Abstract
In mammals, there are five Rad51 paralogs that form two distinct complexes in vivo. One complex is composed of Rad51B-Rad51C-Rad51D-Xrcc2 (BCDX2) and the other Rad51C-Xrcc3 (CX3). We co-expressed and purified human BCDX2 and CX3 protein complexes from insect cells and investigated their binding preferences and structure using transmission electron microscopy (TEM). We visualized the binding of BCDX2 and CX3 to DNA templates containing replication forks and Holliday junctions, intermediates observed during DNA replication and recombination, respectively. We show that both complexes bind with exceptionally high specificity to the DNA junctions with little binding observed elsewhere on the DNAs. Further analysis of the structure of free or DNA-bound BCDX2 and CX3 complexes revealed a multimeric ring structure whose subunits are arranged into a flat disc around a central channel. This work provides the first EM visualization of BCDX2 and CX3 binding to Holliday junctions and forked DNAs and suggests the complexes form ring-shaped structures.
Keywords: DNA, DNA/Damage, DNA/Protein Interaction, DNA/Recombination, DNA/Repair, DNA/Replication, Protein/Binding/DNA
Introduction
Homologous recombination repair (HRR)2 is conserved from bacteria to humans and is a major pathway for the repair of double strand breaks (DSB). It is also involved in re-establishing stalled replication forks and facilitating telomere maintenance in telomerase-negative cells (1).
The eukaryotic Rad51 protein is the structural and functional homolog of the Escherichia coli RecA and a key player in the HRR pathway (2). It forms foci at sites of DNA damage where it binds to single-stranded DNA (ssDNA) forming long nucleoprotein filaments that promote strand exchange with homologous double-stranded DNA (dsDNA) (3). This process is facilitated by a number of accessory proteins including Rad52, Rad54, BRCA1, BRCA2, and the Rad51 paralogs (4–8).
There are five Rad51 paralogs in eukaryotes; Rad51B, Rad51C, Rad51D, Xrcc2, and Xrcc3 that show 20–30% sequence identity to Rad51 (9). Mutations in any of the Rad51 paralogs in chicken or hamster cells leads to increased sensitivity to DNA interstrand cross-linking agents and ionizing radiation (10–12). Furthermore, disruption of Rad51B, Rad51D, or Xrcc2 in mice is embryonically lethal suggesting that the Rad51 paralogs are essential genes (13–15). Yet, the role of the Rad51 paralogs in this process is still unclear. They are thought to facilitate Rad51 at early stages of HRR, as they are involved in the formation of Rad51 DNA damage foci (16–18). They also preferentially bind ssDNA and exhibit homologous pairing or strand annealing activity in vitro (1, 19).
On the other hand, they have also been associated with a role in late stages of HRR. In vivo Rad51 paralogs form two primary complexes, one composed of Rad51B-Rad51C-Rad51D-Xrcc2 (BCDX2), and the other of Rad51C and Xrcc3 (CX3) (20). The Rad51C-Xrcc3 complex has been associated with Holliday junction resolvase activities in mammalian cells (21). In addition, the BCDX2 complex has a preference for binding to branched DNA structures (19, 21–23) and a role for Xrcc2 or Xrcc3 in replication fork progression after DNA damage has also been proposed (24). Thus the role of Rad51 paralogs in HRR is complicated and likely depends on the different functional complexes of the paralogs and their interacting partners.
Understanding the structure-specific DNA binding activity and arrangements of different Rad51 paralog complexes on DNA is key to understanding their role in DNA replication and repair. Therefore, in this study we have examined the two primary Rad51 paralog complexes BCDX2 and CX3, free, and bound to Holliday junctions and replication forks by electron microscopy (EM).
