Abstract
The recent emphasis on soft tissues as the limiting factor in treatment and on soft tissue relationships in establishing the goals of treatment has made 3D analysis of soft tissues more important in diagnosis and treatment planning. It is equally important to be able to detect changes in the facial soft tissues produced by growth and/or treatment. This requires structures of reference for superimposition, and a way to display the changes with quantitative information. This paper outlines a technique for quantifying facial soft tissue changes as viewed in CBCT data, using fully-automated voxel-wise registration of the cranial base surface. The assessment of change of soft tissues is done via calculation of the Euclidean surface distances between the 3D models. Color maps are used for visual assessment of the location and quantification of changes. This methodology allows a detailed examination of soft tissue changes with growth and/or treatment. Because of the lack of stable references with 3D photogrammetry, 3D photography and laser scanning, soft tissue changes cannot be accurately quantified by these methods.
INTRODUCTION
Soft tissues establish the limit to which the orthodontist can alter the dimension of the dental arches and the position of the jaws from both an esthetic and stability standpoint.1–3 Assessment of soft tissue changes produced by growth and/or treatment requires 3D analysis and superimposition because of the complexity of soft tissue behavior and the inability to measure asymmetries in 2D images. Recently, technologies such as 3D photogrammetry 4–8 and laser scanning9–12 of the face have been used for 3D soft tissue superimposition, but their major limitation has been the inability to standardize registration of the images over time. Current procedures to integrate 3D facial images have reported significant errors in head positioning13 and potential errors in facial expression have not been assessed.14
The variability of soft tissue surface appearance has important consequences to the choice of approaches for adequate registration of longitudinal images. A stable reference for superimposition of images is required for a standardized record of the relationship between the facial soft tissues and the underlying skeletal and dental structures. Currently, CBCT technology allows the use of stable reference structures.
No soft tissue structures are stable enough to allow registration between before- and after-treatment images, because the soft tissues change with growth, treatment, head posture, weight gain or loss, aging and facial expression. In 2D cephalometrics, the cranial base often is used for superimpositions to show both hard tissue and soft tissue profile changes because it shows minimal changes after neural growth is completed. While landmark location in 2D is hampered by overlapping multiple structures, locating 3D landmarks on complex curving structures is significantly more difficult and prone to identification errors.14,15 Even though landmark-based geometric morphometric methods16 have been increasingly applied to the study of human form over the last two decades, the use of landmarks is not sufficient because they cannot describe biological forms and patterns.15,17–18 The craniofacial form structural information is represented by surfaces, curves or outlines. The sliding semi-landmark method was proposed to analyze outlines extending the standard Procrustes superimposition procedure.19–21 In addition to translating, scaling, and rotating landmarks optimally, the semi-landmark points are slid along the outline curve until they match as well as possible the positions of corresponding points along an outline in a reference configuration.22 However, semi-landmarks do not include information from the whole curves and surfaces. A workable interpretive system of the biology of craniofacial growth demands the assessment of the complex cause-and-effect interactions among bones growing simultaneously, but with different timing.23
Fortunately, 3D registration can be based on stable surfaces instead of landmarks. The purpose of this paper is to discuss whether 3D imaging technology can quantify soft tissue changes, describe a method to use cranial superimposition of CBCT data to accurately evaluate soft tissue treatment outcomes, and put problems in combining other 3D imaging modalities with CBCT in perspective. Here, we demonstrate the application of a fully automated voxel-wise rigid registration at the cranial base to evaluate 3D soft tissue changes. Establishing this technology has been the focus of several previous studies and our progress to date is described in this paper.
METHODS
The steps in the process of 3-D image acquisition and analysis for evaluation of facial change are:
Image acquisition
Cone-beam CT (CBCT) equipment specialized for maxillofacial imaging now offers a relatively low-dose and convenient way to follow changes in facial morphology in three dimensions for both growing and non-growing subjects. For studies of facial change, the CBCT scans should be acquired with a large field of view so that the entire facial anatomy can be viewed. For the cases presented in this paper, either the iCat (16 × 22 cm field of view) or NewTom 3G (12-inch field of view) scanner was used. The images were reformatted26 to yield a voxel size of 0.5 mm, and then cropped to facilitate image analysis. Experimental protocols were approved by the Institutional Review Board and the radiation safety committee.
Image analysis
Analysis of serial CBCT images to evaluate changes over time is done in a sequence of four steps: (1) model construction, (2) image registration, (3) transparency overlay, and (4) quantitative measurement.
(1) Construction of virtual 3D surface models
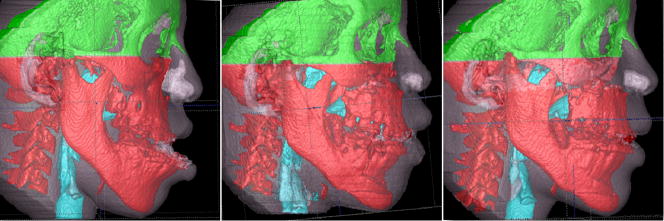
Surface models are created using ITK-SNAP open-source software.22 A surface model of the cranial base is created for the registration in our approach (Figure 1). This construction of surface models requires the generation of an intermediate surface representation (triangular mesh) of the craniofacial structures that is different from the methods used in currently available commercial softwares that create a 3D projected view directly from the volume data (volume rendering). The surface-based method facilitates establishing boundaries between anatomic structures and assigning the proper color label/transparency values to obtain separate display of the mandible, maxilla and cranial base.
Figure 1.
Construction of 3D models of a patient treated with two jaw surgery with visualization of color labeling of different anatomic structures. A, Pre-treatment models. B, 1-week year post-surgery. C, 6 weeks follow-up.
