Abstract
Islam is the second most practiced religion in India, next to Hinduism. It is still unclear whether the spread of Islam in India has been only a cultural transformation or is associated with detectable levels of gene flow. To estimate the contribution of West Asian and Arabian admixture to Indian Muslims, we assessed genetic variation in mtDNA, Y-chromosomal and LCT/MCM6 markers in 472, 431 and 476 samples, respectively, representing six Muslim communities from different geographical regions of India. We found that most of the Indian Muslim populations received their major genetic input from geographically close non-Muslim populations. However, low levels of likely sub-Saharan African, Arabian and West Asian admixture were also observed among Indian Muslims in the form of L0a2a2 mtDNA and E1b1b1a and J*(xJ2) Y-chromosomal lineages. The distinction between Iranian and Arabian sources was difficult to make with mtDNA and the Y chromosome, as the estimates were highly correlated because of similar gene pool compositions in the sources. In contrast, the LCT/MCM6 locus, which shows a clear distinction between the two sources, enabled us to rule out significant gene flow from Arabia. Overall, our results support a model according to which the spread of Islam in India was predominantly cultural conversion associated with minor but still detectable levels of gene flow from outside, primarily from Iran and Central Asia, rather than directly from the Arabian Peninsula.
Keywords: Indian Muslims, mtDNA, Y chromosome, Middle East, sub-Saharan, gene flow
Introduction
Islam was first brought to the Indian Subcontinent in 711 CE, when the Arab military forces conquered Sindh, the lower Indus valley, and incorporated it into the Arabian Empire.1 Subsequently, Sindh not only became an Indo-Muslim state but also an Islamic outpost, where Arabs established trade links with the Middle East and were later joined by mystic teachers, or Sufis. By the end of the tenth century, dramatic changes took place when the Central Asian Turkic tribes accepted both the message and mission of Islam. These aggressively expansive invaders first began to move into Afghanistan and Iran and later into India through the northwest. In the thirteenth century, a Turkic kingdom was established in Delhi, which enabled Persian and Afghan Muslim invaders to further spread across India. Within the next 100 years, the Muslim empire extended its sway east to Bengal and south to the Deccan and remained dominant in the Indian Subcontinent until 1707 CE.1, 2 These last few centuries of expansion of Muslim populations into India were accompanied by extensive religious conversion. Furthermore, the exodus of people from Western Asia, especially from Iran, in the form of mercenaries and businessmen led to significant cultural diffusion of Muslim traditions among the ethnic Indian populations. These Muslim immigrants, who were mostly males, reportedly married local Hindu females and generated a new admixed genetic pool, perhaps with sex-specific differences.2, 3 At present, Islam is the second most practiced religion in India after Hinduism, encompassing 13.4% (138 million) of the total Indian population (Census of India, 2001).
Classical genetic marker studies have revealed that most Indian Muslims are closely related to their neighboring non-Muslim populations, suggesting that they descend primarily from local Hindu converts.4, 5 The exception to this are some Northern and Northwestern Indian Muslims who differ from indigenous Hindu populations, likely because of a higher proportion of genetic lineages of external origin.4, 5, 6, 7 Consistent with historical data, which predict significant local female contributions, the only mitochondrial DNA study that has been reported so far showed that North Indian Muslims exhibit the highest affinity to local Indian regional populations.8 Similarly, yet in contrast to an expectedly higher male contribution of outsiders, the Y-chromosomal evidence that is available so far has revealed predominantly local South Asian-specific lineages among Indian Muslims.8, 9 However, in our recent study based on autosomal STR markers, we have detected genetic signatures characteristic of populations of the Middle East in some of the contemporary Indian Muslim populations.10
According to historical evidence, the Indian Subcontinent has been exposed to several waves of human migrations from the Arabian Peninsula and Iran, the homelands of Indian Muslim rulers.2 The Arabian Peninsula (where Islam was propagated) served as a hub for human migrations, hence the merged genetic signatures of Eurasian and African origin, which has been detected in both maternal11 and paternal12 lineages from the region. Besides Arabia, Iran is a second plausible genetic source for Indian Muslims. It is positioned in the tricontinental nexus and its populations genetically show close proximity to those from the Near East, although with a lesser genetic input from Africa than from the populations of the Arabian Peninsula.13, 14 Besides mtDNA and the Y chromosome, which show relatively low levels of differentiation between these two potential sources, recent studies of lactose tolerance have revealed that Iranian and Arabian populations differ significantly in genetic patterns at this locus.15, 16 The Arabian populations are characterized by a 50–60% frequency of a G−13915 allele, purportedly related to their consumption of camel milk.15 This allele has not been detected so far among Iranian populations who, on the contrary, similar to populations from Europe and the Near East show a moderate frequency of the T−13910 allele, which occurs at a significantly lower frequency in Arabia.15
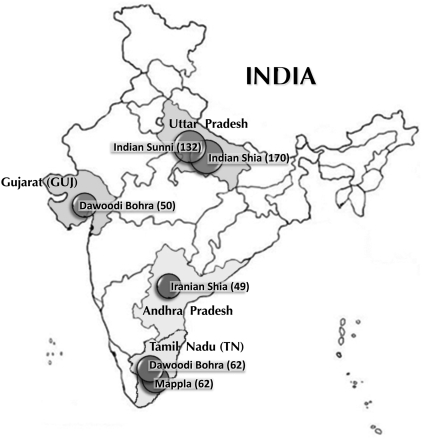
The extent of gene flow associated with the spread of Islam in the Indian Subcontinent is still largely unknown. Two previous studies have assessed mtDNA and the Y-chromosome haplogroup composition of Indian Muslim communities from Uttar Pradesh and Andhra Pradesh and concluded that the spread of Islam in India was mainly a cultural phenomenon and was not accompanied by significant levels of gene flow from West or Central Asia.8, 9 However, a study of more Muslim populations with a wider geographical coverage, larger sample size and high-resolution informative genetic markers would be required to detect signals of minor genetic contribution. To assess the genetic ancestry of contemporary Indian Muslims, we screened six Muslim populations who follow Shia or Sunni faiths from three different geographical regions of India (Figure 1) with ancestry-informative markers from mtDNA, the Y chromosome and the LCT/MCM6 region.
