INTRODUCTION
Librarians are in the recommendation business. Our customers rely on us to recommend what they should read, which database is preferable over another, or which textbook might answer a background question. As digital gate counts increase and outpace traditional face-to-face interactions [1], the need to integrate librarian recommendations into digital systems grows. SmartSearch represents an automated approach to offering digital expert guidance to customers.
The Lane Medical Library & Knowledge Management Center provides information access and knowledge management services for the Stanford University School of Medicine, Stanford Hospital, and the Lucile Packard Children's Hospital. Lane's mission is to get the right knowledge, to the right person, at the right time, in the right context to support translational research, innovative education, and advances in patient care. This is largely accomplished via the LaneConnex web interface [2], a library search platform that performs a metasearch across hundreds of licensed and open access knowledge resources.
Like many academic health libraries, Lane's clinical collection consists of thousands of electronic journals and textbooks. This wealth of knowledge is daunting to users who are often overwhelmed by the sheer quantity of information. Lane's usage statistics show that clinical users consistently overlook expensive clinical resources (e.g., specialty textbooks from AccessMedicine, MDConsult, and Ovid) that librarians have selected for their high value and clinical relevance. SmartSearch addresses this issue.
The goal of the SmartSearch project is to recommend a small number of infrequently consulted, high-value, clinically relevant resources in the context of a standard LaneConnex search. SmartSearch is a resource promotion tool that leverages librarian expertise with the Entrez Programming Utilities (E-Utilities) [3] from the National Center for Biotechnology Information (NCBI) to mimic the information-seeking behavior of a typical reference librarian. System design, development, and an evaluation of its effectiveness will be described.
SMARTSEARCH
The SmartSearch design team consisted of two biomedical librarians, two software developers, a web production specialist, and an interface designer. In designing a recommender system to return “optimally appropriate” clinical resources for a given user query, three general approaches were considered: item-to-item correlation, people-to-people correlation, and attribute-based recommendations [4]. Given an absence of usable user preference data, item-to-item and people-to-people approaches were quickly discarded. Approaches based on popularity alone were also problematic because the resources targeted for recommendation were so lightly used. Given these constraints, an attribute-based recommendation system was selected.
SmartSearch was first deployed in November of 2007. Results appear in the recommendation area of LaneConnex, an area also used for spelling suggestions and exact journal title matches.
Handcrafted Medical Subject Headings (MeSH)-to-resource maps created and maintained by Lane librarians drive SmartSearch recommendations. Recommendations are drawn from a pool of 156 clinical metasearch targets. A metasearch target is a remote resource that Lane's metasearch application searches at either the individual title level (e.g., Abeloff's Clinical Oncology or The American Journal of Bioethics) or across an entire collection (e.g., Clin-eguide, Micromedex). Librarians selected 130 of the 156 metasearch targets to be included in the SmartSearch project. Approximately 85% of these 130 resources are individual clinical textbook, handbook, or atlas titles. To build maps of MeSH terms to recommended resources, Lane librarians were initially aided by the detailed descriptive work done by Lane's cataloging staff. Each resource was described in the Lane catalog using MeSH, and a list of all metasearch targets was then extracted and sorted by MeSH term. The web production specialist then consulted with individual librarians to review and revise maps pertinent to areas of expertise. For example, the librarian for internal medicine added the MeSH term “Nephrology” to two textbook metasearch targets already listed under the heading “Kidney Diseases”: Diseases of the Kidney and Urinary Tract and Brenner and Rector's The Kidney.
Lane librarians, in their role as department liaisons and domain experts, are responsible for the ongoing maintenance and improvement of these subject-specific resource recommendations. At the time of writing, SmartSearch subject maps contained 204 distinct MeSH terms mapped to 130 distinct resources. The other 26 clinical metasearch targets were excluded from the project because they were too general in scope for SmartSearch recommendations, a sufficient number of targets for a given topic were already included, or they were simply overlooked.
SmartSearch maps free-text queries to headings in the MeSH hierarchy. The mapping algorithm automates a common literature search technique known as “pearling.” Pearling is keyword searching in an article database followed by close inspection of the attributes of a handful of relevant and authoritative articles. These attributes are then used to build a more refined search strategy [5]. Three reasons motivate selection of this strategy: the overall strength of the NCBI search engine, the “living” nature of the corpus of PubMed content, and local familiarity with NCBI's application programming interface.
