGTATCTA |
| CYC Terminator | GAATGTAAGCGTGACATAACTAATTACATG |
DNA sequences complementary to the sequences of pLUL and pLHL (S1/S2), pDDB57 (DB1/DB2/DB3/DB4) and pGFP-URA3 (F1/R1) are shown in bold.
The JEN2 locus was also disrupted in C. albicans strain CPK2 (jen1/jen1) to create the double jen1jen2 mutant, CNV4 (Table 1). First, ura3- segregants of CPK2 were selected using 5-fluoroorotic acid (Boeke et al., 1984). The first JEN2 allele was disrupted using a cassette created by PCR amplification of the mini-URA3 blaster (Wilson et al., 2000) with primers CaJEN2-DB1 Fwd and CaJEN2-DB2 Rev (Table 3). Ura3- segregants were selected 5-fluoroorotic acid to create a jen1/jen1, jen2/JEN2 heterozygote. The remaining JEN2 allele in this strain was then disrupted using a second cassette created by PCR amplification of the mini-URA3 blaster (Wilson et al., 2000) with primers CaJEN2-DB3 Fwd and CaJEN2-DB4 (Table 3). Once again, ura3- segregants were selected 5-fluoroorotic acid to create a jen1/jen1, jen2/jen2 double mutant, CNV4 (Table 1). Correct integration of the cassettes and loss of the wild-type JEN2 allele were confirmed by diagnostic PCR with primers DCaJEN2lcFwd and DCaURA3-1Rev, DCaJEN2lcFwd and DminiURA3Rev, URA3-dpl200fwd and URA3-dpl200rev, and CaJEN2A1 and CaJEN2A2 (Table 3).
To reintroduce a functional JEN1 gene into strains CPK2 and CNV4, the JEN1 gene, plus approximately 2000 bp upstream and 600 bp downstream of its coding sequence were PCR amplified using primers CaJEN1Fwd and CaJEN1Rev (Table 3). The resulting PCR fragment was digested with SalI and MluI and ligated into CIp20 (Dennison et al., 2005) to create CIp20-JEN1. This plasmid was then digested with StuI and integrated at the RPS1 locus in C. albicans CPK2 and CNV4 (Murad et al., 2000), thereby generating CNV2-2 and CNV4-2 (Table 1). Correct integration at the RPS1 locus was confirmed by diagnostic PCR, using primers RPS10 and InsCaJEN1 (Table 3), and by Southern blot analysis. As controls the C. albicans strains CPK2, CNV3 and CNV4 were also transformed with the empty CIp20 plasmid. JEN1 reintegration suppressed all jen1/jen1 phenotypes, as expected (not shown). Multiple attempts were made to reintegrate JEN2 into jen2 mutant strains without success. It was not possible to clone the JEN2 locus into several different types of Escherichia coli or C. albicans vectors using distinct PCR strategies or by using the Clonetech Cloning System (In-Fusion™ 2.0 Dry-Down PCR Cloning Kit; http://www.clontech.com). Additionally several attempts to clone PCR-amplified JEN2 directly in S. cerevisiae and into C. albicans genomic locus proved unsuccessful. We conclude that some feature of the JEN2 locus precludes its cloning using a range of standard procedures. This meant that it was not possible to restoring JEN2 in the jen2 mutant. In the absence of a JEN2 reintegrant we compared the phenotypes of five independent jen2 mutants. These jen2 mutants displayed identical phenotypes under all the conditions tested.
Construction of the JEN2-GFP fusion in C. albicans
To tag C. albicans JEN2 at its 3-end, the GFP ORF was PCR amplified from pGFP-URA3 (Gerami-Nejad et al., 2001) using primers CaJEN2GFPFwd and CaJEN2GFPRev (Table 3). The resultant PCR product was used to transform C. albicans RM1000, and transformants were screened for correct integration by diagnostic PCR using primers DJEN2GFPFwd and GFPRev (Table 3).