(2) Image registration
The IMAGINE software developed by NIH (which is available free) 24 was modified at UNC and then used to mask facial structures displaced with growth or treatment and perform a fully automated, voxelwise, rigid registration at the cranial base. The registration of the cranial base utilizes maximization of mutual information to avoid observer-dependent techniques based on overlap of anatomic landmarks. After the software masks the maxillary and mandibular structures, it compares the gray level intensity of each voxel in the cranial base to register the 2 CBCT images.26 These rotation and translation parameters are also applied to register 3D models. After registration, we can assess the overlay of the 3D models.
For subjects in whom cranial base growth is complete, registration is done using the gray level CBCT datasets at the whole surface of the cranial base (Figures 2 and 3). The larger the surface used, the more robust the registration is. For this reason, for adult patients the whole cranial base surface is used for registration. For growing patients, the registration requires two steps. First, an initial head alignment is done using the whole cranial base, and then a finer registration is performed at the stable structure on the anterior cranial base.26
Figure 2.
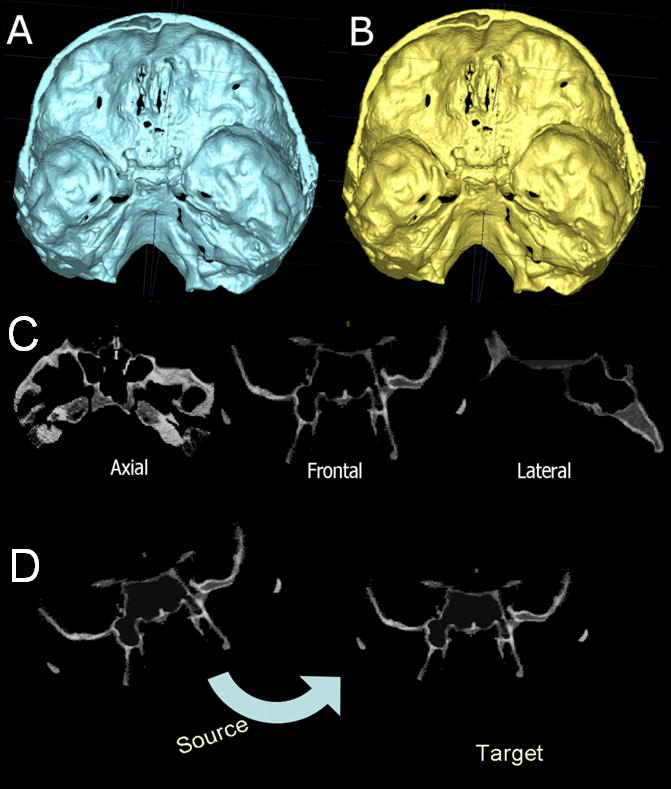
Anatomic structures used for superimpositions of 3D models of non-growing subjects. A, Pretreatment cranial base models, and B, Posttreatment cranial base models. C, The model in B was used to mask all anatomic structures that changed with treatment and generate a gray-level–intensity image containing only the cranial fossa for calculation of registration parameters. D, Fully automated calculation of rotational and translational parameters between the images.
Figure 3.
Cranial base matching. A, Pretreatment cranial base model in white and posttreatment cranial base model in red. B, Pre and posttreatment matching of the cranial base as a result of the voxel-based registration shown in figure 2. C, Color maps of the surface distance between the registered pre and posttreament models shown 0mm surface distances (green color).
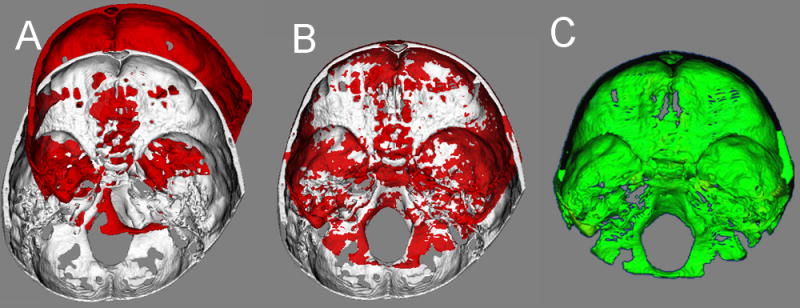
For growing subjects, there is still growth in the sphenooccipital synchondrosis, and the lateral wall of the skull as well as the frontal lobes and sinuses. For this reason, for registration of before- and after-treatment CBCTs of growing subjects, the registration requires two steps.27 First, an initial head alignment is done using the whole cranial base, and then a finer registration with optimal alignment gray level CBCT datasets is performed with subvoxel accuracy at the stable structures on the anterior cranial base (Imagine software, Figure 3).15,28 This registration utilizes a smaller surface area that includes anterior cranial base structures that have completed growth by age 7:25 anterior wall of the sella, anterior clinoid processes, planum sphenoidale, lesser wings of the sphenoid, superior aspect of ethmoid and cribriform plate, cortical ridges on the medial and superior surfaces of the orbital roofs, and inner cortical layer of the frontal bones. (Figure 4)
Figure 4.