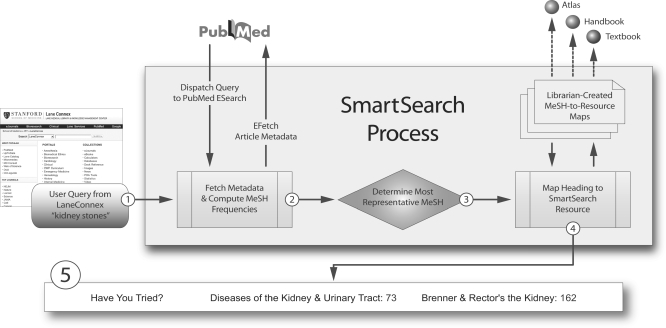
SmartSearch automates this pearling behavior in two phases. First, NCBI's ESearch and EFetch E-Utilities are used to map a user's query to a MeSH term. Second, a locally developed mapping engine matches harvested headings to the librarian-created list of subject-appropriate resources. Each time a user searches LaneConnex, SmartSearch executes a PubMed search using the user's query terms limited to the MEDLINE subset. If PubMed returns results, the top 100 articles are fetched using the EFetch utility. MeSH terms are extracted from each article, and occurrence frequencies are calculated for every unique heading extracted. These frequencies are then compared against a list of known uninformative headings (e.g., “Humans,” “Adult,” “Male”) and weighted accordingly. The heading with the highest weighted frequency score is then selected as the most representative heading for the user's query. This heading is used to search the librarian-created, MeSH-based list of SmartSearch resources, and resources matching the heading are returned to the user interface. For example, the query “kidney stones” maps to the MeSH term “Kidney Diseases,” which in turn produces two high-value clinical textbooks, Diseases of the Kidney and Urinary Tract and Brenner and Rector's The Kidney (Figure 1).
Figure 1.
SmartSearch system flowchart
1. User inputs query terms into LaneConnex: “kidney stones.”
2. SmartSearch dispatches the query to PubMed using ESearch (one of NCBI's Entrez Programming Utilities) and fetches resulting article metadata using EFetch (limited to first 100 articles).
3. Medical Subject Headings (MeSH) terms are extracted and used to compute the most frequently used heading.
4. The algorithm consults a list of known SmartSearch headings and tries to match the heading from step 3. If an exact match is made, resource recommendations for that heading are returned; if no match is made, it traverses up the MeSH hierarchy looking for a known SmartSearch match.
5. One to three “best answer” resources are returned to the LaneConnex user interface if a match in step 4 is made. SmartSearch is silent if no match is found (majority of known-item searches).
If an exact heading-to-resource match cannot be made, the SmartSearch heading-to-resource mapping engine traverses the MeSH tree seeking a broader heading with resource recommendations associated with it. This tree traversal allows the librarian to use broad MeSH terms when associating headings with resource recommendations and keeps the MeSH-to-resource maps manageable in size and complexity. For example, “Kidney Calculi” is the most frequent heading in the “kidney stones” query, but the SmartSearch heading-to-resource mapping engine traverses the MeSH tree until it finds resource recommendations under the heading “Kidney Diseases.” The mapping engine gives priority to heading matches with the greatest specificity by using the structure of MeSH tree numbers: the more specific a MeSH term, the more dots it will have in its associated tree numbers. For example, compare “Kidney Calculi” (C13.351.968.967.500.503) to “Kidney Diseases” (C12.777.419). As SmartSearch examines headings broader than the one associated with a user's query terms, it seeks to return recommendations associated with the heading with the most dots. When a query-to-heading or a heading-to-resource match cannot be made (as is the case in a majority of LaneConnex searches, especially those nonclinical in nature), SmartSearch is silent.