Heterologous expression of C. albicans JEN2 in S. cerevisiae
Candida albicans JEN2 was cloned into the plasmid p416GPD (Mumberg et al., 1995) by gap repair (Orr-Weaver and Szostak, 1983). To achieve this, the JEN2 ORF was PCR amplified with primers JEN2416ForREQ and JEN2416RevREQ (Table 3). Both the plasmid and the PCR product were digested with BamHI and HindIII, purified from an agarose gel, and co-transformed into the S. cerevisiae jen1 mutant (Table 1). Correct clones were identified by colony PCR using primers TDH promoter and CYC terminator (Table 3).
DNA manipulations
Cloning, PCR amplification and Southern analysis were performed as described previously (Sambrook et al., 1989; Dennison et al., 2005).
Transport assays
Cells incubated under derepressing conditions were harvested by centrifugation, washed twice in ice-cold deionized water and resuspended in ice-cold deionized water to a final concentration of about 25–40 mg dry wt. ml−1. 10 µl of yeast cell suspension were mixed in 10 ml of conical tubes with 30 µl of 0.1 M potassium phosphate, pH 5.0. After 2 min of incubation at 30°C in a water bath, the reaction was started by the addition of 10 µl of an aqueous solution of labelled carboxylic acid at the desired concentration and pH value, and stopped by dilution with 5 ml of ice-cold water. The reaction mixtures were filtered immediately through Whatman GF/C membranes, the filters washed with 10 ml of ice-cold water and transferred to scintillation fluid (Opti-phase HiSafe II; LKB FSA Laboratory Supplies, Loughborough, UK). Radioactivity was measured in a Packard Tri-Carb 2200 CA liquid scintillation counter. The following radiolabelled substrates were utilized: D,L-[U-14C] lactic acid, sodium salt (CFB97–Amersham Biosciences); [2,3-14C] succinic acid (NEN Life Science); and L-[1,4(2,3)-14C] malic acid (CFB42-Amersham). Nonspecific 14C adsorption to the filters and to the cells was determined by adding labelled acid after ice-cold water. Background values represented less than 5% of the total incorporated radioactivity. The transport kinetics best fitting the experimental initial uptake rates and the kinetic parameters were determined by a computer-assisted nonlinear regression analysis (GraphPAD Software, San Diego, CA, USA).
Microscopy
Candida albicans and S. cerevisiae cells were examined with a Leica Microsystems DM-5000B epifluorescence microscope with appropriate filter settings. Images were acquired with a Leica DCF350FX digital camera and processed with LAS AF Leica Microsystems software.
RNA isolation and qRT-PCR analysis
Candida albicans wild-type and mutant cells were grown in YNB media supplemented with 2% glucose, till an OD640nm of approximately 0.5, and derepressed for 4 h in media containing different carbon sources, prepared as previously described. Total RNA was then isolated using the standard hot acidic phenol protocol. qRT-PCR was carried out to analyse the expression of JEN1/JEN2 and ACT1 in the conditions tested. qRT-PCR was performed in an LC480 equipment using the standard real-time PCR conditions. Ct values were transformed in expression values using standard curves made on a pool of samples. Both cDNA samples and RT-minus reactions (RNA samples treated in the same way but without the addition of the RT enzyme in the reverse transcription reaction) were analysed, and the level of cDNA was considered as the expression signal of each sample substracting the RT-minus signal. Finally, relative expression was calculated [(Gene of interest cDNA conc)/(calibrator cDNA conc)].
Cell extracts and immunoblotting
Cells were grown in glucose 2% and derepressed for 4 h in media containing different carbon sources. Lactic acid derepressed cells were also subjected to pulses of 0.01, 0.05 and 0.1% glucose. Preparation of total protein extracts followed the NaOHTCA lysis technique (Volland et al., 1994). Sample buffer was added to extracted proteins, heated at 37°C and resolved by SDS polyacrylamide gel electrophoresis in 10% acrylamide gels. The gels were run using a tricine buffer and transferred to PVDF membranes that were probed with monoclonal anti-GFP antiserum (Roche diagnostics) and anti-ACT1 (abcam, Cambrige, UK). Horseradish peroxidase-conjugated anti-mouse immunoglobulin G was used as the secondary antibody (Sigma, St Louis, MO USA) and was detected by enhanced chemiluminescence (ECL).
