Abstract
The dystrobrevin-binding protein 1 (DTNBP1) gene has been one of the most studied and promising schizophrenia susceptibility genes since it was first reported to be associated with schizophrenia in the Irish Study of High Density Schizophrenia Families (ISHDSF). Although many studies have been performed both at the functional level and in association with psychiatric disorders, there has been no systematic review of the features of the DTNBP1 gene, protein or the relationship between function and phenotype. Using a bioinformatics approach, we identified the DTNBP1 gene in 13 vertebrate species. The comparison of these genes revealed a conserved gene structure, protein-coding sequence and dysbindin domain, but a diverse noncoding sequence. The molecular evolutionary analysis suggests the DTNBP1 gene probably originated in chordates and matured in vertebrates. No signature of recent positive selection was seen in any primate lineage. The DTNBP1 gene likely has many more alternative transcripts than the current three major isoforms annotated in the NCBI database. Our examination of risk haplotypes revealed that, although the frequency of a single nucleotide polymorphism (SNP) or haplotype might be significantly different in cases from controls, difference between major geographic populations was even larger. Finally, we constructed the first DTNBP1 interactome and explored its network features. Besides the biogenesis of lysosome-related organelles complex 1 and dystrophin-associated protein complex, several molecules in the DTNBP1 network likely provide insight into the role of DTNBP1 in biological systems: retinoic acid, β-estradiol, calmodulin and tumour necrosis factor. Studies of these subnetworks and pathways may provide opportunities to deepen our understanding of the mechanisms of action of DTNBP1 variants.
Keywords: DTNBP1, schizophrenia, splicing, haplotype, protein–protein interaction, gene network
Introduction
The human dystrobrevin-binding protein 1 (DTNBP1) gene spans ~140 kb on chromosome 6p22.3 and has 10 exons. So far, it has not yet been classified into any known gene family. Dysbindin, a coiled-coil-containing protein encoded by DTNBP1, was initially found to interact with α- and β-dystrobrevin (DTNA and DTNB) in the muscle and brain of mice.1 DTNA and DTNB are members of the dystrophin-associated protein complex (DPC), which links the cytoskeleton to the extracellular matrix and serves as a scaffold for signaling proteins.1,2 Dysbindin is also an essential component of the biogenesis of lysosome-related organelles complex 1 (BLOC-1) and interacts with all seven other components of BLOC-1.3-5
In 2002, Straub et al.6 first identified the DTNBP1 gene as a putative schizophrenia susceptibility gene by undertaking systematic linkage disequilibrium mapping across a linkage region on 6p in the 270 multiply affected pedigrees from the Irish Study of High Density Schizophrenia Families (ISHDSF). Reanalysis of this data identified a single high-risk haplotype containing 8 single nucleotide polymorphisms (SNPs) covering 30 kb.7 As of 19 April 2008, 45 follow-up association studies, 18 of which had positive results, are annotated in the SchizophreniaGene database.8 One meta-analysis has been published.9 So far, DTNBP1 has been one of the most prominent schizophrenia susceptibility genes.10,11 DTNBP1 has been associated with other phenotypes including intelligence, schizoaffective disorder, bipolar disorder and the Hermansky–Pudlak syndrome type 7.3,12,13
DTNBP1 association studies have been frequently cited and reviewed,10,14-16 and will not be discussed here. However, its gene feature, expression and protein’s interactions with other molecules in cellular systems have largely remained speculative.14 For example, there are few amino acid changes (nonsynonymous mutations) observed in the human population, none of which has been reported to be associated with schizophrenia. Recent studies revealed a reduced expression of DTNBP1 in the frontal cortex and hippocampal formation of schizophrenia patients.17-19 DTNBP1 may confer susceptibility to schizophrenia via reduced expression mediated by its high risk haplotype, which might tag one or more cis-acting variants.17,20 These observations call for an investigation of the functional elements or regulatory mechanisms which might affect the DTNBP1 expression and confer the illness. Importantly, protein–protein interaction (PPI) analysis of DISC1 (Disrupted in Schizophrenia 1), another prominent schizophrenia susceptibility gene, suggested that schizophrenia susceptibility genes (for example, DTNBP1 and DISC1) may share common PPIs and affect common biological processes.21 If this is true, network analysis may reveal novel mechanisms of etiology and intervention not easily reached by more conventional approaches (for example, single gene analysis). Here, we apply bioinformatics and systems biology approaches to explore the features of DTNBP1, including its molecular evolution and sequence conservation, gene structure, transcripts, haplotypes and interactome and pathways.
Conserved DTNBP1 gene structure in vertebrates
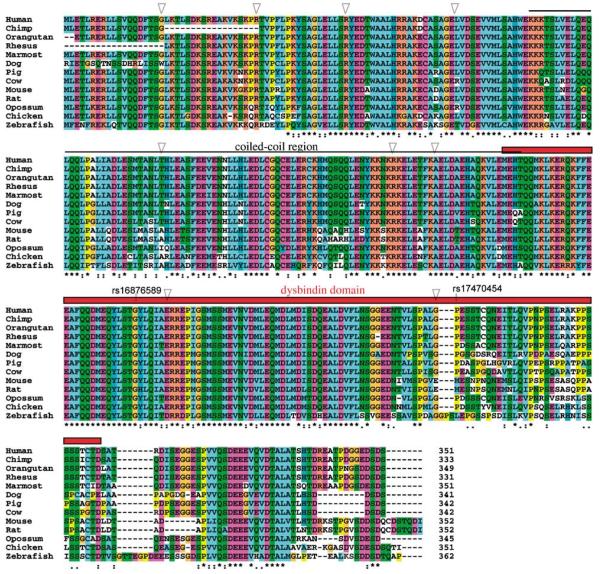
So far, the DTNBP1 gene has only been experimentally verified in mice1 and humans.6 We performed an extensive search for the DTNBP1 gene in several major databases including NCBI Entrez Gene, Ensembl and dbEST, as well as in all the available genomes (see Supplementary Materials and methods). Based on these sources, we identified human DTNBP1 orthologs in 12 other vertebrates, including 4 non-human primates, 6 non-primate mammals, and 2 non-mammals (Table 1). The length of its amino acid sequence is 351 in humans. Similar length and high identity of the amino acids are observed in other species (Table 1). A dysbindin domain is annotated (positions 184–304 in NCBI human dysbindin isoform a, NP_115498). The domain is highly conserved among the vertebrates. For example, the identity is 87.6% between human and chicken dysbindin domains.
Table 1.