Figure 1.
Map of India showing the geographical location of the six Indian Muslim populations included in this study.
Materials and methods
Samples
In total, 472 Indian Muslim mtDNAs, 431 Indian Muslim Y chromosomes and 747 Indian Muslim and non-Muslim MCM6 gene profiles were used in this study. Samples were obtained with informed consent. We compared the mtDNA diversity in Indian Muslims with 15 949 mtDNA profiles from Indian non-Muslims,17, 18, 19 as well as from Pakistan,17 the Middle East,20 Central Asia,13 East Asia21, 22, 23 and Europe.24 We used 3696 previously published Y-chromosomal haplotypes of populations from India,25, 26, 27, 28, 29, 30 Pakistan,26 the Middle East,12, 14, 31 Central Asia,32 East Asia33 and Europe34 to compare with the studied Indian Muslim Y chromosomes. MCM6 gene variants in Indian populations were compared with 581 variants from Pakistan16 and the Middle East.15
mtDNA typing
The first hypervariable segment (HVS-I) of mtDNA was sequenced directly in all samples and variable positions were determined from nps 16 001 to 16 450. The second hypervariable segment (HVS-II) and haplogroup confirmatory diagnostic coding regions were sequenced for 472 samples on the basis of their haplotype information (Supplementary Table 1). In all, 12 samples were selected for whole mtDNA sequencing. The haplotypes defined by control region sequences and coding regions were haplogrouped by their mutational motifs (Supplementary Table 1), following previously published haplogroup trees.35, 36, 37, 38, 39 Complete mtDNA genomes and segments including diagnostic positions were amplified using 24 sets of primers.40 PCRs were carried out with 10 ng of template DNA in a 10 μl reaction volume with 10 p of each primer, 100 μ dNTPs, 1.5 m Mgcl2 and 1 U of Taq DNA polymerase. Thirty-five cycles were performed with 30-s denaturation at 94°C, 30-s annealing at 58°C and 2-min extension at 72°C. The annealing temperature and time were slightly modified for a few sets of primers. PCR products were directly sequenced using the BigDye Terminator cycle sequencing kit and an ABI Prism 3730XL DNA Analyzer (Applied Biosystems, Foster City, CA, USA), following the manufacturer's protocol. The individual mtDNA sequences were compared with rCRS41 using AutoAssembler – ver 2.1 (Applied Biosystems). The sequences generated in this study have been deposited in the GenBank database (accession nos. FJ157366-FJ157837 (mtDNA HVS-I sequences), FJ157838-FJ157849 (complete mtDNA sequences)).
Y-chromosome typing
A total of 431 samples were typed with 23 Y-chromosomal markers (M89, YAP/M145, M96, M35, M78, M130, M356, M9, M45, M304, M172, M410, M69, M82, Apt, M170, M201, M173, M17, M124, M11, M214 and M175). The thermal cycling programs were set up with an initial denaturation at 95°C for 5 min, followed by 30–35 cycles at 94°C for 30 s, at a primer-specific annealing temperature of 52–60°C for 30 s and 72°C for 45 s, followed by a final extension at 72°C for 7 min. PCR products were directly sequenced using the BigDye Terminator cycle sequencing kit (Applied Biosystems) and the ABI Prism 3730XL DNA Analyzer, following the manufacturer's protocol.
LCT/MCM6 gene typing
A 400-bp fragment including the −13.9-kb region of the gene was PCR amplified with primers MCM6i13 and LAC-CL2, as detailed elsewhere.42 PCR products were sequenced using the MCM6i13 or LAC-CL2 primer and the BigDye Terminator cycle sequencing kit (Applied Biosystems) on an ABI Prism 3730XL DNA Analyzer.
Statistical analyses
Phylogenetic trees were constructed using Network 4.2.0.1 (www.fluxus-engineering.com).43, 44 The program Admix 2.0 (http://web.unife.it/progetti/genetica/Isabelle/admix2_0.html)45 was used to calculate the admixture proportions of samples on the basis of the frequency of haplogroups. The age of the L0a2a2 and M52 lineages was estimated on the basis of the molecular clock46, 47 based on synonymous mutation rate, given by Kivisild et al47 and recalibrated by Soares et al46 assuming a mutation rate of one synonymous mutation per 7884 years. PC plots were generated with MVSP 3.1 (http://www.kovcomp.co.uk/mvsp/index.html).48 Arlequin 3.1 (http://cmpg.unibe.ch/software/arlequin3)49 was used to evaluate the genetic structure of the populations by performing analysis of molecular variance (AMOVA), as well as to calculate genetic diversities of mtDNA and the Y chromosome on the basis of haplogroup frequencies.
Results
mtDNA comparisons of Indian Muslim and non-Muslim populations
We analyzed 472 samples for variation in mtDNA control regions and haplogroup-diagnostic coding region sites. Pooled haplogroup frequencies are shown in Table 1 and detailed haplogroup frequencies and definitions are given in Supplementary Tables 1 and 2 and Supplementary Figures 1 and 2. Altogether, haplogroups restricted to the Indian Subcontinent were observed at an average frequency of 63% in Indian Muslim populations as compared with 74% among the non-Muslim neighbors (Table 1). The average contribution of haplogroups of West Eurasian origin to Indian Muslims was 18%, which is not significantly higher than the value observed in non-Muslim populations (14%). In contrast, Iranian Shia Muslims exhibit a high frequency (54%) of West Eurasian lineages. It is interesting that the sub-Saharan African- and Arabian-specific L0a2a2 and R01 lineages were found only in Dawoodi Bohras (TN and GUJ), whereas these lineages were generally absent in Indian non-Muslims, although a related L0a2a2 lineage has been detected previously among the Sindhi population of Pakistan (Figure 2). The Central Asian lineages were found at a lower average frequency of 6% and the haplogroups U7 and W, which exist in similar frequencies in India and Iran, were observed at an average frequency of 6 and 3%, respectively, in Muslim populations. The gene diversity in Muslim populations ranged from 0.80±0.05 to 0.93±0.02, which is slightly higher than that among non-Muslim populations, 0.74±0.02 to 0.86±0.02 (Table 2), and reveals the prevalence of a comparatively high genetic diversity among Indian Muslims. We completely sequenced the mtDNA genome of nine M* samples, which harbor 16223–16275 substitutions in hypervariable segment I (HVS-I), to determine their potential source region. All nine samples were found to share common coding region variants, which enabled us to define a new autochthonous South Asian-specific haplogroup M52, which turned out to share a common origin with one of its sister branches, labeled here as M52a (Figure 3), detected among Indian non-Muslims. The same haplogroup has been recently reported in the Tharus of Nepal and in the Andhra Pradesh population.50 All nine sequences of Muslims are nested within the M52 lineage (Figure 3). Considering this phylogenetic structuring, the newly characterized haplogroup M52 is most likely to have an Indian rather than West Asian or Arabian origin. AMOVA yielded no statistically significant results for any group distinctions on the basis of religion (Indian Muslims and non-Muslims), geography (North India, South India and West India) or other criteria investigated (Supplementary Table 3).