EXPERIMENTAL PROCEDURES
Purification of Human Rad51 Paralog Complexes CX3 and BCDX2
Expression constructs for histidine-tagged human Rad51B, Rad51C, Rad51D, XRCC2, and XRCC3 genes were a generous gift from Dr. Stephen West (London Research Institute, UK). All viruses were generated in Sf21 insect cells (Invitrogen) according to the manufacturer's procedures and had a titer of 1 × 109 pfu/ml. All proteins were expressed as His-tagged proteins and purified according to previously published protocols leaving the His tags on the purified protein complexes (20, 25). Recombinant CX3 and BCDX2 complexes were purified from Sf21 cells grown in spinner flasks in Grace's Insect Media (Invitrogen) supplemented with 10% fetal bovine serum (Sigma). For CX3, 300 ml Sf21 cells (1 × 106 cells/ml) were inoculated with viruses for his-Rad51C (moi, 5) and his-XRCC3 (moi, 5). After 48 h of incubation at 27 °C, the cells were pelleted at 2000 rpm at 4 °C and washed once with cold 1× phosphate-buffered saline. The cell pellet was frozen and kept at −80 °C. For BCDX2, 1 liter of Sf21 cells (1 × 106 cells/ml) was co-infected with his-Rad51B (moi, 5), his-Rad51C (moi, 5), his-Rad51D (moi, 10), and his-XRCC2 (moi, 10) viruses. Both Rad51 paralog complexes were purified by similar procedures. Briefly, the cells were lysed in buffer A (10 mm Tris (pH 7.4), 400 mm NaCl, 8 mm BME, 0.5% Nonidet P-40) for 30 min on ice, and the lysate was dounce homogenized and incubated for 10 min more on ice. The lysate was clarified by spinning at 15,000 rpm for 30 min at 4 °C. The proteins were batch purified with 1 ml of Nickel resin (Qiagen) pre-equilibrated with buffer A, added to the clarified extract and rocked gently at 4 °C. After 1 h, the resin was washed 5 times with 15 ml of buffer A containing 30 mm imidazole (pH 8.0) and 5% glycerol. The bound proteins were eluted from the resin with buffer A containing 250 mm imidazole and 10% glycerol. An AKTA FPLC system equipped with a Superdex 200 10/300 GL column (GE Healthcare) was used to further purify the Rad51 paralog complexes. The column was equilibrated with 10 mm Tris (pH 7.4), 400 mm NaCl, 8 mm BME, and 20% glycerol. Affinity-purified BCDX2 or CX3 complexes were injected, and the column was run at a rate of 0.4 ml/min at 4 °C, and 200-μl fractions were collected. The column was first calibrated using the high molecular mass calibration kit from Amersham Biosciences. The retention volumes of aldolase (158 kDa), ferritin (440 kDa), and thyroglobulin (669 kDa) were determined and fit to the curve y = −0.1732x + 4.37 (R2 = 0.961).
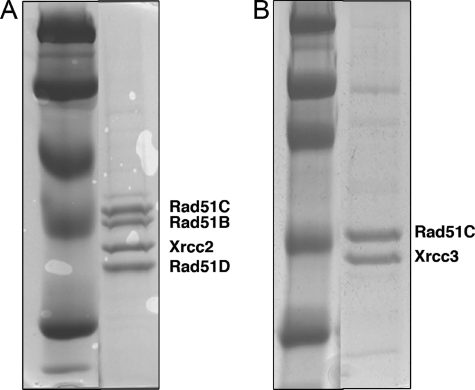
BCDX2 and CX3 were eluted in single peaks from the columns, and their purity was analyzed by SDS-PAGE and visualized using Coomassie Blue stain (Fig. 1). The complexes were determined to be free from contaminating nucleases and all subunits were present in each complex. In agreement with previous studies the apparent stoichiometry of BCDX2 was 1:1:1:1 and CX3 was 1:1 (Fig. 1) (20). The proteins were stored at −80 °C.
FIGURE 1.
Purification of human BCDX2 and CX3 Rad51 paralog complexes. The His-tagged human Rad51B, Rad51C, Rad51D, and Xrcc2 or Rad51C and Xrcc3 complexes were purified from multiply infected Sf21 insect cells as described under “Experimental Procedures.” Protein complexes were separated on a 10% SDS-PAGE gel and visualized by Coomassie Blue staining. A, purified complex containing Rad51B, Rad51C, Rad51D, Xrcc2. B, purified complex containing Rad51C and Xrcc3.
Tungsten-shadowing Electron Microscopy of Bound BCDX2 and CX3 Complexes
The Holliday junction (HJ575) and replication fork (RF25) DNA templates for EM were synthesized as previously described (26–29). For replication forks containing different sized ssDNA gaps at the base of the junction, the pGLGAP plasmid was nicked with N.BbvCIA (NEB) and a 400-nt ssDNA arm was displaced using Klenow fragment (exo-) as previously described (28). To vary the amount of ssDNA at the junction of the fork we used the following primers; primer 1nt (5′-CCTCACTCTCCTCCCCTCCA-3′); primer 5nt (5′-ACTCTCTTCCCCTCCATACC-3′); primer 15nt (5′-CCTCCATACCCTTCCTCCAT-3′); and primer 25nt (5′-CTTCCTCCATCTATACCACC-3′) to convert the ss displaced arm to dsDNA.
