Summary
Our previous microarray expression analysis of the long-lived Little mice (Ghrhrlit/lit) showed a concerted up-regulation of xenobiotic detoxification genes. Here, we show that this up-regulation is associated with a potent increase in resistance against the adverse effects of a variety of xenobiotics, including the hepatotoxins acetaminophen and bromobenzene and the paralyzing agent zoxazolamine. The classic xenobiotic receptors Car (Constitutive Androstane Receptor) and Pxr (Pregnane × Receptor) are considered key regulators of xenobiotic metabolism. Using double and triple knockout/mutant mouse models we found, however, that Car and Pxr are not required for the up-regulation of xenobiotic genes in Little mice. Our results suggest instead that bile acids and the primary bile acid receptor Fxr (farnesoid × receptor) are likely mediators of the up-regulation of xenobiotic detoxification genes in Little mice. Bile acid levels are considerably elevated in the bile, serum, and liver of Little mice. We found that treatment of wild-type animals with cholic acid, one of the major bile acids elevated in Little mice, mimics in large part the up-regulation of xenobiotic detoxification genes observed in Little mice. Additionally, the loss of Fxr had a major effect on the expression of the xenobiotic detoxification genes up-regulated in Little mice. A large fraction of these genes lost or decreased their high expression levels in double mutant mice for Fxr and Ghrhr. The alterations in xenobiotic metabolism in Little mice constitute a form of increased stress resistance and may contribute to the extended longevity of these mice.
Keywords: aging, bile acid metabolism, gene expression, Little mice, mouse models, nuclear hormone receptors, stress resistance, xenobiotic metabolism
Introduction
An increase in various forms of stress resistance has been associated with increased longevity in a variety of long-lived mutants in model organisms such as nematodes, fruit flies, and mice (Larsen, 1993; Lin et al., 1998; Finkel & Holbrook, 2000; Murakami, 2006). In mice, oxidative stress resistance has been one of the most studied systems. For example, in the long-lived Ames dwarf mice several components of the antioxidative defense system, including catalase and superoxide dismutase, are up-regulated and these mice display an enhanced capacity to deal with oxidative stress. After an acute administration of paraquat, a systemic oxidative stressor, the Ames dwarf mice survive significantly longer than wild-type mice (Bartke et al., 2001). Similar observations have been reported for other mouse models of delayed aging. Klotho transgenic mice, p66shc−/− mice and insulin-like growth factor 1 (IGF1)-R+/− mice also show increased resistance against paraquat toxicity (Migliaccio et al., 1999; Napoli et al., 2003; Kurosu et al., 2005). Calorie-restricted mice exhibit increased antioxidative defenses, and they have a slower rate of accumulation of tissue oxidative damage with age (Merry, 2004; Hunt et al., 2006).
The role of other forms of increased stress resistance, such as increased xenobiotic resistance, in long-lived organisms have been started to be explored just recently (Miller et al., 2005; Harper et al., 2006). All organisms are constantly exposed to a wide variety of toxic or potentially harmful compounds of exogenous (environmental contaminants, dietary compounds, drugs) or endogenous (diverse metabolic by-products) origin. Recent evidence suggests that the up-regulation of xenobiotic metabolism, composed of a large number of metabolizing enzymes and transporters that coordinately function in the detoxification and elimination of these potentially harmful compounds, may be another form of enhanced stress resistance shared between long-lived mutants across diverse model organisms. For instance, a recent microarray survey of genes regulated by insulin/IGF-1 signaling in Caenorhabditis elegans suggested a role for the up-regulation of xenobiotic detoxification genes as an important mechanism for longevity assurance (McElwee et al., 2004). Our previous microarray studies in the long-lived Ames dwarf mice and Little mice suggested a similar effect on xenobiotic metabolism (Amador-Noguez et al., 2004). Additional microarray experiments performed in Calorie-restricted mice and Snell dwarf mice have also yielded similar observations (Tsuchiya et al., 2004).
In this paper, we report the alterations in xenobiotic metabolism in the long-lived Little mice (Ghrhrlit/lit). These mutant mice represent an established model of delayed or decelerated aging, and they show an increase in mean lifespan of 23–25% (Flurkey et al., 2001). They are homozygous for a missense mutation (designated as lit) in the growth hormone releasing hormone receptor (Ghrhr) gene (Godfrey et al., 1993). As a result, these mice have substantially reduced levels of circulating growth hormone (1% of the normal level) (Flurkey et al., 2001) and reduced circulating levels of IGF-1 (Donahue & Beamer, 1993). These mice show delayed growth, reaching about 50–60% of the weight of wild-type mice and also have reduced fertility.
Our previous microarray gene expression analysis of Little mice showed a concerted transcriptional up-regulation of xenobiotic detoxification genes (Amador-Noguez et al., 2004). Here, we show that this transcriptional up-regulation is associated with a potent increase in resistance against the adverse effects of a variety of xenobiotic compounds, including the hepatotoxins acetaminophen and bromobenzene and the paralyzing agent zoxazolamine. The classic xenobiotic receptors Car (Constitutive Androstane Receptor) and Pxr (Pregnane × Receptor) are considered key regulators of all phases of xenobiotic metabolism and are known regulators of many of the xenobiotic genes up-regulated in Little mice. Using double and triple knockout/ mutant mouse models we show, however, that Car and Pxr are not required, with the exception of two cytochrome p450s (Cyp2b10 and Cyp2c38), for the up-regulation of xenobiotic detoxification genes in Little mice.
Our results suggest instead that bile acids and the bile acid receptor Fxr (farnesoid × receptor) are likely mediators of the up-regulation of xenobiotic detoxification genes in Little mice. Bile acid levels are significantly elevated in the bile, serum and liver of Little mice. We found that treatment of wild-type animals with cholic acid (CA) mimics in large part the up-regulation of xenobiotic metabolism genes observed in Little mice. Additionally, the loss of Fxr had a major effect on the expression of xenobiotic detoxification genes up-regulated in Little mice. A large fraction of these genes lost or decreased their up-regulation in double mutant mice for Fxr and Ghrhr.
Results
Up-regulation of xenobiotic detoxification genes in the long-lived Little mice
Xenobiotic metabolism, which takes place primarily in the liver, involves four major steps designated as phases 0, 1, 2 and 3. Phase 0 involves the action of transporter genes involved in the uptake of xenobiotics (toxic exogenous compounds) or endobiotics (potentially harmful endogenous metabolites) by the liver. Phase 1 genes participate in the initial chemical modification and activation, by the addition of reactive functional groups, of the typically lipophilic xenobiotic or endobiotic compounds. Phase 1 reactions are frequently, but not always required for phase 2 reactions. Phase 2 conjugation reactions further transform these modified products by the addition of side groups that produce chemically unreactive, soluble products that are then excreted out of the liver by phase 3 transporters (Xu et al., 2005).
Our previous microarray gene expression analysis of Little mice showed a concerted up-regulation of xenobiotic detoxification genes (Amador-Noguez et al., 2004). To expand and corroborate this result and to obtain a testing gene set for the experiments performed in the next sections, we used real-time polymerase chain reaction (PCR) to compare the expression levels of a large set (57 genes) of xenobiotic detoxification genes in the livers of Little mice vs. wild-type mice. The results are summarized in Table 1. The expression of 35 of these genes was significantly different (P < 0.05) in Little mice vs. the wild-type controls, 32 genes were up-regulated in Little mice and 3 genes were down-regulated. In many cases, the increase in the expression levels for these genes was very strong, with 10 genes showing a fold-increase greater than 5.
Table 1.
Up-regulation of xenobiotic detoxification genes in Little mice
| Gene | Description | Phase* | Fold-change† | P-value‡ |
|---|---|---|---|---|
| Abcb1a | ATP-binding cassette, subfamily B (MDR/TAP), member 1 A | 3 | 4.4 | 0.0052 |
| Abcc3 | ATP-binding cassette, subfamily C (CFTR/MRP), member 3 | 3 | 1.5 | 0.3323 |
| Abcd2 | ATP-binding cassette, subfamily D (ALD), member 2 | 3 | 10.0 | 0.1903 |
| Aldh1a1 | Aldehyde dehydrogenase family 1, subfamily A1 | 1 | 2.0 | 0.0005 |
| Bhmt | Betaine-homocysteine S-methyltransferase | 2 | 1.4 | 0.1805 |
| Ces1 | Carboxylesterase 1 | 1 | 1.9 | 0.0402 |
| Ces2 | Carboxylesterase 2 | 1 | 0.7 | 0.4574 |
| Ces3 | Carboxylesterase 3 | 1 | 1.7 | 0.1051 |
| Cyp1a1 | Cytochrome p450, 1a1 | 1 | 1.7 | 0.1084 |
| Cyp1a2 | Cytochrome p450, 1a2 | 1 | 1.1 | 0.8653 |
| Cyp2a4 | Cytochrome p450, 2a4 | 1 | 3.4 | < 0.0001 |
| Cyp2b10 | Cytochrome p450, 2b10 | 1 | 21.0 | 0.0004 |
| Cyp2b13 | Cytochrome p450, 2b13 | 1 | 7715.3 | < 0.0001 |
| Cyp2b9 | Cytochrome p450, 2b9 | 1 | 4126.9 | < 0.0001 |
| Cyp2c37 | Cytochrome p450, 2c37 | 1 | 2.6 | 0.0399 |
| Cyp2c38 | Cytochrome p450, 2c38 | 1 | 2.0 | 0.0142 |
| Cyp2c39 | Cytochrome p450, 2c39 | 1 | 1.6 | 0.0744 |
| Cyp3a11 | Cytochrome p450, 3a11 | 1 | 1.0 | 0.8596 |
| Cyp3a25 | Cytochrome p450, 3a25 | 1 | 1.4 | 0.2590 |
| Cyp4a10 | Cytochrome p450, 4a10 | 1 | 6.8 | 0.0003 |
| Cyp4a12 | Cytochrome p450, 4a12 | 1 | 0.0 | 0.0045 |
| Cyp4a14 | Cytochrome p450, 4a14 | 1 | 26.2 | 0.0003 |
| Cyt19 | Arsenite methyltransferase | 2 | 2.6 | 0.0012 |
| Fmo1 | Flavin containing monooxygenase 1 | 1 | 0.6 | 0.0573 |
| Fmo3 | Flavin containing monooxygenase 3 | 1 | 127.8 | < 0.0001 |
| Fmo4 | Flavin containing monooxygenase 4 | 1 | 3.0 | 0.0221 |
| Gnmt | Glycine N-methyltransferase | 2 | 1.6 | 0.1447 |
| Gsta2 | Glutathione S-transferase, alpha 2 | 2 | 4.3 | 0.0011 |
| Gsta3 | Glutathione S-transferase 3 | 2 | 1.0 | 0.9564 |
| Gsta4 | Glutathione S-transferase, alpha 4 | 2 | 2.1 | 0.0270 |
| Gstk1 | Glutathione S-transferase kappa 1 | 2 | 1.5 | 0.1724 |
| Gstm1 | Glutathione S-transferase, mu 1 | 2 | 1.1 | 0.6268 |
| Gstm2 | Glutathione S-transferase, mu 2 | 2 | 1.6 | 0.0116 |
| Gstm3 | Glutathione S-transferase, mu 3 | 2 | 2.3 | 0.0234 |
| Gstm4 | Glutathione S-transferase, mu 4 | 2 | 1.3 | 0.3302 |
| Gsto1 | Glutathione transferase omega 1 | 2 | 0.6 | 0.1652 |
| Gsto2 | Glutathione transferase omega 2 | 2 | 3.2 | 0.0003 |
| Gstp2 | Glutathione transferase pi 2 | 2 | 0.1 | 0.0017 |
| Gstt1 | Glutathione S-transferase theta 1 | 2 | 1.6 | 0.1219 |
| Gstt2 | Glutathione S-transferase theta 2 | 2 | 2.8 | 0.0069 |
| Mgst3 | Microsomal glutathione S-transferase 3 | 2 | 3.9 | < 0.0001 |
| mOAT6 | Solute carrier family 22 (organic anion transporter), member 20 | 0 | 176.3 | < 0.0001 |
| Mt1 | Metallothionein 1 | 1 | 4.4 | 0.0103 |
| Nnmt | Nicotinamide N-methyltransferase | 2 | 1.4 | 0.4018 |
| Papss2 | 3′-phosphoadenosine 5′-phosphosulfate synthase 2 | 2 | 2.1 | 0.0268 |
| Por | Cytochrome p450 reductase | 1 | 2.4 | 0.0183 |
| Slc21a5 | Solute carrier organic anion transporter family, member 1a5 | 0 | 5.7 | 0.0004 |
| Sth2 | Sulfotransferase family 2A, member 2 | 2 | 30409.2 | < 0.0001 |
| Sult1a1 | Sulfotransferase family 1A, phenol-preferring, member 1 | 2 | 2.4 | 0.0249 |
| Sult1b1 | Sulfotransferase family 1B, member 1 | 2 | 0.9 | 0.5350 |
| Sult1d1 | Sulfotransferase family 1D, member 1 | 2 | 2.8 | 0.0418 |
| Sult1e1 | Sulfotransferase family 1E, member 1 | 2 | 82.8 | 0.0001 |
| Sult2b1 | Sulfotransferase family, cytosolic, 2B, member 1 | 2 | 0.8 | 0.1643 |
| Temt | Indolethylamine N-methyltransferase | 2 | 3.6 | 0.0025 |
| Ugt1a1 | UDP glucuronosyltransferase 1 family, polypeptide A1 | 2 | 4.8 | 0.0004 |
| Ugt1a9 | UDP glycosyltransferase 1 family, polypeptide A9 | 2 | 3.9 | 0.1263 |
| Ugt2b5 | UDP glucuronosyltransferase 2 family, polypeptide B5 | 2 | 0.3 | 0.0068 |
Refers to the phase of xenobiotic metabolism in which each gene participates.
