Synopsis
Pyruvate carboxylase (PC) is a biotin-containing enzyme that catalyses the HCO3−- and MgATP-dependent carboxylation of pyruvate to form oxaloacetate. This is a very important anaplerotic reaction, replenishing oxaloacetate withdrawn from the Krebs cycle for various pivotal biochemical pathways. PC is therefore considered as an enzyme that is crucial for intermediary metabolism, controlling fuel partitioning toward gluconeogenesis, lipogenesis and insulin secretion. The enzyme was discovered in 1959 and over the last decade there has been much progress in understanding its structure and function. PC from most organisms is a tetrameric protein that is allosterically regulated by acetyl CoA and aspartate. High resolution crystal structures of the holoenzyme with various ligands bound have recently been determined, and have revealed details and the relative positions of the biotin carboxylase, carboxyltransferase and biotin carboxyl carrier domains, and also a unique allosteric effector domain. In the presence of the allosteric effector, acetyl CoA, the biotin moiety transfers the carboxyl group intermediate between the biotin carboxylase domain active site on one polypeptide chain and the carboxyltransferase active site on the adjacent antiparallel polypeptide chain. In addition, the bona fide role of PC in the non-gluconeogenic tissues has been studied using a combination of classical biochemistry and genetic approaches. The first cloning of the promoter of the PC gene in mammals and subsequent transcriptional studies reveal some key cognate transcription factors regulating tissue-specific expression. This review summarizes these advances and also offers some prospects in terms of future directions for the study of this important enzyme.
Keywords: pyruvate carboxylase, biotin, structure, kinetics, acetyl CoA, role in metabolism, gene expression
1. Introduction
Pyruvate carboxylase (PC; EC 6.4.1.1) was discovered by Utter & Keech [1] in the course of studies on the intracellular distribution of enzymes involved in the dicarboxylic acid shuttle and its relationship to gluconeogenesis in chicken liver. PC catalyses the HCO3- and MgATP-dependent carboxylation of pyruvate to form oxaloacetate. Oxaloacetate is one of several important intermediates in Krebs cycle (tricarboxylic acid (TCA) cycle) that are withdrawn for use in several biosynthetic pathways. Therefore, this reaction is considered to play an important anaplerotic role in numerous biological processes [2].
2. Metabolic role of PC
2.1 The roles of PC in microorganisms
PC is found in many prokaryotes including Pseudomonas, Bacillus, Rhizobium, Mycobacterium, Staphylococcus and archaebacteria [3], but not in the enteric bacteria which utilize phosphoenolpyruvate carboxylase (PEPC) to convert phosphoenolpyruvate (PEP) directly to oxaloacetate, thus avoiding the need for PC. For a comprehensive review of the enzymes and metabolic fluxes at the PEP-pyruvate-oxaloacetate node in bacteria see Sauer & Eikmanns [4]. In yeast, two metabolic pathways leading to the production of oxaloacetate are the PC-catalysed reaction and the glyoxylate cycle. When yeast are grown on acetate, PC-catalysed oxaloacetate formation is repressed but the glyoxylate cycle is active, and vice versa if grown on glucose minimal media [5]. In E. coli, where PC’s function is replaced by PEPC, overexpression of PC can improve recombinant protein production in this organism. This is thought to be due to improved oxaloacetate production enhancing fuel oxidation [6,7].
2.2 The roles of PC in mammalian tissues
2.2.1 The role of PC in gluconeogenesis
During prolonged fasting, the brain depends substantially, and red blood cells rely entirely for an energy source on glucose synthesised from the non-carbohydrate precursors lactate, alanine, glycerol and glutamine by liver, and to a lesser extent by kidney cortex and small intestine. As shown in Fig. 1, PC catalyses the first committed step in gluconeogenesis, providing oxaloacetate for subsequent conversion to phosphoenolpyruvate by phosphoenolpyruvate carboxykinase (PEPCK). This latter enzyme occurs as two distinct isoforms, one in the mitochondria the other in the cytoplasm, with the relative proportions differing between species. Oxaloacetate may then be converted to PEP by mitochondrial PEPCK [8] and transported through the mitochondrial membranes by the tricarboxylic acid anion carrier system [9] to the cytoplasm where it is converted to glucose via the combination of the reverse pathway of glycolysis, and the other two gluconeogenic enzymes i.e. fructose-1,6-bisphosphatase and glucose-6-phosphatase. However, if the level of cytosolic NADH is low and mitochondrial NADH is high, as in starvation when alanine is the major source of pyruvate in the liver, the oxaloacetate formed by PC can also serve as a carrier of reducing equivalents to the cytoplasm. In this process the oxaloacetate is first converted to malate by mitochondrial malate dehydrogenase (MDH), before leaving the mitochondria via the malate transporter. The cytosolic MDH then converts malate back to oxaloacetate, with reduction of NAD+ to NADH. The oxaloacetate is subsequently converted to PEP by cytosolic PEPCK, and the NADH is used to reduce 1,3-bisphosphoglycerate to glyceraldehyde 3-phosphate. Alterations of nutritional status in mouse and rat have been shown to affect hepatic PC activity [10, 11]. Starvation enhances PC activity, while diabetes also increases gluconeogenesis through enhanced uptake of substrate and increased flux through liver PC in mice and rats [12, 13, 14], indicating the key role of PC in controlling gluconeogenesis. In dairy cows, which absorb little if any hexoses, even when well fed, the expression of PC and PEPCK is markedly elevated during the transition from calving to lactation, suggesting the coordination of these two enzymes in the conversion of lactate and glucogenic amino acids by gluconeogenesis to support lactose synthesis in the mammary gland [15]. Short term treatment with glucagon increased PC mRNA [16] but did not result in an apparent change of PC protein level or activity [17]. Although treatment of lactating dairy cows with dexamethasone did not appear to alter PC expression [28], feed-restriction stimulated PC expression, suggesting this enzyme is under nutritional control [19]. Phlorizin-induced glucose loss in urine of lactating cows stimulates expression of hepatic PC indicating an adaptive response to the demand of hepatic gluconeogenesis [20].
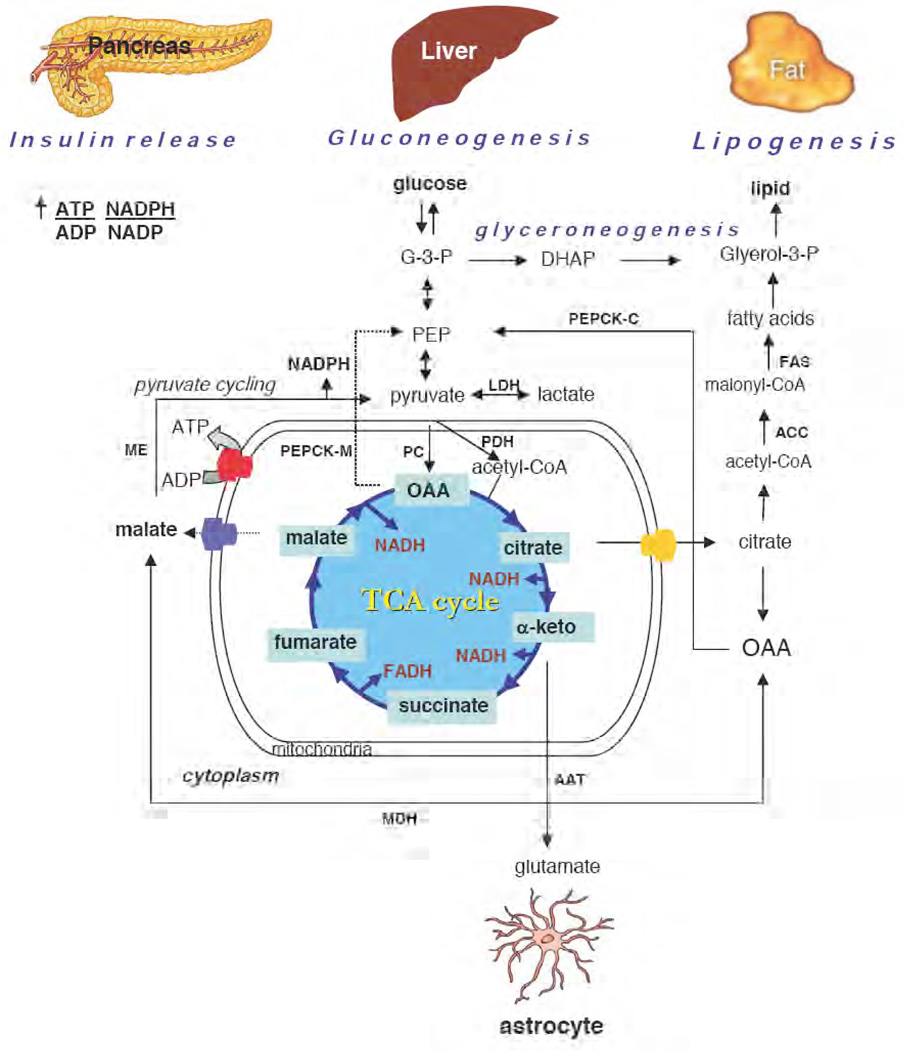
Figure 1. Anaplerotic role of PC in various mammalian tissues.
Glucose is oxidized to pyruvate through the glycolytic pathway. PC replenishes oxaloacetate in the Krebs tricarboxylic acid (TCA) cycle when its intermediates are used for various biosynthetic pathways, depending on the tissues. In liver, oxaloacetate (OAA) is utilized as the precursor for gluconeogenesis whereby OAA can exit the mitochondria as malate before being converted back to OAA by cytosolic malate dehydrogenase (MDH). In some situations OAA is converted to phosphoenolpyruvate (PEP) by mitochondrial phosphoenolpyruvate carboxykinase (PEPCK-M) [8,9]. Cytoplasmic OAA is converted to glucose by the combined activities of cytoplasmic PEPCK (PEPCK-C), fructose-1,6-bisphosphatase, glucose-6-phosphatase and 7 of the 10 glycolytic enzymes. In fat cells, PC provides oxaloacetate to facilitate the export of acetyl CoA in the form of citrate that leaves mitochondria for de novo fatty acid synthesis. Oxaloacetate liberated in the cytoplasm by citrate cleavage enzyme is converted to PEP by PEPCK-C as in the liver [8]. PEP is then converted to glycerol by a pathway known as “glyceroneogenesis”, while acetyl CoA is converted to malonyl-CoA by acetyl CoA carboxylase (ACC). Fatty acid synthase (FAS) catalyses the condensation of two-carbon moieties from malonyl-CoA to produce long chain acyl-CoA that are subsequently esterified with glycerol to form triglycerides. In pancreatic β-cells, PC is involved in a ‘pyruvate cycle’ that involves the exchange of TCA cycle intermediates and cytosolic pyruvate, catalysed by MDH and malic enzyme (ME) [30,31]. This cycle produces a large amount of NADPH which is one of the metabolic coupling factors. In astrocytes, α-ketoglutarate is converted by an aspartate aminotransferase (AAT) to glutamate, one of the neurotransmitter substances of neurons [39]. Key metabolites glyceraldehyde 3-phosphate (G-3-P), dihydroxyacetone phosphate (DHAP), and enzymes lactate dehydrogenase (LDH) and pyruvate dehydrogenase (PDH) are indicated.
2.2.2 The role of PC in adipogenesis
PC is also highly expressed in adipose tissues. As the key gluconeogenic enzymes, glucose-6-phosphatase and fructose-1,6-bisphosphatase, are not present in adipose tissue, Ballard and Hanson [21] proposed the involvement of PC in the pathway of de novo fatty acid synthesis. PC participates in this pathway by providing oxaloacetate for conversion to citrate which is exported from the mitochondria and cleaved in the cytosol to form oxaloacetate and acetyl CoA [21] (see Fig. 1). The latter molecule is a building block for long-chain fatty acid synthesis. In murine 3T3-L1 adipocytes, PC protein, its activity and mRNA are elevated in parallel with other key lipogenic enzymes, viz. ATP-citrate lyase, malic enzyme, acetyl CoA carboxylase (ACC) and fatty acid synthase (FAS), which increase concomitantly with the accumulation of lipid droplets during terminal differentiation of adipocytes [22]. The expression of PC is also regulated tightly with adipocyte differentiation, as treatment of 3T3-L1 preadipocytes with TNFα, a cytokine that inhibits adipocyte differentiation, also suppresses PC expression [23]. However, the inhibition of PC expression may be a secondary effect caused by the suppression of peroxisome proliferator-activated receptor gamma (PPAR-γ) as PPARγ is known to increase PC expression in this cell type [23,24]. In addition to de novo fatty acid synthesis, PC is also involved in glyceroneogenesis, a pathway for synthesizing glycerol required for fatty acid re-esterification [25]. This pathway is important for reducing elevated levels of free fatty acids in the plasma due to lipolysis or to consumption of a high fat diet. PC provides oxaloacetate for conversion to phosphoenolpyruvate by PEPCK, and then to glycerol. Treatment of mice with rosiglitazone or other thiazolidinediones, drugs that reduce free fatty acid levels through re-esterification, enhances expression of PC [26, 27] and PEPCK in adipose tissue, indicating the cooperative role of these two enzymes in the regulation of glyceroneogenesis.
2.2.3 The role of PC in pancreatic islets
The pancreatic islets play a crucial role in glucose homeostasis by secreting insulin in response to fluctuations in plasma glucose levels. Glycolysis plus mitochondrial metabolism are known to be essential for glucose-induced insulin secretion [28]. It is well established that intracellular ATP is required for insulin exocytosis. A higher ATP/ADP ratio is needed for the closure of KATP channels compared with the requirement of the exocytotic process itself. It is known that high TCA cycle activity caused by rapid oxidation of glucose-derived pyruvate produces a large amount of NADH. This supply of reducing equivalents in turn activates oxidative phosphorylation resulting in the formation of ATP. Apart from ATP, other nucleotides, including NADPH, are also known to be important coupling factors required for insulin exocytosis [29]. PC not only provides an anaplerotic input for the TCA cycle by supplying substantial amounts of oxaloacetate, but is also involved in the formation of NADPH. The generation of a high level of NADPH is achieved through a shuttle called “pyruvate cycling” between mitochondria and cytoplasm. The oxaloacetate formed by PC is converted to either malate or citrate, and leaves the mitochondria through the pyruvate/malate [30] or citrate/pyruvate shuttle [31], respectively (see Fig. 1). Malic enzyme [EC 1.1.1.40 malate dehydrogenase (oxaloacetate-decarboxylating) (NADP+)] catalyses the NADPH production in both shuttles. As these two shuttles operate as cycles, the amount of NADPH produced is much higher than that obtained from the pentose phosphate pathway.
Initially, it was estimated that equal proportions of pyruvate enter the TCA cycle through PC and the pyruvate dehydrogenase complex (PDH), and that these two fluxes are equally important for glucose-induced insulin secretion [32]. However, subsequent experiments using NMR analysis showed that pyruvate entering into mitochondria exists in two different pools. In INS1-derived cell lines, the first pool is derived from glycolysis, and from this pyruvate is further oxidized to acetyl CoA by PDH, whilst the second pool is derived by the exchange of TCA cycle intermediates (i.e. pyruvate cycling) involving PC catalytic activity, as described above. It is the latter pool that correlates with insulin secretion [33].
Pharmacological inhibition of PC activity by phenylacetic acid impaired glucose-induced insulin secretion in insulinoma cells, in isolated islets [31, 34], and in intact animals [35]. However, it is still unclear whether this reduced insulin secretion was exclusively caused by specific inhibition of PC as this inhibitor has not been fully characterized [36]. In contrast, partial inhibition of PC expression by siRNA in islets and insulinoma cells did not affect glucose-induced insulin secretion [37]. Specifically, the authors found a 30% increase in the level of acetyl CoA via PDH flux as a key compensatory response to the suppression of PC in islets. As acetyl CoA is a potent allosteric activator of PC, this compensatory mechanism would prevent impairment of anaplerosis, pyruvate cycling, NADPH production and glucose-induced insulin secretion [37]. However, it remains unclear whether this reflects the actual scenario because the PC protein was only reduced to 50% in the β-cells transfected with PC siRNA [37].
A genetic link between PC and type 2 diabetes has recently been revealed. A single nucleotide polymorphism in the PC gene showed evidence of possible association with the acute insulin response phenotype [38].
2.2.4 The role of PC in astrocytes
In both astrocytes and neurons, glucose is metabolized initially via glycolysis. In neurons, glucose-derived pyruvate is mainly converted by PDH to acetyl CoA which subsequently enters the TCA cycle for energy production. Therefore, in fed animals, neurons rely exclusively on exogenous glucose. However, in astrocytes, glucose-derived pyruvate can be either oxidised by PDH to acetyl CoA for energy production or carboxylated by PC to form oxaloacetate for anaplerotic purposes [39]. The need for this anaplerotic reaction is that astrocytes utilize α-ketoglutarate as a precursor for the production of glutamate, some of which is further converted to γ-amino butyric acid, with both being important neurotransmitters [40]. Since a-ketoglutarate is continuously withdrawn from the TCA cycle for this purpose, PC activity is required to replenish this intermediate in astrocytes (see Fig. 1). However, glutamate is not readily diffusible to the neurons and needs to be converted to glutamine by glutamine synthetase [39]. Glutamine is then taken up by neurons for subsequent conversion to glutamate by glutaminase. After being used by neurons, glutamate is taken back by astrocytes. This cycle is known as the glutamine-glutamate cycle that is required for shuffling glutamate between astrocytes and neurons [39]. NMR analysis reveals the increased metabolic flux through PC is concomitant with stimulation of de novo glutamine synthesis in vivo and ex vivo in the brains of hyperammonemic animal models [41, 42]. A recent report also shows that pyruvate flux through PC varies from 20% to 50% of the total pyruvate pool [43], depending on the depletion of glutamate. The presence of PC only in astrocytes explains why neurons cannot directly synthesize glutamate from TCA cycle intermediates [44].