Binding reactions were carried out with CX3 at a molar ratio of 1:5 (DNA to CX3 protein heterodimer) or BCDX2 at a molar ratio of 1:3 (DNA to BCDX2 protein heterotetramer) in 30 μl of binding buffer (20 mm HEPES pH 8.0 (KOH), 10 mm MgCl2, 1 mm dithiothreitol, 10 mm ATP, 60 mm phosphocreatine, and 10 units/ml creatine phosphokinase) for 10 min at 30 °C. Samples were fixed in 0.6% glutaraldehyde (Sigma-Aldrich) and passed over a 2-ml column of 2% agarose beads (Agarose Bead Technology, Colna, Portugal) equilibrated with 10 mm Tris-HCl (pH 7.6), and 0.1 mm EDTA. DNA-rich fractions were mixed with buffer containing 2 mm spermidine and 150 mm NaCl and incubated on glow charged carbon foil grids for 3 min. Carbon grids were washed in water and dehydrated in a series of ethanol washes, air-dried, and rotary shadowcast with tungsten. Samples were visualized on a Tecnai 12 transmission electron microscope at 40 kV. All microscopy images were captured using a Gatan Ultrascan 4000 CCD camera and supporting software (Gatan Inc. Pleasanton CA).
Glycerol Spray Low Voltage Electron Microscopy
Purified BCDX2 (216 μg/ml) or CX3 (65 μg/ml) complexes were diluted 1:1 (to a final volume of 30 μl) into a buffer of 40% glycerol, 0.2 m ammonium bicarbonate (pH 7.5) followed by gentle centrifugation through Sephadex G-50 equilibrated in this buffer. The proteins were sprayed into tiny droplets directly onto very thin, glow discharge-treated carbon foils supported by 400-mesh copper grids using an EFFA atomizer (Earnest Fullam Inc). The samples were placed in an oil-free cryo-pumped evaporator and dried for 18 h at a final vacuum of 1 × 108 torr. Without breaking the vacuum, the sample was rotary shadowcast with a thin layer of tungsten and examined in a Tecnai 12 electron microscope at an accelerating voltage of 16 kV.
Negative Staining of BCDX2 and CX3 Complexes
Purified complexes were adsorbed directly onto glow-charged carbon covered copper mesh grids for 3–5 min followed by staining with 2% (w/v) uranyl acetate in water. Samples were analyzed in a Tecnai 12 EM at 80 kV.
RESULTS
Visualization of BCDX2 and CX3 Binding to DNAs Containing Junctions
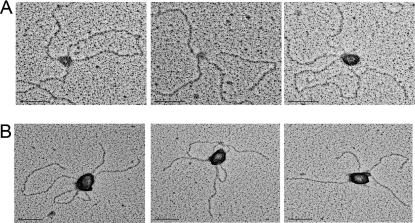
Human Rad51 paralog complexes BCDX2 and CX3 were co-expressed in insect cells, purified, and visualized by Coomassie Blue staining (Fig. 1, A and B). Binding of BCDX2 and CX3 to DNA templates containing a replication fork or Holliday junction structure was examined by EM. The replication fork and Holliday junction templates have been described previously (26, 28). The replication fork template has a 400-bp fork created within a 3.4-kb duplex circle and the Holliday junction contains four 575-bp intersecting arms. BCDX2 or CX3 were incubated with the DNA, fixed, and adsorbed onto carbon grids and then shadowcast with tungsten (Fig. 2). Binding was deliberately kept at subsaturating levels of protein:DNA as incubations at higher ratios resulted in the formation of large protein-DNA aggregates (data not shown) similar to those previously observed by EM (20). Under these conditions, the percentage of replication fork (RF) DNAs with CX3 bound were 60 ± 5% (n = 210, three independent experiments) and 30 ± 5% for BCDX2 (n = 162, two independent experiments).