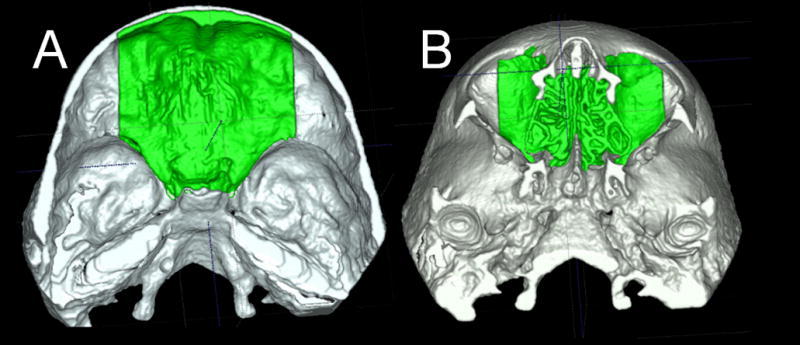
Anatomic structures used for superimpositions of 3D models of growing subjects. The anterior cranial fossa region of the cranial base 3D surface models after treatment was used for registration A shows the superior view and B the inferior view
Validation studies of registration of growing27 and non-growing26 subjects has shown that maximum registrations errors are smaller than the image spatial resolution of 0.5mm.
(3) Transparency overlay
The next step in the analysis involves overlaying the 3D model surfaces that are registered in the same coordinate system. This is done with another tool, CMF software (Maurice Müller Institute, Bern, Switzerland).29 This tool allows different degrees of transparencies to assess visually the boundaries of the soft tissue structures between superimposed models from two different time points. This clearly identifies the location and direction of dental, bone and soft tissue displacements, and allows correlation of hard and soft tissue changes (Figure 5).
Figure 5.
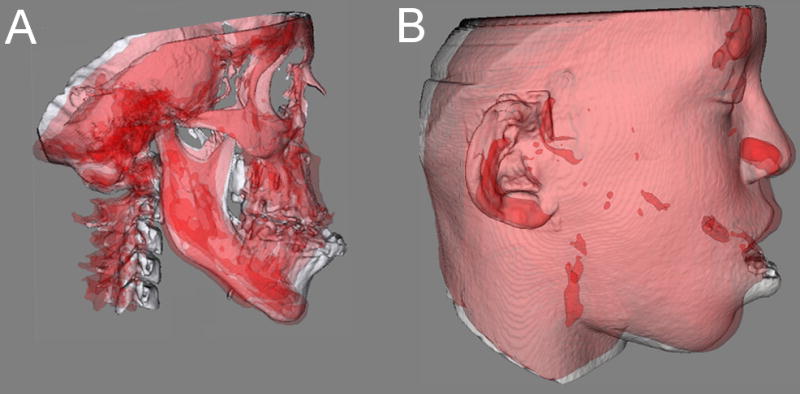
A and B, Transparency overlays of patient in figure 2. Superimposition of pre-surgery (white) and 6 weeks after 2 jaw surgery (red) models of nongrowing patient at the cranial base. A, Hard tissue changes. B, Soft tissue changes.
(4) Quantitative measurements
The CMF application software is then used to measure overall facial changes29 and display of color maps generated from closest-point distances between the surfaces as proposed by Gerig et al.30 The CMF tool calculates thousands of color-coded surface distances in millimeters between before- and after-treatment 3D models by using surface triangles at two different time points, so that the difference between the two surfaces at any location can be quantified. Isolines (contour line tool) are used to delineate and quantify surface changes for specific regions of interest, such as the nose, cheeks, upper and lower lips, and chin (Figure 6). Soft tissue changes are described as displacements relative to the cranial base.
Figure 6.
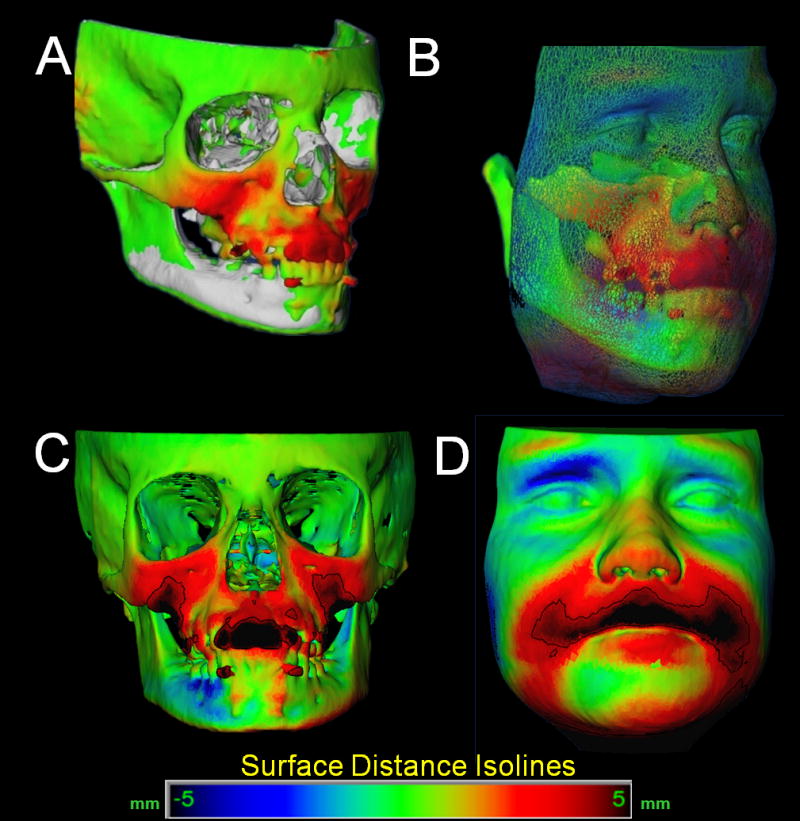
Quantification of soft tissue changes. A- Superimposed Pre-treatment (white) and post-treatment 3D models of (surface distance changes color map of hard tissue changes ). B- Color maps for comparison of hard and soft tissue regional changes. C- Isoline contours adjusted to quantify changes in the upper lip region. D- Isoline contours adjusted to quantify changes in the upper lip region.