Table 1. mtDNA haplogroup frequencies in six Indian Muslims and potential source populations.
| Haplogroups | Indian Shiaa | Indian Sunnia | Dawoodi Bohra (TN)a | Dawoodi Bohra (GUJ)a | Mapplaa | Iranian Shiaa | Indian non-Muslimsb | Arabiac | Iranb |
|---|---|---|---|---|---|---|---|---|---|
| No. of individuals | 120 | 131 | 62 | 50 | 61 | 48 | 598 | 553 | 436 |
| Central Asiand | 0.01 | 0.01 | 0.13 | 0.02 | 0.07 | 0.10 | 0.04 | — | 0.03 |
| West Eurasiane | 0.15 | 0.05 | 0.15 | 0.14 | 0.05 | 0.54 | 0.14 | 0.61 | 0.75 |
| Africanf | — | — | 0.05 | — | — | — | — | 0.13 | — |
| Arabiang | — | — | 0.15 | 0.08 | — | — | 0.01 | 0.18 | 0.03 |
| U7 | 0.06 | 0.06 | 0.05 | 0.10 | 0.05 | 0.04 | 0.05 | 0.01 | 0.09 |
| W | 0.03 | 0.02 | 0.05 | 0.02 | — | 0.04 | 0.02 | 0.01 | 0.03 |
| Indianh | 0.76 | 0.85 | 0.44 | 0.64 | 0.84 | 0.27 | 0.74 | 0.06 | 0.07 |
Present study, (TN, Tamil Nadu and GUJ, Gujarat).
Comparative data are from Metspalu et al.17
Comparative data are from Abu-Amero et al.20
Central Asian haplogroups include A, B, F, MD and MG.
West Eurasian haplogroups include H, HV, I, J, T, U, U1, U2, U2e, U3, U4, U5, U6, N, N1, N2, U9, UK and X.
African haplogroups include L and M1.
Arabian haplogroups include pHV (R01).
Indian haplogroups include M*, M18, M2, M25, M3, M4, M5, M6, M30, M31, M33, M34, M35, M36, M37, M38, M40, M45, M46, M47, M49, M52, R*, R2, R30, R31, R5, R6, R7, R8, U2a, U2b and U2c.
For detailed haplogroup frequencies of all the six Indian Muslim populations see Supplementary Table 2.
Figure 2.
The phylogenetic tree of mtDNA haplogroup L0a2a2. Synonymous positions are marked with ‘s' suffix and highlighted in bold. The age estimate is based on the molecular clock determined by Soares et al,46 assuming a mutation rate of one synonymous mutation per 7884 years. Dawoodi Bohra samples are reported in this study, whereas data for other populations are presented by the phylogenetic tree in Behar et al,39 and by the references to the original sources given therein. Indian denotes the Indian Subcontinent.
Table 2. Genetic diversity based on mtDNA and Y-chromosome analysis of Muslim populations of India.
| Populations | mtDNA haplogroups | Y haplogroups |
|---|---|---|
| Indian Shia | 0.9175±0.0145 | 0.8561±0.0149 |
| Indian Sunni | 0.8833±0.0174 | 0.7806±0.0259 |
| Dawoodi Bohra (TN) | 0.9038±0.0175 | 0.5785±0.0566 |
| Dawoodi Bohra (GUJ) | 0.8996±0.0264 | 0.8147±0.0294 |
| Mappla | 0.8044±0.0487 | 0.8038±0.0367 |
| Iranian Shia | 0.9282±0.0242 | 0.8367±0.0378 |
| North Indiaa | 0.8568±0.0175 | 0.8114±0.0114 |
| West Indiab | 0.8562±0.0284 | 0.7979±0.0235 |
| South Indiac | 0.7437±0.0224 | 0.8680±0.0037 |
North India includes Uttar Pradesh.
West India includes Gujarat.
South India includes Tamil Nadu and Andhra Pradesh.
Figure 3.
Phylogenetic tree reconstruction of the newly defined South Asian-specific haplogroup M52, based on 10 complete mtDNA genomes. This tree was redrawn manually from the output of median joining/reduced network obtained using NETWORK program (version 4.5) http://www.fluxus-engineering.com. The published sequence is shown with the initials of the first author (CS).37 Coalescent times were calculated by a calibration method described elsewhere.46 16182C, 16183C and 16519 polymorphisms were omitted. Suffixes A, C, G and T indicate transversions. Synonymous (s) and nonsynonymous (ns) mutations are distinguished. Recurrent mutations are underlined.
