Abstract
Human hepatocellular carcinoma (HCC) is one of the commonest causes of mortality among solid organ malignancies. The incidence of HCC in the United States is rising. Few proteomic biomarker studies have been done in U.S. populations. Tumor and nonmalignant tissue from three American patients with hepatitis and non-hepatitis-associated HCC were analyzed to find common differences in protein expression. Proteins were separated by 2D electrophoresis (isoelectric focusing followed by 10% SDS-PAGE). Gels were fixed and then stained with Coomassie brilliant blue. Digitization and processing were performed using the PDQuest software. The Student’s t-test was used to detect quantitative protein changes between tumor and nonmalignant liver consistent in all sample pairs with a cutoff made at P < 0.01. This yielded a total of 19 spots with significant (>2 fold) abundance changes. Matrix-assisted laser desorption ionization mass spectrometry analysis was performed using Waters Micomass M@LDI SYSTEM. The proteins were then identified using manual ProFound. Among the 19 spots, 9 showed overexpression and 10 showed underexpression in tumor. Overexpressed proteins included beta-5-tubulin, beta-actin, vimentin, carbamoyl-phosphate synthetase-1, methylenetetrahydrofolate dehydrogenase, serum albumin, catalase, autoimmune regulator, and transcription factor ets. Underexpressed proteins included BiP protein, A-kinase anchoring protein 18 gamma, inorganic pyrophosphatase, keratin 8, repulsive guidance molecule, butyrophilin, superoxide dismutase, TSA, heat-shock 70-kDa protein 9B, and hemoglobin alpha-2. Of particular interest, the protein autoimmune regulator was expressed 14-fold higher in tumor tissue, suggesting it may have a role in HCC. Validation and further investigation of these protein changes may lead to the discovery of new molecular targets for therapy, biomarkers for early detection, and new endpoints for therapeutic efficacy and toxicity.
Keywords: hepatocellular carcinoma, liver, proteomics, cirrhosis
INTRODUCTION
Hepatocellular carcinoma (HCC) is one of the most common causes of mortality from solid organ malignancy in the world, accounting for almost 600,000 deaths a year [1–5]. In the United States the age-adjusted incidence rates have doubled over the past two decades, while prognosis remains poor [2, 3]. Although small improvements have been made in treatment, the median survival rate remains 8 months [2, 3]. At present, the most successful treatment is surgical resection and/or transplantation, which can only be performed in a very select group of patients with earlystage disease [6]. Local-ablative treatment advances in chemoembolization, radiofrequency ablation, and percutaneous ethanol injection have also demonstrated some effectiveness in selected patients. Most patients present with advanced stage disease not amenable to surgical or local-ablative treatment and thus have very limited treatment options outside of clinical trials. Systemic chemotherapy is ineffective [6, 7].
It is estimated that 8500 to 11,500 new cases of HCC occur annually in the United States, with 19,160 new cases projected in the U.S. in 2007 [2, 3, 5]. Although the occurrence of HCC has actually decreased in some high-risk countries, it has increased markedly in low-risk countries such as the United States in recent years [8–10]. Unlike some cancers, for HCC there are well-recognized environmental risk factors. Risk factors of HCC that are relatively common include hepatitis C virus, hepatitis B virus, and heavy alcohol consumption. Other etiologic factors include aflatoxin, oral contraceptives, fluorinated hydrocarbons, tobacco, genetic metabolic enzyme deficiencies (hemochromatosis, α-1-anti-trypsin deficiency, and Wilson’s disease), bile stasis, obesity, and Type 2 diabetes. Interestingly, however, epidemiological studies in the United States of HCC often fail to identify specific risk factors in a large proportion of patients. Between 15 and 50% of patients with HCC in the United States have no readily explicable etiologic factor to account for their disease [11].
Two-dimensional electrophoresis (2-DE), a technique that enables resolution and quantification of hundreds of proteins simultaneously, has stimulated enthusiasm for the identification of proteins expressed in complex disease states. Specifically, this offers new opportunities to characterize complex patterns of tissue protein expression expressed in cancer tissue compared to nonneoplastic tissue. These protein pattern profiles may potentially indicate profiles or signatures of specific host/organ responses to and/or genetic predisposition to cancer [12, 13].
Traditionally, proteins on gels have been visualized as spots stained by dyes and labeled with markers by immunoblotting and by chemical methods (amino acid analysis and Edman sequencing). These techniques are time-consuming, labor-intensive, and expensive and practically only a few proteins can be analyzed within each gel. Mass spectrometry combined with software and automation techniques (MALDI-MS or ESI-MS) has arrived as an important tool to identify and analyze virtually all proteins on a gel [14]. Proteomic tools can be used to discover new molecular targets for therapy, biomarkers for early detection, and new endpoints for therapeutic efficacy and toxicity [15].
Few proteomic biomarker studies have been done in U.S. populations. In this pilot study we sought to examine HCC in patients born and living in the United States with different etiopathologies of HCC. We investigated the protein expression patterns in cancer and nonmalignant adjacent liver in patients with HCC using 2-DE and MALDI-MS techniques. Our rationale was (1) to determine whether there may be common protein profile changes that were present in these patients with HCC regardless of etiopathology, and (2) to compare our results to existing proteomic studies in the literature in predominantly Asian populations of patients.