DTNBP1 gene in 13 species
| Species | Gene ID | DNA |
CDS |
Amino acid |
Dysbindin domain |
Major source |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| Length (kb) |
Repeat a (%) |
Length (bp) |
Identityb (%) |
Length (aa) |
Identityb (%) |
Length (aa) |
Identityb (%) |
|||
| Human | 84062 | 140.2 | 52.8 | 1056 | 351 | 121 | NCBI | |||
| Chimp | ENSPTRG00000017744 | 145.9 | 50.8 | 1002 | 99.0 | 333 | 99.4 | 121 | 99.2 | Ensembl |
| Orangutanc | 143.5 | 49.5 | 1050 | 98.1 | 349 | 97.7 | 121 | 100.0 | Genome | |
| Rhesus | ENSMMUG00000015363 | 147.1 | 52.0 | 996 | 97.3 | 331 | 97.3 | 120 | 97.5 | Ensembl |
| Marmostc | 137.4 | 45.8 | 1056 | 95.1 | 351 | 94.3 | 121 | 94.2 | Genome | |
| Dog | ENSCAFG00000009893 | 115.8 | 40.9 | 1026 | 79.7 | 341 | 78.6 | 121 | 78.5 | Ensembl |
| Pig | 100049697 | 118.4 | 39.0 | 1029 | 83.4 | 342 | 84.6 | 121 | 80.2 | NCBI |
| Cow | 506612 | 89.1 | 38.0 | 1029 | 84.3 | 342 | 85.2 | 121 | 84.3 | NCBI |
| Mouse | 94245 | 80.0 | 28.3 | 1059 | 83.0 | 352 | 77.7 | 121 | 80.2 | NCBI |
| Rat | 641528 | 90.4 | 30.9 | 1059 | 81.8 | 352 | 76.8 | 121 | 81.0 | NCBI |
| Opossum | ENSMODG00000010762 | 207.0 | 1038 | 79.2 | 345 | 82.1 | 120 | 81.8 | Ensembl | |
| Chicken | 420840 | 68.7 | 7.8 | 1056 | 76.4 | 351 | 78.5 | 121 | 87.6 | NCBI |
| Zebrafish | 394109 | 31.1 | 22.5 | 1089 | 67.9 | 362 | 63.2 | 124 | 68.6 | NCBI |
Abbreviations: chimp, chimpanzee; rhesus, rhesus macaque.
The proportion (%) of repetitive sequences identified by the RepeatMasker.
Identity was calculated by comparing with the human DTNBP1 sequence.
Orangutan and marmost DTNBP1 genes were predicted from their draft genomes (see Supplementary Materials and methods).
Gene structure is also similar across species. For example, 10 exons are annotated in the databases or have been consistently predicted in the 13 species. However, gene length varies widely from 31.1 kb in zebrafish to 207.0 kb in opossums, probably as a result of variation in the extent of repeats. For example, human DTNBP1 has 74 088 bp repetitive sequences, accounting for 52.8% of the sequence; conversely, mouse DTNBP1 has 22 648 bp repetitive sequences, accounting for only 28.3% of the sequence (Supplementary Table S1). Repeats account for a smaller proportion in chickens and zebrafish DTNBP1 genes, whose lengths are also shorter.
Figure 1 shows the aligned amino acid sequences and protein features in the 13 species. Overall, the coiled-coil region, dysbindin domain, and some other functional sites are conserved among these species. The N-terminal region is more conserved than the C-terminal region. Phylogenetic trees were reconstructed using the aligned amino acid sequences and CDS sequences, respectively. The protein and CDS trees show similar topology (Supplementary Figure S1).
Figure 1.
Alignment of the 13 DTNBP1 amino acid sequences. The multiple alignment was first generated by computer program Clustal W and then manually checked and refined. Intron positions are indicated by triangles. The predicted coiled-coil region is labeled by a thin line on the top of the alignment and dysbindin domain is labeled by a red thick line. Two nonsynonymous SNPs (rs16876589: G214D, rs17470454: P272S) in isoform a are indicated.
Human and mouse sequences are typically used to identify evolutionary conserved regions and to evaluate sequence conservation.22 Using VISTA tool (http://genome.lbl.gov/vista/), we found that the exons are highly conserved; however, we also identified 41 conserved nongenic regions (CNGs) covering 6729 bp with an average identity of 72.9% (Supplementary Table S2; Supplementary Figure S2). When more genetically distant species (human, dog, mouse, and opossum) were employed in a cross-species sequence comparison, all exons are again highly conserved, but only a few CNGs are present (Figure 2).
Figure 2.
VISTA plot displaying DTNBP1 gene structure and evolutionary conserved regions in a comparison of human, dog, mouse and opossum DNA sequences. Conserved nongenic region (CNG, in red) is defined as an alignment at least 100-bp long and at least 70% identity. The plot used human gene and annotations as reference. In each gene line, a vertical bar denotes an exon, a dashed line denotes a shorter intronic region and a double forward slash denotes a longer intronic region than the human sequence.
DTNBP1 gene likely originated in chordates and matured in vertebrates
Besides the DTNBP1 genes found in these 13 species, an extensive BLAST search of human protein sequence identified hits in 12 other organisms including 5 vertebrates and 4 invertebrates (Supplementary Table S3). Using cutoff values for the BLAST output: identity at the amino acid level of > 30% and requiring the aligned sequences to cover > 50% of the human dysbindin amino acid sequence, significant hits are all from chordates. The most ancient species in our search results is amphioxus (Branchiostoma belcheri tsingtauense, accession no. AY280671), an ancient fish classified as an invertebrate. However, the most significant hits ( > 50% identity in the BLAST output, > 80% coverage of the human amino acids) were only found in vertebrates. Moreover, the complete dysbindin domain was only observed in vertebrates. Thus, the functional DTNBP1 gene probably originated in chordates and the current DTNBP1 gene structure likely matured in vertebrates. Given the simpler nervous system in invertebrate chordates versus vertebrates, this suggests that DTNBP1’s role arose with increasing central nervous system complexity.
Test recent positive selection in the human and other primate lineages
Schizophrenia may be a maladaptive by-product of adaptive cognitive changes during evolution.23 A recent study24 of 76 genes associated with schizophrenia indicated that 28 of these genes may have been under recent positive selection by applying two methods: Haplotter, which tests selective sweeps in human populations using the HapMap allelic data,25 and the ratio of nonsynonymous over synonymous substitution rates (dN/dS ratio), which tests positive selection in a lineage. A dN/dS ratio greater than 1 suggests positive selection at the locus examined.26 The results suggested an enrichment of a positive selection signal in these genes. For DTNBP1, a positive selection signal was found in the European population and a weak signal was also found in the African population in the HapMap data.25 However, no positive selection was detected in the human and chimpanzee lineages.24 Our test using human and four non-human primates further confirmed no positive selection in any primate lineage as the dN/dS ratio in each pairwise primate lineage comparison was consistently smaller than 0.5 (Supplementary Table S4). Furthermore, no positive selection was detected at any single codon site using PAML site models or branchsite models.26 As both the statistical methods tend to be liberal in detecting positive selection and the iHS algorithm implemented in the Haplotter only tests three HapMap subpopulations,25,27 the DTNBP1gene had probably not experienced substantial positive selection in recent evolutionary history.