Y-chromosomal haplogroup profiles of Indian Muslim and non-Muslim populations
We genotyped 23 Y-chromosomal biallelic markers in a total of 431 Indian Muslims. All paternal lineages could be assigned to branches of the major haplogroups C, F and K (Figure 4 and Supplementary Table 4) according to Y-DNA haplogroup tree 2008,51 which are the three founder haplogroups commonly found in all continents outside Africa.52 Among the 17 Y haplogroups observed in Indian Muslims, as among the non-Muslims, R1a1 showed the highest frequency (31%), followed by haplogroup H (20%). The sub-Saharan African- and Arabian-specific paternal lineages E1b1b1a and J*(xJ2) were present in three Muslim populations (Indian Shia, Indian Sunni and Mappla) with an average frequency of 2 and 8%, respectively, whereas they were rare or absent among non-Muslim populations. Haplogroup G, which is common in the Middle East and rare or absent in Indian non-Muslim populations, was also present in three Muslim populations with an average overall frequency of 5%. The Y-chromosomal gene diversity in Muslim populations ranged from 0.58±0.06 to 0.86±0.01 and from 0.80±0.02 to 0.87±0.004 in non-Muslim populations (Table 2). When the paternal genetic structure of Indian Muslims was investigated by AMOVA, the geographical difference between Indian populations (North, South and West) was significant (5.08%, P<0.001), but the differences between religions (Muslims and non-Muslims) within India were not (P=0.08) (Supplementary Table 3). This reflects the large ‘among population within group' variation in the analysis of Indian religious groups. There is a notable variation between different Indian Muslim populations, some being highly similar to local Indian populations and others having similarities with external populations, so that when they are all grouped together as ‘Indian Muslims', the group difference is statistically insignificant from that of non-Muslims.
Figure 4.
Rooted maximum parsimony tree of Y-chromosome haplogroups defined by binary markers, along with their frequency in six Muslim populations of India and their potential source populations. K*=K* (xL,M,NO,P); 3 of the 12 individuals with a K* affiliation were tested for M184 and all three showed the presence of the ancestral allele. Haplogroups E1b1b1a, G and J*(xJ2) are not specific to Indian populations. Markers shown in italics were not genotyped and are included in context for comparison populations. Comparative data are from *Sengupta et al,26 ¶Regueiro et al14 and §Cadenas et al.12
Analysis of the LCT gene
A total of 747 samples of Indian Muslim and non-Muslim populations were sequenced for a 400-bp fragment, which is ∼14 kb upstream of the LCT gene (Table 3). The C/T−13910 variant was widely observed among both the Indian Muslim (Shia 10%, Sunni 10%, Dawoodi Bohra (TN) 14%, Dawoodi Bohra (GUJ) 11%, Mappla 2% and Iranian Shia 4%) and non-Muslim populations (North India 19%, West India 23% and South India 10%). The Iranian population also exhibits the same mutation with 10% frequency.15 The Saudi Arabian-specific T/G−13915 variant15 was completely absent from the Indian population, yet at the same position, we observed a new T/C−13915 variant (Mappla 1% and South India 1%), which is likely to be an Indian-specific mutation.
Table 3. Allele frequencies of LCT/MCM6 variants in South and West Asia.
| Region/population | N | rs4988235 (−13910) C to T mutation T allele frequency | rs41380347 (−13915) T to G G allele frequency | (−13915) T to C C allele frequency | Reference |
|---|---|---|---|---|---|
| Indian Muslims | |||||
| Dawoodi Bohra (GUJ) | 50 | 0.110 | — | — | Present study |
| Indian Sunni | 132 | 0.102 | — | — | Present study |
| Indian Shia | 121 | 0.095 | — | — | Present study |
| Iranian Shia | 49 | 0.041 | — | — | Present study |
| Dawoodi Bohra (TN) | 62 | 0.137 | — | — | Present study |
| Mappla | 62 | 0.016 | — | 0.008 | Present study |
| Indian non-Muslims | |||||
| North Indiaa | 91 | 0.187 | — | — | Our unpublished data |
| West Indiab | 73 | 0.233 | — | — | Our unpublished data |
| South Indiac | 107 | 0.098 | — | 0.009 | Our unpublished data |
| Pakistan | 251 | 0.165 | n.d. | n.d. | Enattah et al, 200716 |
| Arabsd | 40 | 0.130 | 0.105 | — | Enattah et al, 200815 |
| Saudi Arabia | 248 | 0.004 | 0.570 | — | Enattah et al, 200815 |
| Iran | 42 | 0.100 | — | — | Enattah et al, 200815 |
North India includes Uttar Pradesh, Uttaranchal, Jammu and Kashmir.
West India includes Punjab, Gujarat and Rajasthan.
South India includes Tamil Nadu, Andhra Pradesh and Karnataka.
Arabs are from Syria, Iraq, Lebanon and Palestine.
Population affinities and admixture estimates
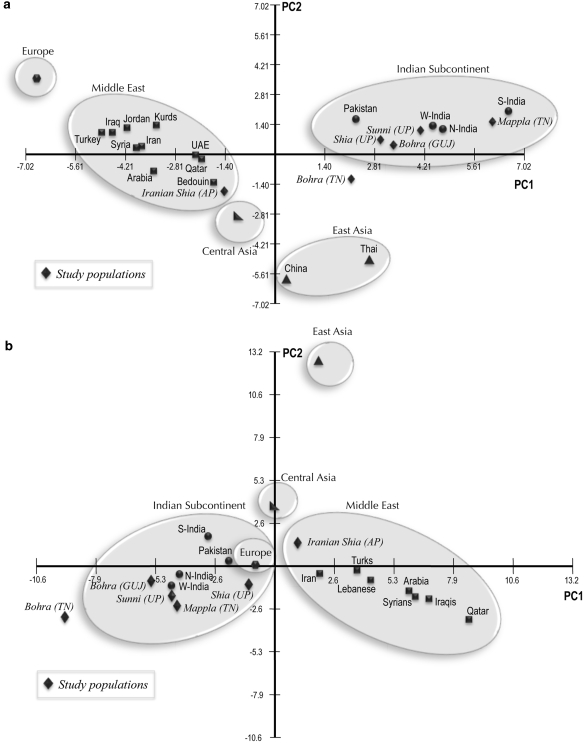
Genetic distance-based PC analyses of Indian Muslim and non-Muslim groups, compared with other world populations for both mtDNA and the Y chromosome, are shown in Figures 5a and b, respectively. In the mtDNA PCA plot (Figure 5a), Shia, Sunni, Dawoodi Bohra (GUJ) and Mappla were found to cluster together with Indian non-Muslim populations, whereas Dawoodi Bohra (TN) seems to be an outlier and Iranian Shia cluster with populations from the Middle East. The East Asian, Central Asian, Middle Eastern and European populations clustered separately according to their geography. In the Y-chromosomal plot (Figure 5b), Shia, Sunni, Dawoodi Bohra (GUJ) and Mappla form a group with their neighboring Indian non-Muslim populations and Europeans, whereas the Dawoodi Bohra (TN), again found as an outlier, and Iranian Shia Muslims seem to be genetically closer to the Middle Eastern group.