METHODS
Usage frequencies of the 156 individual clinical metasearch targets in Lane's clinical metasearch interface were analyzed before and after the introduction of SmartSearch to evaluate its effectiveness as a library resource promotion tool. One hundred thirty of these resources were included in the SmartSearch project; the other 26 were available for use in Lane's clinical metasearch interface but had not been selected for SmartSearch for the reasons mentioned above. The authors hypothesized that resources recommended by the SmartSearch system would be used more heavily than similar resources not included in the project. In statistical terms, the team attempted to disprove the null hypothesis, whereby no statistically significant differences in usage frequency would be observed between resources recommended by SmartSearch (SSR) and resources not recommended by SmartSearch (NSSR) but otherwise similar to SSRs.
User click-through data for 156 resources were extracted from Lane's web analysis tool, WebTrends [6], and divided into 2 sets: August through October of 2007 (pre-SmartSearch data set) and August through October of 2008 (post-SmartSearch). One hundred thirty resources included in the SmartSearch MeSH-to-resource maps were tagged as SSR; the other 26 were tagged NSSR.
RESULTS
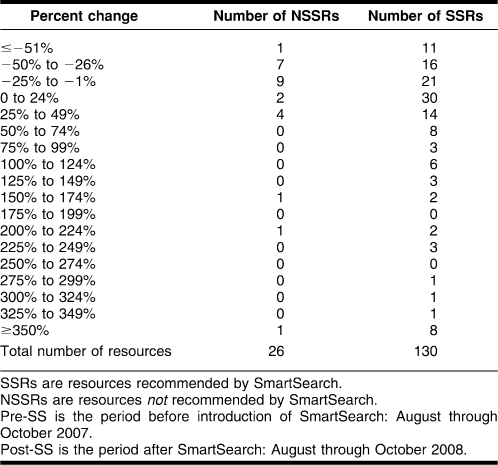
SAS 9.1 (SAS Institute, NC, USA) was used to perform various statistical analyses to determine if the change in use of SSRs was greater than that for NSSRs. The analysis showed that the frequencies were not normally distributed: some resources had large increases skewing the distribution curve. This lack of normality suggests the use of nonparametric measures and procedures, such as looking at median rather than mean. The range in percent change in resource usage between pre-SmartSearch and post-SmartSearch for SSR was from −300% to 1,086%, with a median of 6%. For NSSR, it was −62% to 400%, with a median of −12% (Table 1).
Table 1.
Percent change in resource usage between pre-SmartSearch (SS) and post-SS periods
The percent change values from the 2 groups were compared using the Wilcoxon-Mann-Whitney 2-sample rank-sum test. This nonparametric test determines whether 2 sets of observations come from the same distribution when data are not normally distributed. The result of the test was a 6% chance (P = 0.06) that the 2 sets were from the same distribution. This gave SmartSearch results marginal statistical significance, because the distribution was very close to the commonly accepted value of 5% for disproving a null hypothesis.
As a less stringent evaluation, SmartSearch was also compared to Lane's exact journal title suggestions, another recommendation system in use since 2006. Both occupy the same interface area in LaneConnex search results. During the post-SmartSearch period, approximately 10,160 exact journal title suggestions were made, and users clicked on them 244 times (2.4% user click-through rate). During the same period, SmartSearch made 4,277 recommendations, and users clicked on them 304 times (7.1% user click-through rate). Although statistically inconclusive, these numbers suggest users tend to follow SmartSearch suggestions almost 3 times more frequently than they do exact journal title suggestions.
DISCUSSION
Clear demonstration of statistical significance was hampered by the small size of NSSR and SSR populations. Resources were not randomly assigned to each group, increasing the likelihood that confounding factors (e.g., random use spikes due to irregularly scheduled library classes, updated resource versions, changing resource providers and uniform resource locators) would affect results. Additionally, SmartSearch's marginal influence on resource usage can be partially attributed to low overall use typical of the vast majority of resources accessible via LaneConnex. The average number of clicks per resource for the pre-SmartSearch and post-SmartSearch periods was only 294 and 304, respectively, with medians of 21 and 27 clicks. Tests designed to evaluate much weaker and noisier signals are required for these circumstances.
Yet marginal statistical significance also suggests a need to modify SmartSearch to better leverage this subject-based recommendation system. Potential improvements center around three main areas: interface location, search strategy refinements, and expanded content. None of these enhancements alters the core premise of the automated pearling approach, which the data suggest is strong and sound.