The quantitative changes are visualized using color maps, which can be used to indicate inward (blue) or outward (red) displacement between overlaid structures, registered at the cranial base. An absence of changes is indicated by the green color code. For example, in mandibular advancement surgery, the forward chin and lower lip displacement would be shown in a red color code; in mandibular setback surgery lower lip and chin surfaces would be shown in a blue color code (Figures 7 and 8). This method for showing quantitative changes at multiple locations has been validated and used since 2005.26
Figure 7.
Soft tissue changes 1 year after mandibular advancement surgery. A, Transparency overlays of superimposed pre-surgery (white) and 1 year post-surgery (red). B and C, Surface distance color maps of soft tissue changes in the chin area.
Figure 8.
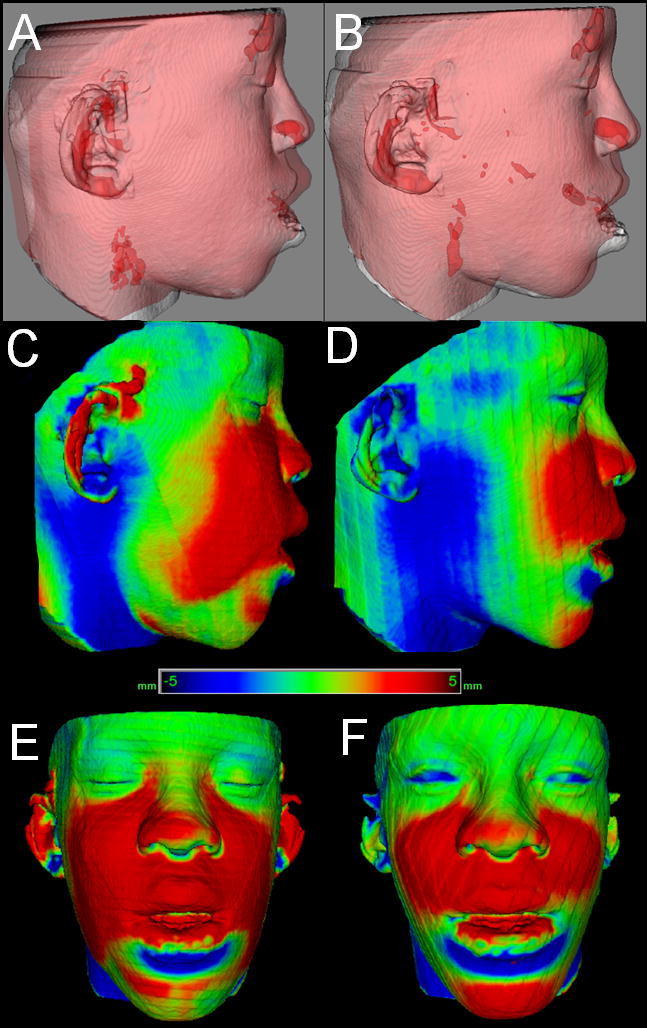
Difference in assessment of soft tissue changes 1 week and 6 weeks after maxillary advancement and mandibular subapical osteotomy. A–B, Transparency overlays. A, Superimposition of pre-surgery (white) and 1 week post-surgery (red). B, Superimposition of pre-surgery (white) and 6 weeks post-surgery (red). C–F, Surface distance color maps of A and B superimpositions. C–D, Lateral views. E–F, Frontal views. Note 1 week post-surgery swelling and that blue surface distances at the neck are artifacts due to differences in cervical positioning between 1 week and 6 week 3D imaging acquisitions.
Discussion
Image registration is a core technology for many imaging tasks. Research efforts over the past 20 years in dentistry, medicine and anthropology have been directed to the development of 3D registration tools for quantitative assessment of facial soft and hard tissues. According to the transformation applied to the images, registration procedures can be classified into two main groups: rigid and nonrigid. The transformation involved in a rigid registration procedure includes translation and rotation, while that of a nonrigid registration includes translation, rotation, scale and affine properties. Rigid registration can be based on landmarks,15–16,31 semi-landmarks,19–22,32–34 curves,35,36 planes,37 surfaces38 or voxel (mutual information).39,40 Non-rigid registration can be based on landmarks, elastic models,42,43 fluid models,44 splines45 and finite element models.46,47 The two obstacles to widespread clinical use of nonrigid (elastic and deformable) registration are computational cost and quantification difficulties as the 3D models are deformed. Nonrigid registration would be required to create a composite of several different jaw shapes to guide the construction of template or standard, normal 3D surface models. To evaluate longitudinal changes, rigid registration is acceptable, and this study utilized voxel based registration on the cranial base of before and after treatment CBCT images.
While CBCT images are lower in contrast than CT, soft and hard tissues are well visualized. Diagnostic benefit and dose detriment tradeoffs are important considerations in choices of radiographic procedures. Concern has recently been raised about increasing numbers of CT examinations in the US and the increased cancer risks, especially in children, which result from these examinations.48 Dental CBCT can be recommended as a dose-sparing technique compared with alternative standard medical CT scans for common oral and maxillofacial radiographic imaging tasks.49 Until we have clear evidence for a threshold dose below which our patients are not at risk, we must assume that radiography involves a small, but real, risk to our patients. CBCT volumes also allow reconstruction of 2D panoramic, lateral, antero-posterior and axial x-rays, eliminating the need for additional radiographic acquisitions.
Although CBCT images show the soft tissue surfaces accurately and therefore are excellent for display of changes due to growth, aging and/or treatment, 3D photographs provide additional information as to color, surface texture, as well as higher resolution of soft tissue surfaces.50 Because of the low radiation dose, the soft tissue visualized in CBCT can have a somewhat roughened texture. Currently-available software programs have tools for superimposition of 3D photographs on landmarks or surface based regions in the soft tissue, but these soft tissue structures are not stable enough to serve as superimposition references. The result is an unknown amount of distortion. Even though the patient “wow” factor with morphed 3D photos may be advantageous from a marketing perspective, no data exist to validate the accuracy of the changes that are displayed to quantify changes over time. It seems a desirable goal to combine CBCT and 3D photography.