Figure 5.
(a) Principal component analysis plot based on mtDNA haplogroup frequencies. UP, Uttar Pradesh; GUJ, Gujarat; AP, Andhra Pradesh; TN, Tamil Nadu. Comparative data references: N-India, W-India and S-India (17–19); Pakistan (17); Middle East (20); Central Asia (13); East Asia (21–23); Europe (24). (b) Principal component analysis plot based on Y haplogroup frequencies. UP, Uttar Pradesh; GUJ, Gujarat; AP, Andhra Pradesh; TN, Tamil Nadu. Comparative data references: N-India, W-India and S-India (25–30); Pakistan (26); Middle East (12, 14, 31); Central Asia (32); East Asia (33); Europe (34).
To obtain quantitative estimates of the Iranian versus Arabian contribution among Indian Muslim groups, admixture analysis was carried out with three putative parental populations, including (i) the geographically closest Indian Hindu population, and a pool of populations from (ii) Arabia or (iii) Iran. With these three putative parental populations, admixture analyses were carried out in two phases. Each phase comprised of two parental populations, that is, (i) the geographically closest Indian non-Muslim population and Arabian population and (ii) the geographically closest Indian non-Muslim population and Iranian population. In the case of Dawoodi Bohra (TN) Muslims, admixture contributions were estimated with local populations from both Tamil Nadu and Gujarat because these Muslims are recent migrants from Gujarat settled in Tamil Nadu. The results of admixture analyses were tabulated (Tables 4 and 5) accordingly.
Table 4. mtDNA – admixture proportions.
| Local Indian closest neighbors (Parental) | ||||||
|---|---|---|---|---|---|---|
| Admixed | Uttar Pradesh | Gujarat | Andhra Pradesh | Tamil Nadu | Arabia (Parental) | Iran (Parental) |
| Indian Shia | 1.15 (0.09) | −0.15 (0.09)a | ||||
| 1.13 (0.06) | −0.13 (0.06)a | |||||
| Indian Sunni | 1.36 (0.08) | −0.36 (0.08)a | ||||
| 1.29 (0.07) | −0.29 (0.07)a | |||||
| Dawoodi Bohra (TN)b | 0.90 (0.11) | 0.10 (0.11) | ||||
| 1.004 (0.12) | −0.004 (0.12)a | |||||
| Dawoodi Bohra (TN)b | 0.70 (0.07) | 0.30 (0.07) | ||||
| 0.77 (0.06) | 0.23 (0.06) | |||||
| Dawoodi Bohra (GUJ) | 0.98 (0.12) | 0.02 (0.12) | ||||
| 1.001 (0.12) | −0.001 (0.12)a | |||||
| Iranian Shia | 0.22 (0.1) | 0.78 (0.1) | ||||
| 0.29 (0.1) | 0.71 (0.1) | |||||
| Mappla | 0.97 (0.06) | 0.03 (0.06) | ||||
| 0.96 (0.05) | 0.04 (0.05) | |||||
Values in parenthesis denote SDs.
Negative values indicate negligible contributions and suggest that the simple admixture model between the given sources is unlikely to be realistic to explain the genetic variation in the given sink population (personal communication, Giorgio Bertorelle).
Admixture contributions were estimated with local populations from both Tamil Nadu and Gujarat because these Muslims are recent migrants from Gujarat settled in Tamil Nadu.
Table 5. Y chromosome – admixture proportions.
| Local Indian closest neighbors (Parental) | ||||||
|---|---|---|---|---|---|---|
| Admixed | Uttar Pradesh | Gujarat | Andhra Pradesh | Tamil Nadu | Arabia (Parental) | Iran (Parental) |
| Indian Shia | 0.68 (0.07) | 0.32 (0.07) | ||||
| 0.50 (0.14) | 0.50 (0.14) | |||||
| Indian Sunni | 0.97 (0.06) | 0.03 (0.06) | ||||
| 0.99 (0.10) | 0.01 (0.10) | |||||
| Dawoodi Bohra (TN)a | 1.18 (0.11) | −0.18 (0.11)b | ||||
| 1.44 (0.26) | −0.44 (0.26)b | |||||
| Dawoodi Bohra (TN)a | 0.96 (0.09) | 0.04 (0.09) | ||||
| 0.94 (0.16) | 0.06 (0.16) | |||||
| Dawoodi Bohra (GUJ) | 0.93 (0.11) | 0.07 (0.11) | ||||
| 0.82 (0.23) | 0.18 (0.23) | |||||
| Iranian Shia | 0.66 (0.1) | 0.34 (0.1) | ||||
| 0.35 (0.13) | 0.65 (0.13) | |||||
| Mappla | 0.73 (0.11) | 0.27 (0.11) | ||||
| 0.71 (0.17) | 0.29 (0.17) | |||||
Values in parenthesis denote SDs.
Admixture contributions were estimated with local populations from both Tamil Nadu and Gujarat because these Muslims are recent migrants from Gujarat settled in Tamil Nadu.
Negative values indicate negligible contributions and suggest that the simple admixture model between the given sources is unlikely to be realistic to explain the genetic variation in the given sink population (personal communication, Giorgio Bertorelle).
Both the maternal and paternal admixture contributions from the closest Hindu parental populations to the respective Shia, Sunni, Dawoodi Bohra (GUJ), Dawoodi Bohra (TN) and Mappla Muslim populations seem to be the highest, with only a minimal contribution from either Iran or Arabia (Tables 4 and 5). The exception is the group of Iranian Shias who show major maternal (71%) and paternal (65%) contribution from Iranian populations (Tables 4 and 5). The sub-Saharan African- and Middle Eastern-specific lineages, such as L0a2a2 (mtDNA) and E1b1b1a (Y haplogroup), were observed among Dawoodi Bohra (TN) and Shia Muslim populations, with a frequency of 5 and 2%, respectively. These significant maternal and paternal lineages, atypical of Indian populations, can be attributed to the nominal Arabian and Iranian admixture contributions. The correlation between the admixture contributions from Arabia and Iran is positive, with significant correlation coefficient values, R2=0.982 for mtDNA and R2=0.939 for Y-chromosome biallelic markers, reflecting the similarity of the genetic composition of the two source pools and thus their poor power to distinguish between the admixture contributions from the two (Figures 6a, b, 7a and b).