Human DTNBP1 putative transcripts and expression
Currently, three major transcripts (isoforms a, b, and c; accession numbers NM_032122, NM_183040 and NM_183041) of human DTNBP1 are annotated in the NCBI Entrez database and more than 160 mRNA or EST sequences are related to human DTNBP1. The transcript isoform a is the reference transcript and has 10 exons. The 10-exon gene structure and sequences are conserved in the 13 species we examined. However, the number of transcripts of human DTNBP1 is likely more than 3. NCBI AceView (version April 2007),28 a comprehensive and well-annotated alternative transcript variants database, annotated 16 human alternative transcripts whose lengths vary from 268 to 1955 bp. Because the transcripts in AceView were annotated in a genomewide fashion, we refined them as follows. We retrieved all the mRNA and EST sequences of the human DTNBP1 gene from the NCBI UniGene database, then performed a clustering analysis of all available mRNA and EST sequences using TGI Clustering Tools (http://compbio.dfci.harvard.edu/tgi/software/), and finally compared the generated transcripts with the AceView transcripts based on the original DTNBP1 mRNA and EST sequences. For the 16 AceView transcripts (named a–p), we deleted transcript j and modified transcript o by deleting the last 200 bp in its 3′ end. In addition, we added EST CD723892 as a partial transcript (details are shown in Supplementary Materials and methods). This resulted in a new set of 16 putative transcripts (Figure 3). Among them, five (a, d, e, g and h) have mRNA evidence and the others have only EST data. Seven transcripts (k, l, m, n, o, p and CD723892) are short, which are likely partial transcripts. According to the expression data in the AceView, the three transcripts a, d and e are detected in many clones from a large number of tissues, whereas short transcripts are shown in a few clones and expressed in only one or a few tissues (Supplementary Table S5). Based on the current data, these alternative transcripts, especially the short ones, must be regarded as putative and require verification. Additional experimental work in our group confirmed six of the eight transcripts assessed (manuscript in preparation).
Figure 3.
Alternative splicing transcripts of the human DTNBP1 gene. Sizes of the exons are approximately scaled. (a) Gene structure. The gray boxes indicate alternative splicing exons. Numbers 1–22 indicate the locations of the 22 SNPs that have been reported to be associated with schizophrenia. (b) Transcripts with available mRNAs. (c) Transcripts with available ESTs. In (b) and (c), black boxes denote coding exons and white boxes denote untranslated exons. For each transcript, NCBI mRNA or EST accession numbers are listed at the left and AceView transcript ID is listed at the right.
SNPs in DTNBP1 gene
There are 558 nonredundant SNPs in the human DTNBP1 gene region in the NCBI dbSNP database (build 129, http://www.ncbi.nlm.nih.gov/SNP/), including 196 HapMap SNPs. Only five SNPs are in protein-coding regions, four of them (rs17470454, rs16876589, rs16876569 and rs16876571) nonsynonymous. The minor allele frequencies of all these five SNPs are very low in CEU and JPT+CHB and 0 in YRI based on the HapMap data, indicating that these mutations occurred recently in non-African population and have not yet been fixed. Twenty-one SNPs are mapped in the CNGs (Supplementary Table S2). There are 22 SNPs that have been reported either individually or within a haplotype that is significantly associated with schizophrenia, including 2 SNPs in CNGs. The locations of the association SNPs are displayed in Figure 3a and their details provided in Supplementary Table S6. In the mouse DTNBP1 gene, there are 385 SNPs, including two nonsynonymous mutations (rs46632574 and rs48618277) and three synonymous mutations (rs51027077, rs50120298 and rs51319012). Overall, few SNPs could be found in the DTNBP1 protein-coding region.
Among the 22 SNPs from the association studies, 16 have allele frequencies from HapMap. The ancestral alleles of the SNPs were inferred based on the maximum parsimony principle using a pipeline in Jiang and Zhao.29 The ancestral alleles of these SNPs are always major alleles with three exceptions: SNPs rs2619538, rs2619539 and rs742106 whose minor alleles in JPT+CHB are ancestral alleles (Supplementary Table S7).
Haplotypes and high-risk haplotypes
We examined haplotype information for DTNBP and compared it to the reported high-risk haplotypes. HapMap SNPs in the DTNBP1 gene were used to infer haplotype using the program PHASE.30 There are 44, 46, 56 haplotypes in CEU, JPT+CHB and YRI, respectively, and 135 haplotypes in the whole HapMap sample. Only a few haplotypes are shared by subpopulations, for example, nine shared by at least two subpopulations and only two shared by three subpopulations. This nonsharing feature of haplotypes between major geographic populations has been reported at other loci.31 Of note, the number of haplotypes in CEU is nearly the same as that in JPT+CHB, though there are twice as many founder chromosomes in JPT+CHB sample. This difference is mainly due to the preascertained SNPs selected in the HapMap project. These SNPs do not directly contain information about the underlying levels of nucleotide diversity.25 Because iHS statistic in Haplotter tests the extended haplotype homozygosity, not the number of haplotypes, the observation of the similar number of haplotypes in CEU and JPT+CHB is not directly related to their different positive selection signals detected by Haplotter.
We re-examined the high-risk haplotype reported in the ISHDSF.7 Eight SNPs were employed in that high-risk haplotype: rs1474605 (P1792), rs1018381 (P1578), rs2619522 (P1763), rs760761 (P1320), rs2005976 (P1757), rs2619528 (P1765), rs1011313 (P1325) and rs3213207 (P1635). Seven of them (excluding rs2005976) are available in the HapMap. These seven SNPs result in the same haplotypes reported in ISHDSF7 (rs2005976 is redundant). We extracted the phased haplotypes and their frequencies from the HapMap and compared haplotype frequencies in the ISHDSF sample (Table 2). Interestingly, for the high-risk haplotype (haplotype 2), its frequency in CEU (0.108) is much higher than that in JPT+CHB (0.005) or YRI (0.025). This confirmed the previous suspicion of lower frequency in other samples.7 A number of haplotypes were not observed in YRI or JPT CHB sample.
Table 2.