METHODS
Tissue Procurement
HCC and nonmalignant adjacent liver tissues were derived from patients undergoing surgery at Indiana University Hospital. HCC and nonmalignant adjacent liver tissues were confirmed grossly and histologically to be consistent with HCC and normal liver, respectively. Tissues were snap-frozen in liquid nitrogen and stored at −80°C until sample preparation as detailed below.
Sample Preparation
Fresh liver samples (n = 6) were weighed, and 250 mg pieces were placed in each of six 50 mL beakers, along with eight volumes of a solution containing 9 M urea, 4% Igepal CA-630 ([octylphenoxy] polyethoxyethanol), 1% DTT, and 0.2% carrier ampholytes (pH 3–10), and thoroughly minced with surgical scissors. The minced samples were then placed in 3 mL DUALL® (Kimble/Kontes, Vineland, NJ) ground-glass tissue grinders and manually homogenized. After complete solubilization at room temperature for 120 min, samples were centrifuged at 100,000 × g for 30 min using a Beckman (Fullerton, CA) TL-100 ultracentrifuge to remove nucleic acid and insoluble materials, and the supernatants were stored at −45°C until 2-DE separation. Protein concentration was determined using amido black 10B [16], an approach that enables the sensitive and accurate assay of solubilized proteins to be performed without interference from constituents of the lysis buffer.
Two-Dimensional Gel Electrophoresis (2DGE) and Image Analysis
Using overnight, passive rehydration at room temperature, 200 µg protein was loaded onto IPG strips (24 cm, nonlinear pH 3–10; Bio-Rad, Hercules, CA). Isoelectric focusing was performed simultaneously on all six IPG strips using a Protean IEF Cell (Bio-Rad) (12 strip capacity), by a program of progressively increasing voltage (150 V for 2 h, 300 V for 4 h, 1500 V for 1 h, 5000 V for 5 h, 7000 V for 6 h, and 10,000 V for 3 h) for a total of 100,000 V/h. A computer-controlled gradient casting system was used to prepare second-dimension SDS gradient slab gels (20 × 25 × 0.15 cm) in which the acrylamide concentration varied linearly from 11 to 17% T. First-dimension IPG strips were loaded directly onto the slab gels following equilibration for 10 min in Equilibration Buffer I and 10 min in Equilibration Buffer II (Equilibration Buffer I: 6 M urea, 2% SDS, 0.375 M Tris-HCl pH 8.8, 20% glycerol, 130 mM DTT; Equilibration Buffer II: 6 M urea, 2% SDS, 0.375 M Tris-HCl pH 8.8, 20% Glycerol, 135 mM iodoacetamide). All 20 second-dimension slab gels were run in parallel at 8°C for 18 h at 160 V and subsequently fixed and stained using a sensitive colloidal Coomassie blue G-250 procedure [17]. After 96 h, gels were washed several times with water and scanned at 95.3 µm/pixel resolution using a GS-800 Calibrated Imaging Densitometer (Bio-Rad). The resulting 12-bit images were analyzed using PDQuest™ software (Bio-Rad, v.7.1). Background was subtracted and protein spot density peaks were detected and counted. Because total spot counts and the total absorbance are directly related to the total protein concentration, individual protein quantities were thus expressed as parts per million of the total integrated absorbance, after normalization against total image density. A reference pattern was constructed and each of the six gels in the match set was matched to the reference gel. Numerous proteins that were uniformly expressed in all patterns were used as landmarks to facilitate rapid gel matching. Raw quantitative data for each protein spot were statistically analyzed and intergroup fold differences were calculated within PDQuest™.
Peptide Mass Fingerprinting
Protein spots were excised manually from the gels and processed automatically using the multifunctional MultiProbe II Station robot (Perkin-Elmer, Concord, ON). The protein spots were de-stained, reduced with dithiothreitol, alkylated with iodoacetamide, and tryptically digested using Promega sequence grade, modified trypsin in preparation for matrix-assisted laser desorption ionization mass spectrometry (MALDI-TOF MS) of the resulting peptides. The tryptic peptides were eluted and manually spotted on the sample target along with α-cyano-4-hydroxycinnamic acid matrix. The target was then analyzed directly using the prOTOF™ 2000 MALDI Orthogonal Time of Flight Mass Spectrometer (Perkin-Elmer/SCIEX) using TOF Works™ software (Perkin-Elmer) for automated batch database searches of the NCBI protein sequence database. Accuracy of monoisotopic peptide mass measurements ranged between 5 and 15 ppm, resulting in high confidence protein identifications. Alternatively, some peptide mass spectra were submitted for online interrogation of the ProFound™ Peptide Mass Database (Proteometrics, LLC, New York, NY). Protein identity was deemed acceptably robust, when the TOF Works™ expectation probability was <0.01 or the Profound™ Z-score exceeded 1.30, corresponding to the 90th percentile.