Figure 6.
(a) The correlation coefficient between admixture contributions from Arabia and Iran to Indian Muslim populations based on mtDNA. (b) mtDNA – admixture proportions: correlation coefficient graph.
Figure 7.
(a) The correlation coefficient between admixture contributions from Arabia and Iran to Indian Muslim populations based on Y chromosome. (b) Y chromosome – admixture proportions: correlation coefficient graph.
Discussion
Historical evidence suggests that Indian Muslims could have originated in two distinct ways: (i) military invasions that led to the establishment of Muslim kingdoms and subsequent immigration of mercenaries, businessmen and political emissaries from Middle Eastern countries, Iran and Arabia, followed by admixture with the local population; and (ii) cultural diffusion as a result of absorption and dominance that resulted in a sizeable population embracing Islam.1, 2, 3 In a nutshell, Indian Muslims could be either the descendants of Iranian and Arabian men who married local Hindu women or the descendants of local converts. We therefore sought to examine contemporary Indian Muslim populations for the occurrence of Middle Eastern genetic signatures, expecting them to be manifested primarily in the male line. For this, we chose six Muslim populations from three different geographical regions of India (Figure 1) that witnessed several human migrations, military invasions from the Middle East and proselytizing of native Hindu populations.1, 2, 3 Despite reported marriages between Muslim males and Hindu females,2, 6 the expected higher Y-chromosomal contribution from the Middle East to contemporary Indian Muslims was not found in this study. Unlike Muslim communities in China and Central Asia,53, 54, 55 which show a marked presence of Western Y chromosomes, Indian Muslims derive most of their Y chromosomes from local neighboring non-Muslim populations, suggesting a regional genetic affinity among Indian Muslim and non-Muslim populations. This suggests that the expansion of Islam in India happened through religious conversions during the implementation of the Muslim faith. In comparison with Indian Muslims following the Shia faith, recent Muslim immigrants from Iran (see Supplementary Text 1 for population history) who also follow Shiism show a genetic proximity to Middle Eastern populations. This shows that this Muslim community maintains its native genetic pool with less genetic affinity to Indian populations. It is interesting that Dawoodi Bohras (TN) were found to exist as a separate genetic entity, with mtDNA lineages L0a2a2 (African specific) and B4ala1 (Polynesian specific), when compared with other Indian Muslim groups. The sub-Saharan African/Arabian mtDNA lineage L0a2a2 can be linked to historical information (Supplementary Text 1) that Dawoodi Bohras belong to a Shia sect of Islam that purportedly migrated to India from Yemen, an area which is known to have a considerable frequency (3%) of African mtDNA lineages, including haplogroup L0a2.56 An alternative interpretation is that L0a2a2 could have persisted in South Asia as the out-of-Africa migration is undermined by the young age estimate of L0a2a2 (Figure 2) and by the absence of this clade among Indian non-Muslim populations. The occurrence of the Polynesian mtDNA lineage B4a1a1 is in accordance with the oral history of the Dawoodi Bohras, which claims that some of their ancestors migrated to India from Thailand. Furthermore, detectable frequencies of other East Asian mtDNA haplogroups, F1a, F1b, F3b, MD, MD5a2 and MG2a, in some contemporary Indian Muslim groups are consistent with historically attested movements of Muslims from Central Asia and contacts with Southeast Asian Muslim communities.55, 57
The paternal haplogroups, E1b1b1a, G and J*(xJ2), frequent widely over Middle East and Arabia,12, 58 from where Islam was propagated, were found to occur at notable frequencies among some of the Indian Muslim groups. Although both maternal and paternal admixture estimates show maximal contribution from the local Indian non-Muslim parental populations, the contribution from Iranian and/or Arabian parental populations cannot be neglected (Tables 4 and 5). The wide spread of the LCT/MCM6 gene C/T−13910 variant among all Indian Muslim populations and the complete absence of the respective Arabian marker in this gene are consistent with gene flow occurring predominantly over Iran than over Arabia. Furthermore, these observations based on uniparental markers are congruent with our recent study on biparental STR markers,10 thus providing a comprehensive view of the genetic heritage of Indian Muslim populations.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgments
We are grateful to all the donors for providing blood samples. We thank Giorgio Bertorelle for his useful advice on the Admixture analysis, Qasim Ayub for comments, AG Reddy for technical support and Mustafa Fakruddin Viramgamwala for help during sample collection. ME, PRM and BD thank the Directorate of Forensic Science, Ministry of Home Affairs, Government of India for the fellowship. TK and KT were supported by the UKIERI Grant RG47772. CTS was supported by the Wellcome Trust.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Schimmel A. Islam in India and Pakistan. Leiden: Brill Academic Publishers; 1982. [Google Scholar]
- Robb P. A History of India. Hampshire: Palgrave Macmillan Publishers; 2002. [Google Scholar]
- Naqvi S. The Iranian Afaquies contributions to the Qutub Shahi and Adil Shahi Kingdoms. Hyderabad: Hussain Book Shop; 2003. [Google Scholar]
- Papiha SS. Genetic variation in India. Hum Biol. 1996;68:607–628. [PubMed] [Google Scholar]
- Lanchbury JS, Agarwal SS, Papiha SS. Genetic differentiation and population structure of some occupational caste groups in Uttar Pradesh, India. Hum Biol. 1996;68:655–678. [PubMed] [Google Scholar]