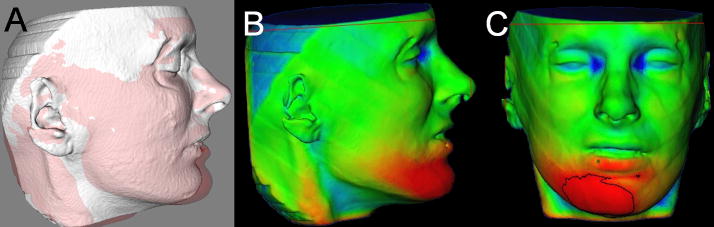
Problems in registering 3D soft tissue photographs to CBCT soft tissue (Figure 9)
Figure 9.
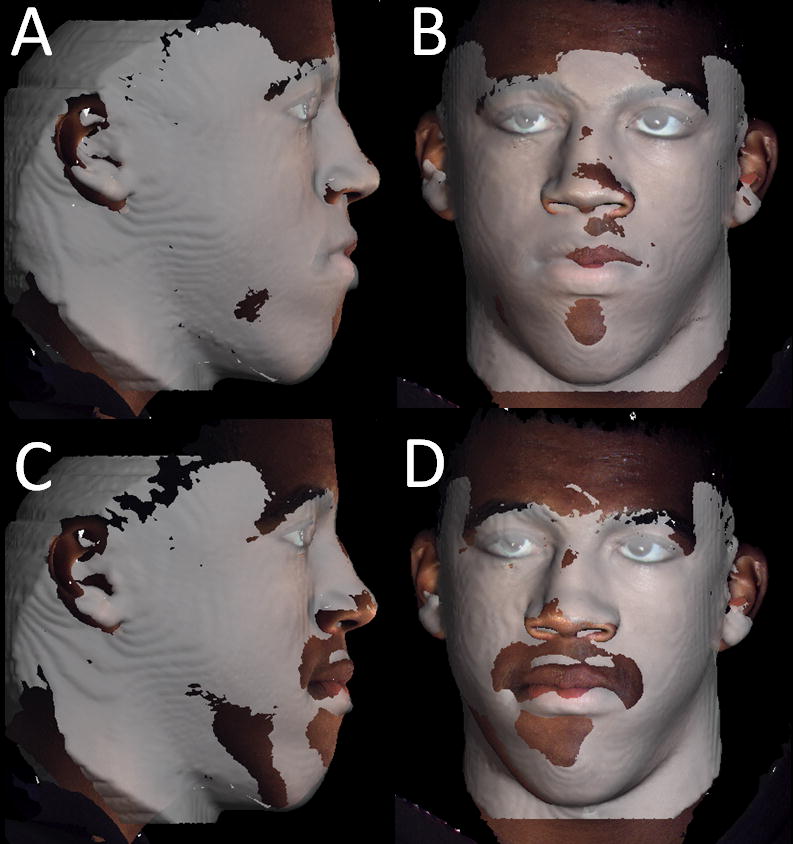
Difference in registration of the 3D photography to two CBCTs taken at the same day. A and B, First CBCT acquisition registered to 3D photograph taken the same day. C and D, Second CBCT acquisition taken the same day. Note that even though it the photograph and the CBCT surface model appear to be registered at the forehead, the contours of the CBCT lower lip, chin and neck do not match the contours of the 3D photograph due to subtle differences in facial expression and head posture.
For 3D photographs and CBCT obtained at close but separate times, Maal et al.50 reported that the registration errors between 3D photographs and CBCT were relatively large at the lateral neck, mouth and around the eyes, with 90–95% of the error in the ± 1.9 mm range. Even after exclusion of artifact regions from the matching process, 90–95% of the error was within ± 1.5 mm. An important step toward overcoming these problems would be simultaneous acquisition of CBCT and 3D photographs, but that is not possible at present and may not be in the near future. Problems that need to be overcome with 3D photograph superimposition include (1) inadequate use of fiducials, (2) head position in acquisition, (3) soft tissue capture errors, and (4) current use of non-rigid registration deformation of soft tissue contours to allow matching of 3D photographs to CBCT soft tissues.
(1) Use of fiducials
Until CBCT and 3D photographs can be acquired simultaneously, the use of fiducials for both CBCT and 3D photograph acquisition can decrease errors due to choice of surface regions or landmarks,13 but fiducials cannot control for soft tissue distortions due to head positioning, different facial expressions and artifacts during image acquisition. For example, if the patient’s head is turned upwards during either the CBCT or the photograph acquisition, the neck and perioral soft tissues are stretched and this cannot be corrected by registration on the fiducials.
(2) Head position in acquisition
For CBCT acquisition, patients are usually held in a fixed position with a strap on the forehead, a chin support or both, or are lying down, depending on the scanner. To minimize deformation of soft tissues around the mouth, the chin support needs to be avoided. As the use of a strap on the forehead causes errors in the forehead region due to small soft-tissue deformations, this should be avoided as well if possible. There is currently no standardization of head position13 during CBCT and 3D photograph acquisition. Differences in head position between the CBCT and 3D photograph acquisitions result in registration errors, which are greatest in the neck region but as Maal et al.50 noted, relatively large elsewhere in the face.