2.2.5 PC in mammalian cell lines
In several mammalian cell lines, PC appears to be important in linking glycolytically-derived pyruvate to the TCA cycle, as those poorly expressing PC are characterized by high lactic acid accumulation. Metabolic engineering of these cells by expression of cytosolic or mitochondrial PC improves the yield of recombinant protein production in these cell lines [45,46]. This improvement in product yield and cell viability is due to an increased flux of pyruvate into the TCA cycle for complete oxidation thereby increasing ATP content while reducing glucose consumption and lactate accumulation [47].
3. PC and metabolic adaptation
As PC is involved in the metabolic crossroad of carbohydrate and lipid metabolism, regulation of its expression in various tissues (gluconeogenic tissue, adipose tissue and pancreatic islets) must be coordinated to achieve an appropriate overall response to various physiological and pathological stimuli. In conditions of over-nutrition (high carbohydrate diet) (Fig. 2A), PC expression is increased in β-cells to enhance pyruvate cycling activity in response to the chronically elevated level of glucose [35]. This condition probably represents an early adaptive response of PC in β-cells. Conversely, gluconeogenic enzymes in liver, including PC, are suppressed by insulin [48,49]. During this period, adipocyte tissue is expanded concomitant with the over-expression of PC and other de novo lipogenic enzymes [50,51]. The excess non-esterified fatty acids and triglycerides are stored in adipose tissue as a result of insulin action. However, chronic exposure of β-cells to an elevated level of glucose due to insulin resistance, as observed in the genetically inherited type 2 diabetic rats, results in decreased PC activity and PC protein [52,53] (see Fig. 2B). The overproduction of hepatic glucose caused by insulin resistance results in a severe hyperglycemia, creating a “glucotoxicity loop” [54]. This condition in turn, causes a global alteration of gene expression in β-cells including down-regulation of PC, concomitant with the failure of β-cell function [55]. In the meantime, adipose tissue develops insulin resistance, causing the accumulation of triglycerides and free fatty acids in the circulation. These free fatty acids and triglycerides further impair β-cell function [54, 56], and decrease PC gene expression [57, 58], creating a “lipotoxicity loop”. Adipose tissue of the ob/ob mice treated with thiazolidinedione shows increased PC expression [27], suggesting that insulin resistance contributes to the down-regulation of PC expression in adipocytes of this obesity-induced diabetic model.
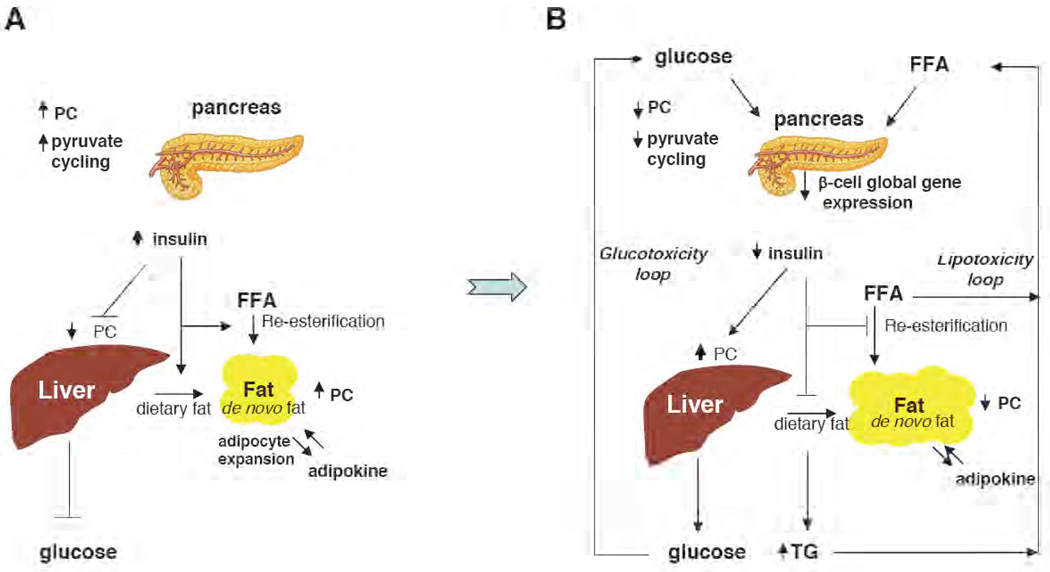
Figure 2. Role of PC during metabolic adaptation.
(A) In an over-nutrition situation, PC activity and pyruvate cycling activity are increased, resulting in increased insulin secretion in response to the chronically elevated level of plasma glucose [43]. Hepatic glucose production is still properly maintained in this condition. Adipocyte mass is expanded concomitant with increased expression of PC and other enzymes of fat synthesis. (B) In obesity-induced type 2 diabetes, chronic exposure of β-cells to an elevated level of glucose due to peripheral insulin resistance impairs β-cell function. Excessive hepatic glucose production results in an elevated level of plasma glucose, creating a “glucotoxicity loop”. Severe hyperglycemia reduces expression of several β-cell specific genes including PC. Mitochondrial metabolism of adipocytes is also impaired, causing down-regulation of some lipogenic enzymes including PC. Triglycerides (TG) and free fatty acids (FFA) released from adipose tissue to plasma further impair β-cell function and PC gene expression, creating a “lipotoxicity loop”. [54]
4. PC deficiency
As PC is involved in numerous metabolic pathways, an inherited deficiency of this enzyme would be expected to cause widespread metabolic disturbances in various tissues. PC deficiency is a rare autosomal recessive inborn error of metabolism with an incidence of 1:250,000 [59]. Three forms of PC deficiency are classified: Type A or the North American phenotype is characterized by a mild lactic acidemia but a normal ratio of plasma lactate to pyruvate, psychomotor retardation and in some, but not all cases, death in the first years of life [59]. The molecular basis of the disorder in type A patients is characterized by the presence of residual PC protein and its diminished activity. There have been four different point mutations of the PC gene associated with this phenotype. These mutations are Val145 to Ala; Arg451 to Cys; Ala610 to Thr and Met743 to Ile [60]. Some of these mutations are located in the active site and may affect binding of substrates to the enzyme (61). The A610T mutation did not appear to affect the enzyme’s Km for pyruvate, but its catalytic activity and steady-state level were markedly decreased, the latter effect being due to its reduced import into the mitochondria and impaired stability [62]. The R451C mutation markedly decreased acetyl CoA dependent activation of the enzyme [61]. The type B or French phenotype is characterized by a severe neonatal lactic acidemia with an elevated lactate to pyruvate ratio, ketoacidosis, hypoglycaemia accompanied with hyperammonemia and hypercitrullinemia [59]. Some type B patients show abnormal brain development [63, 64, 65], and most die within the first few months. The molecular basis of type B patients is characterized by the absence of immunoreactive PC protein or PC mRNA [59]. With the type B phenotype, a complex genotype in which two deletion mutations in both PC alleles was identified, i.e. one allele possesses 2 nucleotide deletions in exon 16, creating a frameshift mutation, while the other allele possesses 4 nucleotide deletions in intron 15, resulting in an aberrant transcript. These two mutations generate premature terminations of the protein [66]. The type C or ‘benign’ phenotype is characterized as a mild lactic acidosis but normal psychomotor development [67].
The above metabolic disturbances caused by PC deficiency can be explained as follows: [i] The lack of PC activity would cause the accumulation of pyruvate which is subsequently converted to lactate by lactate dehydrogenase, causing an elevated level of lactic acid. Considering skeletal muscle and red blood cells are the major organs which utilize glucose via anaerobic glycolysis, the level of lactate released from these two tissues would be extremely high and very toxic for the body without it being excreted or modified; [ii] Lactate and certain glucogenic amino acids derived from protein breakdown cannot be converted to oxaloacetate, and hence there is a limiting amount of oxaloacetate, a starting material for gluconeogenesis. This explains why type B patients face severe hypoglycemia caused by diminished neonatal gluconeogenesis; [iii] Deprivation of oxaloacetate leads to the failure of the liver to oxidize acetyl CoA derived from fatty acids, and this leads to ketoacidosis. The lack of oxaloacetate also impairs TCA cycle activity, affecting various TCA cycle intermediates to be used for various biosynthetic pathways e.g. the argininosuccinic acid cycle in urea. These defects explain why the type B phenotype exhibits metabolic ketoacidosis and elevated levels of citrulline and hyperammonemia; [iv] Delayed mental development with or without brain lesions and impaired psychomotor development may be caused by the low production of neurotransmitter substances [64,65] or an abnormal development of myelin sheath in brain [68]. The latter process relies on the lipogenic role of PC as mentioned earlier. It would be interesting to see whether PC-deficient patients also show impaired insulin secretion in pancreatic β-cells and abnormal adipose tissue development.
Treatment of PC deficiency is varied. Some patients are responsive to biotin treatment, suggesting that they may carry mutations which affect biotinylation. However, most patients do not respond to this treatment [69]. Treatment of patients of the type B phenotype with high doses of citrate or aspartate could potentially regenerate oxaloacetate by non-PC catalysed reactions and thus improve the above metabolic disturbances, but it cannot improve the delayed psychomotor development [64].
5. Gene structure
The PC gene has been conserved throughout evolution as it is found in many bacteria and eukaryotic cells. In fungal species, the PC gene does not contain introns and is encoded by a single copy gene with the exception of Saccharomyces cerevisiae. This yeast contains two functional copies of the PC gene, PYC1 and PYC2, located on different chromosomes [70,71]. Both isozymes are expressed in cytoplasm, unlike PC of higher eukaryotes, where the enzyme is located in the mitochondrial matrix [72]. The expression of the two isozymes is differentially regulated and expressed during different growth conditions [73, 74]. The PYC1 promoter contains two classical TATAA boxes, located −118/−111 and −109/−104 upstream of the translation initiation site [75] while the PYC2 promoter contains one canonical TATAA box, located at −65/−62 and one non-canonical TATA box, located at −290/−285 upstream of the transcription start site [70, 76].
In mammals, the complete genomic structure of PC gene has been reported in rat [95], mouse [78], and human [60]. The organization of the coding exons is highly conserved, i.e. the coding region is split into 19 coding exons, interrupted by 18 introns. Exons 2–10 encode the polypeptide segment that is present in the biotin carboxylation domain, while exons 13–14 encode the polypeptide segment that corresponds to the carboxyl transferase domain. Exons 18–20 encode the C-terminal biotin carboxyl carrier domain. A large [∼10 kb] intron that occurs between exons 10 and 11 is also conserved in these species. The most striking feature of mammalian PC genes is that they are regulated by multiple promoters, implying a complex regulation. In rat, 5 species of PC mRNAs have been reported, each having the same coding sequence but differing in their 5’-untranslated regions [79]. These mRNA variants are the product of alternative splicing of two primary transcripts initiated from two alternative promoters, the proximal and the distal promoters. Neither of these promoters contains a TATA box but both possess multiple GC boxes [77]. These PC mRNAs variants exhibit different translational efficiencies [51]. Furthermore, the production of specific forms of PC mRNA are linked to certain physiological states i.e. development, gluconeogenesis, lipogenesis, [51] which may reflect the differential regulation of each promoter during such conditions. In mouse [78] and human [79], two different transcripts have been identified and are suggested to be the products of two tissue-specific promoters. In cow, up to 6 species of mRNA have been reported from three alternative promoters, i.e. the proximal, middle and distal [80,81]. The proximal and the distal promoter contain multiple GC boxes but lack a TATA box, while the middle promoter contains a TATA and multiple GC boxes. The functional diversity of these PC mRNA variants is unclear. Similar to the rat PC gene, the proximal promoter of the bovine PC gene mediates the mRNA variants that are restricted to gluconeogenic and lipogenic tissues.
6. Transcriptional regulation
6.1 Yeast
In wild type yeast, expression of PYC1 and PYC2 is influenced by both the growth phase and carbon source. When grown in a medium containing glucose or ethanol as the carbon source, PYC1 is constitutively expressed throughout growth phase while PYC2 is only expressed in early log phase [73]. Independent experiments using single null mutants, PYC1 or PYC2, also show that disruption of either PYC gene did not affect the expression pattern of the other PYC gene, indicating the independent regulation of both genes [73]. Also, the type of carbon source influences the PYC1 and PYC2 gene transcription activities. Using PYC1-and PYC2-lacZ chimeric reporter gene constructs, PYC1 promoter activity was shown to be repressed by glucose and glycerol but activated by pyruvate, ethanol and galactose, while the PYC2 promoter is not affected by these carbon sources [82]. Furthermore, the activities of these two promoters are also regulated by nitrogen sources. Among several amino acids that appear to stimulate PYC1 and PYC2 promoter activities, aspartate potently represses the PYC1 promoter but not the PYC2, suggesting PYC1 is more regulatable [74]. Although nutrients certainly regulate PYC1 and PYC2 expression, the cis-acting elements in the promoters of PYC1 and PYC2 genes that mediate transcriptional regulation of both genes are unknown. The only available information is that the minimal promoter of the PYC1 gene is located within the first 330 nucleotides upstream of ATG initiation codon. Among other putative transcription factor binding sites, only one upstream stimulatory sequence located within the −347/−214 region of the PYC1 gene has been identified as a candidate binding site for the transcription factor RTG [82]. This was supported by the 50% down-regulation of PC in the RTG1 and the RTG2 S. cerevisiae mutants [83]. Little information is available for the transcription factors that may regulate PYC2 gene.
6.2 Mammals
The PC genes from mouse and rat are the most extensively studied in terms of transcriptional regulation. The distal promoter of the rat PC gene is active in pancreatic islets while the proximal promoter is active in gluconeogenic tissue (i.e. liver) and in lipogenic tissue, indicating that these two promoters work under different physiological circumstances [51] (see Fig. 3). In insulin-secreting cells, the minimal promoter element of the distal promoter has been located within the first 188 bp upstream of the transcription initiation site and its basal transcription activity is regulated by Sp1/Sp3 and NF-Y [84]. A pancreatic-specific promoter, viz. forkhead transcription factor boxA2 (Foxa2/HNF3β), has been identified as the positive regulator of this promoter [85]. In addition, upstream stimulatory factors (USF1 and USF2) are also involved in the transcription regulation of this promoter.
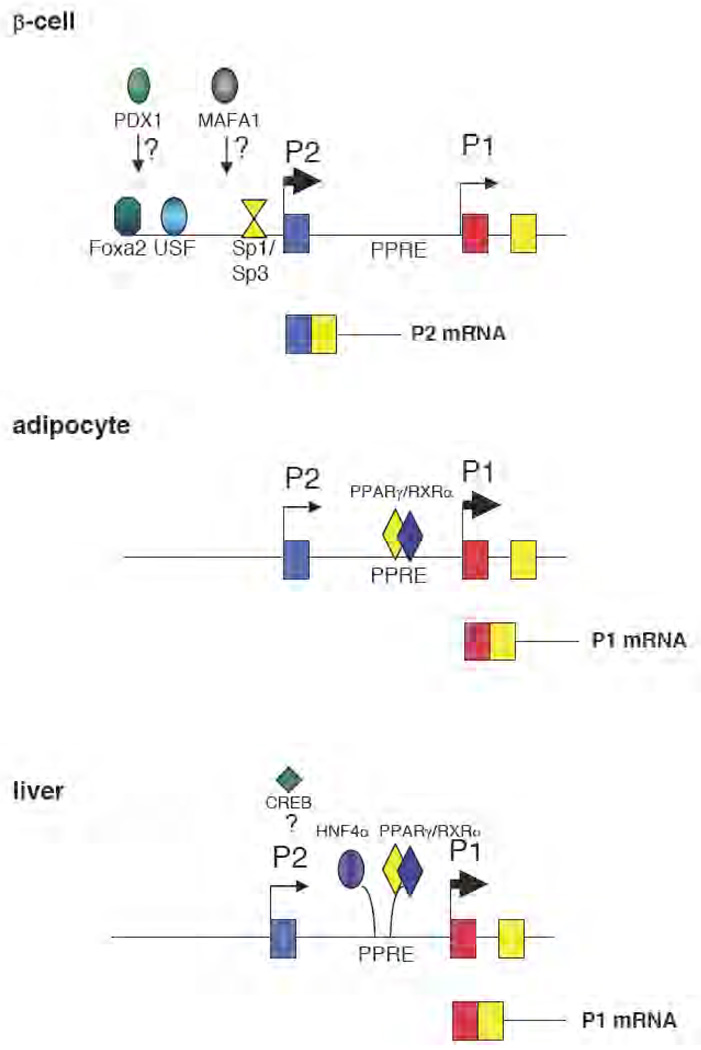
Figure 3. Tissue-specific regulation of PC in mammals.
The PC gene is regulated by two alternative promoters, the proximal (P1) and the distal (P2). In adipose tissue and gluconeogenic tissue (liver), the P1 promoter is active and is responsible for the production of PC mRNA that contains identical 5’-untranslated region exons [77, 78]. In adipocytes, the P1 promoter is regulated by peroxisome proliferator-activated receptor gamma (PPARγ) [23], while in liver it is still unclear whether PPARγ alone or other factors, including hepatocyte nuclear factor 4α (HNF4α) and cAMP-responsive element binding protein (CREB), may also contribute to regulation in this tissue [77, 78, 92]. In pancreatic β-cells PC is regulated by the P2 promoter with an interplay of basal [specific protein (Sp1/Sp3) and upstream stimulatory factors (USFs)] and β-cell specific transcription factors [84, 85]. It is not known whether v-maf musculoaponeurotic fibrosarcoma oncogene homologue A (Mafa1) and pancreatic-duodenal homeobox 1 (Pdx 1) directly regulate P2 activity.
Recent studies have shown that another two pancreatic islet-specific transcription factors, i.e. PDX1 and v-Mafa, are involved in transcriptional regulation of PC in INS1 cells. An adenovirus-mediated suppression of PDX1 affected PC expression concomitantly with impaired insulin secretion in INS1 cells [86]. It remains unclear whether Pdx1 directly regulates PC through the distal promoter because Pdx1 failed to transactivate the distal promoter in INS1 cells [85]. Similarly, overexpression of v-MAFA in INS1 cells causes a 5-fold increase of PC mRNA [87]. Expression of a dominant negative v-MAFA mutant did not stimulate PC expression. However the transcriptional mechanism whereby MAFA regulates promoter of PC gene has not been determined.