Assurances
This study protocol conforms to the ethical guidelines of the “World Medical Association Declaration of Helsinki—Ethical Principles for Medical Research Involving Human Subjects” adopted by the 18th WMA General Assembly, Helsinki, Finland, June 1964, as revised in Tokyo 2004, as reflected in approval by the Indiana University/Purdue University at Indianapolis Institutional Review Board. All patients enrolled in this study gave written informed consent.
RESULTS
Demographics
HCC tissues were derived from patients born and living in the United States with different etiopathologies of HCC. Notably, two patients had non-hepatitis etiologies of HCC without cirrhosis (Table 1).
TABLE 1.
Patient Characteristics
| Age | Gender | Race | Size (cm) | Cirrhosis | Hepatitis | Other risk factors | Survival (mo) |
|---|---|---|---|---|---|---|---|
| 71 | M | Caucasian | 18 | Negative | Negative | Alcohol, DM, | 4 |
| 51 | M | African-American | 8.4 | Negative | Negative | NASH, Obesity | 28 |
| 61 | F | Asian (US born) | 7.7 (m) | Positive | B | Smoking | <1 |
Note. M = male; F = female; DM = diabetes mellitus; NASH = nonalcoholic steatohepatosis; m = multiple.
2DGE of HCC and Adjacent Liver
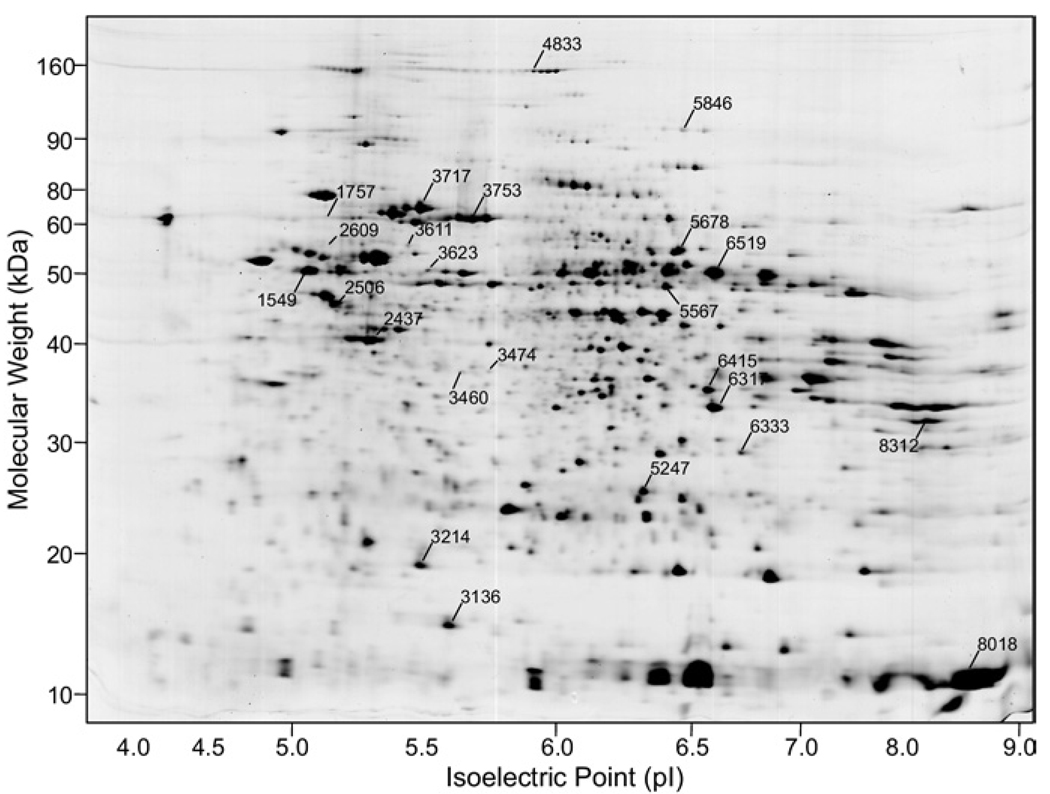
2D proteins isolated from liver sample homogenates were separated via 2-DE and a representative image of the resulting stained gel pattern is shown in Fig. 1. An average of 1693 proteins were resolved in the sample patterns and matched to a reference pattern, and their integrated spot densities (abundances) were compared as described above.
FIG. 1.
Representative Coomassie blue-stained 2-DE gel pattern of human liver homogenates of the six analyzed in this study. Numbered spots indicate proteins that were differentially expressed in nonmalignant versus tumor samples. These were tryptically digested yielding masses subjected to MALDI-MS for submission to a peptide mass database for protein identification. Molecular weight and pI calibrations are estimates based on the calculated MW and pI of protein spots identified in the pattern. Mean protein abundances for the numbered proteins are illustrated graphically in Fig. 2 and additional information is listed in Table 1.