C. albicans morphogenesis
Different environmental signals were used to induce hyphal development in C. albicans. To trigger hyphal development by serum yeast cells were grown in minimal medium containing 0.5% lactic acid, pH 5.0, and then they were incubated with 0, 10 or 20% FBS at 37°C, with agitation, for 3 h (Swoboda et al., 1994). Cells were also platted in YPD agar supplemented with 10% FBS. To monitor the effect of pH, growth was also carried out in minimal medium, but at pH 6.5. The starter culture was divided into two flasks with lactic acid 0.5%, at pH 4.5 and pH 6.5. The culture at pH 6.5 was incubated at 37°C, whereas the one at pH 4.5 at 25°C, for 5 h. Finally, a nutrient limitation stress was imposed by incubating the cells with media containing N-acetylglucosamine (Mattia et al., 1982). The cells were grown in Lee's medium at pH 4.5, 37°C, for several days, and then starved for 24 h, at 37°C. Afterwards, they were ressuspended in BSM medium with or without the addition of 4 mM GlcNAc. All flasks were then incubated for 5 h, at 37°C.
Ex vivo models of C. albicans phagocytosis
Human Blood from several donors was collected by venepuncture using heparinized tubes. For blood killing assays, C. albicans cells were grown in the appropriate carbon source to an OD640 = 0.5, and then incubated with 100 µl of whole blood (100 cells: 100 µl of whole blood/neutrophils) at 37°C for 1 h. Cells were then plated on YPD and colony forming units (CFU) counted after 24 h, at 30°C. Neutrophils were isolated from human blood (Fradin et al., 2005), C. albicans cells were washed with phosphate-buffered saline (PBS) and incubated with neutrophils in a 1:1 ratio. GFP fluorescence was measured after 2.5 h at 37°C (Barelle et al., 2004). Control C. albicans cells were incubated with human plasma. Cultured murine RAW 264.7 macrophage (ECACC, Salisbury, UK), kindly provided by Leanne Clift (University of Aberdeen, UK), were diluted to 1 × 106 cells ml−1 in supplemented DMEM media, plated in six well plates, and grown overnight at 37°C, under 5% CO2. C. albicans CPK20-5 and CNV30-5 (Table 1) were grown also overnight in minimal medium containing 0.5% lactic acid, pH 5.0 to an OD640 = 0.5, and then counted with a haemocytometer. Approximately 3 × 108 cells were then added to the wells containing macrophages to give a C. albicans macrophage ratio of 3:1. Samples were then incubated for 3 h, at 37°C, under 5% CO2. Control cultures of C. albicans strains containing pICL1-GFP or the empty pGFP vector (Barelle et al., 2004) were also incubated with macrophages (3:1). To provide a further control, C. albicans cells were incubated in macrophage growth medium (DMEM containing FBS and glutamine) with no macrophages. Cells were fixed, mounted and GFP quantified as described previously (Barelle et al., 2004; 2006;).
Murine model of systemic candidiasis
Female BALB/c mice (Harlan, UK) were handled and maintained according to the conditions specified by the Home Office (UK) regulations. As described previously (Barelle et al., 2006), mice of approximately 6–8 weeks were infected with 2–6 × 104C. albicans cells/g body weight by lateral tail vein injection. Actual levels of inoculation were assayed by viable plate counting. Fungal burdens and in vivo kidney sections were analysed after 3 or 4 days of infection (Fradin et al., 2005). Kidneys were removed aseptically. Half of each kidney was used for determination of fungal burdens, and the other half was fixed in 4% paraformaldehyde. Fixed kidneys were embedded in Cryo-M-Bed (Bright, Huntingdon, UK) and flash-frozen. Sections (5 µm) were cut and stained with Calcofluor white to identify fungal cells (Barelle et al., 2004). Images were generated at 461 nm (Calcofluor white staining), 516 nm (GFP) and 573 nm (Rhodamine as a control for GFP specificity). For the virulence assay strains were grown for 16 h in NGY medium (0.1% Neopeptone 0.4% glucose, 0.1% yeast extract), washed twice in sterile physiological saline and resuspended in saline to produce inocula of 5 × 104 CFU g−1 bodyweight per mouse. Actual inocula were determined from viable plating of the inocula. Fungal burdens were determined for the kidneys and spleen of all mice. Organs were homogenized in 0.5 ml of saline and dilutions of the resulting homogenate plated onto Saboraud agar. Plates were incubated overnight at 35°C, and then colonies counted. Survival of mice was analysed by log rank/Kaplan–Meier statistics. Organ burdens were compared by the Mann–Whitney U-test.