The fold-change refers to the ratio of the expression values of Little mice over the wild-type mice obtained by real-time PCR analysis.
The P-value refers to a Student’s t-test for the difference in expression between Little and wild-type mice.
The classic xenobiotic receptors Car and Pxr are not required for the up-regulation of xenobiotic detoxification genes in Little mice
The nuclear receptors Car (Constitutive Androstane Receptor) and Pxr (Pregnane × receptor) are widely recognized as key regulators of all phases of xenobiotic metabolism (Kliewer et al., 2002; Kretschmer & Baldwin, 2005). Phase 1 genes regulated by these nuclear receptors include cytochrome p450s, carboxylesterases, and dehydrogenases. Phase 2 regulated genes include glutathione S-transferases, UDP-glucuronosyltransferases, and sulfotransferases. Many of the xenobiotic genes that we found to be up-regulated in Little mice are specifically known to be regulated by Car and/or Pxr. These genes include: Abcb1a (Maglich et al., 2002), Aldh1a1 (Maglich et al., 2002), Cyp2b10 (Zhang et al., 2004), Cyp2a4 (Maglich et al., 2002; Rosenfeld et al., 2003), Gsta2 (Huang et al., 2003), Gsta4 (Rosenfeld et al., 2003), Gstm2 (Maglich et al., 2002), Papss2 (Ueda et al., 2002; Kretschmer & Baldwin, 2005), Por (Ueda et al., 2002), Slc21a5 (Maglich et al., 2002), Sth2 (Rosenfeld et al., 2003), Sult1d1 (Maglich et al., 2002), and Ugt1a1 (Zhang et al., 2004).
We therefore hypothesized that Car and/or Pxr may be involved in the up-regulation of xenobiotic detoxification genes in Little mice. To investigate this idea, we generated double mutant mice for Car and Ghrhr (designated as Car/Little mice) and double mutant mice for Pxr and Ghrhr (designated as Pxr/ Little mice). If the activation of Car or Pxr was responsible for the increase in the expression levels of xenobiotic detoxification genes in Little mice, this up-regulation should be lost in the absence of Car or Pxr in the Car/Little and Pxr/Little double mutant mice. We tested 30 of the xenobiotic detoxification genes up-regulated in Little mice. The results of these experiments are summarized in Table 2. Unexpectedly, our analysis of these genes in the Car/Little mice showed that only Cyp2b10 was up-regulated in a Car-dependent manner. The high level of expression of the rest of these genes was remarkably maintained in the Car/Little double mutant mice. Our analysis of the Pxr/ Little mice showed that the up-regulation of the genes in this set was not dependent on Pxr. Interestingly, a few genes actually showed a stronger up-regulation in the Car/Little mice or Pxr/Little vs. Little mice.
Table 2.
Xenobiotic detoxification genes in the Car/Little, Pxr/Little, Car/Pxr/Little, and Fxr/Little mice
| Little mice |
Car/Little mice |
Pxr/Little mice |
Car/Pxr/Little mice |
Fxr/Little mice |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene* | Fold-change† | P-value‡ | Fold-change† | P-value‡ | Fold-change† | P-value‡ | Fold-change† | P-value‡ | Fold-change† | P-value‡ |
| Abcb1a | 4.4 | 0.0052 | 4.3 | 0.0067 | 8.2 | 0.0004 | 7.4 | 0.0005 | 0.5 | 0.2137 |
| Aldh1a1 | 2.0 | 0.0005 | 2.6 | < 0.0001 | 2.2 | 0.0001 | 2.4 | 0.0007 | 0.4 | 0.1315 |
| Cyp2a4 | 3.4 | < 0.0001 | 4.1 | < 0.0001 | 9.6 | < 0.0001 | 8.5 | < 0.0001 | 4.8 | 0.0026 |
| Cyp2b10 | 21.0 | 0.0004 | 1.2 | 0.7714 | 59.6 | 0.0001 | 1.7 | 0.2841 | 2.3 | 0.1900 |
| Cyp2b13 | 7715.3 | < 0.0001 | 10868.3 | < 0.0001 | 8487.8 | < 0.0001 | 5252.7 | < 0.0001 | 235.0 | 0.0003 |
| Cyp2b9 | 4126.9 | < 0.0001 | 3899.5 | < 0.0001 | 4298.8 | < 0.0001 | 3440.6 | < 0.0001 | 13.7 | 0.0038 |
| Cyp2c38 | 2.0 | 0.0142 | 2.0 | 0.0155 | 2.5 | 0.0022 | 1.2 | 0.3363 | 0.8 | 0.2399 |
| Cyp4a10 | 6.8 | 0.0003 | 14.6 | < 0.0001 | 6.2 | 0.0001 | 7.7 | < 0.0001 | 1.2 | 0.7112 |
| Cyp4a14 | 26.2 | 0.0003 | 112.3 | < 0.0001 | 21.4 | 0.0005 | 37.0 | 0.0001 | 4.2 | 0.0155 |
| Cyt19 | 2.6 | 0.0012 | 3.4 | 0.0021 | 1.8 | 0.0520 | 2.2 | 0.0206 | 1.8 | 0.0153 |
| Fmo3 | 127.8 | < 0.0001 | 145.1 | < 0.0001 | 191.5 | < 0.0001 | 476.2 | 0.0001 | 1.5 | 0.6119 |
| Fmo4 | 3.0 | 0.0221 | 9.6 | 0.0003 | 2.8 | 0.0216 | 7.0 | 0.0008 | 3.0 | 0.0120 |
| Gsta2 | 4.3 | 0.0011 | 5.2 | 0.0006 | 3.1 | 0.0042 | 5.0 | 0.0039 | 7.4 | 0.0005 |
| Gsta4 | 2.1 | 0.0270 | 2.6 | 0.0391 | 3.0 | 0.0052 | 3.9 | 0.0028 | 1.7 | 0.0102 |
| Gstm2 | 1.6 | 0.0116 | 2.0 | 0.0001 | 1.8 | 0.0023 | 1.9 | 0.0159 | 4.5 | 0.0022 |
| Gstm3 | 2.3 | 0.0234 | 2.9 | 0.0093 | 4.2 | 0.0001 | 4.6 | < 0.0001 | 9.8 | 0.0014 |
| Gsto2 | 3.2 | 0.0003 | 5.2 | < 0.0001 | 2.2 | 0.0009 | 2.4 | 0.0142 | 0.3 | 0.0575 |
| Gstt2 | 2.8 | 0.0069 | 3.1 | 0.0009 | 1.6 | 0.0161 | 2.3 | 0.0004 | 0.8 | 0.3889 |
| Mgst3 | 3.9 | < 0.0001 | 4.7 | 0.0138 | 3.0 | 0.0105 | 3.9 | < 0.0001 | 3.2 | 0.0044 |
| mOAT6 | 176.3 | < 0.0001 | 361.4 | < 0.0001 | 273.0 | < 0.0001 | 318.4 | < 0.0001 | 22.2 | 0.0003 |
| Mt1 | 4.4 | 0.0103 | 6.8 | 0.0050 | 4.4 | 0.0125 | 5.5 | 0.0078 | 13.3 | 0.0250 |
| Papss2 | 2.1 | 0.0268 | 4.8 | 0.0004 | 4.0 | 0.0005 | 4.7 | 0.0004 | 0.9 | 0.6981 |
| Por | 2.4 | 0.0183 | 1.8 | 0.0433 | 1.8 | 0.0445 | 3.1 | 0.0020 | 0.8 | 0.2442 |
| Slc21a5 | 5.7 | 0.0004 | 5.2 | 0.0003 | 4.6 | 0.0007 | 2.8 | 0.0152 | 2.6 | 0.0017 |
| Sth2 | 30409.2 | < 0.0001 | 30199.2 | < 0.0001 | 55509.4 | < 0.0001 | 32787.6 | < 0.0001 | 26130.5 | < 0.0001 |
| Sult1a1 | 2.4 | 0.0249 | 3.5 | 0.0121 | 3.1 | 0.0059 | 4.3 | 0.0012 | 1.8 | 0.0005 |
| Sult1d1 | 2.8 | 0.0418 | 2.6 | 0.0282 | 3.2 | 0.0109 | 4.6 | 0.0017 | 0.6 | 0.0438 |
| Sult1e1 | 82.8 | 0.0001 | 53.2 | < 0.0001 | 247.9 | < 0.0001 | 68.8 | 0.0015 | 1900.8 | < 0.0001 |
| Temt | 3.6 | 0.0025 | 4.6 | 0.0012 | 2.4 | 0.0231 | 3.2 | 0.0045 | 1.1 | 0.6197 |
| Ugt1a1 | 4.8 | 0.0004 | 3.7 | < 0.0001 | 2.5 | 0.0001 | 3.4 | < 0.0001 | 1.1 | 0.5335 |
The full names for these genes are provided in Table 1.
The fold-change refers to the ratio of the expression values of each mutant over the wild-type controls obtained by real-time PCR analysis.
The P-value refers to a Student’s t-test for the difference in expression between each mutant and wild-type controls.
Car and Pxr are known to share several common target genes and activators and they play complementary or partially overlapping roles in the protection against xenobiotics (Moore et al., 2000; Maglich et al., 2002; Guo et al., 2003). To rule out the possibility that Car and Pxr could compensate for the absence of each other in the double Car/Little and Pxr/Little mutant mice, we generated triple mutant mice for Car, Pxr, and Ghrhr (designated as Car/Pxr/Little mice). Our analysis showed, however, that the combined loss of Car and Pxr abolished the up-regulation of only one additional gene, Cyp2c38. The up-regulation of the Cyp2b10 gene was, as expected, still lost in the triple mutants. Interestingly, here we also observed that some genes showed a stronger up-regulation in the Car/Pxr/Little mice vs. Little mice.
Together, the results obtained from the Car/Little, Pxr/Little and Car/Pxr/Little mice demonstrate that Car and Pxr, either by themselves or in combination, are not required, with the two exceptions of the Cyp2b10 and Cyp2c38 genes, for the up-regulation of xenobiotic detoxification genes in Little mice.
Increased xenobiotic resistance in Little mice
Xenobiotic metabolism protects the organism against the detrimental effects of potentially toxic xenobiotic and endobiotic compounds. Because of the concerted up-regulation of xenobiotic detoxification genes in Little mice, we hypothesized that these mice may exhibit enhanced resistance to the adverse effects of a variety of xenobiotic compounds. We examined the resistance of these mice against liver damage induced by the hepatotoxins acetaminophen, bromobenzene, and carbon tetrachloride. In addition, we also measured their resistance against the paralyzing agent zoxazolamine.
Zoxazolamine experiments
Zoxazolamine (2-amino-5-chlorobenzoxazole) is a muscle relaxant that has been used extensively as a model substrate for the assessment of changes in hepatic cytochrome p450 activity (Van der Graaff et al., 1986). Increased cytochrome p450 activity results in a more rapid metabolic inactivation of this compound, which is observed as decreased duration of zoxazolamine-induced paralysis (Wei et al., 2000). We compared the ability of Little mice (n = 10) against wild-type mice (n = 10) to recover from paralysis induced by zoxazolamine and found that Little mice recovered significantly faster (P < 0.001) (Fig. 1). Following a single intraperitoneal dose (250 mg kg−1), the wild-type mice recovered after an average time of about 4 h, whereas Little mice recovered after an average time of about 1.5 h.