Peptide Mass Fingerprinting of HCC and Adjacent Liver
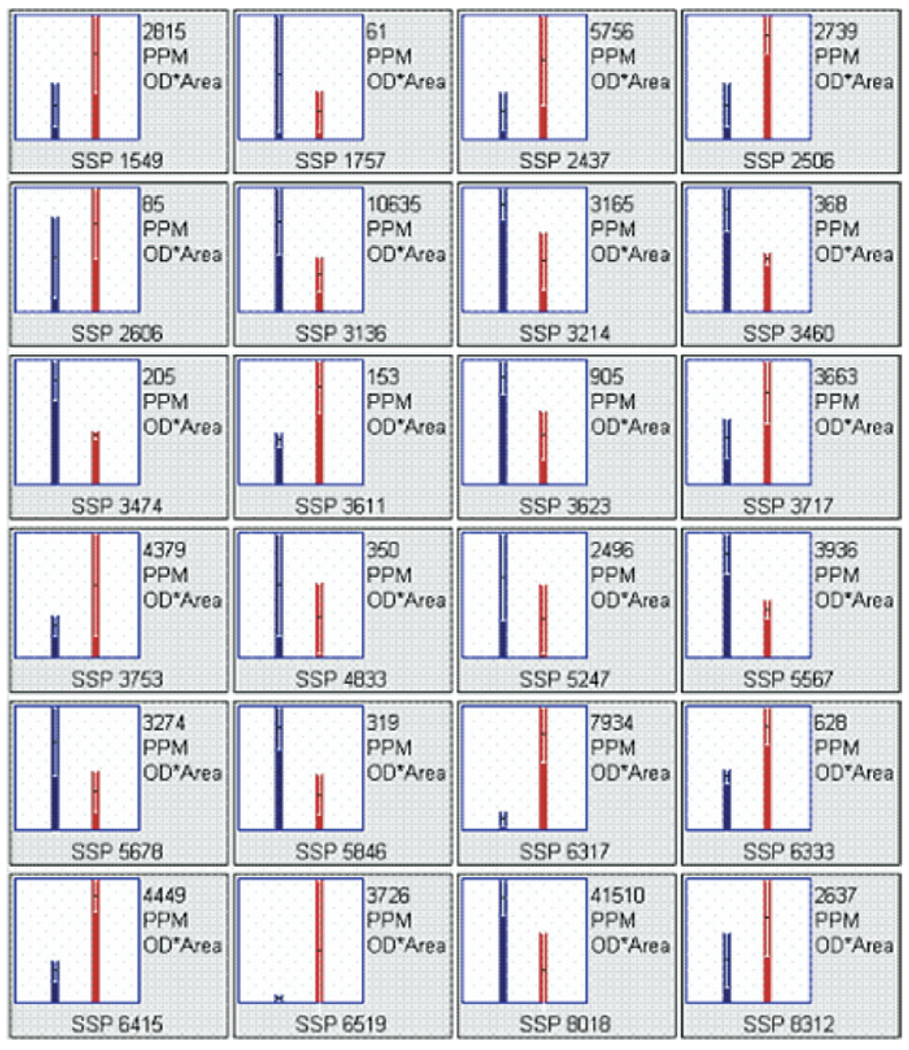
Nineteen proteins, whose expression differed significantly between nonmalignant and tumor samples, had fold differences >2 and/or met an abundance threshold necessary for peptide mass fingerprint identification (>50 ppm), were identified. These proteins (in addition to five others with near significant differences) are illustrated by spot number (SSP) in Fig. 1 and listed in Table 2. Abundance differences are shown graphically in Fig. 2. Overexpressed proteins included beta-5-tubulin, beta-actin, vimentin, carbamoyl-phosphate synthetase-1, methylenetetrahydrofolate dehydrogenase, serum albumin, catalase, AIRE, and transcription factor ets. Underexpressed proteins included BiP protein, A-kinase anchoring protein 18 gamma, inorganic pyrophosphatase, keratin 8, repulsive guidance molecule, butyrophilin, superoxide dismutase, TSA, heat-shock 70-kDa protein 9B, and hemoglobin alpha-2. Of particular interest, the protein autoimmune regulator (AIRE) was expressed 14-fold higher in tumor tissue, suggesting it may have an important role in HCC.
Table 2.