- Aarzoo SS, Afzal M. Gene diversity in some Muslim populations of North India. Hum Biol. 2005;77:343–353. doi: 10.1353/hub.2005.0046. [DOI] [PubMed] [Google Scholar]
- Balgir RS, Sharma JC. Genetic markers in the Hindu and Muslim Gujjars of Northwestern India. Am J Phys Anthropol. 1988;75:391–403. doi: 10.1002/ajpa.1330750310. [DOI] [PubMed] [Google Scholar]
- Terreros MC, Rowold D, Luis JR, Khan F, Agrawal S, Herrera RJ. North Indian Muslims: enclaves of foreign DNA or Hindu converts. Am J Phys Anthropol. 2007;133:1004–1012. doi: 10.1002/ajpa.20600. [DOI] [PubMed] [Google Scholar]
- Gutala R, Carvalho-Silva DR, Jin L, et al. A shared Y-chromosomal heritage between Muslims and Hindus in India. Hum Genet. 2006;120:543–551. doi: 10.1007/s00439-006-0234-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaaswarkhanth M, Dubey B, Meganathan PR, et al. Diverse genetic origin of Indian Muslims: evidence from autosomal STR loci. J Hum Genet. 2009;54:340–348. doi: 10.1038/jhg.2009.38. [DOI] [PubMed] [Google Scholar]
- Abu-Amero KK, González AM, Larruga JM, Bosley TM, Cabrera VM. Eurasian and African mitochondrial DNA influences in the Saudi Arabian population. BMC Evol Biol. 2007;7:32. doi: 10.1186/1471-2148-7-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadenas AM, Zhivotovsky LA, Cavalli-Sforza LL, Underhill PA, Herrera RJ. Y-chromosome diversity characterizes the Gulf of Oman. Eur J Hum Genet. 2008;16:374–386. doi: 10.1038/sj.ejhg.5201934. [DOI] [PubMed] [Google Scholar]
- Quintana-Murci L, Chaix R, Wells RS, et al. Where West meets East: the complex mtDNA landscape of the Southwest and Central Asian corridor. Am J Hum Genet. 2004;74:827–845. doi: 10.1086/383236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regueiro M, Cadenas AM, Gayden T, Underhill PA, Herrera RJ. Iran: tricontinental nexus for Y-chromosome driven migration. Hum Hered. 2006;61:132–143. doi: 10.1159/000093774. [DOI] [PubMed] [Google Scholar]
- Enattah NS, Jensen TG, Nielsen M, et al. Independent introduction of lactase-persistence alleles into human populations reflects different history of adaptation to milk culture. Am J Hum Genet. 2008;82:57–72. doi: 10.1016/j.ajhg.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enattah NS, Trudeau A, Pimenoff V, et al. Evidence for still ongoing convergence evolution of the lactase persistence T-13910 alleles in humans. Am J Hum Genet. 2007;81:615–625. doi: 10.1086/520705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metspalu M, Kivisild T, Metspalu E, et al. Most of the extant mtDNA boundaries in South and Southwest Asia were likely shaped during the initial settlement of Eurasia by anatomically modern humans. BMC Genet. 2004;5:26. doi: 10.1186/1471-2156-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordaux R, Saha N, Bentley GR, Aunger R, Sirajuddin S, Stoneking M. Mitochondrial DNA analysis reveals diverse histories of tribal populations from India. Eur J Hum Genet. 2003;11:253–264. doi: 10.1038/sj.ejhg.5200949. [DOI] [PubMed] [Google Scholar]
- Kivisild T, Bamshad MJ, Kaldma K, et al. Deep common ancestry of Indian and Western-Eurasian mitochondrial DNA lineages. Curr Biol. 1999;9:1331–1334. doi: 10.1016/s0960-9822(00)80057-3. [DOI] [PubMed] [Google Scholar]
- Abu-Amero KK, Larruga JM, Cabrera VM, González AM. Mitochondrial DNA structure in the Arabian Peninsula. BMC Evol Biol. 2008;8:45. doi: 10.1186/1471-2148-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton T, Clifford S, Martinson J, Batzer M, Stoneking M. Genetic evidence for the proto-Austronesian homeland in Asia: mtDNA and nuclear DNA variation in Taiwanese aboriginal tribes. Am J Hum Genet. 1998;63:1807–1823. doi: 10.1086/302131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao YG, Nie L, Harpending H, Fu YX, Yuan ZG, Zhang YP. Genetic relationship of Chinese ethnic populations revealed by mtDNA sequence diversity. Am J Phys Anthropol. 2002a;118:63–76. doi: 10.1002/ajpa.10052. [DOI] [PubMed] [Google Scholar]
- Oota H, Settheetham-Ishida W, Tiwawech D, Ishida T, Stoneking M. Human mtDNA and Y-chromosome variation is correlated with matrilocal versus patrilocal residence. Nat Genet. 2001;29:20–21. doi: 10.1038/ng711. [DOI] [PubMed] [Google Scholar]
- Richard C, Pennarun E, Kivisild T, et al. An mtDNA perspective of French genetic variation. Ann Hum Biol. 2007;34:68–79. doi: 10.1080/03014460601076098. [DOI] [PubMed] [Google Scholar]
- Sahoo S, Singh A, Himabindu G, et al. A prehistory of Indian Y chromosomes: evaluating demic diffusion scenarios. Proc Natl Acad Sci USA. 2006;103:843–848. doi: 10.1073/pnas.0507714103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Zhivotovsky LA, King R, et al. Polarity and temporality of high resolution Y-chromosome distributions in India identify both indigenous and exogenous expansions and reveal minor genetic influence of central Asian pastoralists. Am J Hum Genet. 2006;78:202–221. doi: 10.1086/499411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanseem I, Thangaraj K, Chaubey G, et al. Genetic affinities among the lower castes and tribal groups of India: inference from Y chromosome and mitochondrial DNA. BMC Genet. 2006;7:42. doi: 10.1186/1471-2156-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells RS, Yuldasheva N, Ruzibakiev R, et al. The Eurasian heartland: a continental perspective on Y-chromosome diversity. Proc Natl Acad Sci USA. 2001;98:10244–10249. doi: 10.1073/pnas.171305098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramana GV, Su B, Jin L, et al. Y-chromosome SNP haplotypes suggest evidence of gene flow among caste, tribe, and the migrant Siddi populations of Andhra Pradesh, South India. Eur J Hum Genet. 2001;9:695–700. doi: 10.1038/sj.ejhg.5200708. [DOI] [PubMed] [Google Scholar]