Fig. 1.
Increased resistance to zoxazolamine induced-paralysis in Little mice. Mice were treated with a single intraperitoneal dose of zoxazolamine (250 mg kg−1). The duration of the paralysis was recorded as the time when the mice were able to right themselves repeatedly. The paralysis time was significantly reduced (P < 0.001) in Little mice (n = 10) compared to wild-type mice (n = 10). The increased susceptibility of the Car mice (n = 3) against zoxazolamine induced-paralysis was significantly reversed (P = 0.001) in the Car/Little mice (n = 7), which showed a paralysis time comparable to Little mice. Each bar represents the mean ± (2 × SE) for each group. The P-values refer to a Student’s t-test between the indicated groups.
We also found that the Little mutation reversed the susceptibility of the Car mice against zoxazolamine-induced paralysis. It has been previously shown that the Car mice are highly susceptible to zoxazolamine induced paralysis and they also show high mortality under this treatment (Wei et al., 2000). Wild-type mice pretreated with the xenobiotic activators phenobarbital or TCPOBOP (1,4-bis[2-(3,5-dichloropyridyloxy)] benzene) are protected against zoxazolamine-induced paralysis but this protective effect is lost in Car mice (Wei et al., 2000). Because the increased expression of xenobiotic genes is maintained in the Car/Little mice, we hypothesized that these mice should still be resistant to zoxazolamine-induced paralysis despite the absence of Car. In agreement with this idea, we found that the Car/Little (n = 7) showed a significantly reduced (P = 0.001) paralysis time as compared to Car mice (n = 3). Whereas the Car mice showed an average paralysis time of about 8 h, the Car/Little mice showed an average paralysis time of about 2.5 h (Fig. 1). Although this paralysis time was higher than the one observed in Little mice (around 1.5 h), it was still significantly reduced (P = 0.02) compared to the paralysis time in wild-type mice (about 4 h).
Acetaminophen experiments
Acetaminophen (N-acetyl-4-aminophenol, also known as paracetamol) is a common analgesic that is safe when used at therapeutic doses. However, it can cause severe hepatotoxicity and centrilobular necrosis when given in large doses (Jaeschke et al., 2003; Hinson et al., 2004). To determine whether Little mice are resistant to toxic doses of acetaminophen, we treated wild-type (n = 10) and mutant animals (n = 5) with a single intraperitoneal dose of 375 mg per kg body weight. After 24 h, livers and serum were collected. We observed that the extent of liver damage and centrilobular necrosis was remarkably reduced in Little mice as compared to wild-type mice (Fig. 2A,B). We also measured serum levels of alanine aminotransferase (ALT) and lactate dehydrogenase (LDH) activity as an index of liver toxicity. These enzymes are normally present in the liver and are released into the blood when liver damage occurs. In agreement with the amount of centrilobular necrosis observed in the histological examinations, Little mice had significantly lower ALT and LDH levels than the wild-type mice (Fig. 2C,D).
Fig. 2.
Increased resistance to acetaminophen-induced liver toxicity in Little mice. Mice were administered a 375 mg kg−1 dose of acetaminophen by intraperitoneal injection. (A) After 24 h, liver sections were examined by histological staining (hematoxylin and eosin staining). Liver samples for all treated animals were analyzed and representative histology is shown. While the livers from wild-type animals (n = 10) showed extensive necrosis, only minimal necrosis was observed in Little mice (n = 5). The Car/Pxr double mutants (n = 6) showed even more extensive liver damage than wild-type animals. However, the Car/Pxr/Little (n = 4) mice presented only minimal necrosis that was comparable to the levels seen in Little mice. (B) The necrotic area (as a percentage of total area) was determined for each experimental group as described in the methods section. Little mice and Car/Pxr/Little mice showed a reduced necrotic area as compared to wild-type mice and Car/Pxr mice (P < 0.001 for all comparisons). The Car/Pxr mice showed a substancially larger necrotic area as compared to wild-type mice (P < 0.001). Serum levels of ALT (C) and LDH (D) were measured after 24 h. Little mice and Car/Pxr/Little mice had significantly reduced ALT levels as compared to wild-type mice and Car/Pxr mice (P < 0.02 for all comparisons). Little mice and Car/Pxr/Little mice also had reduced LDH levels as compared to wild-type mice and Car/Pxr mice (P < 0.03 for all comparisons). The Car/Pxr mice showed significantly increased LDH levels as compared to wild-type mice (P < 0.006). Basal Levels of ALT and LDH in Car/Pxr and Car/Pxr/Little mice were indistinguishable from wild-type mice and were not affected by treatment with the PBS vehicle (not shown). In (E), the scatter plot shows the correlation between ALT levels and necrotic areas for the individual mice in all experimental groups. In (B), (C), and (D) bars represents the mean ± (2 × SE) for each group. All P-values refer to a Student’s t-test between the indicated groups.
We also found that the Little mutation reversed the susceptibility of the Car/Pxr mice against acetaminophen-induced liver damage. The Car/Pxr mutants (n = 6) were significantly more susceptible than wild-type mice to acetaminophen-induced liver damage (Fig. 2). However, this susceptibility was rescued in the Car/Pxr/Little mice (n = 4), which were almost as resistant against acetaminophen-induced hepatotoxicity as Little mice. The serum ALT and LDH measurements correlated with the degree of necrosis observed in the histological analysis (Fig. 2E).
Bromobenzene experiments
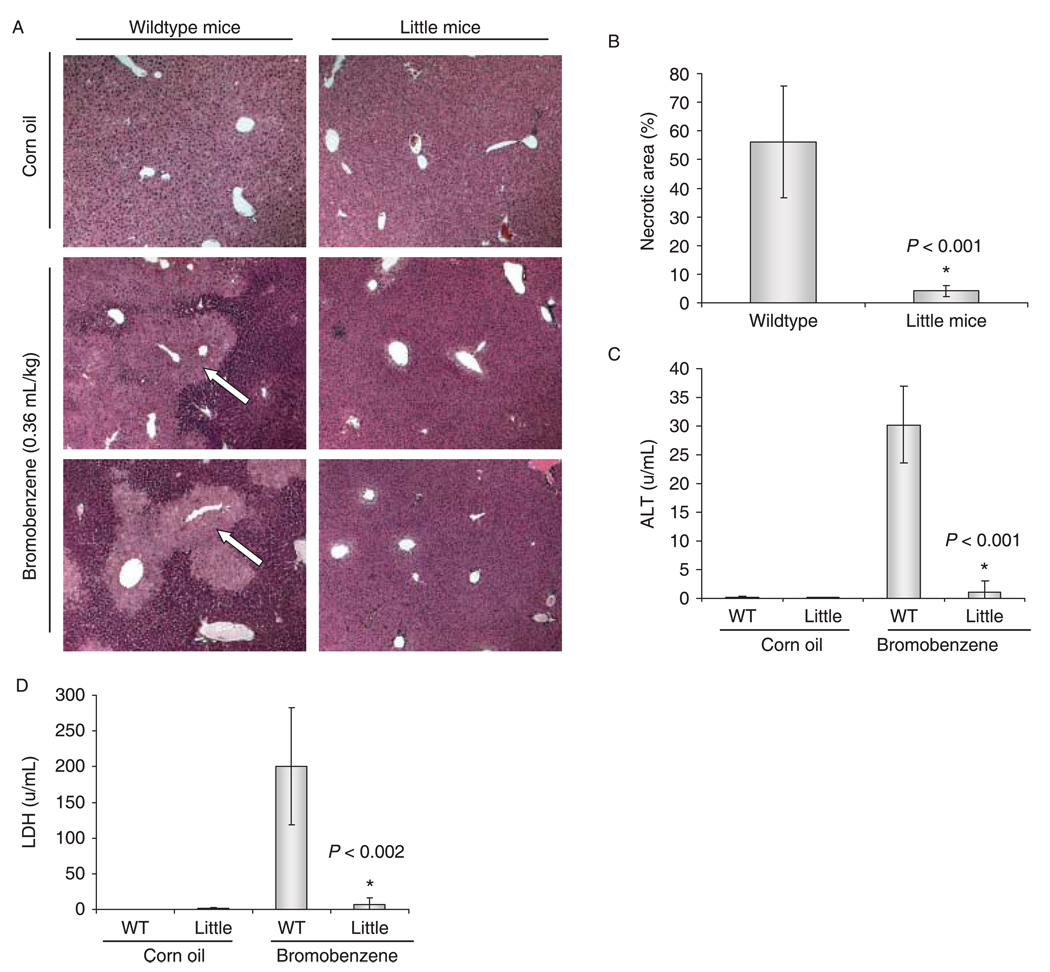
Several halogenated compounds are able to produce centrilobular necrosis in the liver. Bromobenzene has been commonly used as a model of chemical-induced liver injury (Brodie et al., 1971; Zampaglione et al., 1973; Mitchell et al., 1976). To determine if Little mice have increased resistance against bromobenzene-induced liver necrosis, we treated Little mice (n = 5) and wild-type animals (n = 5) with a single intraperitoneal dose of 0.36 mL kg−1. Twenty-four hours after the treatment, Little mice showed only minimal levels of necrosis. However, the wild-type controls presented extensive centrilobular necrosis. In agreement with the amount of centrilobular necrosis, Little mice also showed significantly lower ALT and LDH levels than wild-type mice (Fig. 3).
Fig. 3.
Little mice are resistant to bromobenzene-induced liver toxicity. Mice were treated with a 0.36 mL kg−1 dose of bromobenzene by intraperitoneal injection. (A) After 24 h, liver sections were examined by histological staining (hematoxylin and eosin staining). Liver samples for all treated animals were analyzed and representative histology is shown. The livers from wild-type animals (n = 5) showed extensive necrosis (indicated by arrows), but only minimal necrosis was observed in Little mice (n = 5). (B) The necrotic area (as a percentage of total area) was determined for each experimental group as described in the methods section. Little mice showed a significantly reduced necrotic area as compared to wild-type mice (P < 0.001). Serum levels of ALT (C) and LDH (D) were measured 24 h after treatment. Little mice showed significantly reduced levels of both ALT (P < 0.001) and LDH (P < 0.002). In (B), (C), and (D) bars represents the mean ± (2 × SE) for each group. All P-values refer to a Student’s t-test.
Carbon tetrachloride experiments
Acute CCl4 administration is another widely used model of drug-induced hepatotoxicity (Arosio et al., 2000; Yamazaki et al., 2005; Dwivedi et al., 2006). In this case, we observed that both wild-type (n = 7) and Little (n = 5) mice showed extensive liver damage. However, in contrast with the increased xenobiotic resistance of Little mice against the previous three compounds that we examined, Little mice were actually more susceptible to carbon tetrachloride-induced hepatotoxicity than wild-type mice. Twenty-four hours after an acute dose (50 µL kg−1) Little mice showed more extensive liver damage and significantly higher levels of ALT (Fig. 4).
Fig. 4.
Little mice show increased susceptibility to carbon tetrachloride-induced liver toxicity. Mice were administered a 50 µL kg−1 dose of carbon tetrachloride by intraperitoneal injection. (A) After 24 h, liver sections were examined by histological staining (hematoxylin and eosin staining). Liver samples for all treated animals were analyzed and representative histology is shown. The livers from both wild-type (n = 7) and Little mice (n = 5) showed wide-spread damage, however, the liver damage in Little mice was more extensive. (B) The liver damage areas (as a percentage of total area) were determined for each experimental group as described in the methods section. Little mice showed significantly increased liver damage areas as compared to wild-type mice (P < 0.001). (C) Serum levels of ALT were measured after 24 h and were significantly higher in Little mice (P < 0.02). In (B) and (C) bars represent the mean ± (2 × SE). All P-values shown refer to a Student’s t-test.