Differentially Expressed Proteins From Human Liver Samples (normal vs tumor)
| SSP | Protein name | IPI # | Tumor | Fold (N/T) | pI | kDa | ID Prob | Z-score | % cov |
|---|---|---|---|---|---|---|---|---|---|
| 1757 | BiP protein (grp78; HSPA5) (fragment) | IPI00003362 | < | −2.3 | 5.2 | 71.03 | 0.001 | 2.43 | 43 |
| 3136 | superoxide dismutase, manganese, chain A | IPI00847322 | < | −2.5 | 5.7 | 16.01 | 0.001 | 2.43 | 37 |
| 3214 | peroxiredoxin-2 (Thioredoxin peroxidase 1) | IPI00027350 | < | −2.1 | 5.2 | 18.48 | 0.001 | 1.66 | 24 |
| 3460 | A-kinase anchoring protein 18 gamma | IPI00422254 | < | −1.9 | 5.7 | 37.10 | 0.001 | 2.43 | 24 |
| 3474 | pyrophosphatase (inorganic) | IPI00015018 | < | −2.1 | 5.5 | 33.10 | 0.001 | 2.43 | 10 |
| 3623 | keratin 8, type II cytoskeletal | IPI00554648 | < | −2.2 | 5.4 | 53.73 | 0.001 | 2.43 | 19 |
| 4833 | carbamoyl-phosphate synthetase 1 (charge variant) |
IPI00011062 | < | −1.8 | 6.3 | 166.01 | 0.001 | 2.1 | 12 |
| 5247 | carbonic anhydrase I | IPI00215983 | < | −2.1 | 6.6 | 28.79 | 0.001 | 2.43 | 20 |
| 5567 | repulsive guidance molecule A precursor (RGM domain family member A) |
IPI00015049 | < | −2.1 | 6.7 | 48.42 | 0.001 | 2.43 | 8 |
| 5678 | catalase | IPI00465436 | < | −2.3 | 6.9 | 59.97 | 0.001 | 2.43 | 19 |
| 5846 | C-1-tetrahydrofolate synthase, cytoplasmic (C1-THF synthase) |
IPI00218342 | < | −2.9 | 6.8 | 102.20 | 0.001 | 2.43 | 24 |
| 8018 | hemoglobin, alpha 2 | IPI00410714 | < | −2.2 | 8.7 | 15.30 | 0.001 | 1.57 | 51 |
| 1549 | beta 5 tubulin | IPI00398982 | > | +2.5 | 5.0 | 53.75 | 0.001 | 2.43 | 33 |
| 2437 | actin, beta | IPI00021439 | > | +2.5 | 5.3 | 42.06 | 0.001 | 2.43 | 24 |
| 2506 | vimentin | IPI00418471 | > | +3.1 | 5.1 | 53.72 | 0.001 | 2.43 | 39 |
| 2609 | plectin | IPI00014898 | > | +1.6 | 5.4 | 64.94 | 0.006 | 1.32 | 22 |
| 3611 | hypermethylated in cancer 2 protein (Hic-2) (Hic-3) |
IPI00376570 | > | +2.2 | 5.9 | 60.33 | 0.001 | 1.92 | 6 |
| 3717 | heat shock 70kD protein 9B (grp75; mortalin-2) | IPI00007765 | > | +2.0 | 6.0 | 73.99 | 0.001 | 2.43 | 16 |
| 3753 | serum albumin precursor | IPI00745872 | > | +2.3 | 6.1 | 71.34 | 0.001 | 2.43 | 49 |
| 6317 | 39S ribosomal protein L45, mitochondrial precursor (L45mt) (MRP-L45) |
IPI00185859 | > | +8.8 | 7.9 | 29.05 | 0.01 | 0.78 | 16 |
| 6333 | B-cell receptor-associated protein 31 (6C6-AG tumor-associated antigen) |
IPI00218200 | > | +1.9 | 7.7 | 28.05 | 0.005 | 1.32 | 12 |
| 6415 | butyrophilin | IPI00384734 | > | +3.4 | 7.1 | 33.84 | 0.001 | 1.38 | 11 |
| 6519 | AIRE (autoimmune regulator) | IPI00853285 | > | +14.0 | 7.2 | 55.38 | 0.001 | 2.43 | 11 |
| 8312 | transcription factor ETV7 (TEL-2) | IPI00001714 | > | +2.0 | 8.3 | 38.99 | 0.001 | 2.43 | 24 |
Note. SSP refers to protein spot number from image analysis software PDQuest™; IPI# is the International Protein Index entry (http://www.ebi.ac.uk/IPI/); pI is protein isoelectric point, < is less in tumor vs normal liver, > is greater in tumor vs normal liver; kDa is protein molecular weight; ID Prob and Z-score are derived from the Profound peptide mass database search result and represent robustness of the identification; and % cov refers to the percent of the proteins total amino acid sequence covered by the measure tryptic peptides.
FIG. 2.
Screen capture of protein spot integrated density graphs generated by gel imaging software PDQuest™. Bars represent mean protein abundance ± SD, with nonmalignant on the left and tumor on the right in each frame. Actual mean abundance values (ppm) appear in the top right corner of each frame and the protein spot number (SSP) appears at the bottom of each frame. Protein name and other pertinent data related to those shown here can be found in Table 1.
Protein Classification, Confirmation, and Validation
Identified proteins are classified according to function in Table 3. Several proteins identified in our pilot study have also recently been identified by other investigators (see references in Table 3). Likewise, several proteins have not been previously identified by other investigators as associated with HCC. HCC tissue proteomics has not commonly been studied in U.S. populations, so literature cited is predominantly from Asian investigators outside of the U.S. Table 3 also shows the hepatitis association (hepatitis B or C), direction of the protein difference, mass spectroscopic confirmation according to proteomic technique used, and validation by Western blot or immunohistochemistry according to the recent literature [18–24].
TABLE 3.