The liver X receptor (LXR), a nuclear receptor that acts as a cholesterol sensor, is also found to regulate PC expression in islets. Incubation of mouse insulinoma cells, MIN6 with an LXR agonist, increases glucose-induced insulin secretion concomitantly with increased PC expression [88], whereas treating the cells with a PC inhibitor prevents LXR-mediated glucose-induced insulin secretion. These results suggest that this nuclear receptor regulates insulin secretion via modulation of PC gene activity. However, it is still unclear whether LXR directly stimulates PC gene transcription because an LXR- responsive element in the promoters of the PC gene has not been reported. Another nuclear receptor, PPARα, is also implicated in the regulation of PC transcription. Short term treatment of pancreatic islets with fibrate, a synthetic PPARa ligand, or with free fatty acid, one of PPARα’s natural ligands, increases PPARa expression concomitantly with increased PC expression and glucose-induced insulin secretion, suggesting PC may be regulated by PPARa [89]. Similar to the LXRs study, the molecular mechanism by which PPARα regulates PC expression is unclear because a peroxisome proliferator-activated receptor responsive element (PPRE) has not been reported in the distal promoter which regulates PC expression in pancreatic islets.
In the case of the proximal promoter, a classical PPRE, located −386/−374 upstream of the transcription start site, has been identified (Fig. 3). This site can be transactivated and bound to PPARγ1 and PPARγ2 (adipocyte-specific isoform) equally well [23]. In 3T3-L1 murine adipocytes, disruption of this PPRE abrogates PPAR-γ mediated transcriptional activation [23] while an ectopic expression of PPARγ or treatment of cells with PPARγ agonist increases PC mRNA expression [24]. Heterozygous PPARγ knockout marginally affects PC expression in white adipose tissue [90] while the isoform-specific knockout, PPARγ2 null mice, showed a 50% reduction of PC in white and brown adipose tissue [23]. The remaining 50% of PC mRNA was probably caused by transcriptional compensation by PPARγ1, which was still intact [23]. In another mouse model, where PPARγ1 was over-expressed in the livers of PPARα null mice, it also caused a 6-fold increase of hepatic PC [91]. Collectively, these data clearly show that both isoforms of PPARγ regulate PC expression in vitro and in vivo.
In contrast, little is known about transcriptional regulation of the proximal promoter in liver cells. There is only one report which shows that PC may be regulated by cAMP-responsive element binding protein (CREB) in the liver. Transgenic mice carrying a dominant-negative mutant CREB showed a global reduction of gluconeogenic enzymes including PC, PEPCK, and glucose-6-phosphatase [92]. Although this evidence is consistent with the identification of a putative cAMP-responsive element in the proximal promoter of rat PC gene [93], the molecular mechanism whereby CREB activates PC expression remains to be elucidated.
7. Structure
7.1 Structural overview
PC is found in two forms, the α4 and α4β4 forms, depending on the organism. The α4 form is comprised of 4 identical subunits, each approximately 120–130 kDa in size. Biotin is covalently attached to a specific lysine residue located ∼35 residues from the C-terminus [3]. All three functional domains: biotin carboxylase (BC); carboxyltransferase (CT) and biotin carboxyl carrier protein (BCCP) are located on a single polypeptide chain (see Fig. 4). This α4 type PC is found in most organisms ranging from eubacteria, fungi, invertebrates and vertebrates. In the α4β4 form, each subunit is made up of two polypeptide chains, the ∼55 kDa non-biotinylated polypeptide (α) which possesses the biotin carboxylase activity and the ∼70 kDa polypeptide (β) which carries the biotin and also contains the carboxyl transferase activity. The α4β4 type of PC is found in archaebacteria i.e. Methanobacterium sp, Methanococcus sp and Methanosarcina sp [94, 95, 96] and certain bacteria i.e. Aquifex aeolicus [97], Pseudomonas spp [98, 99]. Interestingly, the majority of α4-type PCs are subject to allosteric regulation. Acetyl CoA is a powerful activator while L-aspartate is an inhibitor of α4-type PC. In contrast, the α4β4-type of PC has an activity that is completely independent of acetyl CoA [94, 95, 100].
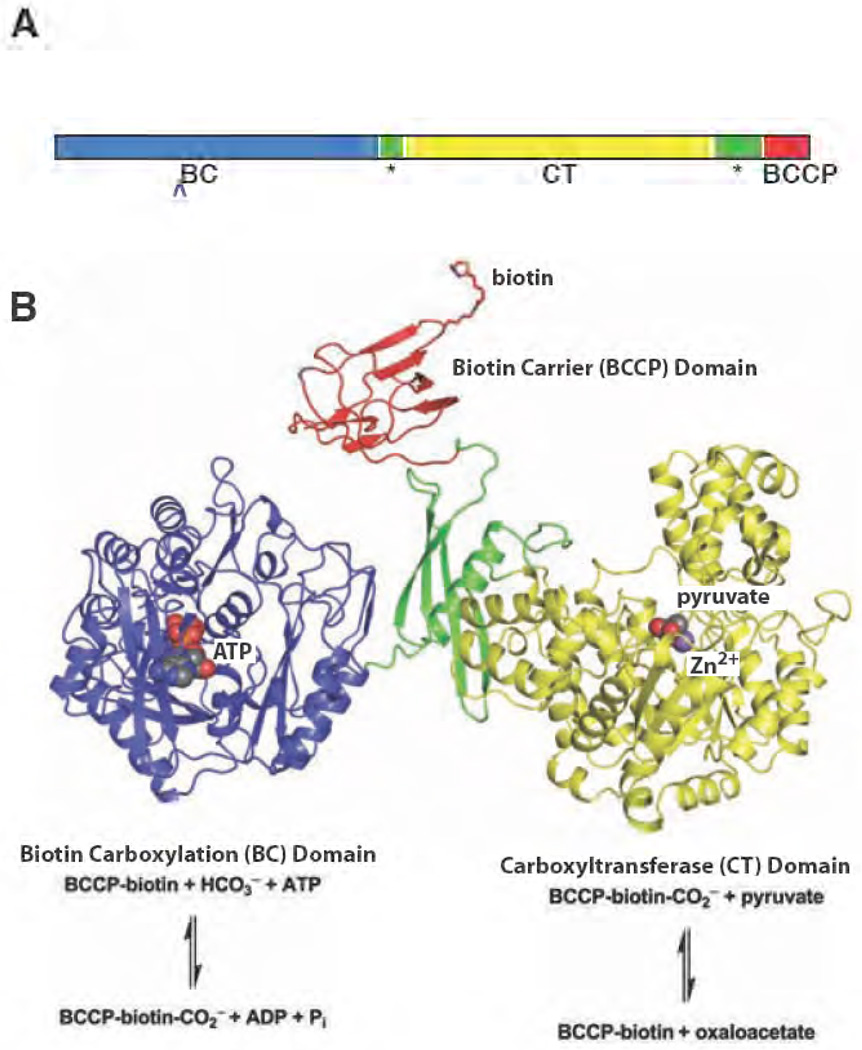
Figure 4. Domain architecture of PC.
(A) Schematic drawing of the primary structure arrangement for the multidomain PC. The central allosteric domain is indicated with a star [103]. (B) The structure of the Staphylococcus aureus PC monomer B [61]. The BC, CT, BCCP and allosteric domains are coloured blue, yellow, red and green, respectively. The chemical reactions catalysed in the individual domains are illustrated below the corresponding domain structure.
Initial structure/function analysis of PC was derived from cDNA and gene cloning studies. Sequence comparison with other biotin-dependent enzymes and limited proteolysis of PC from various sources [75, 101, 102] led to the identification of three functional domains. In α4 PCs, the BC domain is at the N-terminus of the polypeptide chain. Biotin is carboxylated in the BC domain using bicarbonate as a substrate in a reaction that involves MgATP cleavage to form carboxybiotin, MgADP and Pi, and that requires free Mg2+. T he CT domain contains a tightly bound transition metal ion and is the central domain where the carboxy group from carboxybiotin is transferred to pyruvate to form oxaloacetate. The BCCP domain, located at the C-terminus, contains the lysine residue to which the biotin is attached.
7.2. Three dimensional structure
Two recent reports of the complete X-ray crystal structures of α4 PC from the bacteria Rhizobium etli [103] (RePC) and Staphylococcus aureus [61] (SaPC) offer tremendous insight into catalysis and allosteric regulation in PC. The structure of the C-terminal region of human PC (hPC) (comprising all but the BC domain) has also been reported [61]. The structures of RePC and SaPC are surprisingly different in their overall arrangement and each offers a unique perspective on PC catalysis. The structure of RePC reveals the position of the acetyl CoA activator binding site and suggests a possible mechanism for allosteric activation. The structures of both SaPC and hPC, determined in the absence of activator, reveal a tethered biotin bound in a CT active site, offering fresh and surprising insight into the catalytic mechanism in the CT domain. Together, these structures reveal the first complete descriptions of the domain arrangement for any biotin-dependent enzyme, significantly furthering the description of catalysis and allostery in PC at the molecular level. Of particular note in these structures is the revelation of a fourth, central structural domain linking the three functional domains. This domain, termed the allosteric domain, includes the binding site for the allosteric activator acetyl CoA [103]. This domain is structurally unique and had not been predicted on the basis of primary sequence analysis. Chain “A” from the recently published S. aureus PC tetramer provides an example of the complete, intact PC monomer and defines the relative arrangement of the individual domains and active sites (Fig. 4).
7.3. BC Domain Structure
The ATP-dependent carboxylation of biotin from bicarbonate is common among the biotin-dependent carboxylase enzymes. In addition to the complete PC structures determined from R. etli and S. aureus, several structures have been reported for the BC domain in isolation, including the ATP-bound BC subunit of acetyl CoA carboxylase from E. coli [104], the apo BC subunit of PC from Aquifex aeolicus [97] and the apo BC domain of PC from Bacillus thermodenitrificans [105]. The domain consists of three individual subdomains: A, B and C, which are common to all biotin-dependent carboxylase enzymes [106]. The ATP binding pocket of the BC domain consists of an ATP-grasp fold [107] enclosed by a hinged lid (the B-subdomain) that closes over bound ATP [104]. The core of the BC domain structure is very similar among the various enzymes, with root mean square deviations less than 1.2 Å. The position of the B-subdomain, however, changes markedly upon binding ATP. In the presence of ATP, the B-subdomain adopts a closed position over the active site, rotating about a hinge that separates it from the rest of the BC domain [97]. While the overall conformation of the B-subdomain does not change significantly in the presence of ATP, its position is dramatically altered, resulting in a rotation of approximately 45° in the B subdomain and the repositioning of structurally equivalent residues by nearly 20 Å between the apo and ATP-bound states [97,104,105]. In the ATP-bound structure of the BC subunit from E. coli ACC, several backbone amides from the B subdomain interact with the phosphoryl oxygens of ATP to clamp the lid onto the bound ATP substrate, effectively burying 85% of the ATP surface area [106]. In the ATP-bound BC domain of SaPC, the B-subdomain also adopts a closed conformation, but there are large differences relative to that observed for the BC subunit of E. coli ACC [61].
7.4. CT Domain Structure
In the second partial reaction, catalysed at the CT domain of PC, the carboxyl group of carboxybiotin is transferred to pyruvate, generating oxaloacetate. The CT domain of PC shares substantial sequence identity with CT domains from two related biotin-dependent, pyruvate-binding enzymes: the 5S subunit of transcarboxylase (5S) and the α-subunit of the biotin-dependent oxaloacetate decarboxylase (α-OAD). The CT domain structures from both α-OAD [108] and 5S [109] have been determined crystallographically and the overall CT domain structures are structurally homologous to the CT domains from RePC, SaPC and hPC. The CT domain consists of a core, N-terminal α8β8 TIM-barrel with a C-terminal subdomain that forms a funnel leading to a central metal ion bound at the mouth of the TIM-barrel. The CT domain connects from the central hinge domain by a long N-terminal strand that runs the entire exterior length of the CT domain, followed by two short α-helices that lead into the core TIM barrel. In the structure of RePC, a Mg2+ ion is bound at the C-terminal end of this second short helix [103]. This Mg2+ was not observed in the structures of 5S or α-OAD nor was it observed in the CT domains of hPC and SaPC. It is unclear whether this Mg2+ plays a physiological role in PC.
In all reported structures of the homologous CT domains, a catalytically essential metal ion is bound at a conserved position at the mouth of the TIM-barrel, defining the center of the active site for the transcarboxylation reaction in these enzymes [108,109]. In PC, the metal ion is octahedrally coordinated by pyruvate (or a water molecule in the apo structure from R. etli), a conserved Asp, two conserved His residues, and a conserved carbamylated Lys in a manner exactly analogous to the metal ion binding site at the active site of 5S. The identity of the metal ion may vary between species of PC. Metal ion analysis suggests Zn2+ as the active site metal ion in the enzyme from R. etli [103], while the exact identity of the metal ion in S. aureus is unknown [61]. Mutation of several residues ligating this central metal ion results in a significant loss of catalytic activity in PC [103, 110] just as mutations of the analogous residues in α-OAD result in an inactive enzyme [108].
The structures of hPC and SaPC, with both pyruvate and biotin bound in the CT domain, offer new insight into the molecular basis for enzyme dysfunction in mutations currently known to be associated with human PC deficiencies [60, 61, 62, 111]. The A610T mutation is located in the biotin binding site while the M743I mutation is located near the pyruvate binding site [104,110].
7.5. The BCCP domain
The structure and function of the biotin carboxyl carrier domain has been reviewed in the context of swinging domains in multifunctional enzymes [112]. The BCCP domains from hPC, RePC and SaPC share a high degree of structural homology with the BCCP of E. coli acetyl CoA carboxylase [113, 114] and the BCCP domain from the 1.3S subunit of Propionibacterium shermanii transcarboxylase [115].
The structure of S. aureus PC and the structure of the C-terminus of human PC both revealed the enzyme in a tetrameric arrangement with a BCCP domain bound in one of the four CT domains [61]. In this conformation, biotin is projected away from BCCP and positioned in the proximity of pyruvate in the CT active site. While the electron density indicates that the biotin may be partially disordered, it is striking that both the BCCP domain and its biotin adopt a very similar conformation in both the human and S. aureus enzymes, suggesting that BCCP and biotin are bound in a biologically relevant conformation. In human PC, the carbonyl oxygen of biotin is hydrogen bonded to the side chain hydroxyl of conserved S911 (S879 in S. aureus) and the main chain amide of K912 (K880 in S. aureus). These groups may be critical to the enolisation of carboxybiotin in the CT active site. The sulphur atom of biotin is located in a pocket made up of several conserved residues. The N1 atom of biotin is 4.7 Å from the methyl group of pyruvate and is hydrogen bonded to the side-chain hydroxyl group of strictly conserved T908 in human PC (T876 in S. aureus).
7.6. Allosteric Domain Structure
The central structural domain in PC serves to physically connect the three functional domains of the enzyme [61, 103]. It plays a critical role in catalysis through orienting and arranging the individual domains of PC. This central domain has been called the pyruvate tetramerization domain (PT) but its role in tetramerization is obviated by the binding of the acetyl CoA allosteric activator [61]. Since this domain is responsible for binding the allosteric activator, acetyl CoA, and influences the relative position of the other connected domains, it has also been termed the allosteric domain [103]. The allosteric domain displays no sequence similarity with other members of the biotin-dependent enzyme family, suggesting that it is unique to PC. The domain is made up of two separate segments with a topology consisting of four antiparallel β-strands bracketing a long, central α-helix (Fig. 5A). The α-helix consists of residues from the N-terminal segment and serves to connect the BC and CT domains, while the C-terminal segment folds around this helix in a highly twisted, antiparallel (β4)-sheet and connects the CT domain to the BCCP domain. The overall protein fold and the mode of acetyl CoA binding bares some superficial resemblance to the domain organization and acetyl CoA binding site in the GCN-5 related acetyltransferases [116]. The functional domains are connected to the allosteric domain through long, disordered linker arms which presumably allow for the requisite flexibility in domain motions as catalysis proceeds.
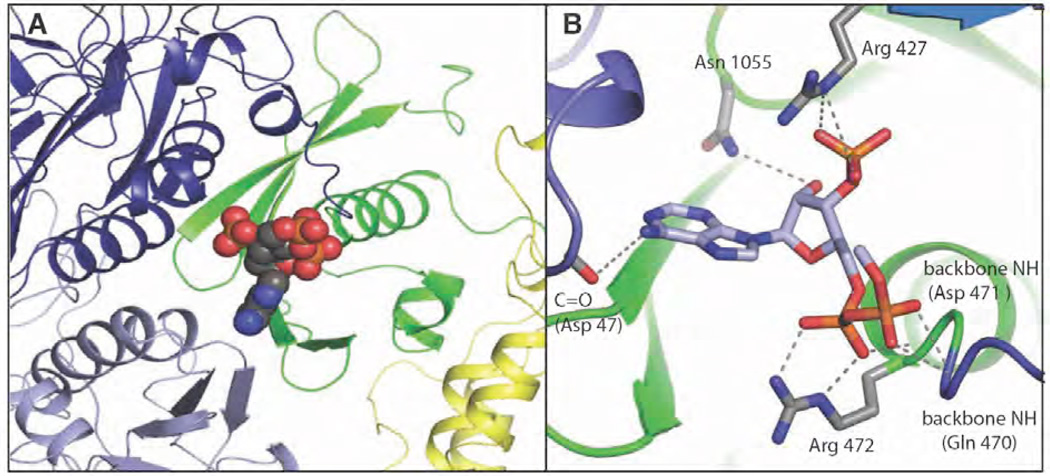
Figure 5. The acetyl CoA binding site in R. etli PC [103].
(A) The nucleotide portion of ethyl-CoA (shown as spheres), a non-hydrolysable analogue of acetyl CoA, is bound at the N-terminal end of the allosteric domain’s central spanning helix and near the dimerization interface for the BC domain. The domains are coloured in dark blue (BC domain 1), light blue (BC domain 2), green (allosteric domain) and yellow (CT domain). (B) Interactions with the nucleotide portion of ethyl-CoA include residues from both BC subunits and from the allosteric domain. Reproduced from [103].