Bile acids as potential endobiotics that up-regulate xenobiotic detoxification genes in Little mice
Xenobiotic detoxification genes are ordinarily up-regulated in response to the exposure to toxic xenobiotics (environmental contaminants, dietary compounds, drugs) or in response to an elevation of potentially harmful endobiotics (bile acids, steroid hormones, fatty acid derivatives, bilirubin). We therefore wondered if there may be an elevation in the levels of some endobiotics in Little mice that triggers the concerted up-regulation of xenobiotic detoxification genes. Although there are a number of different endobiotics that can potentially be involved in the up-regulation of xenobiotic detoxification genes in Little mice, we reasoned that bile acids would be good initial candidates because our previous microarray analysis of long-lived mutant mice hinted at alterations in bile acid homeostasis and metabolism (Amador-Noguez et al., 2004). Bile acids are end products of hepatic cholesterol catabolism that are essential for the absorption of dietary lipids in the intestine. Physiological levels of bile acids are critically regulated, and they can produce toxic effects at high concentrations. Importantly, bile acids also function as signaling molecules that activate nuclear receptors that regulate cholesterol, bile acid, and xenobiotic metabolism (Chiang, 2002; Zhang et al., 2004).
Elevation of bile acid levels in Little mice
We started by measuring total and individual biliary bile acids in Little mice and wild-type mice using gas chromatographymass spectrometry. We found a 70% elevation in the levels of total biliary bile acids in Little mice as compared to wild-type mice. As shown in Table 3, although several individual bile acids were elevated, by far the major contribution to the elevation of total bile acids in Little mice came from the elevation in the levels of CA, one of the main bile acids in mice and other mammals including humans. Biliary CA levels were increased almost four-fold in Little mice (P = 0.005) (Fig. 5A). The proportion (as a percentage of total bile acids) of CA was also increased from 20% in wild-type mice to 41% in Little mice. The analysis of serum bile acids yielded similar observations (Table 4). Little mice showed a four-fold elevation in the levels of total serum bile acids (P < 0.0001). Here, β-muricholic acid and CA were the main contributors to the elevation of total bile acids (Fig. 5B). Little mice also showed a nearly two-fold elevation in the levels of total liver bile acids (P < 0.0001) (Fig. 5C). We measured fecal bile acid excretion rates to determine whether the elevation in bile acid levels in Little mice could be due to an increased rate of synthesis or a reduced rate of excretion. Our analysis showed that Little mice had a ~75% increase in fecal bile acid excretion (Fig. 5D). Because the bile acid excretion rate is directly proportional to the amount synthesized in the liver, these data indicated a generalized increase of bile acid synthesis in Little mice.
Table 3.
Biliary bile acid levels in Little mice and wild-type mice
| Identification | Little mice (µM/L) | Wild-type mice (µM/L) | Mutant/wild-type ratio | P-value* |
|---|---|---|---|---|
| α-muricholic acid | 2274 | 588 | 3.9 | 0.013 |
| β-muricholic acid | 3332 | 1952 | 1.7 | 0.177 |
| Cholic acid | 19040 | 5180 | 3.7 | 0.005 |
| Deoxycholic acid | 1965 | 1701 | 1.2 | 0.564 |
| Iso-deoxycholic acid | 1816 | 3906 | 0.5 | 0.004 |
| Oxodihydroxy bile acid | 3006 | 1688 | 1.8 | 0.005 |
| Unidentified bile acid | 3482 | 2337 | 1.5 | 0.027 |
| ω-muricholic acid | 3969 | 2980 | 1.3 | 0.211 |
| Others | 7030 | 6194 | 1.1 | 0.401 |
| Total | 45914 | 26525 | 1.7 | 0.015 |
The P-value refers to a Student’s t-test for the difference in bile acid levels between Little mice and wild-type controls as determined by gas chromatographymass spectrometry.
Fig. 5.
Elevation of bile acid levels and increased bile acid excretion rate in Little mice. Biliary, serum, and liver bile acids were measured in Little mice as described in the experimental procedures. (A) Little mice (n = 5) showed significantly elevated levels of total biliary bile acids (P < 0.02) and biliary cholic acid (P < 0.005) as compared to wild-type mice (n = 5). (B) Little mice (n = 7) also showed significantly elevated levels of total serum bile acids (P < 0.0001), serum cholic acid (P < 0.002), and serum β-muricholic acid (P < 0.02) as compared to wild-type mice (n = 8). (C) Total liver bile acid levels were also significantly elevated (P < 0.0001) in Little mice (n = 25) as compared to wild-type mice (n = 25). (C) Bile acid excretion rates were determined as described in the experimental procedures. Little mice (n = 5) had a significantly higher bile acid excretion rate (P < 0.0001) than the wild-type mice (n = 7). P-values shown refer to a Student’s t-test, bars represents the mean ± (2 × SE).
Table 4.
Serum bile acid levels in Little mice and wild-type mice
| Identification | Little mice (µM/L) | Wild-type mice (µM/L) | Mutant/wild-type ratio | P-value* |
|---|---|---|---|---|
| Lithocholic acid | 0.37 | 0.10 | 3.7 | 0.0006 |
| Deoxycholic acid | 1.39 | 0.38 | 3.7 | 0.0014 |
| Chenodeoxycholic acid | 2.03 | 0.40 | 5.1 | 0.0532 |
| Cholic acid | 3.56 | 1.03 | 3.5 | 0.0018 |
| Ursodeoxycholic acid | 2.79 | 0.99 | 2.8 | < 0.0001 |
| α-muricholic acid | 0.00 | 0.09 | 0.0 | 0.0049 |
| β-muricholic acid | 10.28 | 1.90 | 5.4 | 0.0228 |
| Total | 20.42 | 4.89 | 4.18 | 0.0018 |
The P-value refers to a Student’s t-test for the difference in bile acid levels between Little mice and wild-type controls as determined by gas chromatographymass spectrometry.
The elevation of bile acid levels in Little mice prompted us to analyze the expression levels of bile acid synthesis genes (Table 5). The main bile acid biosynthetic pathway (neutral) is initiated by Cyp7a1 (cholesterol 7α-hydroxylase). Cyp7a1 is considered to catalyze the rate-limiting step in the classic bile acid synthetic pathway and, like other genes in this pathway, is known to be regulated at the transcriptional level. We did not detect, however, any significant alteration in the expression levels of Cyp7a1 in Little mice. We found, instead, alterations in many other genes in the pathway. For example, Cyp39a1, Akr1d1, Baat, and Slc27a5 were significantly up-regulated. Oxysterol 7α-hydroxylase (Cyp7b1), in the alternative (acidic) pathway, was dramatically down-regulated and the expression level in Little mice was less than 10% of the wild-type level. In addition, Cyp27a1 (sterol 27-hydroxylase) was slightly down-regulated. At this moment, it is not clear how these complex alterations could contribute to the elevation of bile acids in Little mice.
Table 5.
Bile acid synthesis genes in Little mice
| Gene | Description | Fold-change* | P-value† |
|---|---|---|---|
| Acox2 | Acyl-coenzyme A oxidase 2 | 1.1 | 0.6167 |
| Akr1d1 | 3-oxo-5-β-steroid 4-dehydrogenase | 3.1 | 0.0057 |
| Amacr | 2-methylacyl-CoA racemase | 0.8 | 0.5539 |
| Baat | Bile acid coenzyme A: amino acid N-acyltransferase | 1.2 | 0.0389 |
| Ch25h | Cholesterol 25-hydroxylase | 1.2 | 0.6596 |
| Cyp27a1 | Sterol 27-hydroxylase | 0.7 | 0.0322 |
| Cyp39a1 | Cytochrome p450, 39a1 | 8.4 | 0.0010 |
| Cyp46a1 | Cholesterol 24-hydroxylase | 0.8 | 0.5057 |
| Cyp7a1 | Cholesterol 7α-hydroxylase | 0.5 | 0.2224 |
| Cyp7b1 | Oxysterol 7α-hydroxylase | 0.04 | < 0.0001 |
| Cyp8b1 | Sterol 12α-hydroxylase | 0.7 | 0.2508 |
| Dhrs9 | 3α-hydroxysteroid dehydrogenase | 2.3 | 0.1061 |
| Hsd17b4 | D-bifunctional protein | 1.5 | 0.1976 |
| Scp2 | Peroxisomal thiolase 2 | 0.5 | 0.0058 |
| Slc27a5 | Bile acid CoA ligase | 1.6 | 0.0270 |
The fold-change refers to the ratio of the expression values of Little mice over the wild-type mice obtained by real-time PCR analysis.
The P-value refers to a Student’s t-test for the difference in expression between Little and wild-type mice.
Bile acids are synthesized from cholesterol in the liver and alterations in bile acid metabolism usually cause or reflect corresponding alterations in cholesterol metabolism. We observed that Little mice had an increased expression of the genes in the de novo cholesterol synthesis pathway (Table 6). These genes included HMG-CoA synthase, HMG-CoA reductase, mevalonate kinase, mevalonate decarboxylase, squalene synthase, squalene epoxidase, and lanosterol demethylase. Interestingly, however, the cholesterol level in the liver of Little mice was not elevated, and cholesterol plasma levels were actually significantly decreased (P = 0.002) in Little mice (n = 5) to about 70% of that in the normal wild-type mice (n = 5) levels. That Little mice maintain normal liver cholesterol levels despite the up-regulation of cholesterol biosynthesis genes is consistent with an elevation in bile acid biosynthesis.
Table 6.
Cholesterol synthesis genes in Little mice
| Gene | Description | Fold-change* | P-value† |
|---|---|---|---|
| Cyp51 | Lanosterol demethylase | 2.2 | 0.0379 |
| Fdft1 | Squalene synthetase | 2.7 | 0.0008 |
| Fdps | Farnesyl pyrophosphate synthetase | 1.3 | 0.3662 |
| Hmgcr | 3-hydroxy-3-methylglutaryl-coenzyme A reductase | 5.3 | 0.0668 |
| Hmgcs | Hydroxymethylglutaryl-CoA synthase | 3.1 | 0.0166 |
| Mvd | Diphosphomevalonate decarboxylase | 2.3 | 0.0461 |
| Mvk | Mevalonate kinase | 1.9 | 0.0052 |
| Sqle | Squalene epoxidase | 2.4 | 0.0500 |
The fold-change refers to the ratio of the expression values of Little mice over the wild-type mice obtained by real-time PCR analysis.
The P-value refers to a Student’s t-test for the difference in expression between Little and wild-type mice.
Cholic acid treatment in wild-type mice up-regulates a largely overlapping set of xenobiotic detoxification genes
It is possible that the elevation in bile acids, and in particular CA, contributes to the up-regulation of xenobiotic metabolism genes in Little mice. We therefore decided to test if CA treatment of wild-type mice produced an up-regulation of xenobiotic genes similar to Little mice. Wild-type mice were given a diet supplemented with 2% CA for 7 days. All animals tolerated the CA diet well, remained in general good health and displayed no mortality associated with the treatment. We found that out of the testing set of 30 xenobiotic genes shown to be up-regulated in Little mice, 23 genes were significantly up-regulated in wild-type mice after CA treatment (Table 7). An additional three genes had a clear tendency to be up-regulated but did not reach statistical significance at the 0.05 level.
Table 7.
CA treatment in wild-type mice up-regulates a largely overlapping set of detoxification metabolism genes as in Little mice
| Wildtype CA-treated mice |
|||
|---|---|---|---|
| Gene | Description | Fold-change* | P-value† |
| Abcb1a | ATP-binding cassette, subfamily B (MDR/TAP), member 1 A | 15.5 | < 0.0001 |
| Aldh1a1 | Aldehyde dehydrogenase family 1, subfamily A1 | 0.7 | 0.2099 |
| Cyp2a4 | Cytochrome p450, 2a4 | 5.1 | 0.0025 |
| Cyp2b10 | Cytochrome p450, 2b10 | 4.4 | 0.0047 |
| Cyp2b13 | Cytochrome p450, 2b13 | 31.9 | 0.0009 |
| Cyp2b9 | Cytochrome p450, 2b9 | 473.4 | < 0.0001 |
| Cyp2c38 | Cytochrome p450, 2c38 | 0.9 | 0.6353 |
| Cyp4a10 | Cytochrome p450, 4a10 | 5.1 | 0.0148 |
| Cyp4a14 | Cytochrome p450, 4a14 | 71.9 | 0.0001 |
| Cyt19 | Arsenite methyltransferase | 2.2 | 0.0749 |
| Fmo3 | Flavin containing monooxygenase 3 | 124.2 | 0.0001 |
| Fmo4 | Flavin containing monooxygenase 4 | 3.4 | 0.0091 |
| Gsta2 | Glutathione S-transferase, alpha 2 | 17.8 | < 0.0001 |
| Gsta4 | Glutathione S-transferase, alpha 4 | 9.1 | 0.0001 |
| Gstm2 | Glutathione S-transferase, mu 2 | 4.3 | 0.0091 |
| Gstm3 | Glutathione S-transferase, mu 3 | 36.2 | 0.0035 |
| Gsto2 | Glutathione transferase omega 2 | 0.5 | 0.0399 |
| Gstt2 | Glutathione S-transferase theta 2 | 4.6 | 0.0166 |
| Mgst3 | Microsomal glutathione S-transferase 3 | 9.7 | 0.0004 |
| mOAT6 | Solute carrier family 22 (organic anion transporter), member 20 | 8.4 | 0.0004 |
| Mt1 | Metallothionein 1 | 7.5 | 0.0090 |
| Papss2 | 3′-phosphoadenosine 5′-phosphosulfate synthase 2 | 2.4 | 0.0204 |
| Por | Cytochrome p450 reductase | 4.0 | 0.0007 |
| Slc21a5 | Solute carrier organic anion transporter family, member 1a5 | 9.9 | 0.0243 |
| Sth2 | Sulfotransferase family 2A, member 2 | 9.3 | 0.0007 |
| Sult1a1 | Sulfotransferase family 1A, phenol-preferring, member 1 | 4.4 | 0.0005 |
| Sult1d1 | Sulfotransferase family 1D, member 1 | 6.8 | 0.0292 |
| Sult1e1 | Sulfotransferase family 1E, member 1 | 5.8 | 0.0996 |
| Temt | Indolethylamine N-methyltransferase | 0.9 | 0.8868 |
| Ugt1a1 | UDP glucuronosyltransferase 1 family, polypeptide A1 | 4.1 | 0.1843 |
The fold-change refers to the ratio of the expression values of the CA-treated mice over the wild-type mice obtained by real-time PCR analysis.