SmartSearch was introduced as part of the general LaneConnex search interface. Although this interface accounts for the majority of Lane's search traffic, it tends to be used for known item searching more often than question-based clinical searching. SmartSearch should perform better when added to Lane's clinically oriented search portals, where a majority of searches are concept and question based.
Future enhancements may also include modifications to the search algorithm. A stop-words list (“journal,” “book,” “textbook,” etc.) may improve SmartSearch precision and silence it when the user is obviously searching for a known resource. Refining the PubMed search strategy (moving from searching keywords to searching titles or from MeSH terms to major topic MeSH terms) may produce more accurate query-to-heading mappings. Similarly, SmartSearch does not require a minimum number of articles to produce a heading map, and sometimes queries that produce only one or two PubMed articles can produce odd mappings. Simply requiring a minimum article threshold may boost performance.
Exciting future improvements include expanding the number and variety of recommended resources. Although increasing the use of infrequently consulted resources was the original project goal, the SmartSearch experience encourages further examination of this set of clinical textbooks, handbooks, and atlases. Are they the high-value necessities Lane librarians once thought? Do they merit future licensing if usage trends are impervious to a system like SmartSearch? Regardless, Lane is eager to extend SmartSearch suggestions beyond metasearch to include other subject-relevant content, such as portal pages and people. For example, the query “lumbar sympathetic block” might produce the following result:
Have you tried the Anesthesia Portal? Ask Chris Stave, your Lane anesthesia liaison, for more help with your anesthesiology research.
By reducing confounding factors and making minor improvements to the product, the authors are confident they can unleash SmartSearch's full power and further quantify its success.
Acknowledgments
The authors thank Olya Gary for her contributions to interface design work, Pam Murnane for her statistics consulting, and Philip Constantinou for his contributions to development discussions.
Contributor Information
Ryan Max Steinberg, Knowledge Integration Programmer/Architect ryanmax@stanford.edu.
Richard Zwies, Web Production Specialist; Lane Medical Library & Knowledge Management Center, Stanford University Medical Center, 300 Pasteur Drive, Room L109, Stanford, CA 94305 rzwies@stanford.edu.
Charles Yates, System Software Developer, Information Resources and Technology, Building AB, Second Floor - M/C 5569, 301 Ravenswood Avenue, Menlo Park, CA 94025 ceyates@stanford.edu.
Christopher Stave, Instructional and Liaison Program Coordinator cstave@stanford.edu.
Yannick Pouliot, Bioresearch Informationist ypouliot@stanford.edu.
Heidi A Heilemann, Associate Dean for Knowledge Management and Director; Lane Medical Library & Knowledge Management Center, Stanford University Medical Center, 300 Pasteur Drive, Room L109, Stanford, CA 94305 heidi.heilemann@stanford.edu.
REFERENCES
- 1.Carlson S. The deserted library. Chron High Edu. 2001 Nov;48(12):A358. [Google Scholar]
- 2.Ketchell D, Steinberg R.M, Yates C, Heilemann H.A. LaneConnex: an integrated biomedical digital library interface. Inf Technol Libr. 2009 Mar;28(1):31–40. [Google Scholar]
- 3.National Library of Medicine. Entrez programming utilities [Internet] The Library [rev. 16 Feb 2009; cited 17 Aug 2009]. < http://eutils.ncbi.nlm.nih.gov/entrez/query/static/eutils_help.html>.
- 4.Schafer J.B, Konstan J, Riedi J. Proceedings of the 1st ACM Conference on Electronic Commerce; Denver, CO; 3–5 Nov 1999. New York, NY: Electronic Commerce; 1999. Recommender systems in e-commerce; pp. 158–66.http://doi.acm.org/10.1145/336992.337035 [Google Scholar]
- 5.Ramer S.L. Site-ation pearl growing: methods and librarianship history and theory. J Med Libr Assoc. 2005 Jul;93(3):397–400. [PMC free article] [PubMed] [Google Scholar]
- 6.WebTrends [Internet] Portland, OR: WebTrends; 2009 [cited 17 Aug 2009]. < http://www.webtrends.com>. [Google Scholar]