Comparison of DTNBP1 high-risk haplotype in the Irish study and the HapMap samples
 |
1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | Frequency |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ISHDSF | CEU | JPT+CHB | YRI | |||||||||
| Marmost | A | T | G | C | G | A | G | G | ||||
| Rhesus | . | C | . | G | A | . | . | . | ||||
| Orangutan | . | C | . | . | . | . | . | . | ||||
| Chimp | . | C | . | A | . | . | . | A | ||||
| 1 | . | C | T | . | . | G | . | A | 0.733 | 0.750 | 0.705 | 0.633 |
| 2 | G | C | . | T | A | . | . | . | 0.058 | 0.108 | 0.005 | 0.025 |
| 3 | . | C | T | . | . | G | A | A | 0.071 | 0.058 | 0.211 | |
| 4 | . | C | T | T | . | G | . | A | 0.010 | |||
| 5 | G | . | . | T | A | . | . | A | 0.060 | 0.075 | 0.072 | 0.308 |
| 6 | G | C | T | T | . | G | A | . | 0.015 | 0.008 | ||
| Rare | G | . | . | T | A | . | A | . | ||||
Abbreviations: chimp, chimpanzee; rhesus, rhesus macaque; SNP, single nucleotide polymorphism. The SNPs from 1 to 8 are, respectively, rs1474605 (P1792), rs1018381 (P1578), rs2619522 (P1763), rs760761 (P1320), rs2005976 (P1757), rs2619528 (P1765), rs1011313 (P1325) and rs3213207 (P1635). Haplotypes 1 and 2 are the common and high-risk haplotypes in the ISHDSF. Haplotype frequencies were based on the ISHDSF study7 and the HapMap data (CEU, JPT+CHB and YRI). The sum of the haplotype frequencies is less than 1 because some other haplotypes were not included.
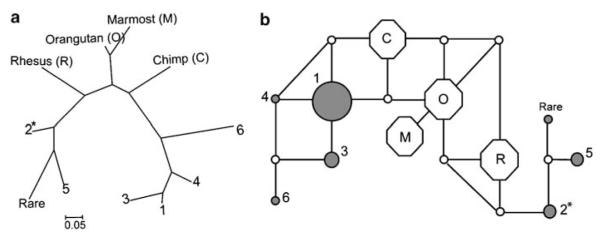
Both van den Oord et al.7 and Mutsuddi et al.32 suggested that the ancestral haplotype was the most common haplotype (haplotype 1, see Figure 1 in van den Oord et al.). We mapped the eight SNPs to four available primates (chimpanzee, orangutan, rhesus macaque and marmost) and identified the corresponding nucleotides at these sites. Figure 4a shows an evolutionary relationship of seven haplotypes and their outgroups (details were provided in the Supplementary Materials and methods). The evolutionary tree indicates that (1) haplotypes 2 and 5 are rare in a branch separated from haplotypes 1, 3, 4 and 6 by primates and (2) the common haplotype is genetically distant from primates. We further drew a haplotype network (Figure 4b), which demonstrates the possible mutational paths based on a median-joining algorithm.33 Both the evolutionary tree and haplotype network indicate that the two haplotype groups are separated by primates. The results suggest that (1) the high-risk and common haplotypes might have undergone different evolutionary paths and, (2) contrary to the previous suggestions, the common haplotype is not ancestral.
Figure 4.
Evolutionary tree and network of DTNBP1 8-SNP haplotypes. (a) The evolutionary tree reconstructed by the neighbor-joining method in MEGA 4.0. (b) The haplotype network generated by the median-joining algorithm in software NETWORK 4.2.01. Seven gray circles denote the seven haplotypes in van den Oord et al.7 Among them, haplotype 2 is the high-risk haplotype and haplotype 1 is the common haplotype. Circle areas are proportional to haplotype frequencies. Four octagon nodes denote the four non-human primates.
We also re-examined the risk haplotype reported in Williams et al.,34 which is based on three markers (rs2619539, rs3213207 and rs2619538), one of which (rs3213207) was in the ISHDSF high-risk haplotype. One risk haplotype (C-A-T) and 2 protective haplotypes (C-A-A and G-G-T) were reported in two independent samples collected in Cardiff and Dublin.34 Table 3 shows the frequencies of haplotypes in these two samples and in the HapMap samples. For the risk haplotype, its frequency in CEU (0.158) is close to that in the control samples (Cardiff: 0.16; Dublin: 0.15). However, the frequencies in JPT+CHB (0.350) and YRI (0.250) are higher than the controls and the cases (0.21 in both the samples). Of note, Numakawa et al.35 failed to detect a significant association between the C-A-T haplotype and schizophrenia in a Japanese sample. The C-A-T risk haplotype was associated with reduced cortical expression of DTNBP1, poor spatial working memory, less severe manic-type symptoms, and early visualprocessing deficits in Irish samples.20,36-38 For the protective haplotype C-A-A, which is an ancestral haplotype, its frequencies in CEU and YRI are high (0.375 and 0.467, respectively), but it absents from the JPT+CHB. This absence is likely due to the low frequency of allele A (0.017) of SNP rs2619538 (Supplementary Table S7). The frequency of the protective haplotype G-G-T, an entirely derived haplotype, is very low (0–0.03) in the Cardiff and Dublin samples and not found in any HapMap sample.
Table 3.
Comparison of haplotypes (rs2619539-rs3213207-rs2619538) in Williams et al.34 and in the HapMap samples
| Haplotype | Cardiff sample |
Dublin sample |
CEU | JPT+CHB | YRI | ||
|---|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | ||||
| GAA | 0.13 | 0.14 | 0.11 | 0.11 | 0.117 | 0.011 | 0.183 |
| CAA a | 0.29 | 0.35 | 0.30 | 0.37 | 0.375 | 0.467 | |
| GGA | 0.10 | 0.08 | 0.10 | 0.10 | 0.117 | 0.006 | 0.025 |
| GAT | 0.26 | 0.24 | 0.28 | 0.26 | 0.233 | 0.633 | 0.075 |
| CAT b | 0.21 | 0.16 | 0.21 | 0.15 | 0.158 | 0.350 | 0.250 |
| GGT a | 0.00 | 0.03 | 0.00 | 0.02 | |||
Protective haplotypes in the two studied samples (Cardiff and Dublin samples).
Risk haplotype in the two studied samples.
The comparisons above suggest that, although a SNP or haplotype might be found at significantly different frequencies in cases and controls, the difference between major geographic populations might be even larger. Whether these population differences impact on differences in the case and control studies requires further examination.
DTNBP1 protein interactions
So far, only one schizophrenia susceptibility gene, DISC1, has had its network extensively investigated.21,39 Here, we present our detailed examination of the network and pathway features for DTNBP1.