The P-value refers to a Student’s t-test for the difference in expression between CA-treated mice and wild-type mice.
As expected (Wang et al., 2002), CA treatment of wild-type mice decreased the expression of bile acid synthesis genes such as Cyp7a1, Cyp8b1, and up-regulated the nuclear receptor Shp (short heterodimer partner) involved in the transcriptional repression of bile acid synthesis genes. Interestingly, CA-treated Little mice also showed these transcriptional changes indicating that these mechanisms of negative feedback regulation of bile acid synthesis are intact in these mice (Table 8).
Table 8.
Selected genes involved in bile acid homeostasis
| Little mice |
CA-treated wild-type mice |
CA-treated Little mice |
Fxr mice |
Fxr/Little mice |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Fold-change* | P-value† | Fold-change* | P-value† | Fold-change* | P-value† | Fold-change* | P-value† | Fold-change* | P-value† |
| Cyp7a1 | 0.5 | 0.2224 | 0.04 | 0.0016 | 0.01 | 0.0010 | 1.4 | 0.5168 | 2.6 | 0.1248 |
| Cyp8b1 | 0.7 | 0.2508 | 0.004 | 0.0020 | 0.003 | < 0.0001 | 2.1 | 0.0239 | 2.0 | 0.0454 |
| Shp | 0.7 | 0.3886 | 4.9 | 0.0073 | 3.9 | 0.0249 | 0.1 | 0.0008 | 0.01 | 0.0096 |
| Slc21a1 | 0.002 | < 0.0001 | 0.03 | 0.0036 | 0.004 | < 0.0001 | 0.9 | 0.5462 | 0.002 | 0.0001 |
| Slc21a5 | 5.7 | 0.0004 | 9.9 | 0.0243 | 5.9 | 0.0263 | 0.8 | 0.2927 | 2.6 | 0.0017 |
| Bsep | 1.1 | 0.8489 | 0.8 | 0.8252 | 0.7 | 0.5674 | 0.2 | 0.0011 | 0.05 | 0.0079 |
| Ntcp | 0.3 | 0.1089 | 0.2 | 0.0422 | 0.2 | 0.0855 | 0.1 | 0.0013 | 0.1 | 0.0014 |
The fold-change refers to the ratio of the expression values of each experimental group over the wild-type mice obtained by real-time PCR analysis.
The P-value refers to a Student’s t-test for the difference in expression between each experimental group and wild-type mice.
Fxr/Little mice
The primary bile acid receptor Fxr (farnesoid × receptor, a nuclear hormone receptor) plays a critical role in the regulation of various steps in bile acid homeostasis, including bile acid synthesis and transport (Chiang, 2004). Loss of Fxr leads to increased susceptibility to bile acid-induced injury. It was previously observed that when Fxr mice were fed a diet supplemented with CA (2%, 7 days) they exhibited severe wasting, hypothermia, drastic reduction of adipose tissue, and a mortality of 30% by day 7 (Sinal et al., 2000).
Because of the overall elevation of bile acid levels in Little mice, we considered that Fxr may have a protective role in these mice and perhaps also contributed to the up-regulation of xenobiotic detoxification genes. To investigate these ideas, we generated double mutant mice for Ghrhr and Fxr (designated as Fxr/Little mice). The Fxr/Little mice showed several signs of poor health: they were generally lethargic, weak, had greatly reduced adipose tissue and noticeable jaundice. An histological examination of livers extracted from these double-mutant mice revealed extensive damage and morphologic alterations, including steatosis, isolated necrosis, prominent and proliferating bile ducts, and focal inflammation (Fig. 6A). We also observed a dramatic elevation of liver and serum bile acid levels in the Fxr/ Little mice (Fig. 6B,C). Serum bile acid levels were over 150-fold elevated in the Fxr/Little mice as compared to wild-type mice (P < 0.0001) and over 50-fold elevated as compared to Little mice or Fxr mice (P < 0.0001 for both comparisons). Liver bile acid levels were over 10-fold elevated in the Fxr/Little mice as compared to wild-type mice (P < 0.0001) and over five-fold elevated as compared to Little mice or Fxr mice (P < 0.0001 for both comparisons). These observations provided further confirmation of a dysregulation of bile acid homeostasis in Little mice.
Fig. 6.
Liver histology and elevation of bile acids in the Fxr/Little mice. (A) An histological examination (hematoxylin and eosin staining) of the livers extracted from these mice revealed extensive damage and morphologic alterations, including steatosis, isolated necrosis, prominent and proliferating bile ducts, and focal inflammation. (B) Liver bile acid levels were over 10-fold elevated in the Fxr/Little (n = 4) mice as compared to wild-type mice (n = 25) (P < 0.0001) and over five-fold elevated as compared to Little mice (n = 25) or Fxr mice (n = 9) (P < 0.0001 for both comparisons). (C) Serum bile acid levels were over 150-fold elevated in the Fxr/Little mice as compared to wild-type mice (P < 0.0001) and over 50-fold elevated as compared to Little mice or Fxr mice (P < 0.0001 for both comparisons). P-values shown refer to a Student’s t-test, bars represents the mean ± (2 × SE).
Importantly, the loss of Fxr had a major effect on the expression of xenobiotic detoxification genes up-regulated in Little mice (Table 2). Several of these genes lost (13 genes) or noticeably decreased (4 genes) their up-regulation in the Fxr/ Little mice. The genes Abcb1a, Aldh1a1, Cyp2b10, Cyp2c38, Cyp4a10, Fmo3, Gsto2, Gstt2, Papp2s, Por, Sult1d1, Temt, and Ugt1a1 were no longer up-regulated in the Fxr/Little mice. The magnitude of the up-regulation in the genes Cyp2b13, Cyp2b9, Cyp4a14, mOat6 was decreased in the Fxr/Little mice as compared to Little mice. Interestingly, a few genes showed an opposite effect and were actually more strongly up-regulated in the Fxr/Little mice than in Little mice. These genes included Gsta2, Gstm2, Gstm3, Mt1, and Sult1e1.
Discussion
Up-regulation of xenobiotic detoxification genes and increased xenobiotic resistance in Little mice
An elevation of xenobiotic metabolism has been recently proposed as a mechanism of extended longevity. The main hypothesis is that the molecular and cellular damage produced by the toxic compounds that this system targets, in particular toxic lipophilic by-products of metabolism, is a major contributor to the aging process (Gems & McElwee, 2005). Here, we investigated the alterations in xenobiotic metabolism in the long-lived Little mice.
These mutant mice showed a remarkably strong and concerted transcriptional up-regulation of xenobiotic detoxification genes. We found that this transcriptional up-regulation was associated with significant in vivo alterations in xenobiotic resistance. Little mice displayed a potent increase in resistance against the adverse effects of the hepatotoxins acetaminophen and bromobenzene and against the paralyzing agent zoxazolamine. The increased resistance against zoxazolamine can be explained by the up-regulation of cytochromes p450s in Little mice, because the rate of zoxazolamine inactivation directly reflects cytochrome p450 activity.
The basis for acetaminophen toxicity resides largely in its conversion to the highly reactive intermediate N-acetyl-p-benzoquinone imine (NAPQI) that covalently binds to cellular macromolecules. NAPQI is normally detoxified by glutathione S-transferases through conjugation with glutathione (GSH). However, NAPQI can accumulate if its rate of production exceeds the rate of GSH-conjugation or when GSH levels are depleted (Hinson et al., 2004). The increased expression of many glutathione S-transferase genes in Little mice, which should rapidly detoxify NAPQI after its formation from acetaminophen, likely contributes to the increased resistance against acetaminophen-toxicity in these mice. This agrees with previous studies that show that pharmacological inducers of phase II enzymes, in particular glutathione S-transferases, protect against acetaminophen hepatotoxicity in mice (Ansher et al., 1983; Seo et al., 2000).
Although Little mice up-regulate cytochrome p450s, and therefore are also expected to produce the toxic intermediate (NAPQI) at a higher rate, the resistance of Little mice against this compound suggests that the up-regulation of glutathione S-transferases increases the rate of conjugation (detoxification) to compensate for increased NAPQI production. This situation could potentially lead to the depletion of glutathione; however, besides conjugation with glutathione, acetaminophen can also be detoxified by sulfation or glucuronidation to form acetaminophen-sulfate and acetaminophen-glucuronide (Brouwer & Jones, 1990; Zamek-Gliszczynski et al., 2006). The increased expression of various sulfotransferases (Sth2, Sult1a1, Sult1d1, and Sult1e1) and the glucuronosyltransferase Ugt1a1 in Little mice could increase their capacity for acetaminophen sulfation and glucuronidation and likely contributes to their enhanced resistance against this compound and would also ameliorate glutathione depletion by enhancing this alternative route of acetaminophen detoxification. Extensive sulfation can cause a depletion of the universal sulfonate donor 3′-phosphoadenosine 5′-phosphosulfate (PAPS). Importantly, Little mice presented a significant up-regulation of Papss2 (3′-phosphoadenosine 5′-phosphosulfate synthase 2), which is responsible for the biosynthesis of the sulfate donor PAPS in mouse liver. Other mouse models of extended longevity have also shown increased resistance against acetaminophen-induced toxicity, such as calorie-restricted mice and methionine-restricted mice (Miller et al., 2005; Harper et al., 2006).
Bromobenzene toxicity depends on its conversion by cytochrome p450s to a toxic intermediate (bromobenzene 3,4-epoxide) that is subsequently detoxified by glutathione S-transferases. The resistance of Little mice against this compound should share a similar basis as with their resistance against acetaminophen.
Interestingly, there is some direct experimental evidence for a role of glutathione S-transferases in longevity. Overexpression of C. elegans gst-10 or mouse Gsta4 increases stress resistance and lifespan in C. elegans (Ayyadevara et al., 2005). Both of these GSTs have high catalytic efficiency toward the conjugation of the lipid peroxidation product 4-hydroxynonenal (4-HNE), which is known to accumulate with age in diverse systems (Sohal & Weindruch, 1996; Levine & Stadtman, 2001; Sohal et al., 2002; Zheng et al., 2005). The up-regulation of Gsta4 in Little mice suggests that the elevation of glutathione S-transferase activity may be a mechanism of longevity assurance shared between nematodes and mice.
In contrast with the increased resistance to acetaminophen and bromobenzene-induced toxicity, we found that Little mice were more sensitive to carbon tetrachloride-induced hepatotoxicity. The toxic effects of CCl4 come from its activation by cytochrome p450s to form trichloromethyl free radical (CCl3), which readily reacts with oxygen to form CCl3O2. These highly reactive species initiate cellular damage by covalent binding to macromolecules and by inducing lipid peroxidation (Weber et al., 2003). Interestingly, although the mechanisms of CCl4 toxicity have been studied extensively, a role for glutathione S-transferases or sulfotransferases in the detoxification of its toxic intermediates has not been described (Weber et al., 2003) and some studies suggest that the CCl4 free radical products are unlikely to be detoxified by a GSH-dependent mechanism (Burk et al., 1984). Therefore, the increased sensitivity of Little mice to CCl4-induced hepatotoxicity must be a consequence of their elevation in cytochrome p450s. In this case, however, in contrast with the two previously discussed hepatotoxins, the elevation in glutathione S-transferases or sulfotransferases cannot provide a protective effect.