In the structure of RePC, ethyl CoA, a non-hydrolyzable analogue of acetyl CoA, is bound at the immediate N-terminal end of the allosteric domain’s central α-helix and near the BC-BC dimer interface. Interactions with the nucleotide portion of ethyl CoA include residues from both BC subunits and the allosteric domain (Fig. 5). Most notably, Arg472 of the hinge domain forms a very tight contact with the α–5’–phosphate of ethyl CoA. This residue lies at the mouth of the central α–helix and, like all of the residues interacting with the nucleotide portion of CoA, it is highly conserved among PC enzymes. A notable exception is the significant lack of conservation among these residues in the sequence of PC from Corynebacterium, where acetyl CoA acts as an inhibitor rather than an activator of the enzyme [117]. Also, subunit type PCs that have mutations or deletions in these residues are not activated by acetyl CoA [3].
7.7. Quaternary Structure of PC and implications for catalysis
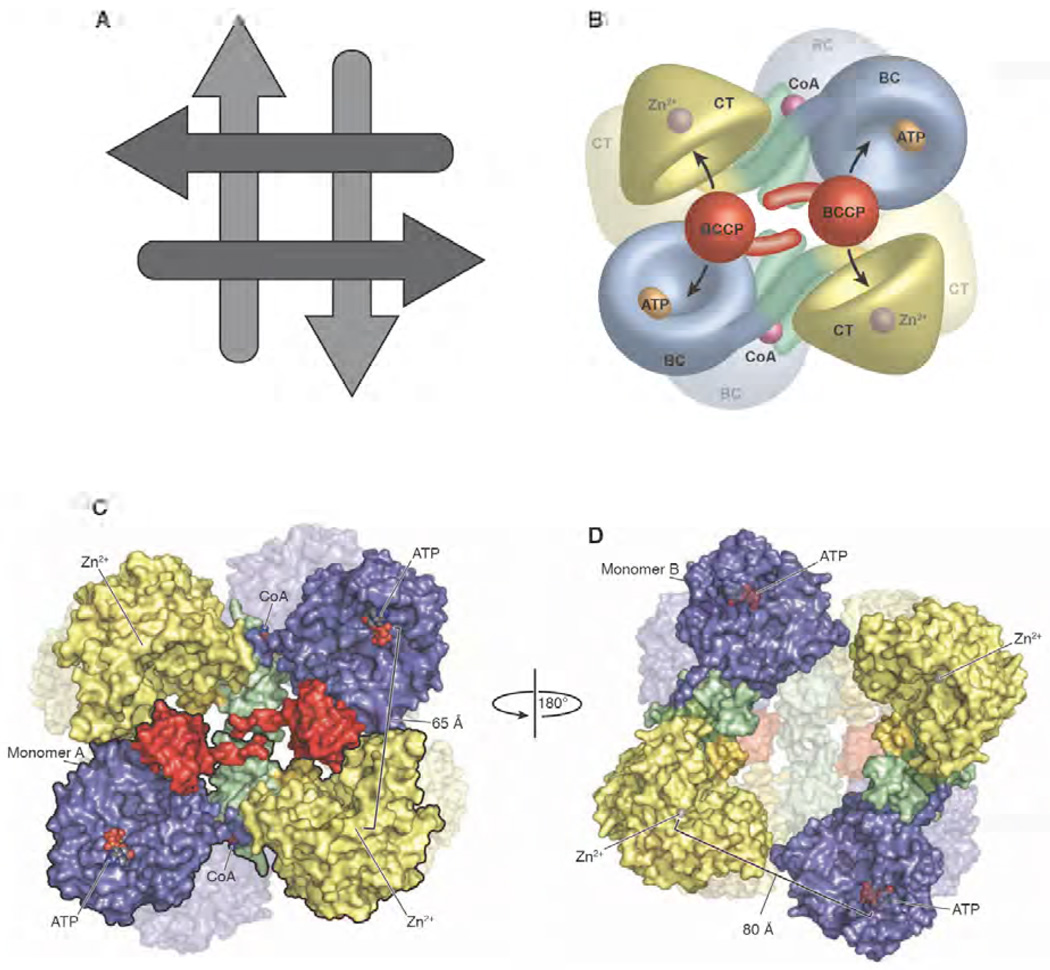
PC exists predominantly as a tetramer in solution and, while it can equilibrate between the tetramer, dimer and monomer, only the tetrameric form of the enzyme catalyses the overall reaction [118, 119]. Early attempts to characterize the subunit arrangement in PC were limited to studies by electron microscopy and were interpreted to suggest that the tetrameric arrangement of PC exhibited either tetrahedral [120, 121] or rhombic [3] geometry. The crystal structures of PC from R. etli and S. aureus confirm that the subunits are arranged in a rhombohedral geometry with the tetramer stabilized at each corner through dimerization interfaces between neighbouring BC domains and neighbouring CT domains (Fig. 6). The dimerization interface is significant for both domains, with a relatively large buried surface area calculated for the BC-BC and CT-CT interface in both SaPC and RePC [77, 126]. Similar dimerization interfaces have been observed for the BC subunit of E. coli acetyl CoA carboxylase [104, 106] and Aquifex aeolicus PC [97] and for the CT subunit of Propionibacterium shermanii transcarboxylase [109] and Vibrio cholerae oxaloacetate decarboxylase [108]. The tetramer consists of a distinct top and bottom face, with the monomers running perpendicularly between these faces (Fig. 6A). On each face, the monomers are arranged in an antiparallel fashion with minimal direct contacts between them. This arrangement facilitates the movement of the biotin carrier domain between neighbouring active site pairs (discussed below).
Figure 6. The quaternary structure of R. etli PC.
(A) Schematic representation of the arrangement of the individual monomers making up the tetramers. The arrows represent the general N-terminal to C-terminal direction of the individual polypeptide chains. The arrangement of the tetramer yields two distinct faces, with the monomers running antiparallel on each face and perpendicularly between the faces. (B) Model of the PC tetramer showing the movement of the BCCP domain between neighbouring active sites on opposing polypeptide chains [103]. (C) Surface representation of the top face of the tetramer with one of the two monomers outlined in black for clarity. The location of the ligand binding sites and the distances between them are indicated [103]. (D) Surface representation of the bottom face of the tetramer after a 180° rotation about the y axis. The distance between opposing active sites increases to 80 Å as a result of the altered orientation of the BC domain. The BCCP domain is disordered on the bottom face of the PC tetramer from R. etli and, consequently, is not modelled into the structure [103]. A,B and C are reproduced from [103].
While the bulk of the intersubunit contacts stabilizing the tetramer occur at the BC and CT domain dimer interfaces, the structures of both hPC and SaPC reveal an additional contribution to tetramer stabilization from the central allosteric domain of PC [126]. Indeed, a single mutation at the allosteric domain interface in both SaPC and hPC is sufficient to fully disrupt tetramerization and eliminate enzyme activity [61]. Interestingly, addition of acetyl CoA restores the tetramer in SaPC and fully recovers enzyme activity [61]. Consistent with this result, the structure of RePC obtained in the presence of an analogue of acetyl CoA does not reveal any interdomain contacts between the allosteric domains [103]. These data suggest that the allosteric domain is involved in stabilizing the PC tetramer only in the absence of acetyl CoA. The binding of the allosteric activator results in a conformational change [see below] that forces apart the allosteric domain interface such that the tetramer is maintained exclusively through contacts at the BC-BC and CT-CT dimer interfaces.
The structures of RePC, SaPC and hPC reveal a notable asymmetry between the top and bottom faces of the tetramer. In RePC, this asymmetry results from substantial differences in the overall spatial arrangement of the domains between the two faces of the tetramer [103]. In the structure of hPC and SaPC, the differences are less dramatic between the two faces. Nevertheless, the domain arrangements in both hPC and SaPC are different between the top and bottom faces of the tetramer, particularly with respect to the position of the BCCP domain [61]. Together, these structures suggest that the opposite faces of the tetramer function independently and raise the question of whether the overall PC-catalysed reaction is subject to half-sites reactivity. Indeed, the BC domain of E. coli acetyl CoA carboxylase exhibits just such half-sites reactivity [122, 123]. It remains to be determined whether the complete reaction catalysed by PC is also subject to half-sites reactivity but the structures are suggestive of such a mechanism.
The arrangement of the individual domains in the PC tetramer from Rhizobium etli revealed a possible intermolecular carboxyl group transfer mechanism between active sites on opposing polypeptide chains (Fig. 6B). In the RePC structure, the BC and CT active sites are much closer between active sites on opposing polypeptide chains than they are between active sites on the same polypeptide chain [103]. To test whether intermolecular transfer was a factor in catalysis by PC, a hybrid tetramer was generated, consisting of two opposing active site mutations whereupon it was demonstrated that activity was recovered in the hybrid tetramers through a fully functional pair of neighbouring active sites [103]. These results were further complemented by the structures of SaPC and hPC which physically revealed the BCCP from one polypeptide chain bound in the CT domain of the neighbouring polypeptide chain [61]. These observations explain why PC is active exclusively as a tetramer: only in the complete tetramer can the domains be arranged to permit carboxybiotin transfer between the BC and neighbouring CT domains on opposing polypeptide chains. The intermolecular transfer of intermediates between active sites on opposing polypeptide chains is an emerging theme of multifunctional enzyme catalysis [124, 125].
In the multidomain structures of PC determined to date, the overall structures of the monomers are quite similar, with all of them showing an overall domain organization similar to what is shown in Fig. 4. However, there are large deviations in the relative positions of the BC and CT domains between the individual monomers of PC in both RePC and SaPC. For example, the four monomers of SaPC show variations ranging from a 6° to 18° rotation around a hinge centred at the BC-allosteric domain interface [61], while the BC domain is rotated 40° and translocated ∼40 Å between the top and bottom faces of the tetramer in RePC [103]. The structure of RePC in the presence of ethyl CoA led to the proposal that the cooperative binding of ethyl CoA at the centre of this hinge on the top face of the monomer was responsible for altering the position from which the BC domain threads [103] [Fig. 6C & Fig. 6D]. Surprisingly, it is the BC domain on the lower face of the activated RePC tetramer, where ethyl CoA is unbound, that deviates most substantially in its orientation relative to the structure of the unactivated SaPC enzyme.
Together, these structures strongly suggest a role for acetyl CoA in influencing the relative orientation of the individual domains. The binding of acetyl CoA results in the repositioning of the neighbouring active site pairs in closer proximity on the top face of the tetramer and further apart on the lower face of the tetramer. This effect now appears to be exerted through a mechanism that is more complex than originally envisioned. It is not yet clear by what mechanism the bottom face of the tetramer is so dramatically altered by the binding of acetyl CoA to the opposite face.
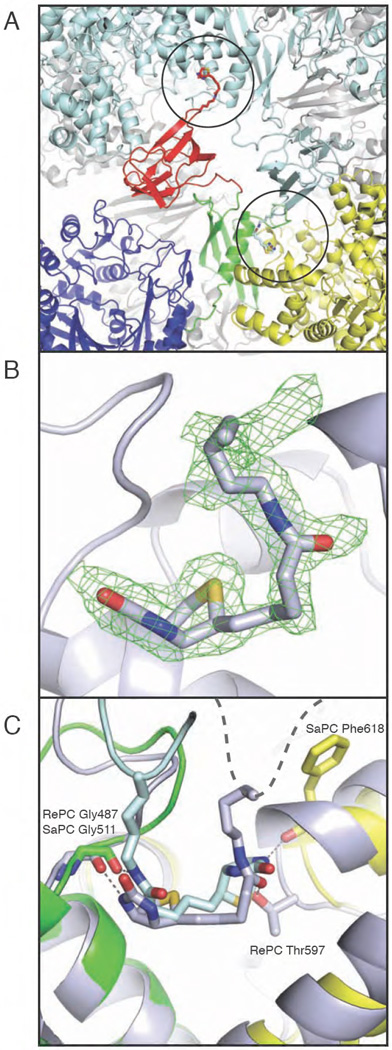
Unexpectedly, the recently reported structure of SaPC revealed three of the four biotinylated BCCP domains bound across the face of the tetramer, with biotin projecting into an exo binding site on the C-terminal side of the allosteric domain, near the interface with the CT domain [61] (Fig. 7A). In contrast, this exo binding site was not observed in the structure of human PC, where a conformational change in the loop at the C-terminal end of the allosteric domain’s central α-helix appears to preclude access to the binding site [61]. The report of an exo binding site in SaPC lead us to re-examine an unidentified lobe of electron density present in the identical region of our original R. etli PC X-ray crystal structure [103]. Indeed, biotin can clearly be modelled into the difference electron density at an exo binding site on the bottom face of the RePC tetramer (Fig. 7B). Thus, PC enzymes from both S. aureus and R. etli include a binding site for biotin that resides outside of the active sites. In SaPC, BCCP-biotin occupies this position in 3 out of 4 cases, the only exception being when BCCP is inserted into the CT domain. In RePC, BCCP-biotin is bound in the exo site only on the bottom face of the tetramer. Furthermore, biotin is bound in a completely opposite orientation in the exo site of RePC than it is in the exo site of SaPC (Fig. 7C). Despite the difference in orientation, the features of the binding sites are similar. Both sites line the binding pocket with hydrophobic residues and, in both cases, the N1 atom of biotin interacts through a hydrogen bond with a main chain carbonyl, indicating that the exo binding site may exclusively accommodate non-carboxylated biotin. It remains to be determined what role this exo site plays in the overall PC reaction mechanism.
Figure 7. The exo biotin binding pocket in bacterial PC.
(A) Cartoon representation of the bottom face of SaPC, where biotin rests in the exo binding site (circled) of the opposing polypeptide chain. One monomer is coloured in blue, green, yellow and red to indicate the BC, allosteric, CT and BCCP domains, respectively. The second monomer is coloured in light blue. The exo binding site is located on the C-terminal end of the allosteric domain’s central spanning helix and at the interface with the CT domain. (B) Biotin in the exo binding site in RePC. The Fo - Fc electron density omit map for biotinylated Lys1114 in RePC was contoured at 2.5σ. (c) Overlay of the allosteric domain in monomer B of SaPC (in colours) and the allosteric domain from the bottom face monomer of RePC (grey). The exo binding pockets are very similar between the two species and each site provides similar contacts with the biotin moiety. However, the binding position of biotin in the exo binding site is completely inverted between RePC and SaPC. The BCCP domain in RePC is disordered, as indicated by the dashed grey line.
8. Mechanism of catalysis and regulation of PC
8.1 The chemical mechanism
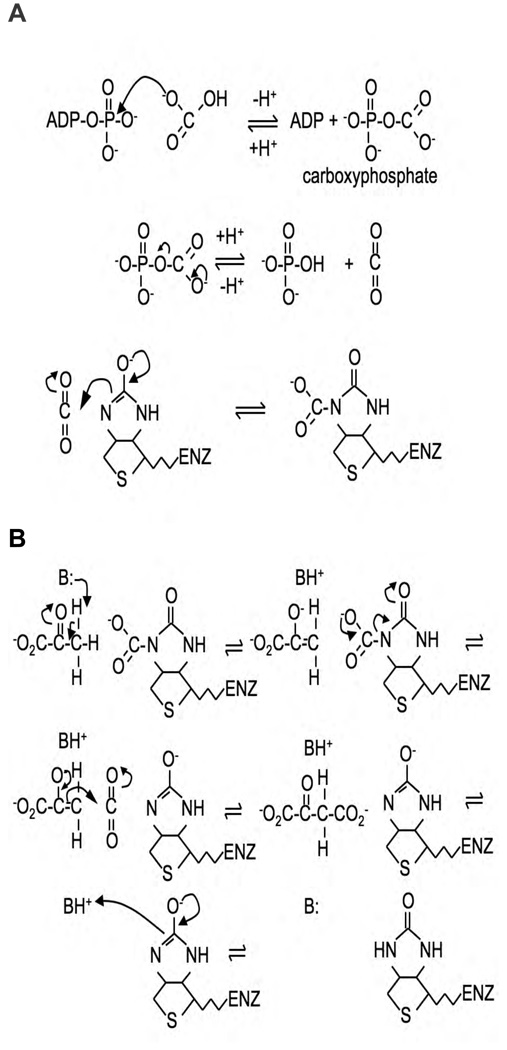
The chemical mechanism of the reaction catalysed by PC has been extensively discussed in reviews by Attwood [126] and Attwood and Wallace [127] and in our view little has changed since these reviews were published in this aspect of the reaction mechanism. Briefly, the first partial reaction, biotin carboxylation, is thought to proceed in two steps with the first step being the activation of bicarbonate by ATP to form a carboxyphosphate intermediate. The second step then involves the carboxylation of the 1’-nitrogen of covalently attached biotin, either directly by carboxyphosphate, or more likely, via decarboxylation of the intermediate to form CO2, which then acts as the carboxylating agent (Fig. 8A). The enol form of biotin, in which the 1’-nitrogen is much more nucleophilic than in the keto form, is the carboxy-group acceptor [126]. The second partial reaction, carboxyltransfer to pyruvate [transcarboxylation], appears to proceed via a stepwise mechanism in which proton transfer steps flank the central carboxyl transfer step between carboxybiotin and the enol form of pyruvate [128] (Fig. 8B).
Figure 8.
(A) Chemical mechanism for the carboxylation of biotin via a carboxyphosphate intermediate. (B) Chemical mechanism for the carboxylation of pyruvate by carboxybiotin. B represents a basic residue in the enzyme active site.