To identify all proteins interacting with DTNBP1, we collected and integrated all the available experimentally verified human PPIs from four major PPI databases (details in Supplementary Materials and methods). These protein pairs were considered as the human protein interactome. A total of 31 proteins were identified to directly interact with DTNBP1. The genes encoding these 31 proteins are provided in Supplementary Table S10. Among them, 3 genes (BLOC1S3, DTNA and SYNE1) have phenotype annotations in the OMIM database and 10 genes (PLDN, CNO, MUTED, BLOC1S1, BLOC1S2, BLOC1S3, DGCR6L, SNAPAP, RANBP5 and ZNF490) have association studies in the SchizophreniaGene database.8 For these 10 genes, only 2 (BLOC1S3 and RANBP5) had positive results. We examined the distribution of characteristic GO terms associated with these genes. Fifty-seven GO terms associated with these 31 genes; 13 terms are associated with at least 5 genes and their hierarchical levels are at least 4 (Supplementary Table S11). The 13 GO terms are ‘cytoplasm’, ‘calcium ion binding’, ‘actin binding’, ‘nucleus’, ‘protein transport’, ‘transcription’, ‘cytoskeleton’, ‘nucleoplasm’, ‘plasma membrane’, ‘intracellular’, ‘signal transduction’, ‘cytosol’ and ‘transport’. Overall, these terms relate to subcellular localization, transport and signal transduction. The most frequently observed GO term (23 genes) is cytoplasm, consistent with the previous report of DTNBP1 subcellular localization.40 The other two frequently observed GO terms are calcium ion binding (21 genes) and actin binding (18 genes). These proteins may be related to synaptic vesicle mobility,41 which is consistent with DTNBP1 being a synaptic and microtubular protein,42 and part of DPC. Note that DTNBP1 and at least five of its interactors (DTNA, PLDN, SNAPAP, RAB11A and SYNE1) have been implicated in neurodevelopmental processes.
Using the Ingenuity Systems Core Analysis, two groups stood out as being highly significant in the human Global Molecular Network (GMN; Figure 5a). Group 1 (P-score = 28) contains 12 proteins. The P-score is defined here as –log10P, where P is calculated using Fisher’s exact test. The top functions of the proteins in group 1 are cell morphology, cellular development and cancer. Group 2 (P-score = 26) has 10 proteins including all of the 8 components in BLOC-1, DTNA and DTNB. The top functions in group 2 are cell morphology, cell-to-cell signaling and interaction, and developmental disorder. For the remaining 10 proteins, 7 could not be mapped to any network and each of the 3 other proteins was mapped to a small network with P-score≤3. Interestingly, cancer is annotated as a functional feature in the DTNBP1 interactors. This is likely because cancer genes are widely annotated in the human genome and have been associated with schizophrenia (for example, TP53,43 AKT144).
Figure 5.
DTNBP1 interactome and extracted networks/pathways in the Ingenuity Global Molecular Network (GMN). (a) DTNBP1 interactome contains 31 proteins that directly interact with DTNBP1 in the human interactome. Using the Ingenuity System Core Analysis, these 31 proteins could be classified into group 1 (green, 12 proteins), group 2 (red, 9 proteins) and the others (10 proteins). Nodes in red indicate available phenotype annotation in the OMIM database and nodes in diamond indicate neurodevelopment annotation in the Gene Ontology database. (b) Subnetwork for the proteins in group 2 and their direct interactors in GMN. Nodes in brown denote the proteins directly linked to the BLOC-1 and DPC complexes and node in gray denote the proteins in group 2 and their direct interactors. A solid line indicates a physical interaction, a dashed line with an arrow indicates a regulation relationship, and a solid line with an arrow indicates both a physical interaction and a regulation relationship.
As expected, frequent connections between proteins in group 2 were observed because most proteins are in the BLOC-1 complex. Interestingly, no direct interaction was found between any pair of proteins within group 1 or between any pair of the remaining 10 proteins (we broadly named these 10 proteins as group 3). When DTNBP1 is removed, there is no direct interaction between any two groups, with one exception (a connection between ABI3 and KIAA0408). To further examine this feature, we added proteins that directly interact with the 31 DTNBP1 interactors (distance 1), that is, DTNBP1 interactors with distance 2. The network shows that DTNBP1 serves as a superhub (a network node with more than 15 links). Conversely, when DTNBP1 is intentionally removed, the DTNBP1 interactors are only loosely connected (Supplementary Figure S4).
DTNBP1’s networks/pathways
We further explored the features of DTNBP1 pathways and protein interaction networks in the Ingenuity GMN. We described the networks for the proteins in group 2 because the analysis of proteins in group 1 produced little additional information. Using the Ingenuity Network Analysis Tool, we extracted a complex subnetwork (details in Supplementary Materials and methods), which contains 140 molecules including 125 single proteins, 4 protein complexes, 9 protein groups and 2 other molecules (Supplementary Figure S5; Supplementary Table S12). Moreover, in the network, 30 genes have phenotype annotation in the OMIM database and 19 genes have been collected in the SchizophreniaGene database including 5 genes with at least 2 positive results. As expected, the network includes two protein complexes, BLOC-1 and DPC, which involve DTNBP1. Fourteen molecules directly link to the components of these two complexes: retinoic acid (RA), PRKACA, calmodulin (CaM), Nos, F-actin, ABCA1, SNAP25, SNAP23, Ap1, tumour necrosis factor (TNF), PRX, actin, STX12 and EBAG9. To interpret the results efficiently, we simplified the network by limiting: (1) components of the BLOC-1 and DPC complexes, (2) molecules directly linked to the components of these two complexes (14 molecules), (3) the molecules directly linked these 14 molecules, and (4) any molecules having at least 15 links (that is, degree≥15) under the assumption that highly connected molecules tend to have important function.45 The simplified network is shown in Figure 5b. The features are described below.
Dysbindin in the BLOC-1 complex
Dysbindin is an essential component of the BLOC-1 complex.3 BLOC-1 is a 200-kDa ubiquitously expressed soluble algometric protein complex known to contain at least eight components: DTNBP1, MUTED, PLDN, CNO, SNAPAP, BLOC1S1, BLOC1S2 and BLOC1S3. A number of the genes (DTNBP1, MUTED, PLDN, CNO and BLOC1S3) encoding these proteins are defective in inbred mouse strains serving as models of the Hermansky–Pudlak syndrome.3,5 Figure 5b shows eight molecules that directly interact with the BLOC-1 components: DTNA and DTNB that interact with DTNBP1, EBAG9, SNAP23, PRKACA and SNAP25 that interact with SNAPAP, and STX12 and F-actin that interact with PLDN. Six proteins (STX12, SNAP23, PRKACA, EBAG9, SNAPAP, SNAP25) are involved in membrane fusion of the synaptic vesicle.46-49 In the fusion process, the SNARE complex plays a central role,46 which decreases the release of glutamate from astrocytes in mice.50 A number of protein- and gene-expression studies have presented evidence of DTNBP1 involvement in glutamate neurotransmission.19,35,42 Therefore, the role of DTNBP1 and its interactors in the membrane fusion process of the synaptic vesicle is a plausible mechanism by which risk of schizophrenia is increased through disruption of synaptic glutamate neurotransmission.