Protein Classification and Literature Confirmation
| Literature |
||||||
|---|---|---|---|---|---|---|
| Class | Protein name | T | Hep | MS confirmation | Valid | Ref |
| Acid base balance | Carbonic anhydrase I | < | B | T; LC; MALDI | 20, 21 | |
| Amino acid metab. | Carbamoyl-phosphate synthetase 1 (charge variant) |
< | C | LC; MALDI | 22 | |
| Amino acid metab. | C-1-tetrahydrofolate synthase, cytoplasmic (C1-THF synthase) |
< | T; MALDI | 21 | ||
| Calcium regulation | A-kinase anchoring protein 18 gamma | < | ||||
| Chaperone | BiP protein (grp78; HSPA5) (fragment) | >a | T | WB, IHC | 23 | |
| Chaperone | Heat shock 70-kD protein 9B (grp75; mortalin-2) | > | B | LC; MALDI | 20 | |
| Cytoskeleton | beta 5 tubulin | > | LC | 19 | ||
| Cytoskeleton | Actin, beta | > | T | 19 | ||
| Cytoskeleton | Vimentin | <a | B | LC; MALDI | 20 | |
| Cytoskeleton | Plectin | <a | IHC | 24 | ||
| Cytoskeleton | Keratin 8, type II cytoskeletal | < | C | LC; MALDI | 20 | |
| Detox, redox | Superoxide dismutase, manganese, chain A | < | B | LC; MALDI | 20 | |
| Detox, redox | Peroxiredoxin-2 (Thioredoxin peroxidase 1) | >a | C | LC; MALDI | 19, 22 | |
| Detox, redox | Catalase | < | T; MALDI | 21 | ||
| Immunity | Butyrophilin | > | ||||
| Immunity | AIRE (autoimmune regulator) | > | ||||
| Mitochondrial protein synthesis |
39S-ribosomal protein L45, mitochondrial precursor (L45 mt) (MRP-L45) |
> | ||||
| Neuro-axonal guidance |
Repulsive guidance molecule A precursor (RGM domain family) |
< | ||||
| Oxygen transport | Hemoglobin, alpha 2 | < | ||||
| Phosphate regulation | Pyrophosphatase (inorganic) | < | ||||
| Protein transport | B-cell receptor-associated protein 31 (6C6-AG tumor-associated antigen) |
> | ||||
| Protein transport | Serum albumin precursor | > | ||||
| Transcription Factor | Transcription factor ETV7 (TEL-2) | > | ||||
| Unknown (tumor suppressor?) |
Hypermethylated in cancer 2 protein (Hic-2) (Hic-3) |
> | ||||
Note. < = less in tumor versus normal; > = greater in tumor versus normal; metab. = metabolism; MS = mass spectroscopy; T = tandem MS/MS; LC = LC-MS/MS; MALDI = MALDI-ToF/MS; T = tumor; Hep = hepatitis; Valid = validation; Ref = references.
Opposing direction compared to current study.
DISCUSSION
HCC is a deadly cancer that is on the rise in the U.S. The treatment options for HCC are suboptimal. Multiple environmental risk factors, which lead to HCC development, complicate discovery of viable treatments, but there may be a common HCC etiology that links them. Proteomic studies have recently been used outside of the U.S. to help promote discovery of diagnostic, prognostic, and therapeutic biomarkers. In our pilot study, we sought to determine shared protein profile changes in HCC from American patients with various HCC risk factors. Our study demonstrated multiple proteins with known associations previously identified by investigators in predominantly Asian populations of patients. A few of these proteins had differences that were opposite that seen in our pilot study. This may be a reflection of our different population of patients or small sample size (due simply to genetic variability) or may indicate that they are not as relevant to HCC. Increasing our sample size and subsequent validation of our samples will help to determine the reasons for these discrepancies.
In addition, our study found HCC protein association not previously characterized in the literature. This may be a reflection of our different population of patients and could represent an opportunity to discover novel HCC associations. Alternatively, even though some of these proteins are not mentioned in the Asian literature, it is certainly possible that they may have been detected or discovered and simply not reported.
Some of the previously unreported molecules detected in this study could theoretically have significant roles in HCC tumorigenesis. For example, butyrophilin, up-regulated in HCC, has been shown to be up-regulated in the tumor microenvironment where it may decrease T-cell immune activity, assisting the tumor’s ability to evade immune detection and may thus serve as a potential therapeutic molecular target [25, 26]. The expression of AIRE (autoimmune regulator) has also been shown in the tumor microenvironment to be altered and appears to play a role in negative selection of autoreactive lymphocytes [27]. Repulsive guidance molecule A, down-regulated in HCC, has been shown to be a pro-apoptotic in embryonic systems, suggesting that it may have a role in suppressing tumorigenicity [28]. Similarly, Bap31 has been shown to regulate apoptosis [29]. Transcription factor ETV7 (Tel-2), up-regulated in HCC, may promote chromosomal translocations in human cancers [30]. Moreover, TEL-FGFR3 transformants have been shown to lead to leukemia in syngeneic mice [31]. Finally, hypermethylated in cancer (Hic) protein isoforms, e.g., Hic-1, have recently been shown to be up-regulated in hepatitis and HCC, but the Hic-3 isoform has not previously been reported in association with HCC or other cancers [32].