Acknowledgments
We thank Alexandra Rodaki, Brice Enjalbert and Susan Nicholls for their helpful advice, and all the members of the Aberdeen Fungal Group for their support. We also acknowledge Drs Bernard Hube and Mike Lorenz for fruitful discussions. This study was supported by the Portuguese grant POCI/BIA-BCM/57812/2004 (Eixo 2, Medida 2.3, QCAIII – FEDER). N. V. received FCT PhD fellowship (SFRH/BD/23503/2005). The work performed in Aberdeen was funded by the BBSRC (BB/C510391/1), and by the Wellcome Trust (080088).
References
- Akita O, Nishimori C, Shimamoto T, Fujii T, Iefuji H. Transport of pyruvate in Saccharomyces cerevisiae and cloning of the gene encoded pyruvate permease. Biosci Biotechnol Biochem. 2000;64:980–984. doi: 10.1271/bbb.64.980. [DOI] [PubMed] [Google Scholar]
- Alonso-Monge R, Navarro-Garcia F, Molero G, Diez-Orejas R, Gustin M, Pla J, et al. Role of the mitogen-activated protein kinase Hog1p in morphogenesis and virulence of Candida albicans. J Bacteriol. 1999;181:3058–3068. doi: 10.1128/jb.181.10.3058-3068.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade RP, Casal M. Expression of the lactate permease gene JEN1 from the yeast Saccharomyces cerevisiae. Fungal Genet Biol. 2001;32:105–111. doi: 10.1006/fgbi.2001.1254. [DOI] [PubMed] [Google Scholar]
- Barelle CJ, Manson CL, MacCallum DM, Odds FC, Gow NA, Brown AJ. GFP as a quantitative reporter of gene regulation in Candida albicans. Yeast. 2004;21:333–340. doi: 10.1002/yea.1099. [DOI] [PubMed] [Google Scholar]
- Barelle CJ, Priest CL, Maccallum DM, Gow NA, Odds FC, Brown AJ. Niche-specific regulation of central metabolic pathways in a fungal pathogen. Cell Microbiol. 2006;8:961–971. doi: 10.1111/j.1462-5822.2005.00676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensen ES, Martin SJ, Li M, Berman J, Davis DA. Transcriptional profiling in Candida albicans reveals new adaptive responses to extracellular pH and functions for Rim101p. Mol Microbiol. 2004;54:1335–1351. doi: 10.1111/j.1365-2958.2004.04350.x. [DOI] [PubMed] [Google Scholar]
- Berman J, Sudbery PE. Candida albicans: a molecular revolution built on lessons from budding yeast. Nat Rev Genet. 2002;3:918–930. doi: 10.1038/nrg948. [DOI] [PubMed] [Google Scholar]
- Boeke JD, LaCroute F, Fink GR. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Boles E, de Jong-Gubbels P, Pronk JT. Identification and characterization of MAE1, the Saccharomyces cerevisiae structural gene encoding mitochondrial malic enzyme. J Bacteriol. 1998;180:2875–2882. doi: 10.1128/jb.180.11.2875-2882.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AJ, Gow NA. Regulatory networks controlling Candida albicans morphogenesis. Trends Microbiol. 1999;7:333–338. doi: 10.1016/s0966-842x(99)01556-5. [DOI] [PubMed] [Google Scholar]
- Brown V, Sexton JA, Johnston M. A glucose sensor in Candida albicans. Eukaryot Cell. 2006;5:1726–1737. doi: 10.1128/EC.00186-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffo J, Herman MA, Soll DR. A characterization of pH-regulated dimorphism in Candida albicans. Mycopathologia. 1984;85:21–30. doi: 10.1007/BF00436698. [DOI] [PubMed] [Google Scholar]
- Calderone RA. Candida and Candidiasis. Washington, DC: American Society for Micobiology Press; 2002. [Google Scholar]
- Calderone RA, Fonzi WA. Virulence factors of Candida albicans. Trends Microbiol. 2001;9:327–335. doi: 10.1016/s0966-842x(01)02094-7. [DOI] [PubMed] [Google Scholar]