Dysbindin in the DPC complex
Dysbindin interacts with DTNA in muscle and DTNB in the brain, both components of DPC.1 DPC is a multifunctional protein complex that has been studied extensively both genetically and biochemically in muscle. Mutations in genes involved in DPC cause several types of muscular dystrophies.51,52 DPC is a dynamic and strategically located cellular signaling complex.53 In Figure 5b, most of the DPC components are shown including dystrophin (DMD) and it related proteins (UTRN, DTNA, DTNB and DRP2), dystroglycans (DAG1), syntrophins (SNTA1, SNTB1, SNTB2, SNTG1 and SNTG2), one intercellular binding partner (F-actin) and two signaling molecules associated with the complex: CaM and Nos (a group of proteins including nNOS). Sarcoglycan complex was not present, as previously reported.53 DTNA is highly expressed in skeletal muscle whereas DTNB, which is abundantly expressed in the brain, kidney, lung and liver, is often considered a non-muscle protein.51 Furthermore, DTNB is expressed at significant levels within the cerebral cortex and hippocampus.54 However, in brain, some of the individual components of DPC are expressed presynaptically and others postsynaptically. These data argue that the functions of DPC protein components in brain are different from those in muscle.55 Interestingly, to our knowledge, no other gene in the DPC complex has been studied for its association with schizophrenia.
Dysbindin and other molecules
Besides BLOC-1 and DPC, several molecules in the network are noteworthy: RA, β-estradiol and CaM. RA is a metabolic product of vitamin A (retinol) and an established signaling molecule involved in neuronal patterning, neural differentiation and axon out-growth.56 Disruption of RA signaling has been implicated in the development of Alzheimer’s disease and possibly, Parkinson’s disease and depression.56,57 Retinoids may play an etiological role in schizophrenia.58 Figure 5b depicts multiple paths from RA to DTNBP1; most are via regulation of expression.
Figure 5b also shows β-estradiol, which link to multiple molecules including Akt, TNF, insulin and CaM. Estrogens may be protective in schizophrenia as men develop schizophrenia at an earlier age and with greater severity than women.58,59 A recent study showed estrogen directly induces expression of RA biosynthetic enzymes in rats, suggesting a coordinated role of estrogens and RA.60
CaM, a calcium-binding protein, and PRKACA, a cAMP-dependent protein kinase, are both involved in long-term potentiation (LTP) of synaptic transmission.61 In nervous tissue, Nos is involved in synaptic long-term depression (LTD). LTP and its counterpart LTD have long been considered as a potential mechanism for memory formation and learning.62 Upregulation of human CaM mRNA(s) in dorsolateral prefrontal cortex was reported in schizophrenics.63
Conclusion
The DTNBP1 gene is highly conserved in its structure and functional domains, but not in its noncoding sequences. This gene probably originated in chordates and matured in vertebrates, consistent with its role in complex nervous systems. No strong evidence of recent positive selection was found at the gene locus. The human DTNBP1 gene likely has many more alternative transcripts than the current three major isoforms annotated in the NCBI database. Our examination of risk haplotypes revealed large difference between major geographic populations, which need to be taken into account in interpreting association studies. Specifically, for the haplotypes based on the eight SNPs reported in the ISHDSF, we found that (1) the seven haplotypes were separated as two groups by primate lineages, indicating different evolutionary processes of the high-risk haplotype from the common haplotype and (2) contrary to prior suggestion, the most common haplotype is not the ancestral haplotype. Finally, we constructed the first DTNBP1 interactome, which contains 31 interactors, and explored its network features in the Ingenuity GMN. The networks indicate that DTNBP1 is involved in both muscle and neuronal functions. DTNBP1 may confer its susceptibility to schizophrenia through its impact on glutamate neurotransmission, RA signaling pathway, and synaptic LTP and LTD, which link to other biological pathways.
Supplementary Material
Acknowledgments
We regret that many DTNBP1 studies, especially association studies, have not been cited in this review because of our focus on gene feature and network analysis. This work was supported by a NARSAD Young Investigator Award and a Jeffress Trust grant to ZZ and a research grant (R01MH41953) to KSK/BPR.
Footnotes
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
References
- 1.Benson MA, Newey SE, Martin-Rendon E, Hawkes R, Blake DJ. Dysbindin, a novel coiled-coil-containing protein that interacts with the dystrobrevins in muscle and brain. J Biol Chem. 2001;276:24232–24241. doi: 10.1074/jbc.M010418200. [DOI] [PubMed] [Google Scholar]
- 2.Veroni C, Grasso M, Macchia G, Ramoni C, Ceccarini M, Petrucci TC, et al. beta-dystrobrevin, a kinesin-binding receptor, interacts with the extracellular matrix components pancortins. J Neurosci Res. 2007;85:2631–2639. doi: 10.1002/jnr.21186. [DOI] [PubMed] [Google Scholar]
- 3.Li W, Zhang Q, Oiso N, Novak EK, Gautam R, O’Brien EP, et al. Hermansky–Pudlak syndrome type 7 (HPS-7) results from mutant dysbindin, a member of the biogenesis of lysosome-related organelles complex 1 (BLOC-1) Nat Genet. 2003;35:84–89. doi: 10.1038/ng1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nazarian R, Starcevic M, Spencer MJ, Dell’Angelica EC. Reinvestigation of the dysbindin subunit of BLOC-1 (biogenesis of lysosome-related organelles complex-1) as a dystrobrevin-binding protein. Biochem J. 2006;395:587–598. doi: 10.1042/BJ20051965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Starcevic M, Dell’Angelica EC. Identification of snapin and three novel proteins (BLOS1, BLOS2, and BLOS3/reduced pigmentation) as subunits of biogenesis of lysosome-related organelles complex-1 (BLOC-1) J Biol Chem. 2004;279:28393–28401. doi: 10.1074/jbc.M402513200. [DOI] [PubMed] [Google Scholar]