The classic xenobiotic receptors Car and Pxr are key regulators of all phases of xenobiotic metabolism and several of their target genes are up-regulated in Little mice. However, the results of our experiments using the double mutants Car/Little and Pxr/ Little and the triple mutant Car/Pxr/Little demonstrate that, with the exception of Cyp2b10 and Cyp2c38, these xenobiotic nuclear receptors are not required for the up-regulation of xenobiotic genes in Little mice. There are a number of other nuclear receptors and transcription factors that could be involved in this up-regulation, including Aryl hydrocarbon receptor (Ahr), Peroxisome proliferator activated receptors (Ppar), Vitamin D receptor (Vdr), NF-E2-related factor-2 (Nrf2), and others (Wang & LeCluyse, 2003; Xu et al., 2005). The Nrf2 transcription factor is an interesting candidate. Hepatocyte-specific deletion of the Nrf2-repressor Keap1 constitutively activates Nrf2 and up-regulates a set of xenobiotic genes that partially overlaps with the genes up-regulated in Little mice. These overlapping genes include Sth2, Cyp2b13, Fmo3, Cyp2b9, Gstm3, Mgst3, and Gsta2 (Okawa et al., 2006). Additionally, like Little mice, the Keap1 mice have increased resistance against acetaminophen-induced hepatotoxicity (Okawa et al., 2006).
We found that the Little mutation reverses the susceptibility of the Car mice against zoxazolamine induced-paralysis, and the susceptibility of the Car/Pxr mice against acetaminophen-induced liver necrosis. It has been previously reported that loss of Car function significantly increases zoxazolamine sensitivity (Wei et al., 2000). The constitutive up-regulation of cytochrome p450s introduced by the Little mutation is likely to be responsible for the loss of sensitivity against zoxazolamine-induced paralysis in the Car/Little double mutant mice. We found that the combined loss of Car and Pxr results in considerably increased susceptibility to acetaminophen-induced hepatotoxicity. Although at this time the precise mechanisms for the increased toxicity in Car/Pxr mice are unclear, they may be related to the failure to activate key phase I or phase II xenobiotic detoxification genes. Most likely, the constitutive up-regulation of xenobiotic detoxification genes associated with the introduction of the Little mutation is responsible for reversing this susceptibility in the Car/Pxr/Little mice.
Bile acids as potential mediators of the xenobiotic response in Little mice
Little mice showed significantly elevated bile acid levels in bile, liver, and plasma. Our data indicated that this elevation was primarily a consequence of a generalized increase of bile acid synthesis, because the fecal bile acid excretion rate, which is directly proportional to the amount synthesized in the liver, was considerably elevated in Little mice. We found that the expression of the canalicular bile salt export pump (Bsep), the major bile salt export pump in rodents, and Mrp2, another major bile salt export pump, were not affected in Little mice implying that bile acid export was not compromised. However, we observed some alterations in genes involved in bile acid uptake by the liver. The Na+-independent organic anion-transporting polypeptide Slc21a1, important for bile salt uptake into hepatocytes, was greatly down-regulated in Little mice and this may contribute to the elevation in plasma bile acids levels. This down-regulation may constitute a defense response against the elevation of liver bile acids because Slc21a1 was also down-regulated in the CA-treated wild-type mice. Interestingly, Slc21a5, which shows overlapping substrate specificities to Slc21a1, is highly up-regulated in Little mice and in the CA-treated wild-type mice. It has been proposed that Slc21a5 plays an important role in situations in which the expression of Slc21a1 is compromised and its up-regulation may represent an important response in the hepatic detoxification of bile acids (Meier & Stieger, 2002). Although Little mice could be regarded as mildly cholestatic, the elevation of bile acids in Little mice does not induce a pathological state of the liver. Little mice do not show histological signs of liver damage and their ALT and LDH serum levels are normal.
The analysis of biliary bile acids showed an almost four-fold elevation in the levels of CA, which also resulted in an altered bile acid composition in Little mice with a two-fold increase in the proportion (as a percentage of total bile acids) of this bile acid. We found that CA treatment of wild-type mice up-regulated a largely overlapping set (close to 80%) of the xenobiotic genes up-regulated in Little mice. These observations support the idea that bile acids, in particular CA, play a role in the up-regulation of these genes in Little mice. Previously, CA has been shown to up-regulate Cyp2b10, Cyp3a11 and affect the expression of bile acid transporters (Fickert et al., 2001; Zollner et al., 2006), but such a broad response on the expression of phase I and phase II detoxification genes has not, to our knowledge, been documented before.
Some of the genes up-regulated in CA-treated wild-type mice do not reach the levels seen in Little mice. This may be due to the fact that the elevation in bile acids in Little mice is a chronic state vs. the acute nature of CA treatment or it may be an indication that other mechanisms (including the elevation in other bile acids) also play a role in the induction of these genes in Little mice. Treatment of Little mice with CA yielded some interesting observations. Little mice, just like wild-type mice, decreased the expression of the bile acid synthesis genes Cyp7a1 and Cyp8b1, and up-regulated the nuclear receptor Shp after CA treatment. This indicated that the transcriptional mechanisms of negative feedback regulation of bile acid synthesis, which are largely dependent on Fxr, are functional in Little mice. Because the basal expression levels of these genes are not affected in Little mice, it is possible that they have a higher threshold for the activation of these negative feedback mechanisms than normal mice. The two- or three-fold elevation of bile acids in these mutant mice may not be enough to activate this response, but when there is a very high or sudden elevation in bile acid levels, as with cholic acid treatment, the response can be triggered.
We are proposing that the elevation of bile acid levels, and in particular CA levels, is a likely contributor to the increased expression of xenobiotic detoxification genes and increased xenobiotic resistance in Little mice. However, the elevation of other endobiotics (some of them may be other bile acids) could also contribute. Furthermore, in addition to the proposed contribution of bile acids to the elevation of xenobiotic detoxification genes, it is possible that the simple increase in bile acid excretion rates, by itself, may be an important contributor to the increased xenobiotic resistance in Little mice. Besides renal excretion, biliary excretion is a major mechanism involved in the elimination of xenobiotics, and an increase in the rate at which conjugated xenobiotics are excreted into bile is expected to decrease their toxicity. Of particular relevance to the increased resistance against acetaminophen and bromobenzene in Little mice, sulfate and glucuronide metabolites of acetaminophen are known to be excreted into bile (Brouwer & Jones, 1990; Zamek-Gliszczynski et al., 2006), as well as bromobenzeneglutathione conjugates (Madhu & Klaassen, 1992).
Our analysis of the Fxr/Little double mutant mice further confirmed a dysregulation of bile acid homeostasis in Little mice. We observed a remarkable synergistic effect in the elevation of bile acid levels in the Fxr/Little mice. This dramatic elevation in bile acid levels occurred without a significant up-regulation of Cyp7a1 in the Fxr/Little mice as compared to either wild-type or Fxr mice (Table 8). However, the expression of canalicular Bsep is severely decreased in Fxr mice, and this down-regulation, not present in Little mice, is maintained in the Fxr/Little mice (Table 8). It is likely that the combination of the down-regulation of Bsep in Fxr mice with the increased bile acid biosythesis in Little mice is a primary cause for the accumulation and elevation of bile acid levels in these double mutants. The liver damage seen in the Fxr/Little mice is very likely a consequence of bile acid accumulation and toxicity. In particular, the prominent bile ducts seen in the livers of the Fxr/Little mice are likely to be related to the elevation in bile acid levels, because it has been previously shown that bile acid feeding enhances bile duct proliferation while external bile drainage decreases proliferation (Alpini et al., 1999; Alvaro et al., 2000).
Importantly, many of the xenobiotic detoxification genes up-regulated in Little mice lost completely, or to some extent, their elevated levels of expression in the Fxr/Little mice. The majority of these genes are not known targets, direct or indirect, of Fxr. It is possible that these genes represent previously unidentified Fxr targets, and that the loss of their up-regulation contributes to the accumulation of bile acids and to the liver damage phenotype of the Fxr/Little mice. These observations support a model in which bile acids, through the activation of Fxr, up-regulate a large portion of the xenobiotic detoxification genes in Little mice. As mentioned before, Fxr plays a central role in bile acid homeostasis by regulating bile acid synthesis genes and bile acid transporters. The regulation by Fxr of xenobiotic genes involved in bile acid detoxification would be consistent with this role and would add another level to the regulation of bile acid homeostasis by this nuclear receptor. However, there is a possibility that the down-regulation of these genes in the Fxr/ Little mice could be a nonspecific consequence of liver damage and hepatotoxicity and further investigations are required to clarify this issue.
Our results support the hypothesis that alterations in xenobiotic metabolism and increased xenobiotic resistance is a trait associated with longevity in mice. Our analysis leads us to propose that bile acids, possibly acting through Fxr, may be important mediators of this response. The next step to further these hypotheses would be to test the effects that a chronic (possibly lifelong) elevation of xenobiotic detoxification genes would have on aging and longevity. This could be achieved through chemical intervention by using drugs (we would propose the use of bile acid-like compounds) that induce expression of xenobiotic detoxification genes, through the use of additional mouse models with a constitutive elevation of xenobiotic detoxification genes such as the Keap1 knockout mice (Okawa et al., 2006) or by genetic interventions that involve the overexpression of constitutively active forms of nuclear receptors (we would propose Fxr as an interesting candidate) that regulate the xenobiotic response.
Experimental procedures
Mice
Little mice breeders were purchased from Jackson laboratories. Ghrhrlit/Ghrhrlit females and Ghrhrlit/+ males were crossed to produce the Ghrhrlit/+ and Ghrhrlit/Ghrhrlit male mice used in these experiments. Heterozygote Ghrhr+/lit or wild-type Ghrhr+/+ littermates were used as controls for Little mice. The generation of Car/Little was performed as follows: Car−/− males were crossed with Ghrhr+/lit females. The resulting male and female Car+/−; Ghrhr+/lit double heterozygote mice were crossed to generate Car−/−; Ghrhr+/lit breeders from which the Car−/−; Ghrh lit/lit (Car/Little) double mutants were obtained. The Pxr/Little and Fxr/Little double mutants and the Car/Pxr/Little triple mutants were generated similarly. The Car, Pxr, and Fxr mice were obtained from Moore’s laboratory. Car, Pxr, and Fxr knockout mice were generated as previously described (Sinal et al., 2000; Wei et al., 2000; Staudinger et al., 2001). Animals were weaned at the age of 21–30 days and housed in groups of two to four animals of the same gender. Animals were housed in a pathogen-free animal facility with controlled photoperiod of 12 h light:12 h darkness (lights on from 07:00 to 19:00 hours) and a temperature of 22 ± 2 °C. Animals were given free access to water and pelleted diet (5010 rodent diet, LabDiet, PMI Nutrition International, Brentwood, MO, USA). All the animals used in these experiments were 2–4 months old. All animal protocols followed the provisions of the Animal Welfare Act and were approved by the Institutional Animal Care and Use Committee at Baylor College of Medicine.
Xenobiotic resistance experiments and cholic acid feeding
Mice were given a single intraperitoneal dose of acetaminophen (375 mg kg−1), CCl4 (50 µL kg−1), bromobenzene (0.36 mL kg−1), or zoxazolamine (250 mg kg−1). CCl4, bromobenzene, and zoxazolamine were dissolved in corn oil at a concentration of 5 µL mL−1, 36 µL mL−1, and 25 mg mL−1, respectively. Acetaminophen was dissolved in phosphate-buffered saline at a concentration of 37.5 mg mL−1. All chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA). At the end of the experiments, the animals were anesthetized with isoflurane and sacrificed by cervical dislocation, and the liver and blood were collected. For histological examination, the major lobe of the livers was fixed in 10% buffered formalin, embedded in paraffin, sectioned at 5 µm, and stained with hematoxylin and eosin. Samples were examined under a light microscope. For RNA isolation the rest of the liver was flash frozen in liquid nitrogen and stored at −80 °C until use. Serum was obtained from whole blood by centrifugation at 20 800 g for 10 min at 4 °C, using ethylenediaminetetraacetic acid coated tubes. ALT, LDH, and cholesterol measurements were performed in the clinical chemistry laboratory at the Texas Children’s Hospital, Houston, TX, USA. The quantification of the necrotic areas was performed in three random fields (at a magnification of × 100) from each animal in the different experimental groups using the NIH ImageJ software (Bethesda, MD, USA; http://rsb.info.nih.gov/ij/). The levels of necrosis were reported as a percentage of the total area. All chemicals were purchased from Sigma. For the cholic acid treatment experiments, wild-type and Little mice were fed mouse diet supplemented with 2% CA (LabDiet, PMI Nutrition International) for 7 days. At the end of this period, the serum and liver from these animals were collected for analysis.