(3) Soft tissue capture errors
Registration errors also result from errors in capture of soft-tissue surfaces, in both CBCT and stereophotogrammetry imaging. With CBCT, the soft-tissue surface can appear roughened due to the low radiation dose. With stereophotogrammetry, it is not possible to capture the eye region correctly, because the light pattern used to reconstruct a 3D photograph interferes with light reflection in the lenses of the eyes.50
(4) Effects of rigid vs non-rigid deformation on soft tissue contours
In a rigid registration algorithm, only translational and rotational movements are allowed as the different data sets are fused. In order to register the textured surface of a 3D photograph to the untextured surface of a cone-beam CT, rigid registration of the surfaces frequently is not sufficient. This can be due to the rougher surface of the CBCT, different facial expressions during the two acquisitions that are done at separate times, and/or acquisition artifacts.
A possible solution is the use of non-rigid registration algorithms, which allow deformational movements of the surface as well. Unfortunately, these algorithms deform the images and contribute to errors rather than remove them. At present they should be avoided.
3D soft tissue analysis in the future
Registration tools using “best fit” between 3D renderings, landmarks or surfaces that have changed with time does not allow quantification of local changes, and this can lead to misleading interpretation of changes (Figure 7). The superimposition methods presented in this paper not only allows visualization, but also provide precise localization of the soft tissue growth and adaptation to skeletal changes.
Although it seems reasonable that a combination of data from 3D photographs and CBCT would be better than either method alone, the added value of 3D photographs still needs to be assessed in carefully controlled studies. The superimposition methodology presented here allows quantification of soft tissue surface changes from any 3D data set, but its application to other imaging modalities such as laser scanners and 3D cameras requires registration to the CBCT data sets. This would require either simultaneous acquisition of the photograph and CBCT, or standardization of head position with calibration of the CBTC and 3D camera acquisition parameters. Due to recent technological advances in imaging, there is now the promise that many if not most of the criteria for an ideal standardized record of the relationship between the soft tissue facial mask and the underlying skeletal and dental structures can be met. The potential in the future for melding 3D facial photography with Cone Beam CT promises to provide a record, which is three-dimensional, easily obtained, able to capture facial and dental display, is measurable and can be used as a longitudinal record. From a clinical diagnostic standpoint, the record will depict all of the soft and hard tissue structures with six degrees of freedom.
We are applying the methodology presented in this paper to research in progress. Currently, superimposition of 3D surface models is still too time-consuming and computing-intensive to apply these methods in routine clinical use. Our current focus is on developing a simplified analysis so that in the near future these methods can be used clinically. This approach to 3D image analysis methods has been streamlined and continuously updated with new methods for quantification, with collaboration from the Maurice Müller Institute, the UNC medical image analysis group, the neuroimaging laboratory, and the statistical modeling group in the UNC Biomedical Research Imaging Center.
Acknowledgments
Supported by National Institute for Dental and Craniofacial Research (NIDCR) DE017727, DE018962, DE005215 and CAPES 3827-05-4.
Footnotes
Conflict of Interest Statement
There is no conflict of interest for any of the contributing authors.
References
- 1.Tessier P. Subperiosteal face-lift. Ann Chir Plast Esthet. 1989;34(3):193–7. [PubMed] [Google Scholar]
- 2.Ackerman JL, Proffit WR. Soft tissue limitations in orthodontics: treatment planning guidelines. Angle Orthod. 1997;67(5):327–36. doi: 10.1043/0003-3219(1997)067<0327:STLIOT>2.3.CO;2. [DOI] [PubMed] [Google Scholar]
- 3.Ackerman JL, Proffit WR, Sarver DM, Ackerman MB, Kean MR. Pitch, roll, and yaw: describing the spatial orientation of dentofacial traits. Am J Orthod Dentofacial Orthop. 2007 Mar;131(3):305–10. doi: 10.1016/j.ajodo.2006.05.032. [DOI] [PubMed] [Google Scholar]
- 4.Aldridge K, Boyadjiev SA, Capone GT, DeLeon VB, Richtsmeier JT. Precision and error of three-dimensional phenotypic measures acquired from 3dMD photogrammetric images. Am J Med Genet A. 2005 Oct 15;138A(3):247–53. doi: 10.1002/ajmg.a.30959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ayoub AF, Xiao Y, Khambay B, Siebert JP, Hadley D. Towards building a photo-realistic virtual human face for craniomaxillofacial diagnosis and treatment planning. Int J Oral Maxillofac Surg. 2007 May;36(5):423–8. doi: 10.1016/j.ijom.2007.02.003. Epub 2007 Apr 10. [DOI] [PubMed] [Google Scholar]