- 6.Straub RE, Jiang Y, MacLean CJ, Ma Y, Webb BT, Myakishev MV, et al. Genetic variation in the 6p22.3 gene DTNBP1, the human ortholog of the mouse dysbindin gene, is associated with schizophrenia. Am J Hum Genet. 2002;71:337–348. doi: 10.1086/341750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van den Oord EJ, Sullivan PF, Jiang Y, Walsh D, O’Neill FA, Kendler KS, et al. Identification of a high-risk haplotype for the dystrobrevin binding protein 1 (DTNBP1) gene in the Irish study of high-density schizophrenia families. Mol Psychiatry. 2003;8:499–510. doi: 10.1038/sj.mp.4001263. [DOI] [PubMed] [Google Scholar]
- 8.Allen NC, Bagade S, McQueen MB, Ioannidis JPA, Kavvoura FK, Khoury MJ, et al. Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database. Nat Genet. 2008;40:827–834. doi: 10.1038/ng.171. [DOI] [PubMed] [Google Scholar]
- 9.Li D, He L. Association study between the dystrobrevin binding protein 1 gene (DTNBP1) and schizophrenia: a meta-analysis. Schizophr Res. 2007;96:112–118. doi: 10.1016/j.schres.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 10.Riley B, Kendler KS. Molecular genetic studies of schizophrenia. Eur J Hum Genet. 2006;14:669–680. doi: 10.1038/sj.ejhg.5201571. [DOI] [PubMed] [Google Scholar]
- 11.Sun J, Kuo P-H, Riley BP, Kendler KS, Zhao Z. Candidate genes for schizophrenia: a survey of association studies and gene ranking. Am J Med Genet B Neuropsychiatr Genet. 2008 doi: 10.1002/ajmg.b.30743. e-pub ahead of print. [DOI] [PubMed] [Google Scholar]
- 12.Breen G, Prata D, Osborne S, Munro J, Sinclair M, Li T, et al. Association of the dysbindin gene with bipolar affective disorder. Am J Psychiatry. 2006;163:1636–1638. doi: 10.1176/ajp.2006.163.9.1636. [DOI] [PubMed] [Google Scholar]
- 13.Zinkstok JR, de Wilde O, van Amelsvoort TA, Tanck MW, Baas F, Linszen DH. Association between the DTNBP1 gene and intelligence: a case–control study in young patients with schizophrenia and related disorders and unaffected siblings. Behav Brain Funct. 2007;3:19. doi: 10.1186/1744-9081-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Donovan MC, Williams NM, Owen MJ. Recent advances in the genetics of schizophrenia. Hum Mol Genet. 2003;12, Spec No 2:R125–R133. doi: 10.1093/hmg/ddg302. [DOI] [PubMed] [Google Scholar]
- 15.Owen MJ, Williams NM, O’Donovan MC. The molecular genetics of schizophrenia: new findings promise new insights. Mol Psychiatry. 2004;9:14–27. doi: 10.1038/sj.mp.4001444. [DOI] [PubMed] [Google Scholar]
- 16.Ross CA, Margolis RL, Reading SA, Pletnikov M, Coyle JT. Neurobiology of schizophrenia. Neuron. 2006;52:139–153. doi: 10.1016/j.neuron.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 17.Weickert CS, Straub RE, McClintock BW, Matsumoto M, Hashimoto R, Hyde TM, et al. Human dysbindin (DTNBP1) gene expression in normal brain and in schizophrenic prefrontal cortex and midbrain. Arch Gen Psychiatry. 2004;61:544–555. doi: 10.1001/archpsyc.61.6.544. [DOI] [PubMed] [Google Scholar]
- 18.Weickert CS, Rothmond DA, Hyde TM, Kleinman JE, Straub RE. Reduced DTNBP1 (dysbindin-1) mRNA in the hippocampal formation of schizophrenia patients. Schizophr Res. 2008;98:105–110. doi: 10.1016/j.schres.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Talbot K, Eidem WL, Tinsley CL, Benson MA, Thompson EW, Smith RJ, et al. Dysbindin-1 is reduced in intrinsic, glutamatergic terminals of the hippocampal formation in schizophrenia. J Clin Invest. 2004;113:1353–1363. doi: 10.1172/JCI20425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bray NJ, Preece A, Williams NM, Moskvina V, Buckland PR, Owen MJ, et al. Haplotypes at the dystrobrevin binding protein 1 (DTNBP1) gene locus mediate risk for schizophrenia through reduced DTNBP1 expression. Hum Mol Genet. 2005;14:1947–1954. doi: 10.1093/hmg/ddi199. [DOI] [PubMed] [Google Scholar]
- 21.Camargo LM, Collura V, Rain JC, Mizuguchi K, Hermjakob H, Kerrien S, et al. Disrupted in Schizophrenia 1 Interactome: evidence for the close connectivity of risk genes and a potential synaptic basis for schizophrenia. Mol Psychiatry. 2007;12:74–86. doi: 10.1038/sj.mp.4001880. [DOI] [PubMed] [Google Scholar]
- 22.Dermitzakis ET, Reymond A, Antonarakis SE. Conserved nongenic sequences–an unexpected feature of mammalian genomes. Nat Rev Genet. 2005;6:151–157. doi: 10.1038/nrg1527. [DOI] [PubMed] [Google Scholar]
- 23.Brune M. Schizophrenia–an evolutionary enigma? Neurosci Biobehav Rev. 2004;28:41–53. doi: 10.1016/j.neubiorev.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 24.Crespi B, Summers K, Dorus S. Adaptive evolution of genes underlying schizophrenia. Proc Biol Sci. 2007;274:2801–2810. doi: 10.1098/rspb.2007.0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 27.MacCallum C, Hill E. Being positive about selection. PLoS Biol. 2006;4:e87. doi: 10.1371/journal.pbio.0040087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thierry-Mieg D, Thierry-Mieg J. AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 2006;7(Suppl 1: S12):11–14. doi: 10.1186/gb-2006-7-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang C, Zhao Z. Mutational spectrum in the recent human genome inferred by single nucleotide polymorphisms. Genomics. 2006;88:527–534. doi: 10.1016/j.ygeno.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 30.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao Z, Yu N, Fu Y-X, Li W-H. Nucleotide variation and haplotype diversity in a 10-kb noncoding region in three continental human populations. Genetics. 2006;174:399–409. doi: 10.1534/genetics.106.060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mutsuddi M, Morris DW, Waggoner SG, Daly MJ, Scolnick EM, Sklar P. Analysis of high-resolution HapMap of DTNBP1 (Dysbindin) suggests no consistency between reported common variant associations and schizophrenia. Am J Hum Genet. 2006;79:903–909. doi: 10.1086/508942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 34.Williams NM, Preece A, Morris DW, Spurlock G, Bray NJ, Stephens M, et al. Identification in 2 independent samples of a novel schizophrenia risk haplotype of the dystrobrevin binding protein gene (DTNBP1) Arch Gen Psychiatry. 2004;61:336–344. doi: 10.1001/archpsyc.61.4.336. [DOI] [PubMed] [Google Scholar]
- 35.Numakawa T, Yagasaki Y, Ishimoto T, Okada T, Suzuki T, Iwata N, et al. Evidence of novel neuronal functions of dysbindin, a susceptibility gene for schizophrenia. Hum Mol Genet. 2004;13:2699–2708. doi: 10.1093/hmg/ddh280. [DOI] [PubMed] [Google Scholar]