8.2 The catalytic mechanism of the biotin carboxylation reaction
There has been a concentration of studies of putative acid/base catalysts involved in the enolisation of the biotin in each of the partial reactions. Earlier pH profile [129], kinetic isotope effect [128, 130] and chemical modification [130,131,132] studies had suggested that a cysteine-lysine ion pair may be involved. A scheme was proposed in which the lysine ε-amino group deprotonates the sulphydryl of the cysteine. The thiolate then deprotonates the 1’-N of biotin whilst the ε-NH3+ stabilises the enolate oxygen of the enol form of biotin [149, 128, 130]. A pair of lysine and cysteine residues was sought in the biotin carboxylase subunit from E. coli acetyl CoA carboxylase, that was conserved in other biotin-dependent carboxylases and was close enough to each other in the structure of the subunit to be able to act as an ion pair. Such a pair of residues were C230 and K238 and Levert et al. [133] mutated these residues and examined the effects of these mutations on catalysis. Both of the mutations, K238Q and C230A increased the Km for ATP by about 50–80 fold in the bicarbonate-dependent ATP cleavage reaction, suggesting an involvement of both residues in ATP binding [133]. Kondo et al. [134] also mutated K238 (K238A, K238Q, K238R) and also found that the mutations also caused many fold increases in the Km for ATP (124 fold, 51 fold and 76 fold respectively). The K238Q mutant also lacked the ability to carboxylate free biotin, whilst there was little effect on this reaction in the C230A mutant [133]. Thus, while K238 may be involved in stabilisation of the enolate oxygen of biotin, C230 does not appear to play a role in this process.
The role of the corresponding cysteine residue in the yeast isozyme of PC, Pyc1 (C249), was investigated by site-directed mutagenesis of this residue [135]. The C249A mutation had little effect on the Km for ATP but rather, its main effect was to increase the Ka of activation of the enzyme by acetyl CoA by about 5 fold and that of K+ by about 4 fold [135]. Branson et al. [135] suggested that the reason for this apparent difference in function between C230 in biotin carboxylase and C249 in Pyc1 was that the reaction and conformation of the biotin carboxylase would be more similar to that in PC in the context of the acetyl CoA holoenzyme. From the structure of the R. etli enzyme, it is apparent that the corresponding cysteine (C237) is located 30–40 Å from the acetyl CoA binding site and so does not participate directly in the binding of acetyl CoA [103]. This suggests that this cysteine is playing a role in the allosteric activation of the enzyme by acetyl CoA, however, comparison of the position of C237, relative to ATPγS and other close-by residues, in the acetyl CoA-bound subunit with that in the subunit without acetyl CoA shows only small differences. Thus the relationship between this cysteine and acetyl CoA activation remains unclear.
The biotin carboxylation and ATPase reactions catalysed by biotin carboxylase [130 133] and the full reaction of Pyc1 [158] exhibit large inverse solvent deuterium isotope effects. From proton inventory analyses, these effects have been attributed to a single proton transfer step [130, 135]. Originally, Tipton and Cleland [130] had attributed these effects to proton transfer from a cysteine sulphydryl to the ε-amino of a lysine on the basis of the low fractionation factor of sulphydryl groups and experiments with sulphydryl-group modifying reagents [136]. However, as pointed out by Branson et al. [135], at this time it was not known that low deuterium isotope fractionation factors can also be produced as a result of the presence of low-barrier hydrogen bonds [137]. Branson et al. [135] suggested the possibility that such is the case in both biotin carboxylase and PC and that such a low-barrier hydrogen bond is associated with proton rearrangements, leading to biotin enolisation. However, at the moment it is not clear which amino acid(s) might be involved in this and the lack of an experimentally determined structure in which the biotin is clearly located in this part of the enzyme active site hampers the search for such a residue(s).
8.3 The catalytic mechanism of the transcarboxylation reaction
As regards the second partial reaction, pH profile [129] and kinetic isotope effect [128] studies on the oxamate-induced decarboxylation of oxaloacetate had suggested the involvement of a cysteine-lysine ion pair in enolisation of biotin at the site of the second partial reaction. However, in this part of the amino acid sequences of PC, there are no highly conserved cysteine residues.
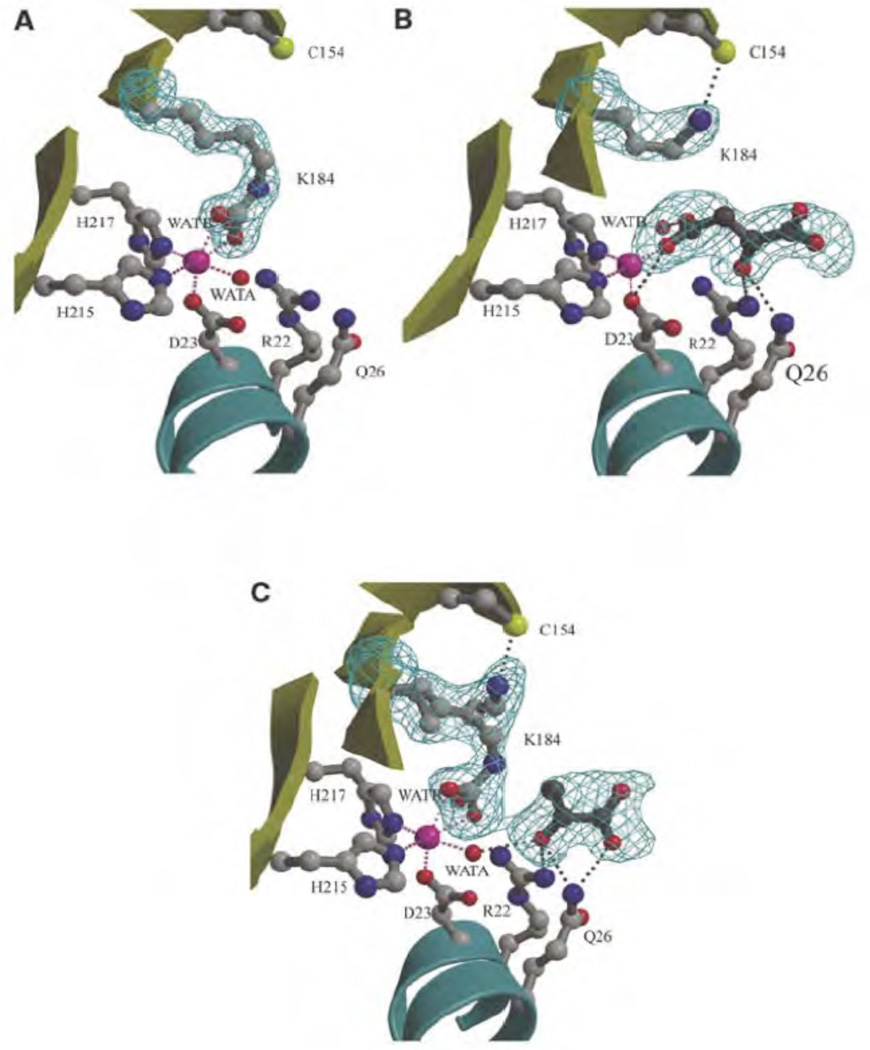
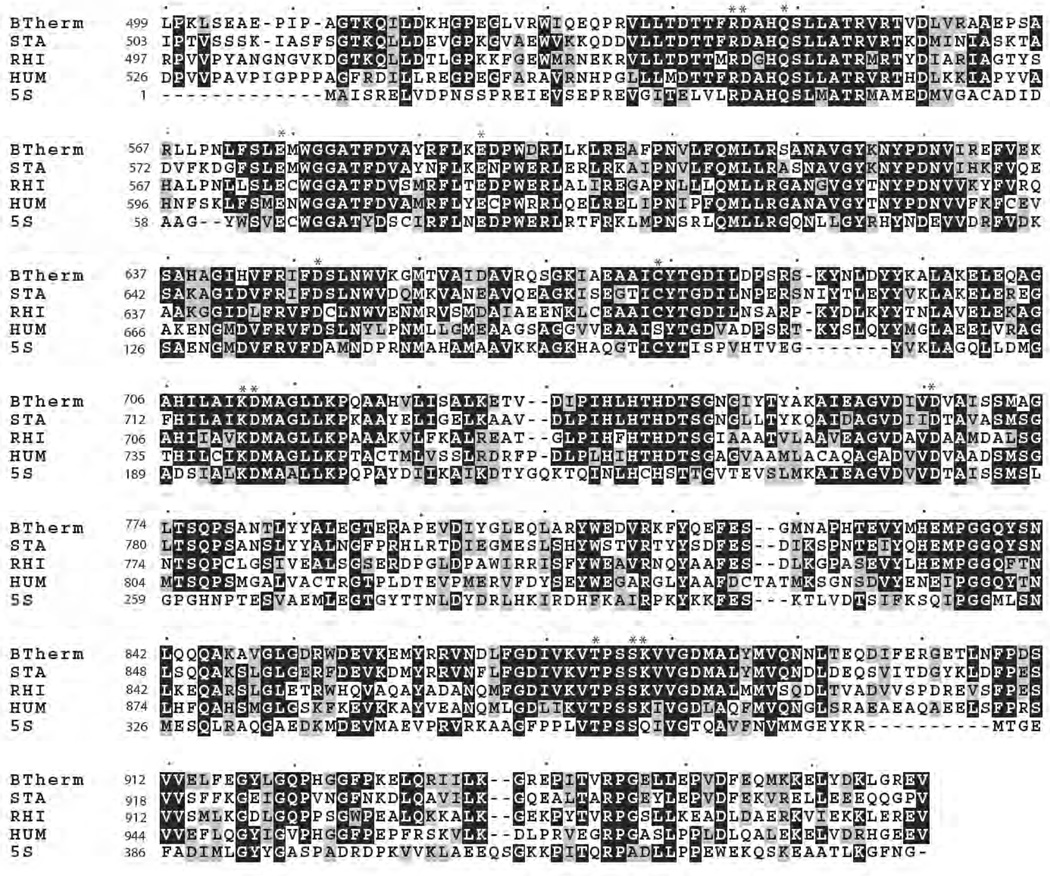
Recently, the structure of 5S, which shares homology with the CT domain of PC, was determined [109]. One of the most interesting features of the structure was the presence of a carbamylated lysine (K184) in close proximity to the binding site of pyruvate and with the CO2 from this residue coordinated to the active site cobalt in the free 5S structure [109] (Fig. 9A). The corresponding lysines in R. etli PC (K718)(see Fig. 10) [103] and hPC (K741), but not SaPC (K712) [61], were also found to be carbamylated and coordinated with the bound metal ion. In the 5S-oxaloacetate complex, formed by soaking crystals of free 5S with oxaloacetate, K184 was not carbamylated and the 4-carboxy group of oxaloacetate had replaced the carbamyl carboxy group of carbamylated K184 as a ligand for the cobalt [109]. In this complex, the ε-amino group of K184 was displaced and formed a hydrogen bond with C154 [109] (Fig. 9B). In the 5S-pyruvate complex there appeared to be two chemical and conformational states of K184, one carbamylated form, as in the free 5S subunit and one in the non-carbamylated form as in the 5S-oxaloacetate complex [109] (Fig. 9C). Based on these structures, Hall et al. [109] suggested that K184 might be carbamylated as part of the catalytic cycle to transfer the carboxy group from carboxybiotin to pyruvate. Mutation of this lysine (K184E, K184A) resulted in catalytically inactive forms of transcarboxylase which did suggest that K184 does play an essential role in catalysis. Mutation of the corresponding lysine in PC from B. thermodenitrificans (K712) (see Fig. 10) [110] resulted in an enzyme with less than 0.02% of the specific activity of the wild type enzyme. However, mutation of the corresponding lysine in PC from R. etli (K718) [103] resulted in an enzyme that still retained 4% of the specific activity of the wild type enzyme. This suggests that this lysine is not absolutely essential for catalysis in PC and hence that carbamyl-lysine is not a catalytic intermediate in the carboxy transfer reaction.
Figure 9.
The active site of 5S showing the bound cobalt ion (pink sphere), its water ligands, and side chains of residues which interact with either the cobalt ion or substrate (ball and stick) are shown. Potential interactions involving the metal ion and active site ligands are represented by pink and grey dashed lines, respectively. (A) Free 5S, showing carbamylated lysine (K184) coordinating the cobalt ion. (B) 5S–oxaloacetate complex, showing the noncarbamylated K184 interacting with cysteine-154 (C154) and oxaloacetate interacting with the cobalt ion. (C) 5S-pyruvate complex and two conformations of carbamylated/noncarbamylated K184 (both refined at half occupancy). WATA and WATB are water molecules. Adapted with permission from Macmillan Publishers Ltd: (EMBO Journal) [132], copyright (2004).
Figure 10.
Part of a sequence alignment (ClustalW) showing the sequences of pyruvate carboxylase from B. thermodenitrificans (BTHERM)[146], R. etli (RHI)[103], S. aureus (STA)[61] and humans (HUM)[61] and that of the transcarboxylase 5S subunit from P. shermanii (5S)[109]. Identical residues are outlined in grey residues. Asterisks indicate conserved residues for which functions are described in Section 8.3. Single dots indicate every 10th residue.
A possible catalytic role of C154 in 5S was also tested by mutation (C154A), however the mutant retained about 50% of the activity of the wild type enzyme [109]. In any case, this residue is not conserved in all PC sequences and is replaced by serine in many sequences and alanine in others. The corresponding residue in R. etli PC is C682, which is the closest cysteine to K718 and is positioned similarly to C154 relative to K184 in 5S.
In the structure of the complexes of 5S with pyruvate and oxaloacetate R22 and Q26 both form hydrogen bonds with the carbonyl oxygen [109], suggesting a role in promoting enolisation of pyruvate as a prelude to carboxylation in the forward reaction and promoting decarboxylation of oxaloacetate in the reverse reaction. The corresponding residues are highly conserved in PCs (see Fig. 10).
Yong-Biao et al. [110] mutated a number of highly conserved residues in the transcarboxylation domain of B. thermodenitrificans PC and investigated the effects of these mutations on the catalytic activity and quaternary structure of the enzyme. As described above, mutation of K712 (K712Q) resulted in a very great, but not complete, reduction in catalytic activity, whilst the mutant K712R retained about 1% of the specific activity of the wild type and had little effect on the Km for pyruvate.
Yong-Biao et al. [110] also mutated four aspartate and two glutamate residues in the CT domain: D543; D649; D713; D762; E576; E592 all of which are highly conserved in PCs and in 5S of transcarboxylase (see Fig. 10). The mutation D762N had only a small effect on the transcarboxylation reaction. The mutations D713N, E576Q and E592Q did have effects on the overall reaction, but these effects were attributed to their marked destabilisation of the native tetrameric structure of the enzyme inducing a high degree of dissociation into dimers and monomers [110].
The conservative mutation D543E resulted in a mutant that retained about 4.7% of the specific activity of the wild type enzyme and that had a Km for pyruvate that was 54 fold higher than that of the wild type. The mutation D649N resulted in a mutant that retained about 2% the specific activity of the wild type enzyme and that had a Km for pyruvate that was 4 fold higher than that of the wild type. The effects of these mutations [D543E and D649N] on the oxamate-induced oxaloacetate decarboxylation reaction were less with 4.5 and 14 fold reductions in specific activity respectively and only small changes in the Kms for oxaloacetate and oxamate [110]. Both mutations did however reduce the rate of (3H) pyruvate detritiation by about 40 fold [110]. On the basis of the effects of these mutations, Yong-Biao et al. [110] suggested a mechanism in which D543 initially acts as a base to deprotonate the pyruvate to form the enolate. The positioning of the ε-NH3+ of K712 adjacent to the ureido oxygen of carboxybiotin then induces its decarboxylation and subsequent carboxylation of the enol form of pyruvate. The protonated D543 then protonates the 1’-N of biotin to return it to the keto form. The carboxyl group of D649 acts as a ligand to the metal ion, positioning it adjacent to the carbonyl oxygen of pyruvate to facilitate enolisation. The corresponding aspartate residues in transcarboxylase 5S are D23 and D127 respectively (see Fig. 10) and those in R. etli PC are D549 and D655. The carboxyl group of D23 is shown to be a ligand of the cobalt ion in 5S (Fig. 9) [109] and is 5.6Å from the methyl carbon of pyruvate. D549 in R. etli PC is also a ligand of the bound metal ion (zinc) [103]. The carboxyl groups of D127 and D655 are about 16Å from the bound metal ion and D127 is about 12Å from the methylene carbon of pyruvate. Thus the putative roles of D543 and D469 in the mechanism proposed by Yong-Biao et al. [110] are not supported by the structures of transcarboxylase 5S and R. etli PC.
In the recent structures of hPC and SaPC, in some of the subunits, biotin was bound at the active site of the CT domain [61]. Here the 1’-N of biotin interacts with the hydroxyl of T908, which is highly conserved in PCs and in 5S (see Fig. 10) whilst the ureido oxygen interacts with the amide of K912. It remains unclear whether these interactions would be sufficient to induce decarboxylation of carboxybiotin in the forward reaction or enolisation of the biotin in the reverse reaction.
8.4 The allosteric regulation of PC by acetyl CoA
Acetyl CoA is a positive regulator of PCs from many, but not all species and the degree of dependency of the activity of the enzyme on it also varies [138]. Previous work had indicated that the locus of action of acetyl CoA in stimulating the activity of PC was the biotin carboxylation reaction [118, 139, 140, 141, 142, 143] and not the transcarboxylation reaction [144]. Acetyl CoA was shown to enhance the steady state rate of ATP cleavage [139, 140, 141] in the absence of pyruvate and also the rate constant for the first order approach to steady state in the chicken enzyme [141], but not the yeast enzyme [140]. The rate constants for the approach to steady state of enzyme-carboxybiotin complex formation in the absence of pyruvate were also enhanced by acetyl CoA [140, 141]. Acetyl CoA was also shown to reduce the affinity of nucleoside triphosphate binding in stopped-flow experiments with the fluorescent ATP analogue, formycin A 5’-triphosphate [145].
More recently, a mutant form of B. thermodenitrificans PC was produced in which the lysine to which the biotin is normally covalently bound was mutated to an alanine residue. This resulted in the production of an unbiotinylated apoenzyme, which could however carboxylate free biotin in a reaction that proceeded 8-fold faster in the presence of acetyl CoA than its absence [146].