- 6.Goos MI, Alberink IB, Ruifrok AC. 2D/3D image (facial) comparison using camera matching. Forensic Sci Int. 2006 Nov 10;163(1–2):10–7. doi: 10.1016/j.forsciint.2005.11.004. Epub 2005 Dec 6. [DOI] [PubMed] [Google Scholar]
- 7.Sawyer AR, See M, Nduka C. 3D Stereophotogrammetry Quantitative Lip Analysis. Aesthetic Plast Surg. 2008 Jun 27; doi: 10.1007/s00266-008-9191-1. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 8.Weinberg SM, Naidoo S, Govier DP, Martin RA, Kane AA, Marazita ML. Anthropometric precision and accuracy of digital three-dimensional photogrammetry: comparing the Genex and 3dMD imaging systems with one another and with direct anthropometry. J Craniofac Surg. 2006 May;17(3):477–83. doi: 10.1097/00001665-200605000-00015. [DOI] [PubMed] [Google Scholar]
- 9.Nishii Y, Nojima K, Takane Y, et al. Integration of the maxillofacial three-dimensional CT image and the threedimensional dental surface image. J Japan Orthod Soc. 1998;57:189–94. [Google Scholar]
- 10.Terai H, Shimahara M, Sakinaka Y, Tajima S. Accuracy of integration of dental casts in three-dimensional models. J Oral Maxillofac Surg. 1999;57:662–5. doi: 10.1016/s0278-2391(99)90425-1. [DOI] [PubMed] [Google Scholar]
- 11.Kau CH, Cronin AJ, Richmond S. A three-dimensional evaluation of postoperative swelling following orthognathic surgery at 6 months. Plast Reconstr Surg. 2007 Jun;119(7):2192–9. doi: 10.1097/01.prs.0000260707.99001.79. [DOI] [PubMed] [Google Scholar]
- 12.Kau CH, Richmond S, Savio C, Mallorie C. Measuring adult facial morphology in three dimensions. Angle Orthod. 2006 Sep;76(5):773–8. doi: 10.1043/0003-3219(2006)076[0773:MAFMIT]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 13.Hajeer MY, Millett DT, Ayoub AF, Siebert JP. Applications of 3D imaging in orthodontics: part II. J Orthod. 2004 Mar;31(1):155–62. doi: 10.1179/146531204225011346. [DOI] [PubMed] [Google Scholar]
- 14.Curry S, Baumrind S, Carlson S, Beers A, Boyd R. Integrated three-dimensional craniofacial mapping at the Craniofacial Research Instrumentation Laboratory/University of the Pacific. Semin Orthod. 2001;7:258–65. [Google Scholar]
- 15.Cevidanes LH, Franco AA, Gerig G, Proffit WR, Slice DE, Enlow DH, Yamashita HK, Kim YJ, Scanavini MA, Vigorito JW. Assessment of mandibular growth and response to orthopedic treatment with 3-dimensional magnetic resonance images. Am J Orthod Dentofac Orthop. 2005 Jul;128(1):16–26. doi: 10.1016/j.ajodo.2004.03.032. [DOI] [PubMed] [Google Scholar]
- 16.Rohlf FJ, Marcus LE. A revolution in morphometrics. Tree. 1993;8:129–132. doi: 10.1016/0169-5347(93)90024-J. [DOI] [PubMed] [Google Scholar]
- 17.Oxnard CE. One biologist’s view of morphometrics. Ann Rev Ecol Syst. 1978;9:219–241. [Google Scholar]
- 18.Moyers RE, Bookstein FL. The inappropriateness of conventional cephalometrics. Am J orth od. 1979;75:599–617. doi: 10.1016/0002-9416(79)90093-9. [DOI] [PubMed] [Google Scholar]
- 19.Green WDK. The thin-plate spline and images with curving features. In: Mardia KV, Gill CA, Dryden IL, editors. Image Fusion and Shape Variability. Leeds: University of Leeds Press; 1966. pp. 79–87. [Google Scholar]
- 20.Bookstein FL. Applying landmark methods to biological outline data. In: Mardia KV, Gill CA, Dryden IL, editors. Image Fusion and Shape Variability. Leeds: University of Leeds Press; 1966c. pp. 59–70. [Google Scholar]
- 21.Bookstein FL. Landmark methods for forms without landmarks: localizing group differences in outline shape. Med Image Anal. 1997;1:225–243. doi: 10.1016/s1361-8415(97)85012-8. [DOI] [PubMed] [Google Scholar]
- 22.Adams DC, Rohlf FJ, Slice DE. Geometric morphometrics: ten years of progress following the ‘revolution’. Ital J Zool. 2004;71:5–16. [Google Scholar]
- 23.Enlow D. Discussion. Am J Orthod Dentofac Orthop. 2000;117:147. [Google Scholar]
- 24.Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G. User guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage. 2006 Jul 1;31(3):1116–28. doi: 10.1016/j.neuroimage.2006.01.015. Epub 2006 Mar 20. [DOI] [PubMed] [Google Scholar]
- 25.Melsen B The cranial base. Acta Odontol Scand. Suppl 62. Thesis, Aarbus; 1974. 32, The postnatal development of the cranial base studied histologically on human autopsy material. [Google Scholar]
- 26.Cevidanes LH, Bailey LJ, Tucker GR, Jr, Styner MA, Mol A, Phillips CL, Proffit WR, Turvey T. Superimposition of 3D cone-beam CT models of orthognathic surgery patients. Dentomaxillofac Radiol. 2005;34(6):369–375. doi: 10.1259/dmfr/17102411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cevidanes LHS, Heymann G, Cornelis MA, DeClerck HJ. Superimposition of 3D Cone-Beam CT models of growing patients. Am J Orthod Dentofac Orthop. 2009 doi: 10.1016/j.ajodo.2009.01.018. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rueckert D, Sonoda LI, Hayes C, Hill DLG, Leach MO, Hawkes DJ. Nonrigid registration using free-form deformations: application to breast MR images. IEEE Trans Med Imaging. 1999;18:712–21. doi: 10.1109/42.796284. [DOI] [PubMed] [Google Scholar]