- 36.Donohoe G, Morris DW, Clarke S, McGhee KA, Schwaiger S, Nangle JM, et al. Variance in neurocognitive performance is associated with dysbindin-1 in schizophrenia: a preliminary study. Neuropsychologia. 2007;45:454–458. doi: 10.1016/j.neuropsychologia.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 37.Donohoe G, Morris DW, De Sanctis P, Magno E, Montesi JL, Garavan HP, et al. Early visual processing deficits in dysbindin-associated schizophrenia. Biol Psychiatry. 2008;63:484–489. doi: 10.1016/j.biopsych.2007.07.022. [DOI] [PubMed] [Google Scholar]
- 38.Corvin A, Donohoe G, Nangle JM, Schwaiger S, Morris D, Gill M. A dysbindin risk haplotype associated with less severe manic-type symptoms in psychosis. Neurosci Lett. 2008;431:146–149. doi: 10.1016/j.neulet.2007.11.031. [DOI] [PubMed] [Google Scholar]
- 39.Chubb JE, Bradshaw NJ, Soares DC, Porteous DJ, Millar JK. The DISC locus in psychiatric illness. Mol Psychiatry. 2008;13:36–64. doi: 10.1038/sj.mp.4002106. [DOI] [PubMed] [Google Scholar]
- 40.Di Pietro SM, Falcon-Perez JM, Tenza D, Setty SR, Marks MS, Raposo G, et al. BLOC-1 interacts with BLOC-2 and the AP-3 complex to facilitate protein trafficking on endosomes. Mol Biol Cell. 2006;17:4027–4038. doi: 10.1091/mbc.E06-05-0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schweizer FE, Ryan TA. The synaptic vesicle: cycle of exocytosis and endocytosis. Curr Opin Neurobiol. 2006;16:298–304. doi: 10.1016/j.conb.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 42.Talbot K, Cho DS, Ong WY, Benson MA, Han LY, Kazi HA, et al. Dysbindin-1 is a synaptic and microtubular protein that binds brain snapin. Hum Mol Genet. 2006;15:3041–3054. doi: 10.1093/hmg/ddl246. [DOI] [PubMed] [Google Scholar]
- 43.Ni X, Trakalo J, Valente J, Azevedo MH, Pato MT, Pato CN, et al. Human p53 tumor suppressor gene (TP53) and schizophrenia: case–control and family studies. Neurosci Lett. 2005;388:173–178. doi: 10.1016/j.neulet.2005.06.050. [DOI] [PubMed] [Google Scholar]
- 44.Thiselton DL, Vladimirov VI, Kuo PH, McClay J, Wormley B, Fanous A, et al. AKT1 is associated with schizophrenia across multiple symptom dimensions in the Irish study of high density schizophrenia families. Biol Psychiatry. 2008;63:449–457. doi: 10.1016/j.biopsych.2007.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jeong H, Mason SP, Barabasi AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 46.Rizo J, Chen X, Arac D. Unraveling the mechanisms of synaptotagmin and SNARE function in neurotransmitter release. Trends Cell Biol. 2006;16:339–350. doi: 10.1016/j.tcb.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 47.Ruder C, Reimer T, Delgado-Martinez I, Hermosilla R, Engelsberg A, Nehring R, et al. EBAG9 adds a new layer of control on large densecore vesicle exocytosis via interaction with Snapin. Mol Biol Cell. 2005;16:1245–1257. doi: 10.1091/mbc.E04-09-0817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ilardi JM, Mochida S, Sheng ZH. Snapin: a SNARE-associated protein implicated in synaptic transmission. Nat Neurosci. 1999;2:119–124. doi: 10.1038/5673. [DOI] [PubMed] [Google Scholar]
- 49.Sudhof TC. The synaptic vesicle cycle: a cascade of protein-protein interactions. Nature. 1995;375:645–653. doi: 10.1038/375645a0. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Q, Pangrsic T, Kreft M, Krzan M, Li N, Sul JY, et al. Fusion-related release of glutamate from astrocytes. J Biol Chem. 2004;279:12724–12733. doi: 10.1074/jbc.M312845200. [DOI] [PubMed] [Google Scholar]
- 51.Rees ML, Lien CF, Gorecki DC. Dystrobrevins in muscle and non-muscle tissues. Neuromuscul Disord. 2007;17:123–134. doi: 10.1016/j.nmd.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 52.Rando TA. The dystrophin-glycoprotein complex, cellular signaling, and the regulation of cell survival in the muscular dystrophies. Muscle Nerve. 2001;24:1575–1594. doi: 10.1002/mus.1192. [DOI] [PubMed] [Google Scholar]
- 53.Mehler MF. Brain dystrophin, neurogenetics and mental retardation. Brain Res Rev. 2000;32:277–307. doi: 10.1016/s0165-0173(99)00090-9. [DOI] [PubMed] [Google Scholar]
- 54.Blake DJ, Nawrotzki R, Loh NY, Gorecki DC, Davies KE. beta-dystrobrevin, a member of the dystrophin-related protein family. Proc Natl Acad Sci USA. 1998;95:241–246. doi: 10.1073/pnas.95.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Culligan K, Ohlendieck K. Diversity of the brain dystrophin-glycoprotein complex. J Biomed Biotechnol. 2002;2:31–36. doi: 10.1155/S1110724302000347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maden M. Retinoic acid in the development, regeneration and maintenance of the nervous system. Nat Rev Neurosci. 2007;8:755–765. doi: 10.1038/nrn2212. [DOI] [PubMed] [Google Scholar]
- 57.Mey J, McCaffery P. Retinoic acid signaling in the nervous system of adult vertebrates. Neuroscientist. 2004;10:409–421. doi: 10.1177/1073858404263520. [DOI] [PubMed] [Google Scholar]
- 58.Palha JA, Goodman AB. Thyroid hormones and retinoids: a possible link between genes and environment in schizophrenia. Brain Res Rev. 2006;51:61–71. doi: 10.1016/j.brainresrev.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 59.Rao ML, Kolsch H. Effects of estrogen on brain development and neuroprotection—implications for negative symptoms in schizophrenia. Psychoneuroendocrinology. 2003;28(Suppl 2):83–96. doi: 10.1016/s0306-4530(02)00126-9. [DOI] [PubMed] [Google Scholar]
- 60.Li XH, Kakkad B, Ong DE. Estrogen directly induces expression of retinoic acid biosynthetic enzymes, compartmentalized between the epithelium and underlying stromal cells in rat uterus. Endocrinology. 2004;145:4756–4762. doi: 10.1210/en.2004-0514. [DOI] [PubMed] [Google Scholar]
- 61.Blitzer RD, Iyengar R, Landau EM. Postsynaptic signaling networks: cellular cogwheels underlying long-term plasticity. Biol Psychiatry. 2005;57:113–119. doi: 10.1016/j.biopsych.2004.02.031. [DOI] [PubMed] [Google Scholar]
- 62.Sullivan JM. Cellular and molecular mechanisms underlying learning and memory impairments produced by cannabinoids. Learn Mem. 2000;7:132–139. doi: 10.1101/lm.7.3.132. [DOI] [PubMed] [Google Scholar]
- 63.Hakak Y, Walker JR, Li C, Wong WH, Davis KL, Buxbaum JD, et al. Genome-wide expression analysis reveals dysregulation of myelination-related genes in chronic schizophrenia. Proc Natl Acad Sci USA. 2001;98:4746–4751. doi: 10.1073/pnas.081071198. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.