Bile acids measurements
For the collection of bile, mice were put under general anesthesia (avertin). After distal ligation of the common bile duct, the gall bladder was cannulated and bile was collected for a period of 15 min. The amount of bile collected was roughly proportional to the weight of the animals, about 25–30 µL for wild-type mice and about 15–20 µL for Little mice. Bile samples were immediately frozen at −20 °C.
Biliary and serum bile acid analysis in Little mice (total and individual bile acids) was performed by gas chromatographymass spectrometry of the methyl ester-trimethylsilyl ethers after liquid-solid extraction, solvolysis and enzymic hydrolysis of the bile acid conjugates, and anion exchange chromatographic separation and purification of the hydrolyzed bile acids. The methods for bile acid analysis have been extensively described previously (Setchell et al., 1998). Bile acid concentrations were quantified by gas chromatography from a comparison of their peak area response with the peak area response obtained for the known amount of the added internal standard, nordeoxycholic acid (Setchell & Matsui, 1983). The identification of a bile acid was based upon its gas chromatography retention index relative to a homologous series of n-alkanes (methylene-unit value) and the electron ionization mass spectrum compared to a published library of reference bile acids (Lawson & Setchell, 1988).
Bile acid extraction from liver was performed by homogenization in 75% ethanol (1.5 mL per 100 mg of liver) followed by incubation at 50 °C for 2 h with intermittent shaking. After clearance by centrifugation the supernatant was used for bile acid measurements. To determine fecal bile acid excretion mice were housed individually and feces were collected over a period of 3 days. Feces were dried, weighed, finely ground using a mortar and pestle and 50 mg were extracted with 1 mL of 75% ethanol and incubation at 50 °C for 2 h with intermittent shaking. The rate of bile acid excretion was calculated from the fecal bile acid content (µM g −1) and the daily feces output (g day −1 per 100 g of body weight). Liver and fecal bile acid content was measured enzymatically using a kit from Diagnostic Chemical Limited (Carlottetown, PE Canada) according to the manufacturer’s protocol. Total bile acid serum levels were also determined by this method. Liver cholesterol measurements were performed using a cholesterol quantitation kit from Biovision (Mountain View, CA, USA) according to the manufacturer’s protocol.
Real-time PCR
Total RNA was extracted from frozen livers using the Quiagen RNeasy purification kit (Qiagen, Valencia, CA, USA) in accordance with manufacturer’s protocols. DNase-treated total liver RNA was reverse transcribed using SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA, USA) according to the vendor’s protocol. Real-time PCR was performed using SYBR Green PCR Master Mix and the ABI Prism 7000 Sequence Detection System (Applied Biosystems, Foster city, CA, USA). B-actin was used as a normalizer. Amplification specificity of all targets was confirmed by melting curve analysis. Thermal cycling conditions consisted of an initial step at 95 °C for 10 min to activate the Taq DNA polymerase and 40 cycles of sequential denaturation at 95 °C for 15 s and annealing/extension at 60 °C for 60 s. Data analysis was performed using the ABI Prism 7000 SDS software (Applied Biosystems). The primers used are listed in the Supplementary Material. The real-time PCR analysis was performed according to the comparative CT method. The fold changes were calculated according to the expression: 2−ΔΔCT For example: ΔΔCT = ΔCT, experimental − ΔCT, wild-type. Where, ΔCT, experimental = Average CT, target gene − Average CT, β-actin and ΔCT, wild-type = Average CT, target gene − Average CT, β-actin. The averages CT values for experimental and control wild-type mice were based on three to six biological replicates. The CT value for each biological replicate represents the average of three technical replicates. The P-values reported for these changes refer to a two-tailed T-test (assuming homoscedasticity) between the normalized CT values (CT, target gene – CT, β-actin) in experimental vs. control mice.
Supplementary Material
Acknowledgments
We would like to thank Dorene M. Rudmad for her help in the preparation of histological samples and Dr Ching-Nan Ou for his help in the measurement of ALT, LDH, and cholesterol serum levels. We also thank Dr Milton Finegold for his expertise in the histological analysis of liver samples. This work was supported by AG 20752, DK 56338, and The Ellison Medical Foundation.
Footnotes
Supplementary material
The following supplementary material is available for this article:
This material is available as part of the online article from: http://www.blackwell-synergy.com/doi/abs/10.1111/j.1474-9726.2007.00300.x
(This link will take you to the article abstract).
Please note: Blackwell Publishing are not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Alpini G, Glaser SS, Ueno Y, Rodgers R, Phinizy JL, Francis H, Baiocchi L, Holcomb LA, Caligiuri A, LeSage GD. Bile acid feeding induces cholangiocyte proliferation and secretion: evidence for bile acid-regulated ductal secretion. Gastroenterology. 1999;116:179–186. doi: 10.1016/s0016-5085(99)70242-8. [DOI] [PubMed] [Google Scholar]
- Alvaro D, Gigliozzi A, Attili AF. Regulation and deregulation of cholangiocyte proliferation. J. Hepatol. 2000;33:333–340. doi: 10.1016/s0168-8278(00)80377-3. [DOI] [PubMed] [Google Scholar]
- Amador-Noguez D, Yagi K, Venable S, Darlington G. Gene expression profile of long-lived Ames dwarf mice and Little mice. Aging Cell. 2004;3:423–441. doi: 10.1111/j.1474-9728.2004.00125.x. [DOI] [PubMed] [Google Scholar]
- Ansher SS, Dolan P, Bueding E. Chemoprotective effects of two dithiolthiones and of butylhydroxyanisole against carbon tetrachloride and acetaminophen toxicity. Hepatology. 1983;3:932–935. doi: 10.1002/hep.1840030608. [DOI] [PubMed] [Google Scholar]
- Arosio B, Gagliano N, Fusaro LM, Parmeggiani L, Tagliabue J, Galetti P, De Castri D, Moscheni C, Annoni G. Aloe-Emodin quinone pretreatment reduces acute liver injury induced by carbon tetrachloride. Pharmacol. Toxicol. 2000;87:229–233. doi: 10.1034/j.1600-0773.2000.d01-79.x. [DOI] [PubMed] [Google Scholar]
- Ayyadevara S, Engle MR, Singh SP, Dandapat A, Lichti CF, Benes H, Shmookler Reis RJ, Liebau E, Zimniak P. Lifespan and stress resistance of Caenorhabditis elegans are increased by expression of glutathione transferases capable of metabolizing the lipid peroxidation product 4-hydroxynonenal. Aging Cell. 2005;4:257–271. doi: 10.1111/j.1474-9726.2005.00168.x. [DOI] [PubMed] [Google Scholar]
- Bartke A, Brown-Borg H, Mattison J, Kinney B, Hauck S, Wright C. Prolonged longevity of hypopituitary dwarf mice. Exp. Gerontol. 2001;36:21–28. doi: 10.1016/s0531-5565(00)00205-9. [DOI] [PubMed] [Google Scholar]
- Brodie BB, Reid WD, Cho AK, Sipes G, Krishna G, Gillette JR. Possible mechanism of liver necrosis caused by aromatic organic compounds. Proc. Natl Acad. Sci. USA. 1971;68:160–164. doi: 10.1073/pnas.68.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer KL, Jones JA. Altered hepatobiliary disposition of acetaminophen metabolites after phenobarbital pretreatment and renal ligation: evidence for impaired biliary excretion and a diffusional barrier. J. Pharmacol. Exp. Ther. 1990;252:657–664. [PubMed] [Google Scholar]
- Burk RF, Lane JM, Patel K. Relationship of oxygen and glutathione in protection against carbon tetrachloride-induced hepatic microsomal lipid peroxidation and covalent binding in the rat. Rationale for the use of hyperbaric oxygen to treat carbon tetrachloride ingestion. J. Clin. Invest. 1984;74:1996–2001. doi: 10.1172/JCI111621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang JY. Bile acid regulation of gene expression: roles of nuclear hormone receptors. Endocr. Rev. 2002;23:443–463. doi: 10.1210/er.2000-0035. [DOI] [PubMed] [Google Scholar]
- Chiang JY. Regulation of bile acid synthesis: pathways, nuclear receptors, and mechanisms. J. Hepatol. 2004;40:539–551. doi: 10.1016/j.jhep.2003.11.006. [DOI] [PubMed] [Google Scholar]
- Donahue LR, Beamer WG. Growth hormone deficiency in ‘little’ mice results in aberrant body composition, reduced insulin-like growth factor-I and insulin-like growth factor-binding protein-3 (IGFBP-3), but does not affect IGFBP-2-1 or −4. J. Endocrinol. 1993;136:91–104. doi: 10.1677/joe.0.1360091. [DOI] [PubMed] [Google Scholar]
- Dwivedi S, Sharma R, Sharma A, Zimniak P, Ceci JD, Awasthi YC, Boor PJ. The course of CCl4 induced hepatotoxicity is altered in mGSTA4-4 null (−/−) mice. Toxicology. 2006;218:58–66. doi: 10.1016/j.tox.2005.10.012. [DOI] [PubMed] [Google Scholar]
- Fickert P, Zollner G, Fuchsbichler A, Stumptner C, Pojer C, Zenz R, Lammert F, Stieger B, Meier PJ, Zatloukal K, Denk H, Trauner M. Effects of ursodeoxycholic and cholic acid feeding on hepatocellular transporter expression in mouse liver. Gastroenterology. 2001;121:170–183. doi: 10.1053/gast.2001.25542. [DOI] [PubMed] [Google Scholar]
- Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- Flurkey K, Papaconstantinou J, Miller RA, Harrison DE. Lifespan extension and delayed immune and collagen aging in mutant mice with defects in growth hormone production. Proc. Natl Acad. Sci. USA. 2001;98:6736–6741. doi: 10.1073/pnas.111158898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gems D, McElwee JJ. Broad spectrum detoxification: the major longevity assurance process regulated by insulin/IGF-1 signaling? Mech. Ageing Dev. 2005;126:381–387. doi: 10.1016/j.mad.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Godfrey P, Rahal JO, Beamer WG, Copeland NG, Jenkins NA, Mayo KE. GHRH receptor of little mice contains a missense mutation in the extracellular domain that disrupts receptor function. Nat. Genet. 1993;4:227–232. doi: 10.1038/ng0793-227. [DOI] [PubMed] [Google Scholar]
- Guo GL, Lambert G, Negishi M, Ward JM, Brewer HB, Jr, Kliewer SA, Gonzalez FJ, Sinal CJ. Complementary roles of farnesoid × receptor, pregnane × receptor, and constitutive androstane receptor in protection against bile acid toxicity. J. Biol. Chem. 2003;278:45062–45071. doi: 10.1074/jbc.M307145200. [DOI] [PubMed] [Google Scholar]
- Harper JM, Salmon AB, Chang Y, Bonkowski M, Bartke A, Miller RA. Stress resistance and aging: influence of genes and nutrition. Mech. Ageing Dev. 2006;127:687–694. doi: 10.1016/j.mad.2006.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinson JA, Reid AB, McCullough SS, James LP. Acetaminophen-induced hepatotoxicity: role of metabolic activation, reactive oxygen/ nitrogen species, and mitochondrial permeability transition. Drug Metab. Rev. 2004;36:805–822. doi: 10.1081/dmr-200033494. [DOI] [PubMed] [Google Scholar]