Validation and further investigation of these protein changes may lead to the discovery of new molecular targets for therapy, biomarkers for early detection, and new endpoints for therapeutic efficacy and toxicity.
The proteins that we have identified will be organized into a protein database that integrates the spots on the 2-DE map as well as the MS data and the table of identified proteins. The literature searches conducted on the identified proteins to explore and establish possible new relationships of the proteins with the process of tumorigenesis coupled with the implementation of a workflow system will be used to form a basis for a bioinformatics approach to find patterns of biological significance. We will advance this pilot study forward by increasing the overall sample numbers and subgrouping patients with common etiologies. Furthermore, newer proteomic techniques (Laser capture microdissection, ICAT, SILAC, ITRAQ, and MudPIT) will be used to complement these studies by enhancing sample purity, protein quantification, and characterizing posttranslational modifications, and assisting in determining a protein’s functional significance to HCC.
In summary, our pilot study demonstrates multiple significant protein differences in a small sample of American patients with HCC when comparing tumor to normal liver. The individual protein associations discovered are in many cases confirmatory of previous proteomic studies in HCC, validating our technique, but also lead us to hypothesize whether the American population of patients with HCC may have a unique proteomic profile. The magnitude of the problem in the U.S. is small compared to countries where hepatocellular carcinoma is the major cause of death from solid organ malignancy. The threat to the U.S., however, is growing, so it remains a worthwhile pursuit to determine if there is a unique proteomic profile that would be clinically relevant to the discovery of diagnostic, prognostic, and therapeutic biomarkers to advance the knowledge and treatment of HCC in this country.
ACKNOWLEDGMENT
This work was supported in part by a grant from the NIH-NIAAA (P50 AA07611-16-20).
REFERENCES
- 1.Bosch FX, et al. Epidemiology of hepatocellular carcinoma. Clin Liver Dis. 2005;9:191. doi: 10.1016/j.cld.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 2.El-Serag HBR, Lenhard K. Hepatocellular carcinoma: Epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 3.El-Serag HB. Hepatocellular carcinoma: Recent trends in the United States. Gastroenterology. 2004;127(5 Suppl 1):S27. doi: 10.1053/j.gastro.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 4.Marrero JA. Hepatocellular carcinoma. Curr Opin Gastroenterol. 2005;21:308. doi: 10.1097/01.mog.0000159817.55661.ca. [DOI] [PubMed] [Google Scholar]
- 5.Jemal A, et al. Cancer statistics, 2005. CA Cancer J Clin. 2005;55:10. doi: 10.3322/canjclin.55.1.10. [DOI] [PubMed] [Google Scholar]
- 6.Olthoff KM. Surgical options for hepatocellular carcinoma: Resection and transplantation. Liver Transpl Surg. 1998;4(5 Suppl 1):S98. [PubMed] [Google Scholar]
- 7.Llovet JM. Updated treatment approach to hepatocellular carcinoma. J Gastroenterol. 2005;40:225. doi: 10.1007/s00535-005-1566-3. [DOI] [PubMed] [Google Scholar]
- 8.McGlynn KA, London WT. Epidemiology and natural history of hepatocellular carcinoma. Best Pract Res Clin Gastroenterol. 2005;19:3. doi: 10.1016/j.bpg.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 9.Tanaka Y, Hanada K, Mizokami M, et al. Inaugural article: A comparison of the molecular clock of hepatitis C virus in the United States and Japan predicts that hepatocellular carcinoma incidence in the United States will increase over the next two decades. Proc Natl Acad Sci USA. 2002;99:15584. doi: 10.1073/pnas.242608099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Srivatanakul P, Sriplung H, Deerasamee S. Epidemiology of liver cancer: An overview. Asian Pac J Cancer Prev. 2004;5:118. [PubMed] [Google Scholar]
- 11.Di Bisceglie AMO, Klein SE, Waggoner JL, et al. The role of chronic viral hepatitis in hepatocellular carcinoma in the United States. Am J Gastroenterol. 1991;86:335. [PubMed] [Google Scholar]
- 12.Aebersold R, Leavitt J. Sequence analysis of proteins separated by polyacrylamide gel electrophoresis: Towards an integrated protein database. Electrophoresis. 1990;11:517. doi: 10.1002/elps.1150110702. [DOI] [PubMed] [Google Scholar]