Thus it is clear that the main action of acetyl CoA is to enhance the rate of the carboxylation of biotin in the overall reaction. Islam et al. [100] found that a chimeric form of PC, comprising the biotin carboxylase domain of PC from A. aeolicus and the transcarboxylation and biotin carboxyl carrier domain from B. thermodenitrificans PC, had activity that was independent of acetyl CoA, a characteristic of the A. aeolicus enzyme and not the B. thermodentrificans enzyme. In addition, in a recent work by Jitrapakdee et al. [147] the acetyl CoA-activation characteristics of the two yeast isozymes of PC (Pyc1 and Pyc2) and chimeric constructs of these isozymes were studied. Pyc1 and Pyc2 have different degrees of dependency on acetyl CoA for activity, have different Kas and exhibit different degrees of cooperativity for the activation [147]. From studying different chimeric constructs of these isozymes, the characteristics of acetyl CoA activation were shown to mainly reside in the BC domains of the isozymes. However, there were determinants of the Ka for acetyl CoA that lay outside this domain [147]. Kondo and coworkers [100, 148] showed that when the BC domain alone of B. thermodenitrificans PC was expressed, although it possessed biotin-dependent ATPase activity, this activity was not stimulated by acetyl CoA, with free biotin as a substrate. Similarly the CT + BCCP domains, expressed as a single protein, had oxaloacetate decarboxylating activity that was also not activated by acetyl CoA [148]. When ATP cleavage by the expressed BC domain of B. thermodenitrificans PC was measured with the biotinylated biotin carboxyl carrier protein as a substrate, the activity was stimulated by acetyl CoA, although to a lesser extent than in the complete enzyme. Thus it appeared that the acetyl CoA binding site and its main focus of action are not completely co-located in the BC domain of PC and that there may be contributions to binding from other parts of the enzyme.
The recently published structure of R. etli PC complexed with the acetyl CoA analogue, ethyl CoA, revealed the activator binding site to lie between the BC domain and the CT domain and between the CT domain and the BCCP domain [103]. As discussed in Section 7, acetyl CoA appears to have a marked effect on the conformation of a pair of subunits on one side of the tetramer to allow the binding of the biotin from one subunit to the CT domain of its partner [103]. Thus, it appears that biotin of one subunit is carboxylated in its own subunit’s BC domain and then transfers its carboxyl group to pyruvate in its partner’s CT domain [103]. However, the conformation of the pair of subunits on the other side of the tetramer, which do not have acetyl CoA bound to them, does not allow this interaction to occur [103]. As pointed out by St Maurice et al. [103] and described in Section 7, this interconnection between acetyl CoA binding, subunit pair conformation and the ability of the biotin to reach its partner subunit’s transcarboxylation site are very suggestive of half-sites reactivity. The recent studies on dimeric biotin carboxylase which show that this enzyme displays half-sites reactivity, with the strong likelihood that there is obligate activity switching between the subunits, so catalysis alternates between the two [122, 123].
Before the structure of PC was determined, it had been assumed that the entire catalytic cycle took place within the confines of a single subunit and that each subunit within the tetramer acted independently of the others. Thus, much of the data that have accumulated over a number of years to investigate the role of acetyl CoA has been interpreted in this context. In view of the interaction between pairs of subunits, the effect of acetyl CoA on the structure and the likelihood of half-sites reactivity described above, this interpretation will have to be re-evaluated.
Some PCs are not activated by acetyl CoA and most of those that are activated are capable of catalysing the carboxylation of pyruvate in the absence of acetyl CoA, albeit at a considerably reduced rate [138]. If we assume that all PCs share the same mechanism, namely a half-sites reactivity system with obligate activity switching between the pairs of subunits on either side of the tetramer (see above), then a major a role of acetyl CoA may be to facilitate this switching process. In order to more fully understand the role of acetyl CoA, it would be very useful to have a structure of R. etli PC in the absence of ethyl CoA and in addition the structure of a PC that is not activated by acetyl CoA. In addition, to understand the connection between acetyl CoA binding and half-sites reactivity, this aspect of the function of the enzyme must be further investigated.
9. Interacting proteins
Apart from biotin protein ligase or holocarboxylase [EC 6.3.4.15] which biotinylates newly synthesized PC protein, several other proteins have been reported to interact with PC in vivo and in vitro. In the yeast, Hansenula polymorpha, PC is associated with the peroxisomal alcohol oxidase (AO) [149]. This flavoenzyme catalyses the oxidation of methanol to formaldehyde and hydrogen peroxide.
Deletion of the PC gene in this yeast impairs AO activity, causing the accumulation of inactive AO in the cytosol. In vitro studies reveal that PC protein but not its activity is required to mediate AO assembly in the peroxisome whereby PC assists in the binding of AO’s cofactor, FAD, to newly synthesized AO monomer. This interaction is mediated through the transcarboxylation domain and the linking peptide between the biotin carboxylation and the transcarboxylation domains [150]. A similar requirement for PC in AO assembly has also been reported in the yeast Pichia pastoris [149].
In 3T3-L1 adipocytes, PC was shown to interact with prohibitin, a protein involved in mitochondrial biogenesis [151]. The mechanism by which prohibitin interacts with PC is not well understood, although it was demonstrated that exogenous prohibitin inhibits partially purified PC from adipocytes [151].
Acknowledgements
This work was supported by the Career Development Grant from Faculty of Science, Mahidol University and the Thailand Research Fund to S.J., and the National Institutes of Health grants GM070455 to W.W.C, J.C.W and P.V.A, and AR35186 to I. R. M.St.M. was supported in part by a fellowship from the Natural Science and Engineering Research Council of Canada. The authors apologize in advance for any inadvertent oversight in the selection of relevant work due a limitation of space.
Abbreviations
- 5S
5S subunit of transcarboxylase
- α-OAD
α-subunit of the biotin-dependent oxaloacetate decarboxylase
- ACC
acetyl CoA carboxylase
- acetyl CoA
acetyl coenzyme A
- AO
peroxisomal alcohol oxidase
- BC
biotin carboxylase
- BCCP
biotin carboxyl carrier protein
- CREB
cAMP-responsive element binding protein
- CT
carboxyltransferase
- FAS
fatty acid synthase
- hPC
human pyruvate carboxylase
- LXR
liver X receptor
- MDH
malate dehydrogenase
- PC
pyruvate carboxylase
- PDH
pyruvate dehydrogenase complex
- PEP
phosphoenolpyruvate
- PEPC
phosphoenolpyruvate carboxylase
- PEPCK
phosphoenolpyruvate carboxykinase
- PPAR
peroxisome proliferator-activated receptor
- PPRE
peroxisome proliferator-activated receptor
- PPRE
peroxisome proliferator-activated receptor responsive element
- RePC
Rhizobium etli pyruvate carboxylase
- SaPC
Staphylococcus aureus pyruvate carboxylase
- TCA
tricarboxylic acid
References
- 1.Utter MF, Keech DB. Formation of oxaloacetate from pyruvate and carbon dioxide. J. Biol. Chem. 1960;235:17–18. [PubMed] [Google Scholar]
- 2.Owen OE, Kalhan SC, Hanson RW. The key role of anaplerosis and cataplerosis for citric acid cycle function. J. Biol. Chem. 2002;277:30409–30412. doi: 10.1074/jbc.R200006200. [DOI] [PubMed] [Google Scholar]
- 3.Jitrapakdee S, Wallace JC. Structure, function and regulation of pyruvate carboxylase. Biochem J. 1999;340:1–16. doi: 10.1042/bj3400001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sauer U, Eikmanns BJ. The PEP-pyruvate-oxaloacetate node as the switch point for carbon flux distribution in bacteria. FEMS Microbiol. Rev. 2005;29:765–794. doi: 10.1016/j.femsre.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 5.de Jong-Gubbels P, Bauer J, Niederberger P, Stückrath I, Kötter P, van Dijken JP, Pronk JT. Physiological characterisation of a pyruvate-carboxylase- negative Saccharomyces cerevisiae mutant in batch and chemostat cultures. Antonie Van Leeuwenhoek. 1998;74:253–263. doi: 10.1023/a:1001772613615. [DOI] [PubMed] [Google Scholar]
- 6.Gokarn RR, Eiteman MA, Altman E. Metabolic analysis of Escherichia coli in the presence and absence of carboxylating enzymes phosphoenolpyruvate carboxykinase and pyruvate carboxylase. Appl. Environ. Microbiol. 2000;66:1844–1850. doi: 10.1128/aem.66.5.1844-1850.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.March JC, Eiteman MA, Altman E. Expression of an anaplerotic enzyme, pyruvate carboxylase, improves recombinant protein production in Escherichia coli. Appl. Environ. Microbiol. 2002;68:5620–5624. doi: 10.1128/AEM.68.11.5620-5624.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanson RW, Patel YM. Phosphoenolpyruvate carboxykinase (GTP): The gene and the enzyme. Adv. Enzymol. 1994;69:203–281. doi: 10.1002/9780470123157.ch6. [DOI] [PubMed] [Google Scholar]
- 9.Robinson BH. Transport of phosphoenolpyruvate by the tricarboxylate transporting system in mammalian mitochondria. FEBS Lett. 1971;14:309–312. doi: 10.1016/0014-5793(71)80287-9. [DOI] [PubMed] [Google Scholar]
- 10.Bizeau ME, Short C, Thresher JS, Commerford SR, Willis WT, Pagliassotti MJ. Increased pyruvate flux capacities account for diet-induced increases in gluconeogenesis in vitro. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2001;281:R427–R433. doi: 10.1152/ajpregu.2001.281.2.R427. [DOI] [PubMed] [Google Scholar]
- 11.Hagopian K, Ramsey JJ, Weindruch R. Krebs cycle enzymes from livers of old mice are differentially regulated by caloric restriction. Exp Gerontol. 2004;39:1145–1154. doi: 10.1016/j.exger.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 12.Salto R, Sola M, Oliver FJ, Vargas AM. Effects of starvation, diabetes and carbon tetrachloride intoxication on rat kidney cortex and liver pyruvate carboxylase levels. Arch. Physiol. Biochem. 1996;104:845–850. doi: 10.1076/apab.104.7.845.13111. [DOI] [PubMed] [Google Scholar]
- 13.Andrikopoulos S, Proietto J. The biochemical basis of increased hepatic glucose-production in a mouse model of type-2 (noninsulin-dependent) diabetes-mellitus. Diabetologia. 1995;38:1389–1396. doi: 10.1007/BF00400598. [DOI] [PubMed] [Google Scholar]
- 14.Large V, Beylot M. Modifications of citric acid cycle activity and gluconeogenesis in streptozotocin-induced diabetes and effects of metformin. Diabetes. 1999;48:1251–1257. doi: 10.2337/diabetes.48.6.1251. [DOI] [PubMed] [Google Scholar]
- 15.Greenfield RB, Cecava MJ, Donkin SS. Changes in mRNA expression for gluconeogenic enzymes in liver of dairy cattle during the transition to lactation. J Dairy Sci. 2002;83:1228–1236. doi: 10.3168/jds.S0022-0302(00)74989-7. [DOI] [PubMed] [Google Scholar]
- 16.She P, Linberg GL, Hippen AR, Beitz DC, Young JW. Regulation of messenger ribonucleic acid expression for gluconeogenic enzymes during glucagons infusions into lactating cows. J. Dairy Sci. 1999;82:1153–1163. doi: 10.3168/jds.S0022-0302(99)75338-5. [DOI] [PubMed] [Google Scholar]
- 17.Williams EL, Rodriguez SM, Beitz DC, Donkin SS. Effects of short-term Glucagons administration on gluconeogenic enzymes in the liver of midlactation dairy cows. J. Dairy Sci. 2006;89:693–703. doi: 10.3168/jds.S0022-0302(06)72132-4. [DOI] [PubMed] [Google Scholar]
- 18.Hammon HM, Philipona C, Zbinden Y, Blum JW, Donkin SS. Effects of dexamethasone and growth hormone treatment on hepatic gluconeogenic enzymes in calves. J. Dairy Sci. 2005;88:2107–2116. doi: 10.3168/jds.S0022-0302(05)72887-3. [DOI] [PubMed] [Google Scholar]
- 19.Velez JC, Donkin SS. Feed restriction induces pyruvate carboxylase but not phosphoenolpyruvate carboxykinase in dairy cows. J Dairy Sci. 2005;88:2938–4298. doi: 10.3168/jds.S0022-0302(05)72974-X. [DOI] [PubMed] [Google Scholar]
- 20.Bradford BJ, Allen MS. Phlorizin administration increases hepatic gluconeogenic enzyme mRNA abundance but not feed intake in late-lactation dairy cows. J. Nutrition. 2005;135:2206–2211. doi: 10.1093/jn/135.9.2206. [DOI] [PubMed] [Google Scholar]
- 21.Ballard FJ, Hanson RW. The citrate cleavage pathway and lipogenesis in rat adipose tissue: replenishment of oxaloacetate. J. Lipid Res. 1967;8:73–79. [PubMed] [Google Scholar]
- 22.Wise LS, Sul HS, Rubin CS. Coordinate regulation of the biosynthesis of ATP-citrate lyase and malic enzyme during adipocyte differentiation. Studies on 3T3-L1 cells. J. Biol. Chem. 1984;259:4827–4832. [PubMed] [Google Scholar]
- 23.Jitrapakdee S, Slawik M, Medina-Gomez G, Campbell M, Wallace JC, Sethi JK, O’Rahilly S, Vidal-Puig A. The peroxisome proliferator-activated receptor-γ regulates murine pyruvate carboxylase gene expression in vivo and in vitro. J. Biol. Chem. 2005;280:27466–27476. doi: 10.1074/jbc.M503836200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, Lazar MA. Differential Gene Regulation by PPARγ Agonist and Constitutively Active PPARγ2. Mol. Endocrinol. 2002;16:1040–1048. doi: 10.1210/mend.16.5.0825. [DOI] [PubMed] [Google Scholar]
- 25.Reshef L, Olswang Y, Cassuto H, Blum B, Croniger CM, Kalhan SC, Tilghman SM, Hanson RW. Glyceroneogenesis and the triglyceride/fatty acid cycle. J. Biol Chem. 2003;278:30413–30416. doi: 10.1074/jbc.R300017200. [DOI] [PubMed] [Google Scholar]
- 26.Wellen KE, Uysal KT, Wiesbrock S, Yang Q, Chen H, Hotamisligil GS. Interaction of Tumor Necrosis Factor-α- and Thiazolidinedione-Regulated Pathways in Obesity. Endocrinology. 2004;145:2214–2220. doi: 10.1210/en.2003-1580. [DOI] [PubMed] [Google Scholar]
- 27.Wilson-Fritch L, Nicoloro S, Chouinard M, Lazar MA, Chui P.C LeszykJ, Straubhaar J, et al. Mitochondrial remodeling in adipose tissue associated with obesity and treatment with rosiglitazone. J. Clin. Invest. 2004;114:1281–128938. doi: 10.1172/JCI21752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wiederkehr A, Wollheim CB. Implication of mitochondria in insulin secretion and action. Endocrinology. 2006;147:2643–2649. doi: 10.1210/en.2006-0057. [DOI] [PubMed] [Google Scholar]
- 29.MacDonald MJ, Fahein LA, Brown LJ, Hasan NM, Buss JD, Kendrick MA. Perspective: emerging evidence for signaling roles of mitochondrial anaplerotic products in insulin secretion. Am J. Physiol. Endocrinol. Metab. 2005;288:E1–E15. doi: 10.1152/ajpendo.00218.2004. [DOI] [PubMed] [Google Scholar]
- 30.MacDonald MJ. Feasibility of a mitochondrial pyruvate malate shuttle in pancreatic islets. Further implication of cytosolic NADPH in insulin secretion. J. Biol. Chem. 1995;270:20051–20058. [PubMed] [Google Scholar]
- 31.Farfari S, Schulz V, Corkey B, Prentki B. Glucose regulated anaplerosis and cataplerosis in pancreatic β-cells: possible implication of a pyruvate/citrate shuttle in insulin secretion. Diabetes. 2000;49:718–726. doi: 10.2337/diabetes.49.5.718. [DOI] [PubMed] [Google Scholar]