- 29.Chapuis J, Schramm A, Pappas I, Hallermann W, Schwenzer-Zimmerer K, Langlotz F, Caversaccio M. A new system for computer-aided preoperative planning and intraoperative navigation during corrective jaw surgery. IEEE Trans Inf Technol Biomed. 2007 May;11(3):274–87. doi: 10.1109/titb.2006.884372. [DOI] [PubMed] [Google Scholar]
- 30.Gerig G, Jomier M, Chakos M. Valmet: a new validation tool for assessing and improving 3D object segmentation. Med Image Comput Comput Assist Interv Int Conf Med Image Comput Comput Assist Interv. 2001;2208:516–528. [Google Scholar]
- 31.Rohr K. Computational Imaging and Vision Series. Vol. 21 Dordrecht, Boston, London: Kluwer Academic Publishers; 2001. Landmark-based image analysis: using geometric and intensity models. [Google Scholar]
- 32.Andresen R, Bookstein FL, Conradsen K, Ersboll BK, Marsh JL, Kreiborg S. Surface-bounded growth modeling applied to human mandibles. IEEE Trans Med Imaging. 2000;19:1053–63. doi: 10.1109/42.896780. [DOI] [PubMed] [Google Scholar]
- 33.Gunz P, Mitteroecker P, Bookstein FL. Semilandmarks in three dimensions. In: Slice DL, editor. Modern morphometrics in physical anthropology. New York: Kluwer Academic/Plenum Publishers; 2005. [Google Scholar]
- 34.Perez SI, Bernal V, Gonzales PN. Differences between sliding semi-landmark me hods in geometric morphometrics, with an application to human craniofacial and dental variation. J Anat. 2006;208:769–784. doi: 10.1111/j.1469-7580.2006.00576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Subsol G, Thirion JP, Ayache N. A scheme for automatically building three-dimensional morphometric anatomic atlases: application to a skull atlas. Med Image Anal. 1998;2:37–60. doi: 10.1016/s1361-8415(01)80027-x. [DOI] [PubMed] [Google Scholar]
- 36.Subsol G, Roberts N, Doran M, Thirion JP, Whitehouse GH. Automatic analysis of cerebral atrophy. Mag Reson Imaging. 1997;15:917–927. doi: 10.1016/s0730-725x(97)00002-7. [DOI] [PubMed] [Google Scholar]
- 37.Baumrind S, Moffitt FH, Curry S. The geometry of three-dimensional measurement from paired coplanar x-ray images. Am J Orthod. 1983;84:313–322. doi: 10.1016/s0002-9416(83)90347-0. [DOI] [PubMed] [Google Scholar]
- 38.Thompson PM, MacDonald D, Mega MS, Holmes CJ, Evans AC, toga Aw. Detection and mapping of abnormal brain structure with a probabilistic atlas of cortical surfaces. J Comput Assist Tomogr. 1997;21(4):567–581. doi: 10.1097/00004728-199707000-00008. [DOI] [PubMed] [Google Scholar]
- 39.Maes F, Collignon A, Vandermeulen D, Marchal G, Suetens P. Multimodality image registration by maximization of mutual information. IEEE Trans Med Imaging. 1997;16:187–198. doi: 10.1109/42.563664. [DOI] [PubMed] [Google Scholar]
- 40.Swennen GR, Mollemans W, De Clercq C, Abeloos J, Lamoral P, Lippens F, Neyt N, Casselman J, Schutyser F. A Cone-Beam Computed Tomography Triple Scan Procedure to Obtain a Three-Dimensional Augmented Virtual Skull Model proper for Orthognathic Surgery Planning. J Craniofac Surg. 2009 Mar 5; doi: 10.1097/SCS.0b013e3181996803. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 41.Crum WR, Hartkens T, Hill Dl. Non-rigid image registration: theory and practice. Br J Radiol. 2004;77(Sp2):S140–153. doi: 10.1259/bjr/25329214. [DOI] [PubMed] [Google Scholar]
- 42.Rohr K, Stiehl HS, Sprengel R, Beil W, Buzug TM, Weese J, et al. Point-based elastic registration of medical image data using approximating thin-plate splines. Visualization in Biomedical Computing, Lecture Notes in Computer Science. 1996;1131:297–306. [Google Scholar]
- 43.Bajcsy R, Kovacic S. Multiresolution elastic matching. Comp Vision Graphics Image Processing. 1989;46:1–21. [Google Scholar]
- 44.Christensen GE, Rabbitt RD, Miller MI. Deformable templates using large deformation kinematics. IEEE Trans Image Processing. 1996;5:1435–1447. doi: 10.1109/83.536892. [DOI] [PubMed] [Google Scholar]
- 45.Rueckert D, Sonoda LI, Hayes C, Hill DLG, Leach MO, Hawkes DJ. Nonrigid registration using free-form deformations: application to breast MR images. IEEE Trans Med Imaging. 1999;18:712–721. doi: 10.1109/42.796284. [DOI] [PubMed] [Google Scholar]
- 46.Hajnal JV, Hill DLG, Hawkes DJ, editors. Medical image restoration. Boca Raton: CRC Press; 2001. [Google Scholar]
- 47.Shenton ME, Gerig G, McCarley RW, Szekely G, Kininis R. Amygdala-hippocampal shape differences in schizophreia: the application of 3D shape models to volumetric MR data. Psychiatry Res. 2002;115:15–35. doi: 10.1016/s0925-4927(02)00025-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brenner DJ, Hall EJ. Computed tomography--an increasing source of radiation exposure. N Eng J Med. 2007;357:2277–2278. doi: 10.1056/NEJMra072149. [DOI] [PubMed] [Google Scholar]
- 49.Ludlow J, Ivanovic M. comparative dosimetry of dental CBCT devices and 64-slice CT for oral and maxillofac radiology. Oral Surg, Oral Med, Oral Pathol, Oral Radiol, Endodontol. 2008;106:106–114. doi: 10.1016/j.tripleo.2008.03.018. [DOI] [PubMed] [Google Scholar]
- 50.Maal TJ, Plooij JM, Rangel FA, Mollemans W, Schutyser FA, Bergé SJ. The accuracy of matching three-dimensional photographs with skin surfaces derived from cone-beam computed tomography. Int J Oral Maxillofac Surg. 2008 Jul;37(7):641–6. doi: 10.1016/j.ijom.2008.04.012. [DOI] [PubMed] [Google Scholar]