- Carman AJ, Vylkova S, Lorenz MC. Role of acetyl coenzyme A synthesis and breakdown in alternative carbon source utilization in Candida albicans. Eukaryot Cell. 2008;7:1733–1741. doi: 10.1128/EC.00253-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal M, Paiva S, Andrade RP, Gancedo C, Leao C. The lactate-proton symport of Saccharomyces cerevisiae is encoded by JEN1. J Bacteriol. 1999;181:2620–2623. doi: 10.1128/jb.181.8.2620-2623.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal M, Paiva S, Queiros O, Soares-Silva I. Transport of carboxylic acids in yeasts. FEMS Microbiol Rev. 2008;32:974–994. doi: 10.1111/j.1574-6976.2008.00128.x. [DOI] [PubMed] [Google Scholar]
- Cormack BP, Bertram G, Egerton M, Gow NA, Falkow S, Brown AJ. Yeast-enhanced green fluorescent protein (yEGFP) a reporter of gene expression in Candida albicans. Microbiology. 1997;143:303–311. doi: 10.1099/00221287-143-2-303. [DOI] [PubMed] [Google Scholar]
- Dennison PM, Ramsdale M, Manson CL, Brown AJ. Gene disruption in Candida albicans using a synthetic, codon-optimised Cre-loxP system. Fungal Genet Biol. 2005;42:737–748. doi: 10.1016/j.fgb.2005.05.006. [DOI] [PubMed] [Google Scholar]
- El Barkani A, Kurzai O, Fonzi WA, Ramon A, Porta A, Frosch M, Muhlschlegel FA. Dominant active alleles of RIM101 (PRR2) bypass the pH restriction on filamentation of Candida albicans. Mol Cell Biol. 2000;20:4635–4647. doi: 10.1128/mcb.20.13.4635-4647.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradin C, Kretschmar M, Nichterlein T, Gaillardin C, d'Enfert C, Hube B. Stage-specific gene expression of Candida albicans in human blood. Mol Microbiol. 2003;47:1523–1543. doi: 10.1046/j.1365-2958.2003.03396.x. [DOI] [PubMed] [Google Scholar]
- Fradin C, De Groot P, MacCallum D, Schaller M, Klis F, Odds FC, Hube B. Granulocytes govern the transcriptional response, morphology and proliferation of Candida albicans in human blood. Mol Microbiol. 2005;56:397–415. doi: 10.1111/j.1365-2958.2005.04557.x. [DOI] [PubMed] [Google Scholar]
- Gerami-Nejad M, Berman J, Gale CA. Cassettes for PCR-mediated construction of green, yellow, and cyan fluorescent protein fusions in Candida albicans. Yeast. 2001;18:859–864. doi: 10.1002/yea.738. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Woods RA. Transformation of yeast by the lithium acetate/single-stranded carrier DNA/PEG method. In: Brown AJP, Tuite MF, editors. Methods in Microbiology: Yeast Gene Analysis. London: Academic Stress; 1998. pp. 53–66. [Google Scholar]
- Gow NA, Gooday GW. Growth kinetics and morphology of colonies of the filamentous form of Candida albicans. J Gen Microbiol. 1982;128:2187–2194. doi: 10.1099/00221287-128-9-2187. [DOI] [PubMed] [Google Scholar]
- Gow NA, Brown AJ, Odds FC. Fungal morphogenesis and host invasion. Curr Opin Microbiol. 2002;5:366–371. doi: 10.1016/s1369-5274(02)00338-7. [DOI] [PubMed] [Google Scholar]
- Grobler J, Bauer F, Subden RE, Van Vuuren HJ. The mae1 gene of Schizosaccharomyces pombe encodes a permease for malate and other C4 dicarboxylic acids. Yeast. 1995;11:1485–1491. doi: 10.1002/yea.320111503. [DOI] [PubMed] [Google Scholar]
- Hwang CS, Rhie GE, Oh JH, Huh WK, Yim HS, Kang SO. Copper- and zinc-containing superoxide dismutase (Cu/ZnSOD) is required for the protection of Candida albicans against oxidative stresses and the expression of its full virulence. Microbiology. 2002;148:3705–3713. doi: 10.1099/00221287-148-11-3705. [DOI] [PubMed] [Google Scholar]
- Kaur S, Mishra P. Differential increase in cytoplasmic pH at bud and germ tube formation in Candida albicans: studies of a nongerminative variant. Can J Microbiol. 1994;40:720–723. doi: 10.1139/m94-114. [DOI] [PubMed] [Google Scholar]