- 32.MacDonald MJ. Estimates of glycolysis, pyruvate (de)carboxylation, pentose phosphate pathway, and methyl succinate metabolism in incapacitated pancreatic islets. Arch. Biochem. Biophys. 1993;305:205–214. doi: 10.1006/abbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 33.Lu D, Mulder H, Zhao P, Burgess SC, Jensen MV, Kamzolova S, et al. 13C NMR isotopomer analysis reveals a connection between pyruvate cycling and glucose-stimulated insulin secretion (GSIS) Proc. Natl. Acad. Sci. U.S.A. 2002;99:2708–2713. doi: 10.1073/pnas.052005699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fransson U, Rosengren AH, Schuit FC, Renstöm E, Mulder H. Anaplerosis via pyruvate carboxylase is required for the fuel-induced rise in the ATP:ADP ratio in rat pancreatic islets. Diabetologia. 2006;49:1578–1586. doi: 10.1007/s00125-006-0263-y. [DOI] [PubMed] [Google Scholar]
- 35.Liu YQ, Han J, Epstein PN, Long YS. Enhanced rat beta-cell proliferation in 60% pancreatectomized islets by increased glucose metabolic flux through pyruvate carboxylase pathway. Am. J. Physiol. Endocrinol. Metab. 2005;2889:E471–E478. doi: 10.1152/ajpendo.00427.2004. [DOI] [PubMed] [Google Scholar]
- 36.Bahl JJ, Matsuda M, DeFronzo RA, Bressler R. In vitro and in vivo suppression of gluconeogenesis by inhibition of pyruvate carboxylase. Biochem. Pharmacol. 1997;53:67–74. doi: 10.1016/s0006-2952(96)00660-0. [DOI] [PubMed] [Google Scholar]
- 37.Jensen MV, Joseph JW, Ilkayeva O, Burgess S, Lu D, Ronnebaum SM, Odegaard M, Becker TC, Sherry AD, Newgard CB. Compensatory response to pyruvate carboxylase suppression in islets β-cells. Preservation of glucose-stimulated insulin secretion. J. Biol. Chem. 2006;281:22541–22351. doi: 10.1074/jbc.M604350200. [DOI] [PubMed] [Google Scholar]
- 38.Palmer ND, Lagefeld CD, Campbell JK, Williams AH, Saad M, Norris JM, Haffner SM, Rotter JI, Wagenknecht LE, Bergman RN, Rich SS, Bowden DW. Genetic mapping of disposition index and acute insulin response to loci on chromosome 11q. Diabetes. 2006;55:911–918. doi: 10.2337/diabetes.55.04.06.db05-0813. [DOI] [PubMed] [Google Scholar]
- 39.Hertz L, Peng L, Dienel GA. Energy metabolism in astrocytes: high rate of oxidative metabolism and spatiotemporal dependence on glycolysis/glycogenolysis. J. Cereb. Blood. Flow. Metab. 2007;27:219–249. doi: 10.1038/sj.jcbfm.9600343. [DOI] [PubMed] [Google Scholar]
- 40.Gamberino WC, Berkich DA, Lynch CJ, Xu B, LaNoue KF. Role of pyruvate carboxylase in facilitation of synthesis of glutamate and glutamine in cultured astrocytes. J. Neurochem. 1997;69:2312–2325. doi: 10.1046/j.1471-4159.1997.69062312.x. [DOI] [PubMed] [Google Scholar]
- 41.Lapidot A, Gopher A. Cerebral metabolic compartmentation. Estimation of glucose flux via pyruvate carboxylase/pyruvate dehydrogenase by 13C NMR isotopomer analysis of D-(U-13C)glucose metabolites. J.Biol. Chem. 1994;269:27198–27208. [PubMed] [Google Scholar]
- 42.Kanamatsu T, Tsukada Y. Effects of ammonia on the anaplerotic pathway and amino acid metabolism in the brain: an ex vivo 13C NMR spectroscopic study of rats after administering (2-13C)) glucose with or without ammonium acetate. Brain Res. 1999;841:11–19. doi: 10.1016/s0006-8993(99)01772-2. [DOI] [PubMed] [Google Scholar]
- 43.Teixeira AP, Santos SS, Carinhas N, Oliveira R, Alves PM. Combining metabolic flux analysis tools and 31C NMR to estimate intracellular fluxes of cultured astrocytes. Neurochem. Int. 2007;52:478–486. doi: 10.1016/j.neuint.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 44.Miller KE, Richards BA, Kriebel RM. Glutamine-, glutamine synthetase-,glutamate dehydrogenase- and pyruvate carboxylase-immunoreactivities in the rat dorsal root ganglion and peripheral nerve. Brain Res. 2002;945:202–21163. doi: 10.1016/s0006-8993(02)02802-0. [DOI] [PubMed] [Google Scholar]
- 45.Elias CB, Carpentier E, Durocher Y, Bisson L, Wagner R, Kamen R. Improving glucose and glutamine metabolism of human HEK 293 and Trichoplusia ni insect cells engineered to express a cytosolic pyruvate carboxylase enzyme. Biotechnol. Prog. 2003;19:90–97. doi: 10.1021/bp025572x. [DOI] [PubMed] [Google Scholar]
- 46.Kim SH, Lee GM. Functional expression of human pyruvate carboxylase for reduced lactic acid formation of Chinese hamster ovary cells (DG44) Appl. Microbiol. Biotechnol. 2007;76:659–665. doi: 10.1007/s00253-007-1041-6. [DOI] [PubMed] [Google Scholar]
- 47.Irani N, Wirth M, Heuvel JD, Wagner R. Improvement of the primary metabolism of cell cultures by introducing a new cytoplasmic pyruvate carboxylase reaction. Biotechnol. Bioeng. 1999;66:238–246. doi: 10.1002/(sici)1097-0290(1999)66:4<238::aid-bit5>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 48.Weinberg MB, utter MF. Effect of streptozotocin-induced diabetes mellitus on the turnover of rat liver pyruvate carboxylase and pyruvate dehydrogenase. Biochem. J. 1980;188:601–608. doi: 10.1042/bj1880601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Desvergne B, Michalik L, Wahli W. Transcriptional regulation of metabolism. Physiol. Rev. 2006;86:465–514. doi: 10.1152/physrev.00025.2005. [DOI] [PubMed] [Google Scholar]
- 50.Lynch CJ, McCall KM, Billingsley ML, Bohlen LM, Hreniuk SP, Martin LF, et al. Pyruvate carboxylase in genetic obesity. Am. J. Physiol. 1992;262:E608–E618. doi: 10.1152/ajpendo.1992.262.5.E608. [DOI] [PubMed] [Google Scholar]
- 51.Jitrapakdee S, Gong Q, MacDonald MJ, Wallace JC. Regulation of pyruvate carboxylase gene expression by alternate promoters during development, in genetically obese rats and insulin-secreting cells. Multiple transcripts with 5’-end heterogeneity modulate translation. J. Biol. Chem. 1998;273:34422–34428. doi: 10.1074/jbc.273.51.34422. [DOI] [PubMed] [Google Scholar]
- 52.MacDonald MJ, Tang J, Polonsky KS. Low mitochondrial glycerol phosphate dehydrogenase and pyruvate carboxylase in pancreatic islets of Zucker diabetic fatty rats. Diabetes. 1996;45:1626–1630. doi: 10.2337/diab.45.11.1626. [DOI] [PubMed] [Google Scholar]
- 53.McDonald MJ, Efendić S, Ostenson CG. Normalization by insulin treatment of low mitochondrial glycerol phosphate dehydrogenase and pyruvate carboxylase in pancreatic islets of the GK rat. Diabetes. 1996;45:886–890. doi: 10.2337/diab.45.7.886. [DOI] [PubMed] [Google Scholar]
- 54.Poitout V, Robertson RP. Secondary beta-cell failure in type 2 diabetes-a convergence of glucotoxicity and lipotoxicity. Endocrinology. 2002;143:339–342. doi: 10.1210/endo.143.2.8623. [DOI] [PubMed] [Google Scholar]
- 55.Laybutt DR, Glandt M, Xu G, Ahn YB, Trivedi N, Boner-Weeir S, et al. Critical reduction in β-cell mass results in two distinct outcomes over time. Adaptation with impaired glucose tolerance or decompensated diabetes. J. Biol. Chem. 2003;278:2997–3005. doi: 10.1074/jbc.M210581200. [DOI] [PubMed] [Google Scholar]
- 56.Boucher A, Lu D, Burgess SC, Telemaque-Potts S, Jensen MV, Mulder H, et al. Biochemical mechanism of lipid-induced impairment of glucose-stimulated insulin secretion and reversal with a malate analogue. J. Biol. Chem. 2004;279:27263–27271. doi: 10.1074/jbc.M401167200. [DOI] [PubMed] [Google Scholar]
- 57.Busch AK, Cordery D, Denyer GS, Biden TJ. Expression profiling of palmitate- and oleate-regulated genes provides novel insights into the effects of chronic exposure on pancreatic β-cell function. Diabetes. 2002;51:977–987. doi: 10.2337/diabetes.51.4.977. [DOI] [PubMed] [Google Scholar]
- 58.Iizuka K, Nakajima H, Namb aM, Miyagawa J, Miyazaki J, Hanafusa T, et al. Metabolic consequence of long-term exposure of pancreatic β cells to free fatty acid with special reference to glucose insensitivity. Biochim. Biophys. Acta. 2002;1586:23–31. doi: 10.1016/s0925-4439(01)00082-5. [DOI] [PubMed] [Google Scholar]
- 59.Robinson BH. Lactic acidemia and mitochondrial disease. Mol. Genet. Metab. 2006;89:3–13. doi: 10.1016/j.ymgme.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 60.Carbone MA, MacKay N, Ling M, Cole DE, Douglas C, Rigat B, et al. Amerindian pyruvate carboxylase deficiency is associated with two distinct missense mutations. Am. J. Hum. Genet. 1998;62:1312–1319. doi: 10.1086/301884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xiang S, Tong L. Crystal structures of human and S aureus pyruvate carboxylase and molecular insights into the carboxyltransfer reaction. Nat. Struct. Mol. Biol. 2008;15:295–302. doi: 10.1038/nsmb.1393. [DOI] [PubMed] [Google Scholar]
- 62.Carbone MA, Robinson BH. Expression and characterization of a human pyruvate carboxylase variant by retroviral gene transfer. Biochem J. 2003;370:275–282. doi: 10.1042/BJ20021397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pineda M, Campistol J, Vilaseca MA, Briones P, Ribes A, Temudo T, Pons M, Cusi V, Rolland M. An atypical French form of pyruvate carboxylase deficiency. Brain and Development. 1995;17:276–279. doi: 10.1016/0387-7604(95)00057-i. [DOI] [PubMed] [Google Scholar]
- 64.Ahmad A, Kahler SG, Kishnani PS, Artigas-Lopez M, Pappu AS, Steiner R, Millington DS, van Hove JLK. Treatment of pyruvate carboxylase deficiency with high doses of citrate and aspartate. Am. J. Med. Genet. 1999;87:331–338. doi: 10.1002/(sici)1096-8628(19991203)87:4<331::aid-ajmg10>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 65.García-Cazorla A, Rabier D, Touati G, Chadefaux-Vekemans B, Marsac C, de Lonlay P, Saudubray J-M. Pyruvate carboxylase deficiency: Metabolic characteristics and new neurological aspects. Ann. Neurol. 2006;59:121–127. doi: 10.1002/ana.20709. [DOI] [PubMed] [Google Scholar]
- 66.Carbone MA, Applegarth DA, Robinson BH. Intron retention and frameshift mutations result in severe pyruvate carboxylase deficiency in two male siblings. Hum. Mut. 2002;20:48–56. doi: 10.1002/humu.10093. [DOI] [PubMed] [Google Scholar]
- 67.Hamilton J, Rae MD, Logan RW, Robinson PH. A case of benign pyruvate carboxylase deficiency with normal development. J. Inher. Metab. Dis. 1997;20:401–403. doi: 10.1023/a:1005350600278. [DOI] [PubMed] [Google Scholar]
- 68.Schiff M, Levart V, Acquaviva C, Vianey-Saban C, Rolland M-O, Guffon N. A case of pyruvate carboxylase deficiency with a typical clinical and neuroradiological presentation. Mol. Genet. Metab. 2006;87:175–177. doi: 10.1016/j.ymgme.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 69.Higgins JJ, Ide SE, Oghalai JS, Polymeropoulos MH. Lack of mutations in the biotin-binding region of the pyruvate carboxylase (PC) in a family with partial PC deficiency. Clin. Biochem. 1997;30:79–81. doi: 10.1016/s0009-9120(96)00125-7. [DOI] [PubMed] [Google Scholar]
- 70.Stucka R, Dequin S, Salmon JM, Gancedo C. DNA sequences in chromosomes II and VII code for pyruvate carboxylase isoenzymes in Saccharomyces cerevisiae: analysis of pyruvate carboxylase-deficient strains. Mol. Gen. Genet. 1991;229:307–315. doi: 10.1007/BF00272171. [DOI] [PubMed] [Google Scholar]
- 71.Walker ME, Val DL, Rohde M, Devenish RJ, Wallace JC. Yeast pyruvate carboxylase: identification of two genes encoding isoenzymes. Biochem Biophys Res Commun. 1991;176:1210–1217. doi: 10.1016/0006-291x(91)90414-3. [DOI] [PubMed] [Google Scholar]
- 72.Rohde M, Lim F, Wallace JC. Electron microscopic localization of pyruvate carboxylase in rat liver and Saccharomyces cerevisiae by immunogold procedures. Arch Biochem Biophys. 1991;290:197–201. doi: 10.1016/0003-9861(91)90608-l. [DOI] [PubMed] [Google Scholar]
- 73.Brewster NK, Val DL, Walker ME, Wallace JC. Regulation of pyruvate carboxylase isozyme (PYC1, PYC2) gene expression in Saccharomyces cerevisiae during fermentative and nonfermentative growth. Arch. Biochem. Biophys. 1994;311:62–71. doi: 10.1006/abbi.1994.1209. [DOI] [PubMed] [Google Scholar]
- 74.Huet C, Menendez J, Gancedo C, Francois JM. Regulation of pyc1 encoding pyruvate carboxylase isozyme I by nitrogen sources in Saccharomyces cerevisiae. Eur. J. Biochem. 2000;267:6817–6823. doi: 10.1046/j.1432-1033.2000.01779.x. [DOI] [PubMed] [Google Scholar]
- 75.Lim F, Morris CP, Occhiodoro F, Wallace JC. Sequence and domain structure of yeast pyruvate carboxylase. J. Biol. Chem. 1988;263:11493–11497. [PubMed] [Google Scholar]
- 76.Val DL, Chapman-Smith A, Walker ME, Cronan JE, Jr, Wallace JC. Polymorphism of the yeast pyruvate carboxylase 2 gene and protein: effects on protein biotinylation. Biochem J. 1995;312:817–825. doi: 10.1042/bj3120817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jitrapakdee S, Booker GW, Cassady AI, Wallace JC. The rat pyruvate carboxylase gene structure. Alternate promoters generate multiple transcripts with the 5’-end heterogeneity. J. Biol. Chem. 1997;272:20522–20530. doi: 10.1074/jbc.272.33.20522. [DOI] [PubMed] [Google Scholar]
- 78.Jitrapakdee S, Petchamphai N, Sunyakumthorn P, Wallace JC, Boonsaeng V. Structural and promoter regions of the murine pyruvate carboxylase gene. Biochem. Biophys. Res. Commun. 2001;287:411–417. doi: 10.1006/bbrc.2001.5599. [DOI] [PubMed] [Google Scholar]
- 79.Jitrapakdee S, Walker ME, Wallace JC. Identification of novel alternatively spliced pyruvate carboxylase mRNAs with divergent 5’-untranslated regions which are expressed in a tissue-specific manner. Biochem. Biophys. Res. Communs. 1996;223:695–700. doi: 10.1006/bbrc.1996.0958. [DOI] [PubMed] [Google Scholar]
- 80.Agca C, Bidwell CA, Donkin SS. Cloning of bovine pyruvate carboxylase and 5’-untranslated region variants. Anim. Biotechnol. 2004;15:47–66. doi: 10.1081/ABIO-120037897. [DOI] [PubMed] [Google Scholar]
- 81.Hazelton SR, Spurlock DM, Bidwell CA, Donkin SS. Cloning the genomic sequence and identification of promoter regions of bovine pyruvate carboxylase. J. Dairy Sci. 2008;91:91–99. doi: 10.3168/jds.2007-0542. [DOI] [PubMed] [Google Scholar]
- 82.Menéndez J, Gancedo C. Regulatory regions in the promoters of the Saccharomyces cerevisiae PYC1 and PYC2 genes encoding isoenzymes of pyruvate carboxylase. FEMS Microbiol. Lett. 1998;164:345–352. doi: 10.1111/j.1574-6968.1998.tb13108.x. [DOI] [PubMed] [Google Scholar]
- 83.Small WC, Brodeur RD, Sandor A, Federova, Li G, Butow RA, Srere P. Enzymatic and metabolic studies on retrograde regulation mutants of yeast. Biochemistry. 1995;34:5569–5576. doi: 10.1021/bi00016a031. [DOI] [PubMed] [Google Scholar]
- 84.Sunyakumthorn P, Boonsaen T, Boonsaeng V, Wallace JC, Jitrapakdee S. Involvement of Specific Proteins (Sp1/Sp3) and Nuclear Factor Y (NF-Y) in Basal Transcription of the Distal Promoter of the Rat Pyruvate Carboxylase Gene in β-cells. Biochem. Biophys. Res. Communs. 2005;329:188–196. doi: 10.1016/j.bbrc.2005.01.108. [DOI] [PubMed] [Google Scholar]
- 85.Boonsaen T, Rojvirat P, Surinya KH, Wallace JC, Jitrapakdee S. Transcriptional regulation of the distal promoter of the rat pyruvate carboxylase gene by hepatocyte nuclear factor 3β/Foxa2 and upstream stimulatory factors in insulinoma cells. Biochem J. 2007;405:359–367. doi: 10.1042/BJ20070276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gauthier BR, Brun T, Sarret EJ, Ishihara H, Schaad O, Descombes P, Wollheim CB. Oligonucleotide microarray analysis reveals PDX1 as an essential regulator of mitochondrial metabolism in rat islets. J. Biol. Chem. 2004;279:31121–31130. doi: 10.1074/jbc.M405030200. [DOI] [PubMed] [Google Scholar]
- 87.Wang H, Brun T, Kataoka K, Sharma AJ, Wollheim CB. MAFA controls genes implicated in insulin biosynthesis and secretion. Diabetologia. 2007;50:348–358. doi: 10.1007/s00125-006-0490-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Efanov AM, Sewing S, Bokvist K, Gromada J. Liver X receptor activation stimulates insulin secretion via modulation of glucose and lipid metabolism in pancreatic β-cells. Diabetes. 2004;53 suppl:S75–S78. doi: 10.2337/diabetes.53.suppl_3.s75. [DOI] [PubMed] [Google Scholar]
- 89.Yoshikawa H, Tajiri Y, Sako Y, Hashimoto T, Umeda F, Nawata H. Effects of free fatty acids on beta-cell functions: a possible involvement of peroxisome proliferator-activated receptors alpha or pancreatic/duodenal homeobox. Metabolism. 2001a;50:613–618. doi: 10.1053/meta.2001.22565. [DOI] [PubMed] [Google Scholar]