- Kivisild T, Rootsi S, Metspalu M, et al. The genetic heritage of the earliest settlers persists both in Indian tribal and caste populations. Am J Hum Genet. 2003;72:313–332. doi: 10.1086/346068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Zahery N, Semino O, Benuzzi G, et al. Y-chromosome and mtDNA polymorphisms in Iraq, a crossroad of the early human dispersal and of post-Neolithic migrations. Mol Phylogenet Evol. 2003;28:458–472. doi: 10.1016/s1055-7903(03)00039-3. [DOI] [PubMed] [Google Scholar]
- Karafet TM, Osipova LP, Gubina MA, Posukh OL, Zegura SL, Hammer MF. High levels of Y-chromosome differentiation among native Siberian populations and the genetic signature of a boreal hunter-gatherer way of life. Hum Biol. 2002;74:761–789. doi: 10.1353/hub.2003.0006. [DOI] [PubMed] [Google Scholar]
- Xue Y, Zerjal T, Bao W, et al. Male demography in East Asia: a north-south contrast in human population expansion times. Genetics. 2006;172:2431–2439. doi: 10.1534/genetics.105.054270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semino O, Passarino G, Oefner PJ, et al. The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: a Y chromosome perspective. Science. 2000;290:1155–1159. doi: 10.1126/science.290.5494.1155. [DOI] [PubMed] [Google Scholar]
- Thangaraj K, Chaubey G, Singh VK, et al. In situ origin of deep rooting lineages of mitochondrial Macrohaplogroup ‘M' in India. BMC Genomics. 2006;7:151. doi: 10.1186/1471-2164-7-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong QP, Bandelt HJ, Sun C, et al. Updating the East Asian mtDNA phylogeny: a prerequisite for the identification of pathogenic mutations. Hum Mol Genet. 2006;15:2076–2086. doi: 10.1093/hmg/ddl130. [DOI] [PubMed] [Google Scholar]
- Sun C, Kong QP, Palanichamy MG, et al. The dazzling array of basal branches in the mtDNA macrohaplogroup M from India as inferred from complete genomes. Mol Biol Evol. 2006;23:683–690. doi: 10.1093/molbev/msj078. [DOI] [PubMed] [Google Scholar]
- Palanichamy MG, Sun C, Agrawal S, et al. Phylogeny of mtDNA macrohaplogroup N in India, based on complete sequencing: implications for the peopling of South Asia. Am J Hum Genet. 2004;75:966–978. doi: 10.1086/425871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behar DM, Villems R, Soodyall H, et al. The dawn of human matrilineal diversity. Am J Hum Genet. 2008;82:1130–1140. doi: 10.1016/j.ajhg.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder MJ, Taylor SL, Tobe VO, Nickerson DA. Automating the identification of DNA variations using quality-based fluorescence re-sequencing: analysis of the human mitochondrial genome. Nucleic Acids Res. 1998;26:967–973. doi: 10.1093/nar/26.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- Ingram CJ, Elamin MF, Mulcare CA, et al. A novel polymorphism associated with lactose tolerance in Africa: multiple causes for lactase persistence. Hum Genet. 2007;120:779–788. doi: 10.1007/s00439-006-0291-1. [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Sykes BC, Richards MB. Mitochondrial portraits of human populations using median networks. Genetics. 1995;141:743–753. doi: 10.1093/genetics/141.2.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Dupanloup I, Bertorelle G. Inferring admixture proportions from molecular data: extension to any number of parental populations. Mol Biol Evol. 2001;18:672–675. doi: 10.1093/oxfordjournals.molbev.a003847. [DOI] [PubMed] [Google Scholar]
- Soares P, Ermini L, Thomson N, et al. Correcting for purifying selection: an improved human mitochondrial molecular clock. Am J Hum Genet. 2009;84:740–759. doi: 10.1016/j.ajhg.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivisild T, Shen P, Wall DP, et al. The role of selection in the evolution of human mitochondrial genomes. Genetics. 2006;172:373–387. doi: 10.1534/genetics.105.043901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach WL, Services KC. MVSP – a multi-variate statistical package for Windows ver 3.1, 2004
- Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Fornarino S, Pala M, Battaglia V, et al. Mitochondrial and Y-chromosome diversity of the Tharus (Nepal): a reservoir of genetic variation. BMC Evol Biol. 2009;9:154. doi: 10.1186/1471-2148-9-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karafet TM, Mendez FL, Meilerman MB, Underhill PA, Zegura SL, Hammer MF. New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res. 2008;18:830–838. doi: 10.1101/gr.7172008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobling MA, Tyler-Smith C. The human Y chromosome: an evolutionary marker comes of age. Nat Rev Genet. 2003;4:598–612. doi: 10.1038/nrg1124. [DOI] [PubMed] [Google Scholar]
- Wang W, Wise C, Baric T, Black ML, Bittles AH. The origins and genetic structure of three co-resident Chinese Muslim populations: the Salar, Bo'an and Dongxiang. Hum Genet. 2003;113:244–252. doi: 10.1007/s00439-003-0948-y. [DOI] [PubMed] [Google Scholar]
- Zerjal T, Wells RS, Yuldasheva N, Ruzibakiev R, Tyler-Smith C. A genetic landscape reshaped by recent events: Y-chromosomal insights into central Asia. Am J Hum Genet. 2002;71:466–482. doi: 10.1086/342096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comas D, Calafell F, Mateu E, et al. Trading genes along the Silk Road: mtDNA sequences and the origin of Central Asian populations. Am J Hum Genet. 1998;63:1824–1838. doi: 10.1086/302133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivisild T, Reidla M, Metspalu E, et al. Ethiopian mitochondrial DNA heritage: tracking gene flow across and around the gate of tears. Am J Hum Genet. 2004;75:752–770. doi: 10.1086/425161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao YG, Lü XM, Luo HR, Li WH, Zhang YP. Gene admixture in the Silk Road region of China: evidence from mtDNA and melanocortin 1 receptor polymorphism. Genes Genet Syst. 2000a;75:173–178. doi: 10.1266/ggs.75.173. [DOI] [PubMed] [Google Scholar]
- Zalloua PA, Xue Y, Khalife J, et al. Y-chromosomal diversity in Lebanon is structured by recent historical events. Am J Hum Genet. 2008;82:873–882. doi: 10.1016/j.ajhg.2008.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.