- Kuczynski JT, Radler F. The anaerobic metabolism of malate of Saccharomyces bailii and the partial purification and characterization of malic enzyme. Arch Microbiol. 1982;131:266–270. doi: 10.1007/BF00405891. [DOI] [PubMed] [Google Scholar]
- Lodi T, Fontanesi F, Ferrero I, Donnini C. Carboxylic acids permeases in yeast: two genes in Kluyveromyces lactis. Gene. 2004;339:111–119. doi: 10.1016/j.gene.2004.06.019. [DOI] [PubMed] [Google Scholar]
- Lodi T, Diffels J, Goffeau A, Baret PV. Evolution of the carboxylate Jen transporters in fungi. FEMS Yeast Res. 2007;7:646–656. doi: 10.1111/j.1567-1364.2007.00245.x. [DOI] [PubMed] [Google Scholar]
- Lorenz MC, Fink GR. The glyoxylate cycle is required for fungal virulence. Nature. 2001;412:83–86. doi: 10.1038/35083594. [DOI] [PubMed] [Google Scholar]
- Lorenz MC, Fink GR. Life and death in a macrophage: role of the glyoxylate cycle in virulence. Eukaryot Cell. 2002;1:657–662. doi: 10.1128/EC.1.5.657-662.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz MC, Bender JA, Fink GR. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot Cell. 2004;3:1076–1087. doi: 10.1128/EC.3.5.1076-1087.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCallum DM, Odds FC. Temporal events in the intravenous challenge model for experimental Candida albicans infections in female mice. Mycoses. 2005;48:151–161. doi: 10.1111/j.1439-0507.2005.01121.x. [DOI] [PubMed] [Google Scholar]
- Mattia E, Carruba G, Angiolella L, Cassone A. Induction of germ tube formation by N-acetyl-D-glucosamine in Candida albicans: uptake of inducer and germinative response. J Bacteriol. 1982;152:555–562. doi: 10.1128/jb.152.2.555-562.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollapour M, Piper PW. Hog1 mitogen-activated protein kinase phosphorylation targets the yeast Fps1 aquaglyceroporin for endocytosis, thereby rendering cells resistant to acetic acid. Mol Cell Biol. 2007;27:6446–6456. doi: 10.1128/MCB.02205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- Murad AM, Lee PR, Broadbent ID, Barelle CJ, Brown AJ. CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast. 2000;16:325–327. doi: 10.1002/1097-0061(20000315)16:4<325::AID-YEA538>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Negredo A, Monteoliva L, Gil C, Pla J, Nombela C. Cloning, analysis and one-step disruption of the ARG5,6 gene of Candida albicans. Microbiology. 1997;143:297–302. doi: 10.1099/00221287-143-2-297. [DOI] [PubMed] [Google Scholar]
- Odds FC. Candida and Candidosis. 2nd. London: Bailliere Tindall; 1988. [Google Scholar]
- Orr-Weaver TL, Szostak JW. Multiple, tandem plasmid integration in Saccharomyces cerevisiae. Mol Cell Biol. 1983;3:747–749. doi: 10.1128/mcb.3.4.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiva S, Kruckeberg AL, Casal M. Utilization of green fluorescent protein as a marker for studying the expression and turnover of the monocarboxylate permease Jen1p of Saccharomyces cerevisiae. Biochem J. 2002;363:737–744. doi: 10.1042/0264-6021:3630737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiva S, Devaux F, Barbosa S, Jacq C, Casal M. Ady2p is essential for the acetate permease activity in the yeast Saccharomyces cerevisiae. Yeast. 2004;21:201–210. doi: 10.1002/yea.1056. [DOI] [PubMed] [Google Scholar]