- 90.Anghel SI, Bedu E, Vivier CD, Descombes P, Desvergne B, Wahli W. Adipose tissue integrity as a prerequisite for systemic energy balance: a critical role for peroxisome proliferator-activated receptor gamma. J Biol Chem. 2007;282:29946–29957. doi: 10.1074/jbc.M702490200. [DOI] [PubMed] [Google Scholar]
- 91.Yu S, Matsusue K, Kashireddy P, Cao WQ, Yeldandi V, Yeldandi AV, Rao MS, Gonzalez FJ, Reddy JK. Adipocyte-specific gene expression and adipogenic steatosis in the mouse liver due to peroxisome proliferator-activated receptor gamma1 (PPARgamma1) overexpression. J. Biol. Chem. 2003;278:498–505. doi: 10.1074/jbc.M210062200. [DOI] [PubMed] [Google Scholar]
- 92.Herzig S, Long F, Jhala US, Hedrick S, Quinn R, Bauer A, et al. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001;413:179–183. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- 93.Impey S, McCorkle SR, Cha-Molstad H, Dwyer JM, Yochum GS, Boss JM, McWeeney S, et al. Defining the CREB Regulon: A Genome-Wide Analysis of Transcription Factor Regulatory Regions. Cell. 2004;119:1041–1054. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 94.Mukhopadhyay B, Stoddard SF, Wolfe RS. Purification, regulation and biochemical characterization of pyruvate carboxylase from Methanobacterium thermoautotrophicum strains ΔH. J. Biol. Chem. 1998;273:5155–5166. doi: 10.1074/jbc.273.9.5155. [DOI] [PubMed] [Google Scholar]
- 95.Mukhopadhyay B, Patel VJ, Wolfe RS. A stable archael pyruvate carboxylase from the hyperthermophile Methanococcus jannaschii. Arch. Microbiol. 2000;174:406414. doi: 10.1007/s002030000225. [DOI] [PubMed] [Google Scholar]
- 96.Mukhopadhyay B, Purwantini E, Kreder CL, Wolfe RS. Oxaloacetate synthesis in the Methanarchaeon Methanosarcina barkeri: pyruvate carboxylase genes and a putative Escherichia coli-type bifunctional biotin protein ligase gene (bpl/birA) exhibit a unique organization. J. Bacteriol. 2001;183:3804–3810. doi: 10.1128/JB.183.12.3804-3810.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kondo S, Nakajima Y, Sugio S, Yong-Biao J, Sueda S, Kondo H. Structure of the biotin carboxylase subunit of pyruvate carboxylase from Aquifex aeolicus at 2.2 A resolution. Acta Crystallogr D Biol Crystallogr. 2004;60:486–492. doi: 10.1107/S0907444904000423. [DOI] [PubMed] [Google Scholar]
- 98.Goss JA, Cohen ND, Utter MF. Characterization of the subunit structure of pyruvate carboxylase from Pseudomonas citronellolis. J. Biol. Chem. 1981;256:11819–11825. [PubMed] [Google Scholar]
- 99.Lai H, Kraszewski JL, Parwantini E, Mukhopadhyay B. Identiification of pyruvate carboxylase genes in Pseudomonas aeruginosa PAO1 and development of a P. aeruginosa-based overexpression system for α4- and α4β4-type pyruvate carboxylase. Appl. Environ. Microbiol. 2006;72:7785–7792. doi: 10.1128/AEM.01564-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Islam MN, Sueda S, Kondo H. Construction of new forms of pyruvate carboxylase to assess the allosteric regulation by acetyl CoA. Protein Eng. Des. Sel. 2005;18:71–78. doi: 10.1093/protein/gzi011. [DOI] [PubMed] [Google Scholar]
- 101.Jitrapakdee S, Booker GW, Cassady AI, Wallace JC. Cloning, sequencing and expression of rat liver pyruvate carboxylase. Biochem. J. 1996;316:631–637. doi: 10.1042/bj3160631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jitrapakdee S, Nezic M, Cassady AI, Khew-Goodall Y, Wallace JC. Molecular cloning and domain structure of chicken pyruvate carboxylase. Biochem. Biophys. Res. Communs. 2002;295:387–393. doi: 10.1016/s0006-291x(02)00651-4. [DOI] [PubMed] [Google Scholar]
- 103.St.Maurice M, Reinhardt L, Surinya KH, Attwood PV, Wallace JC, Cleland WW, Rayment I. Domain architecture of pyruvate carboxylase, a biotin-dependent multifunctional enzyme. Science. 2007;317:1076–1079. doi: 10.1126/science.1144504. [DOI] [PubMed] [Google Scholar]
- 104.Thoden JB, Blanchard CZ, Holden HM, Waldrop GL. Movement of the biotin carboxylase B-domain as a result of ATP binding. J Biol Chem. 2000;275:16183–16190. doi: 10.1074/jbc.275.21.16183. [DOI] [PubMed] [Google Scholar]
- 105.Kondo S, Nakajima Y, Sugio S, Sueda S, Islam MN, Kondo H. Structure of the biotin carboxylase domain of pyruvate carboxylase from Bacillus thermodenitrificans. Acta Crystallographica. 2007;63:885–890. doi: 10.1107/S0907444907029423. [DOI] [PubMed] [Google Scholar]
- 106.Waldrop GL, Rayment I, Holden HM. Three-dimensional structure of the biotin carboxylase subunit of acetyl CoA carboxylase. Biochemistry. 1994;33:10249–10256. doi: 10.1021/bi00200a004. [DOI] [PubMed] [Google Scholar]
- 107.Galperin MY, Koonin EV. A diverse superfamily of enzymes with ATP-dependent carboxylate-amine/thiol ligase activity. Protein Sci. 1997;6:2639–2643. doi: 10.1002/pro.5560061218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Studer R, Dahinden P, Wang W-W, Auchli Y, Li X-D, Dimroth P. Crystal structure of the carboxyltransferase domain of the oxaloacetate decarboxylase Na+ pump from Vibrio cholerae. J. Mol. Biol. 2006;367:547–547. doi: 10.1016/j.jmb.2006.12.035. [DOI] [PubMed] [Google Scholar]
- 109.Hall PR, Zheng R, Antony L, Pusztai-Carey M, Carey PR, Yee VC. Transcarboxylase 5S structures: assembly and catalytic mechanism of a multienzyme complex subunit. EMBO J. 2004;23:3621–3631. doi: 10.1038/sj.emboj.7600373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yong-Biao J, Islam MN, Sueda S, Kondo H. Identification of the catalytic residues involved in the carboxyl transfer of pyruvate carboxylase. Biochemistry. 2004;43:5912–5920. doi: 10.1021/bi035783q. [DOI] [PubMed] [Google Scholar]
- 111.Wexler ID, Kerr DS, Du Y, Kaung MM, Stephenson W, Lusk MM, Wappner RS, Higgins JJ. Molecular characterization of pyruvate carboxylase deficiency in two consanguineous families. Pediatr Res. 1998;43:579–584. doi: 10.1203/00006450-199805000-00004. [DOI] [PubMed] [Google Scholar]
- 112.Perham RN. Swinging arms and swinging domains in multifunctional enzymes: catalytic machines for multistep reactions. Annu. Rev. Biochem. 2000;69:961–1004. doi: 10.1146/annurev.biochem.69.1.961. [DOI] [PubMed] [Google Scholar]
- 113.Athappilly FK, Hendrickson WA. Structure of the biotinyl domain of acetyl CoA carboxylase determined b MAD phasing. Structure. 1995;3:1407–1419. doi: 10.1016/s0969-2126(01)00277-5. [DOI] [PubMed] [Google Scholar]
- 114.Roberts EL, Shu N, Howard MJ, Broadhurst RW, Chapman-Smith A, Wallace JC, Morris T, Cronan JE, Jr, Perham RN. Solution structures of apo-and holo-biotinyl domains from acetyl coenzyme A carboxylase of Escherichia coli determined by triple-resonance NMR spectroscopy. Biochemistry. 1999;38:5045–5053. doi: 10.1021/bi982466o. [DOI] [PubMed] [Google Scholar]
- 115.Reddy DV, Rothemund S, Shenoy BC, Carey PR, Sonnichsen FD. Structural characterization of the entire 1.3S subunit of transcarboxylase from Propiobacterium shermanii. Protein Sci. 1998;7:2156–2163. doi: 10.1002/pro.5560071013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dyda F, Klein DC, Hickman AB. GCN5-related N-acetyltransferases: a structural overview. Annu. Rev. Biophys. Biomol. Struct. 2000;29:81–103. doi: 10.1146/annurev.biophys.29.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Peters-Wendisch PG, Wendisch VF, Paul S, Eikmanns BJ, Sahm H. Pyruvate carboxylase as an anaplerotic enzyme in Corynebacterium glutamicum. Microbiology. 1997;143:1095–1103. doi: 10.1099/00221287-143-4-1095. [DOI] [PubMed] [Google Scholar]
- 118.Attwood PV. Locus of action of acetyl CoA in the biotin-carboxylation reaction of pyruvate carboxylase. Biochemistry. 1993;32:12736–12742. doi: 10.1021/bi00210a024. [DOI] [PubMed] [Google Scholar]
- 119.Khew-Goodall YS, Johannssen W, Attwood PV, Wallace JC, Keech DB. Studies on dilution inactivation of sheep liver pyruvate carboxylase. Arch. Biochem. Biophys. 1991;284:98–105. doi: 10.1016/0003-9861(91)90269-o. [DOI] [PubMed] [Google Scholar]
- 120.Johannssen W, Attwood PV, Wallace JC, Keech DB. Localisation of the active site of pyruvate carboxylase by electron microscopic examination of avidin-enzyme complexes. Eur. J. Biochem. 1983;133:201–206. doi: 10.1111/j.1432-1033.1983.tb07448.x. [DOI] [PubMed] [Google Scholar]
- 121.Mayer F, Wallace JC, Keech DB. Further electron microscope studies onpyruvate carboxylase. Eur. J. Biochem. 1980;112:265–272. doi: 10.1111/j.1432-1033.1980.tb07202.x. [DOI] [PubMed] [Google Scholar]
- 122.de Queiroz MS, Waldrop GL. Modeling and numerical simulation of biotin carboxylase kinetics: Implications for half-sites reactivity. J. Theor. Biol. 2007;246:167–175. doi: 10.1016/j.jtbi.2006.12.025. [DOI] [PubMed] [Google Scholar]
- 123.Janiyani K, Bordelon T, Waldrop GL, Cronan JE., Jr Function of Escherichia coli biotin carboxylase requires catalytic activity of both subunits of the homodimer. J. Biol. Chem. 2001;276:29864–29870. doi: 10.1074/jbc.M104102200. [DOI] [PubMed] [Google Scholar]
- 124.Rangan VS, Joshi AK, Smith S. Fatty acid synthase dimers containing catalytically active beta-ketoacyl synthase or malonyl/acetyltransferase domains in only one subunit can support fatty acid synthesis at the acyl carrier protein domains of both subunits. J. Biol. Chem. 1998;273:34949–39953. doi: 10.1074/jbc.273.52.34949. [DOI] [PubMed] [Google Scholar]
- 125.Tang Y, Kim CY, Mathews II, Cane DE, Khosla C. The 2.7-Angstrom crystal structure of a 194-kDa homodimeric fragment of the 6-deoxyerythronolide B synthase. Proc. Natl. Acad. Sci. USA. 2006;103:11124–11129. doi: 10.1073/pnas.0601924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Attwood PV. The structure and the mechanism of action of pyruvate carboxylase. Int. J. Biochem. Cell. Biol. 1995;27:231–249. doi: 10.1016/1357-2725(94)00087-r. [DOI] [PubMed] [Google Scholar]
- 127.Attwood PV, Wallace JC. Chemical and catalytic mechanisms of carboxyl transfer reactions in biotin-dependent enzymes. Acc. Chem. Res. 2002;35:113–120. doi: 10.1021/ar000049+. [DOI] [PubMed] [Google Scholar]
- 128.Attwood PV, Tipton PA, Cleland WW. Carbon-13 and deuterium isotope effects on oxalacetate decarboxylation by pyruvate carboxylase. Biochemistry. 1986;25:8197–8205. doi: 10.1021/bi00373a012. [DOI] [PubMed] [Google Scholar]
- 129.Attwood PV, Cleland WW. Decarboxylation of oxalacetate by pyruvate carboxylase. Biochemistry. 1986;25:8191–8196. doi: 10.1021/bi00373a011. [DOI] [PubMed] [Google Scholar]
- 130.Tipton PA, Cleland WW. Carbon-13 and deuterium isotope effects on the catalytic reactions of biotin carboxylase. Biochemistry. 1988;27:4325–4331. doi: 10.1021/bi00412a020. [DOI] [PubMed] [Google Scholar]
- 131.Headlam MJ, Attwood PV. Cysteine-lysine ion pairs in yeast pyruvate carboxylase. Biochem. Soc.Trans. 1998;26:S 74. doi: 10.1042/bst026s074. [DOI] [PubMed] [Google Scholar]
- 132.Werneburg BG, Ash DE. Chemical modifications of chicken liver pyruvate carboxylase: evidence for essential cysteine-lysine pairs and a reactive sulfhydryl group. Arch. Biochem. Biophys. 1993;303:214–221. doi: 10.1006/abbi.1993.1275. [DOI] [PubMed] [Google Scholar]
- 133.Levert KL, Lloyd RB, Waldrop GL. Do cysteine 230 and lysine 238 of biotin carboxylase play a role in the activation of biotin? Biochemistry. 2000;39:4122–4128. doi: 10.1021/bi992662a. [DOI] [PubMed] [Google Scholar]
- 134.Kazuta Y, Tokunaga E, Aramaki E, Kondo H. Identification of Lysine-238 of Escherichia coli Biotin Carboxylase as an ATP-Binding Residue. FEBS Letters. 1998;427:377–380. doi: 10.1016/s0014-5793(98)00472-4. [DOI] [PubMed] [Google Scholar]
- 135.Branson JP, Nezic M, Jitrapakdee S, Wallace JC, Attwood PV. Kinetic Characterization of Yeast Pyruvate Carboxylase Isozyme Pyc1 and the Pyc1 Mutant, C249A. Biochemistry. 2004;43:1075–1081. doi: 10.1021/bi035575y. [DOI] [PubMed] [Google Scholar]
- 136.Cleland WW. The use of isotope effects in the detailed analysis of catalytic mechanisms of enzymes. Bioorg. Chem. 1987;15:283–302. [Google Scholar]
- 137.Cleland WW. Low-barrier hydrogen bonds and enzymatic catalysis. Arch. Biochem. Biophys. 2000;382:1–5. doi: 10.1006/abbi.2000.2011. [DOI] [PubMed] [Google Scholar]
- 138.Wallace J. Distribution and biological functions of pyruvate carboxylase in nature. In: Keech D, Wallace J, editors. Pyruvate Carboxylase. Boca Raton: CRC Press; 1985. pp. 5–64. [Google Scholar]
- 139.Attwood PV, Graneri BD. Bicarbonate-dependent ATP cleavage catalysed by pyruvate carboxylase in the absence of pyruvate. Biochem. J. 1992;287:1011–1017. doi: 10.1042/bj2871011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Branson JP, Nezic M, Wallace JC, Attwood PV. Kinetic characterization of yeast pyruvate carboxylase isozyme pyc1. Biochemistry. 2002;41:4459–4466. doi: 10.1021/bi011888m. [DOI] [PubMed] [Google Scholar]
- 141.Legge GB, Branson JP, Attwood PV. Effects of acetyl CoA on the pre-steady-state kinetics of the biotin carboxylation reaction of pyruvate carboxylase. Biochemistry. 1996;35:3849–3856. doi: 10.1021/bi952797q. [DOI] [PubMed] [Google Scholar]
- 142.Phillips NF, Snoswell MA, Chapman-Smith A, Keech DB, Wallace JC. Isolation of a carboxyphosphate intermediate and the locus of acetyl CoA action in the pyruvate carboxylase reaction. Biochemistry. 1992;31:9445–9450. doi: 10.1021/bi00154a017. [DOI] [PubMed] [Google Scholar]
- 143.Scrutton MC, Keech DB, Utter MF. Pyruvate carboxylase IV. Partial reactions and the locus of activation by acetyl coenzyme A. J. Biol. Chem. 1965;240:574–581. [PubMed] [Google Scholar]
- 144.Attwood PV, Wallace JC. The carboxybiotin complex of chicken liver pyruvate carboxylase. A kinetic analysis of the effects of acetyl CoA, Mg2+ ions and temperature on its stability and on its reaction with 2-oxobutyrate. Biochem. J. 1986;235:359–364. doi: 10.1042/bj2350359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Geeves MA, Branson JP, Attwood PV. Kinetics of nucleotide binding to pyruvate carboxylase. Biochemistry. 1995;34:11846–11854. doi: 10.1021/bi00037a024. [DOI] [PubMed] [Google Scholar]
- 146.Adina-Zada A, Jitrapakdee S, Surinya KH, McIldowie MJ, Piggott MJ, Cleland WW, Wallace WW, Attwood PV. Insights into the mechanism and regulation of pyruvate carboxylase by characterisation of a biotin-deficient mutant of the Bacillus thermodenitrificans enzyme. Int. J. Biochem. Cell Biol. 2008 doi: 10.1016/j.biocel.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 147.Jitrapakdee S, Adina-Zada A, Besant PG, Surinya KH, Cleland WW, Wallace JC, Attwood PV. Differential regulation of the yeast isozymes of pyruvate carboxylase and the locus of action of acetyl CoA. Int. J. Biochem. Cell Biol. 2007;39:1211–1223. doi: 10.1016/j.biocel.2007.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sueda S, Islam MN, Kondo H. Protein engineering of pyruvate carboxylase: investigation on the function of acetyl CoA and the quaternary structure. Eur. J. Biochem. 2004;271:1391–1400. doi: 10.1111/j.1432-1033.2004.04051.x. [DOI] [PubMed] [Google Scholar]
- 149.Ozimek P, van Dijk R, Latchev K, Gancedo C, Wang DY, van der Klei IJ, Veenhuis M. Pyruvate carboxylase is an essential protein in the assembly of yeast peroxisomal oligomeric alcohol oxidase. Mol. Biol. Cell. 2003;14:786–797. doi: 10.1091/mbc.E02-07-0417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ozimek P, Klompmaker SH, Visser N, Veenhuis M, van der Klei IJ. The transcarboxylase domain of pyruvate carboxylase is essential for assembly of the peroxisomal flavoenzyme alcohol oxidase. FEMS Yeast Res. 2007;7:1082–1092. doi: 10.1111/j.1567-1364.2007.00214.x. [DOI] [PubMed] [Google Scholar]
- 151.Vessal M, Mishra S, Moulik S, Murphy LJ. Prohibitin attenuates insulin-stimulated glucose and fatty acid oxidation in adipose tissue by inhibiting pyruvate carboxylase. FEBS J. 2006;273:568–576. doi: 10.1111/j.1742-4658.2005.05090.x. [DOI] [PubMed] [Google Scholar]