- Huang W, Zhang J, Chua SS, Qatanani M, Han Y, Granata R, Moore DD. Induction of bilirubin clearance by the constitutive androstane receptor (CAR) Proc. Natl Acad. Sci. USA. 2003;100:4156–4161. doi: 10.1073/pnas.0630614100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt ND, Hyun DH, Allard JS, Minor RK, Mattson MP, Ingram DK, de Cabo R. Bioenergetics of aging and calorie restriction. Ageing Res. Rev. 2006;5:125–143. doi: 10.1016/j.arr.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Jaeschke H, Knight TR, Bajt ML. The role of oxidant stress and reactive nitrogen species in acetaminophen hepatotoxicity. Toxicol. Lett. 2003;144:279–288. doi: 10.1016/s0378-4274(03)00239-x. [DOI] [PubMed] [Google Scholar]
- Kliewer SA, Goodwin B, Willson TM. The nuclear pregnane × receptor: a key regulator of xenobiotic metabolism. Endocr. Rev. 2002;23:687–702. doi: 10.1210/er.2001-0038. [DOI] [PubMed] [Google Scholar]
- Kretschmer XC, Baldwin WS. CAR and PXR: xenosensors of endocrine disrupters? Chem. Biol. Interact. 2005;155:111–128. doi: 10.1016/j.cbi.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Kurosu H, Yamamoto M, Clark JD, Pastor JV, Nandi A, Gurnani P, McGuinness OP, Chikuda H, Yamaguchi M, Kawaguchi H, Shimomura I, Takayama Y, Herz J, Kahn CR, Rosenblatt KP, Kuro OM. Suppression of aging in mice by the hormone Klotho. Science. 2005;309:1829–1833. doi: 10.1126/science.1112766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen PL. Aging and resistance to oxidative damage in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA. 1993;90:8905–8909. doi: 10.1073/pnas.90.19.8905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson AM, Setchell KD. Mass spectrometry of bile acids. In: Setchell KD, Kritchevsky D, Nair PP, editors. The Bile Acids. Vol. 4. New York: Plenum Press; 1988. pp. 167–267. [Google Scholar]
- Levine RL, Stadtman ER. Oxidative modification of proteins during aging. Exp. Gerontol. 2001;36:1495–1502. doi: 10.1016/s0531-5565(01)00135-8. [DOI] [PubMed] [Google Scholar]
- Lin YJ, Seroude L, Benzer S. Extended life-span and stress resistance in the Drosophila mutant Methuselah. Science. 1998;282:943–946. doi: 10.1126/science.282.5390.943. [DOI] [PubMed] [Google Scholar]
- Madhu C, Klaassen CD. Bromobenzene-glutathione excretion into bile reflects toxic activation of bromobenzene in rats. Toxicol. Lett. 1992;60:227–236. doi: 10.1016/0378-4274(92)90278-r. [DOI] [PubMed] [Google Scholar]
- Maglich JM, Stoltz CM, Goodwin B, Hawkins-Brown D, Moore JT, Kliewer SA. Nuclear pregnane × receptor and constitutive androstane receptor regulate overlapping but distinct sets of genes involved in xenobiotic detoxification. Mol. Pharmacol. 2002;62:638–646. doi: 10.1124/mol.62.3.638. [DOI] [PubMed] [Google Scholar]
- McElwee JJ, Schuster E, Blanc E, Thomas JH, Gems D. Shared transcriptional signature in Caenorhabditis elegans Dauer larvae and long-lived daf-2 mutants implicates detoxification system in longevity assurance. J. Biol. Chem. 2004;279:44533–44543. doi: 10.1074/jbc.M406207200. [DOI] [PubMed] [Google Scholar]
- Meier PJ, Stieger B. Bile salt transporters. Annu. Rev. Physiol. 2002;64:635–661. doi: 10.1146/annurev.physiol.64.082201.100300. [DOI] [PubMed] [Google Scholar]
- Merry BJ. Oxidative stress and mitochondrial function with aging – the effects of calorie restriction. Aging Cell. 2004;3:7–12. doi: 10.1046/j.1474-9728.2003.00074.x. [DOI] [PubMed] [Google Scholar]
- Migliaccio E, Giorgio M, Mele S, Pelicci G, Reboldi P, Pandolfi PP, Lanfrancone L, Pelicci PG. The p66shc adaptor protein controls oxidative stress response and lifespan in mammals. Nature. 1999;402:309–313. doi: 10.1038/46311. [DOI] [PubMed] [Google Scholar]
- Miller RA, Buehner G, Chang Y, Harper JM, Sigler R, Smith-Wheelock M. Methionine-deficient diet extends mouse lifespan, slows immune and lens aging, alters glucose, T4, IGF-I and insulin levels, and increases hepatocyte MIF levels and stress resistance. Aging Cell. 2005;4:119–125. doi: 10.1111/j.1474-9726.2005.00152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JR, Snodgrass WR, Gillette JR. The role of biotransformation in chemical-induced liver injury. Environ. Health Perspect. 1976;15:27–38. doi: 10.1289/ehp.761527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore LB, Parks DJ, Jones SA, Bledsoe RK, Consler TG, Stimmel JB, Goodwin B, Liddle C, Blanchard SG, Willson TM, Collins JL, Kliewer SA. Orphan nuclear receptors constitutive androstane receptor and pregnane × receptor share xenobiotic and steroid ligands. J. Biol. Chem. 2000;275:15122–15127. doi: 10.1074/jbc.M001215200. [DOI] [PubMed] [Google Scholar]
- Murakami S. Stress resistance in long-lived mouse models. Exp. Gerontol. 2006;41:1014–1019. doi: 10.1016/j.exger.2006.06.061. [DOI] [PubMed] [Google Scholar]
- Napoli C, Martin-Padura I, de Nigris F, Giorgio M, Mansueto G, Somma P, Condorelli M, Sica G, De Rosa G, Pelicci P. Deletion of the p66Shc longevity gene reduces systemic and tissue oxidative stress, vascular cell apoptosis, and early atherogenesis in mice fed a high-fat diet. Proc. Natl Acad. Sci. USA. 2003;100:2112–2116. doi: 10.1073/pnas.0336359100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okawa H, Motohashi H, Kobayashi A, Aburatani H, Kensler TW, Yamamoto M. Hepatocyte-specific deletion of the keap1 gene activates Nrf2 and confers potent resistance against acute drug toxicity. Biochem. Biophys. Res. Commun. 2006;339:79–88. doi: 10.1016/j.bbrc.2005.10.185. [DOI] [PubMed] [Google Scholar]
- Rosenfeld JM, Vargas R, Jr, Xie W, Evans RM. Genetic profiling defines the xenobiotic gene network controlled by the nuclear receptor pregnane × receptor. Mol. Endocrinol. 2003;17:1268–1282. doi: 10.1210/me.2002-0421. [DOI] [PubMed] [Google Scholar]
- Seo KW, Kim JG, Park M, Kim TW, Kim HJ. Effects of phenethyl-isothiocyanate on the expression of glutathione S-transferases and hepatotoxicity induced by acetaminophen. Xenobiotica. 2000;30:535–545. doi: 10.1080/004982500237532. [DOI] [PubMed] [Google Scholar]
- Setchell KD, Matsui A. Serum bile acid analysis. Clin. Chim. Acta. 1983;127:1–17. doi: 10.1016/0009-8981(83)90070-0. [DOI] [PubMed] [Google Scholar]
- Setchell KD, Schwarz M, O’Connell NC, Lund EG, Davis DL, Lathe R, Thompson HR, Weslie Tyson R, Sokol RJ, Russell DW. Identification of a new inborn error in bile acid synthesis: mutation of the oxysterol 7alpha-hydroxylase gene causes severe neonatal liver disease. J. Clin. Invest. 1998;102:1690–1703. doi: 10.1172/JCI2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinal CJ, Tohkin M, Miyata M, Ward JM, Lambert G, Gonzalez FJ. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell. 2000;102:731–744. doi: 10.1016/s0092-8674(00)00062-3. [DOI] [PubMed] [Google Scholar]
- Sohal RS, Mockett RJ, Orr WC. Mechanisms of aging: an appraisal of the oxidative stress hypothesis. Free Radic. Biol. Med. 2002;33:575–586. doi: 10.1016/s0891-5849(02)00886-9. [DOI] [PubMed] [Google Scholar]
- Sohal RS, Weindruch R. Oxidative stress, caloric restriction, and aging. Science. 1996;273:59–63. doi: 10.1126/science.273.5271.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudinger JL, Goodwin B, Jones SA, Hawkins-Brown D, MacKenzie KI, LaTour A, Liu Y, Klaassen CD, Brown KK, Reinhard J, Willson TM, Koller BH, Kliewer SA. The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc. Natl Acad. Sci. USA. 2001;98:3369–3374. doi: 10.1073/pnas.051551698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya T, Dhahbi JM, Cui X, Mote PL, Bartke A, Spindler SR. Additive regulation of hepatic gene expression by dwarfism and caloric restriction. Physiol. Genomics. 2004;17:307–315. doi: 10.1152/physiolgenomics.00039.2004. [DOI] [PubMed] [Google Scholar]
- Ueda A, Hamadeh HK, Webb HK, Yamamoto Y, Sueyoshi T, Afshari CA, Lehmann JM, Negishi M. Diverse roles of the nuclear orphan receptor CAR in regulating hepatic genes in response to phenobarbital. Mol. Pharmacol. 2002;61:1–6. doi: 10.1124/mol.61.1.1. [DOI] [PubMed] [Google Scholar]
- Van der Graaff M, Vermeulen NP, Crul IE, Breimer DD. Dose-dependent pharmacokinetics of zoxazolamine in the rat. Drug Metab. Dispos. 1986;14:331–335. [PubMed] [Google Scholar]
- Wang H, LeCluyse EL. Role of orphan nuclear receptors in the regulation of drug-metabolising enzymes. Clin. Pharmacokinet. 2003;42:1331–1357. doi: 10.2165/00003088-200342150-00003. [DOI] [PubMed] [Google Scholar]
- Wang L, Lee YK, Bundman D, Han Y, Thevananther S, Kim CS, Chua SS, Wei P, Heyman RA, Karin M, Moore DD. Redundant pathways for negative feedback regulation of bile acid production. Dev. Cell. 2002;2:721–731. doi: 10.1016/s1534-5807(02)00187-9. [DOI] [PubMed] [Google Scholar]
- Weber LW, Boll M, Stampfl A. Hepatotoxicity and mechanism of action of haloalkanes: carbon tetrachloride as a toxicological model. Crit. Rev. Toxicol. 2003;33:105–136. doi: 10.1080/713611034. [DOI] [PubMed] [Google Scholar]
- Wei P, Zhang J, Egan-Hafley M, Liang S, Moore DD. The nuclear receptor CAR mediates specific xenobiotic induction of drug metabolism. Nature. 2000;407:920–923. doi: 10.1038/35038112. [DOI] [PubMed] [Google Scholar]
- Xu C, Li CY, Kong AN. Induction of phase I, II and III drug metabolism/transport by xenobiotics. Arch. Pharm. Res. 2005;28:249–268. doi: 10.1007/BF02977789. [DOI] [PubMed] [Google Scholar]
- Yamazaki Y, Kakizaki S, Horiguchi N, Takagi H, Mori M, Negishi M. Role of nuclear receptor CAR in carbon tetrachloride-induced hepatotoxicity. World J. Gastroenterol. 2005;11:5966–5972. doi: 10.3748/wjg.v11.i38.5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamek-Gliszczynski MJ, Nezasa K, Tian X, Kalvass JC, Patel NJ, Raub TJ, Brouwer KL. The important role of bcrp (abcg2) in the biliary excretion of sulfate and glucuronide metabolites of acetaminophen, 4-methylumbelliferone, and harmol in mice. Mol. Pharmacol. 2006;70:2127–2133. doi: 10.1124/mol.106.026955. [DOI] [PubMed] [Google Scholar]
- Zampaglione N, Jollow DJ, Mitchell JR, Stripp B, Hamrick M, Gillette JR. Role of detoxifying enzymes in bromobenzene-induced liver necrosis. J. Pharmacol. Exp. Ther. 1973;187:218–227. [PubMed] [Google Scholar]
- Zhang J, Huang W, Qatanani M, Evans RM, Moore DD. The constitutive androstane receptor and pregnane × receptor function coordinately to prevent bile acid-induced hepatotoxicity. J. Biol. Chem. 2004;279:49517–49522. doi: 10.1074/jbc.M409041200. [DOI] [PubMed] [Google Scholar]
- Zheng J, Mutcherson R, 2nd, Helfand SL. Calorie restriction delays lipid oxidative damage in Drosophila melanogaster. Aging Cell. 2005;4:209–216. doi: 10.1111/j.1474-9726.2005.00159.x. [DOI] [PubMed] [Google Scholar]
- Zollner G, Wagner M, Moustafa T, Fickert P, Silbert D, Gumhold J, Fuchsbichler A, Halilbasic E, Denk H, Marschall HU, Trauner M. Coordinated induction of bile acid detoxification and alternative elimination in mice: role of FXR-regulated organic solute transporter-alpha/beta in the adaptive response to bile acids. Am. J. Physiol. Gastrointest. Liver Physiol. 2006;290:G923–G932. doi: 10.1152/ajpgi.00490.2005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.