- Paiva S, Vieira N, Nondier I, Haguenauer-Tsapis R, Casal M, Urban-Grimal D. Glucose-induced ubiquitylation and endocytosis of the yeast Jen1 transporter: role of lysine 63-linked ubiquitin chains. J Biol Chem. 2009;284:19228–19236. doi: 10.1074/jbc.M109.008318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlroth J, Choi B, Spellberg B. Nosocomial fungal infections: epidemiology, diagnosis, and treatment. Med Mycol. 2007;45:321–346. doi: 10.1080/13693780701218689. [DOI] [PubMed] [Google Scholar]
- Piekarska K, Mol E, van den Berg M, Hardy G, van den Burg J, van Roermund C, et al. Peroxisomal fatty acid beta-oxidation is not essential for virulence of Candida albicans. Eukaryot Cell. 2006;5:1847–1856. doi: 10.1128/EC.00093-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekarska K, Hardy G, Mol E, van den Burg J, Strijbis K, van Roermund C, et al. The activity of the glyoxylate cycle in peroxisomes of Candida albicans depends on a functional beta-oxidation pathway: evidence for reduced metabolite transport across the peroxisomal membrane. Microbiology. 2008;154:3061–3072. doi: 10.1099/mic.0.2008/020289-0. [DOI] [PubMed] [Google Scholar]
- Prigneau O, Porta A, Poudrier JA, Colonna-Romano S, Noel T, Maresca B. Genes involved in beta-oxidation, energy metabolism and glyoxylate cycle are induced by Candida albicans during macrophage infection. Yeast. 2003;20:723–730. doi: 10.1002/yea.998. [DOI] [PubMed] [Google Scholar]
- Queirós O, Paiva S, Moradas-Ferreira P, Casal M. Molecular and physiological characterization of monocarboxylic acids permeases in the yeast Kluyveromyces lactis. Yeast. 2003;20:S237. [Google Scholar]
- Queiros O, Pereira L, Paiva S, Moradas-Ferreira P, Casal M. Functional analysis of Kluyveromyces lactis carboxylic acids permeases: heterologous expression of KlJEN1 and KlJEN2 genes. Curr Genet. 2007;51:161–169. doi: 10.1007/s00294-006-0107-9. [DOI] [PubMed] [Google Scholar]
- Ramirez MA, Lorenz MC. Mutations in alternative carbon utilization pathways in Candida albicans attenuate virulence and confer pleiotropic phenotypes. Eukaryot Cell. 2007;6:280–290. doi: 10.1128/EC.00372-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez MA, Lorenz MC. The transcription factor homolog CTF1 regulates β-oxidation in Candida albicans. Eukaryot Cell. 2009;8:1604–1614. doi: 10.1128/EC.00206-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao GR, Sirsi M, Ramakrishnan T. Enzymes in Candida albicans. II. Tricarboxylic acid cycle and related enzymes. J Bacteriol. 1962;84:778–783. doi: 10.1128/jb.84.4.778-783.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodaki A, Enjalbert B, Young T, Odds FC, Gow NAR, Brown AJP. Glucose promotes stress resistance in the fungal pathogen, Candida albicans. Mol Biol Cell. 2009;20:4845–4855. doi: 10.1091/mbc.E09-01-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez SB, Thornton RJ. Factors influencing the utilisation of l-malate by yeasts. FEMS Microbiol Lett. 1990;60:17–22. doi: 10.1016/0378-1097(90)90337-p. [DOI] [PubMed] [Google Scholar]
- Rubin-Bejerano I, Fraser I, Grisafi P, Fink GR. Phagocytosis by neutrophils induces an amino acid deprivation response in Saccharomyces cerevisiae and Candida albicans. Proc Natl Acad Sci USA. 2003;100:11007–11012. doi: 10.1073/pnas.1834481100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon JM. l-malic-acid permeation in resting cells of anaerobically grown Saccharomyces cerevisiae. Biochim Biophys Acta. 1987;901:30–34. doi: 10.1016/0005-2736(87)90253-7. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Scannell DR, Butler G, Wolfe KH. Yeast genome evolution – the origin of the species. Yeast. 2007;24:929–942. doi: 10.1002/yea.1515. [DOI] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Soares-Silva I, Paiva S, Kotter P, Entian KD, Casal M. The disruption of JEN1 from Candida albicans impairs the transport of lactate. Mol Membr Biol. 2004;21:403–411. doi: 10.1080/09687860400011373. [DOI] [PubMed] [Google Scholar]