- 13.Steiner S, Witzmann FA. Proteomics: Applications and opportunities in preclinical drug development. Electrophoresis. 2000;21:2099. doi: 10.1002/1522-2683(20000601)21:11<2099::AID-ELPS2099>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 14.Lahm HW, Langen H. Mass spectrometry: A tool for the identification of proteins separated by gels. Electrophoresis. 2000;21:2105. doi: 10.1002/1522-2683(20000601)21:11<2105::AID-ELPS2105>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 15.Petricoin EF, Zoon KC, Kohn EC, et al. Clinical proteomics: Translating benchside promise into bedside reality. Nat Rev Drug Discov. 2002;1:683. doi: 10.1038/nrd891. [DOI] [PubMed] [Google Scholar]
- 16.Kaplan RS, Pedersen PL. Determination of microgram quantities of protein in the presence of milligram levels of lipid with amido black 10B. Anal Biochem. 1985;150:97. doi: 10.1016/0003-2697(85)90445-2. [DOI] [PubMed] [Google Scholar]
- 17.Candiano G, et al. Blue silver: A very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis. 2004;25:1327. doi: 10.1002/elps.200305844. [DOI] [PubMed] [Google Scholar]
- 18.Sun S, Lee NPY, Poon RTP, et al. Oncoproteomics of hepatocellular carcinoma: From cancer markers’ discovery to functional pathways. Liver Intl. 2007:1021. doi: 10.1111/j.1478-3231.2007.01533.x. [DOI] [PubMed] [Google Scholar]
- 19.Lee IN, Chen CH, Sheu JC, et al. Identification of human hepatocellular carcinoma-related biomarkers by two-dimensional difference gel electrophoresis and mass spectrometry. J Proteome Res. 2005;4:2062. doi: 10.1021/pr0502018. [DOI] [PubMed] [Google Scholar]
- 20.Li C, Tan YX, Zhou H, et al. Proteomic analysis of hepatitis B virus-associated hepatocellular carcinoma: Identification of potential tumor markers. Proteomics. 2005;5:1125. doi: 10.1002/pmic.200401141. [DOI] [PubMed] [Google Scholar]
- 21.Liang CR, Leow CK, Neo JC, et al. Proteome analysis of human hepatocellular carcinoma tissues by two-dimensional difference gel electrophoresis and mass spectrometry. Proteomics. 2005;5:2258. doi: 10.1002/pmic.200401256. [DOI] [PubMed] [Google Scholar]
- 22.Blanc JF, Lalanne C, Plomion C, et al. Proteomic analysis of differentially expressed proteins in hepatocellular carcinoma developed in patients with chronic viral hepatitis C. Proteomics. 2005;5:3778. doi: 10.1002/pmic.200401194. [DOI] [PubMed] [Google Scholar]
- 23.Luk JM, Lam CT, Siu AF, et al. Proteomic profiling of hepatocellular carcinoma in Chinese cohort reveals heatshock proteins (Hsp27, Hsp70, GRP78) up-regulation and their associated prognostic values. Proteomics. 2006;6:1049. doi: 10.1002/pmic.200500306. [DOI] [PubMed] [Google Scholar]
- 24.Cheng CC, Liu YH, Ho CC, et al. The influence of plectin deficiency on stability of cytokeratin 18 in hepatocellular carcinoma. J Mol Histol. 2007 doi: 10.1007/s10735-007-9155-9. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 25.Martin-Orozco N, Dong C. Inhibitory costimulation and antitumor immunity. Semin Cancer Biol. 2007;17:288. doi: 10.1016/j.semcancer.2007.06.003. [Epub 2007 Jun 23] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor MR, Peterson JA, Ceriani RL, et al. Cloning and sequence analysis of human butyrophilin reveals a potential receptor function. Biochim Biophys Acta. 1996;1306:1. doi: 10.1016/0167-4781(19)60000-x. [DOI] [PubMed] [Google Scholar]
- 27.Scarpino S, Di Napoli A, Stoppacciaro A, et al. Expression of autoimmune regulator gene (AIRE) and T regulatory cells in human thymomas. Clin Exp Immunol. 2007;149:504. doi: 10.1111/j.1365-2249.2007.03442.x. [Epub 2007 Jun 22] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shin GJ, Wilson NH. Overexpression of repulsive guidance molecule (RGM) a induces cell death through Neogenin in early vertebrate development. J Mol Histol. 2008;39:105. doi: 10.1007/s10735-007-9138-x. [Epub 2007 Sep 7] [DOI] [PubMed] [Google Scholar]
- 29.Nieto-Miguel T, Fonteriz RI, Vay L, et al. Endoplasmic reticulum stress in the proapoptotic action of edelfosine in solid tumor cells. Cancer Res. 2007;67:10368. doi: 10.1158/0008-5472.CAN-07-0278. [DOI] [PubMed] [Google Scholar]
- 30.Gu X, Shin BH, Akbarali Y, et al. Tel-2 is a novel transcriptional. J Biol Chem. 2001;276:9421. doi: 10.1074/jbc.M010070200. [Epub 2000 Dec 6] [DOI] [PubMed] [Google Scholar]
- 31.Maeda T, Yagasaki F, Ishikawa M, et al. Transforming property of TEL-FGFR3 mediated through PI3-K in a T-cell lymphoma that subsequently progressed to AML. Blood. 2005;105:2115. doi: 10.1182/blood-2003-12-4290. [Epub 2004 Oct 28] [DOI] [PubMed] [Google Scholar]
- 32.Nishida N, Nagasaka T, Nishimura T, et al. Aberrant methylation of multiple tumor suppressor genes in aging liver, chronic hepatitis, and hepatocellular carcinoma. Hepatology. 2008;47:908. doi: 10.1002/hep.22110. [DOI] [PMC free article] [PubMed] [Google Scholar]