Recent work from our laboratory has demonstrated that the phosphoinositide PtdIns(3,5)P2 has an essential role in autophagy in the mammalian nervous system. This low abundance, signaling lipid is synthesized by an enzyme complex that is localized on the vacuole membrane in yeast and in the endosome/lysosome compartment in mammalian cells. In mice with mutations in FIG4 and VAC14, two components of the PtdIns(3,5)P2 regulatory complex, autophagy intermediates accumulate in brain and spinal cord. The data indicate that PtdIns(3,5)P2 is required for completion of basal autophagy in mammalian cells.
PtdIns(3,5)P2 is synthesized from the precursor PtdIns(3)P by a protein complex that contains three major components, the 5-kinase FAB1/PIP5K3/PIKfyve, the 5-phosphatase FIG4/SAC3, and the scaffold protein VAC14, as well as several minor proteins. Mutations of the three major genes in yeast result in a greatly enlarged, single-lobed vacuole, but do not appear to affect autophagy. However, mutation of FAB1 in Drosophila and in C. elegans results in accumulation of LC3 homologs, suggesting a connection between PtdIns(3,5)P2 and autophagy in multicellular organisms.
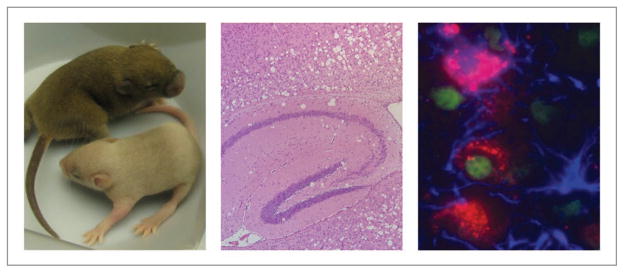
We identified spontaneous mouse mutants affecting two of the major components of the PtdIns(3,5)P2 regulatory complex, a null mutant of Fig4 in the “pale tremor” mouse and a missense mutant of Vac14 in the “infantile gliosis” mouse. The level of PtdIns(3,5)P2 is reduced by more than 50% in cultured fibroblasts from these mutants, demonstrating the conserved role of mouse Fig4 and Vac14 in biosynthesis of PtdIns(3,5)P2. The major in vivo effect in both mutant mice is severe neurodegeneration, including spongiform degeneration of the brain (Fig. 1), reduced number of myelinated axons in sciatic nerve, loss of neurons from sensory and autonomic ganglia, and lethality by 6 weeks of age. The cytoplasm of cultured fibroblasts and neurons becomes filled with large vacuoles whose limiting membrane contains the lysosomal membrane protein LAMP-2.
Figure 1.
The Fig4 mouse mutant “pale tremor” exhibits diluted pigmentation, spongiform degeneration of the brain, and accumulation of autophagy intermediates in neurons and astrocytes. Left, wild-type littermate above, mutant below. Middle, cerebral cortex overlying hippocampal region of mutant brain. Right, accumulation of autophagy substrate p62 (red) in neurons (green, nuclear NeuN) and astrocytes (blue, GFAP); pink, colocalization of p62 and GFAP in astrocytes. (Representative examples of previously described phenotypes).
Our investigation of autophagy in the PtdIns(3,5)P2-deficient mouse mutants was motivated by three considerations. First, the corresponding yeast mutants have abnormal vacuoles, and some functions of the yeast vacuole are carried out by the autophagy pathway in mammalian cells. Second, autophagosome membranes are known to contain PtdIns(3)P, the substrate for biosynthesis of PtdIns(3,5)P2. Third, mutations of the autophagy proteins ATG5 and ATG7 result in neurodegeneration in the mouse. We therefore hypothesized that neurodegeneration in Fig4 and Vac14 mutant mice might be a consequence of a direct role for PtdIns(3,5)P2 in autophagy. To test this hypothesis, we examined autophagy in brain and spinal cord of the mutant mice. Four proteins with a role in autophagy, p62, ubiquitin, LC3-II and LAMP-2, were found to be highly elevated in the mutant tissue. The p62 and ubiquinated proteins were colocalized in cytoplasmic inclusion bodies, predominantly in astrocytes and to a lesser extent in neurons (Fig. 1). Ultrastructural analysis revealed the accumulation of electron-dense vesicles in mutant brain; however, upregulation of autophagy was not detected. The colocalization of p62 and LAMP-2 in vesicles indicated that p62 was correctly sequestered in autophagosomes, and that the autophagosomes could fuse with lysosomes. Autophagy thus appears to be blocked at a subsequent step, indicating that PtdIns(3,5)P2 is required for completion of basal autophagy, resolution of the autolysosome and regeneration of lysosomes.
We suggest the following model to explain the abnormalities in PtdIns(3,5)P2-deficient brain: (1) PtdIns(3,5)P2 is generated at the cytoplasmic surface of the autolysosome as a transient signal that mediates resolution of the autolysosome, possibly by membrane budding. (2) Close physical association of the kinase and phosphatase at the surface of the autolysosome facilitates dynamic regulation of PtdIns(3,5)P2 concentration and the rate of autophagy. (3) The mammalian lysosome and the yeast vacuole undergo repeated cycles of fusion and resolution that are initiated by cycles of PtdIns(3,5)P2 synthesis and breakdown.
The nervous system of Fig4 and Vac14 mutant mice is highly sensitive to deficiency of PtdIns(3,5)P2, with neuronal loss that begins before birth. This sensitivity may be related to the inability of post-mitotic neurons to reduce the abundance of accumulated proteins via cell division. Defects in the response of astrocytes to neuronal injury in the mutant mice may exacerbate neuronal loss. Increased excitotoxicity may also contribute to neurodegeneration, as suggested by recent evidence from the Dolmetsch laboratory that endosomal internalization of the calcium channel CaV1.2 is dependent on the activity of FAB1.
Based on the movement disorder in the mutant mice, we tested human FIG4 as a candidate gene for Charcot-Marie-Tooth disease and ALS. We identified recessive loss-of-function mutations of FIG4 in four families with Charcot-Marie-Tooth disease type 4J (OMIM#611228), a severe form of the disease. We also observed heterozygosity for deleterious mutations of FIG4 in six patients with ALS (OMIM# 6122577).
The number of neurological disorders associated with impaired autophagy is rapidly growing. In the best-known examples, mutation of an autophagy substrate generates a toxic protein that is resistant to autophagic degradation and accumulates in p62-positive inclusion bodies like those in the mutant mice. Examples include Parkinson disease and SOD1-based ALS. We propose that the FIG4 mutations in CMT4J and ALS represent another, distinct type of autophagy-related disease that is caused by mutations in the autophagy machinery itself. Growing recognition that impaired autophagy may be a common pathway for many types of neurodegeneration raises the possibility that induction of alternative pathways of protein degradation could provide a common treatment for these genetically heterogeneous disorders.