- Soares-Silva I, Paiva S, Diallinas G, Casal M. The conserved sequence NXX[S/T]HX[S/T]QDXXXT of the lactate/pyruvate:H(+) symporter subfamily defines the function of the substrate translocation pathway. Mol Membr Biol. 2007;24:464–474. doi: 10.1080/09687680701342669. [DOI] [PubMed] [Google Scholar]
- Stewart E, Gow NA, Bowen DV. Cytoplasmic alkalinization during germ tube formation in Candida albicans. J Gen Microbiol. 1988;134:1079–1087. doi: 10.1099/00221287-134-5-1079. [DOI] [PubMed] [Google Scholar]
- Stewart E, Hawser S, Gow NA. Changes in internal and external pH accompanying growth of Candida albicans: studies of non-dimorphic variants. Arch Microbiol. 1989;151:149–153. doi: 10.1007/BF00414430. [DOI] [PubMed] [Google Scholar]
- Strijbis K, van Roermund CW, Visser WF, Mol EC, van den Burg J, MacCallum DM, et al. Carnitine-dependent transport of acetyl coenzyme A in Candida albicans is essential for growth on nonfermentable carbon sources and contributes to biofilm formation. Eukaryot Cell. 2008;7:610–618. doi: 10.1128/EC.00017-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swoboda RK, Bertram G, Delbruck S, Ernst JF, Gow NA, Gooday GW, Brown AJ. Fluctuations in glycolytic mRNA levels during morphogenesis in Candida albicans reflect underlying changes in growth and are not a response to cellular dimorphism. Mol Microbiol. 1994;13:663–672. doi: 10.1111/j.1365-2958.1994.tb00460.x. [DOI] [PubMed] [Google Scholar]
- Thomas BJ, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- Tournu H, Serneels J, Van Dijck P. Fungal pathogens research: novel and improved molecular approaches for the discovery of antifungal drug targets. Curr Drug Targets. 2005;6:909–922. doi: 10.2174/138945005774912690. [DOI] [PubMed] [Google Scholar]
- Viljoen M, Subden RE, Krizus A, Van Vuuren HJ. Molecular analysis of the malic enzyme gene (mae2) of Schizosaccharomyces pombe. Yeast. 1994;10:613–624. doi: 10.1002/yea.320100506. [DOI] [PubMed] [Google Scholar]
- Volland C, Urban-Grimal D, Geraud G, Haguenauer-Tsapis R. Endocytosis and degradation of the yeast uracil permease under adverse conditions. J Biol Chem. 1994;269:9833–9841. [PubMed] [Google Scholar]
- Wilson RB, Davis D, Mitchell AP. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J Bacteriol. 1999;181:1868–1874. doi: 10.1128/jb.181.6.1868-1874.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RB, Davis D, Enloe BM, Mitchell AP. A recyclable Candida albicans URA3 cassette for PCR product-directed gene disruptions. Yeast. 2000;16:65–70. doi: 10.1002/(SICI)1097-0061(20000115)16:1<65::AID-YEA508>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Wysong DR, Christin L, Sugar AM, Robbins PW, Diamond RD. Cloning and sequencing of a Candida albicans catalase gene and effects of disruption of this gene. Infect Immun. 1998;66:1953–1961. doi: 10.1128/iai.66.5.1953-1961.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Lorenz MC. Carnitine acetyltransferases are required for growth on non-fermentable carbon sources but not for pathogenesis in Candida albicans. Microbiology. 2008;154:500–509. doi: 10.1099/mic.0.2007/014555-0. [DOI] [PubMed] [Google Scholar]