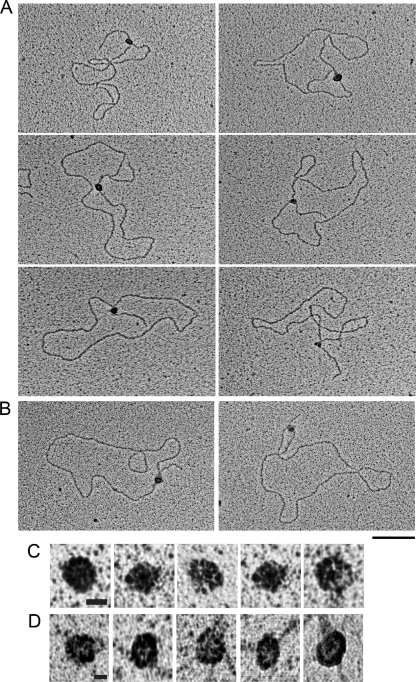
FIGURE 2.
BCDX2 and CX3 binding to replication fork DNAs as seen by EM. Replication fork DNAs were incubated with A, BCDX2 or B, CX3, and the products were visualized by EM. Samples were mounted onto charged carbon-coated copper grids and shadowed with tungsten. Representative bound replication forks are shown. The bar is equivalent to 50 nm. C, CX3 and D, BCDX2 protein complexes at higher magnification. The bar is equivalent to 9 nm.
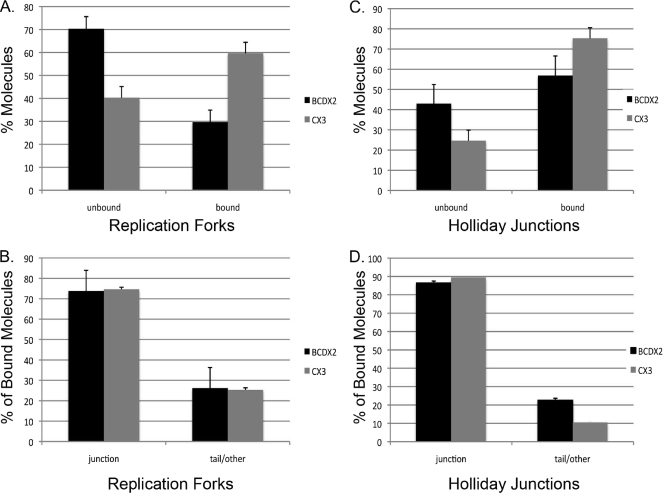
Examination of the binding preferences of these CX3 complexes on RF DNAs (n = 210) revealed that 75 ± 4% were localized exclusively at the forked junction (Figs. 2 and 3B). The remaining CX3 bound molecules were bound at sites other than the three-way junction, for example along the tail or along the circular portion of the DNA template (25 ± 4%). Similarly, BCDX2 revealed a high specificity for the fork junction. Of the 162 RF DNA molecules scored, 74 ± 10% showed the BCDX2 complex localized to the fork, with the remaining complexes bound at other sites on the DNA (26 ± 10%) (Figs. 2 and 3B).
FIGURE 3.
Quantitative analysis of BCDX2 and CX3 binding to replication forks and Holliday junctions. Binding was characterized as either unbound or bound and expressed as a percentage of molecules analyzed. A, the percentage of replication forks bound by BCDX2 (n = 162) or CX3 (n = 210). C, the percentage of Holliday junctions bound by BCDX2 (n = 258) or CX3 (n = 182). The percentage of bound molecules was further categorized as either bound at the junction or bound elsewhere (tail/other) on the DNA and expressed as the percentage of total bound molecules. B, percentage of bound replication forks with BCDX2 or CX3 at the junction. D, percentage of bound Holliday junctions with BCDX2 and CX3 at the junction. Error bars represent ± S.D.
We next examined binding of CX3 and BCDX2 to Holliday junctions (HJ) by EM under conditions in which 75 ± 5% of the HJ DNAs were bound by CX3 and 57 ± 10% were bound by BCDX2 (Fig. 3C and 5). Of the 182 CX3 molecules bound to HJ DNA, 89 ± 0.5% was localized to the junction. Similarly, 86 ± 19% (n = 258) of the bound BCDX2 complexes were localized to the junction (Fig. 3D). The remaining molecules were bound nonspecifically on the HJ DNA template either at the end of one arm, or rarely along the arm itself. Given the large number of possible binding sites for BCDX2 or CX3 on Holliday junctions and replication forks these findings suggest a strong preference for binding to the junctions relative to the much longer regions of DNA along the arms, or at the ends of the dsDNA.
Kinetics of Rad51 Paralog Complex Binding
It is possible that Rad51 paralog complexes bind directly to DNA junctions using a bind and release mechanism; or alternatively, they may bind to duplex DNA and slide until they reach a junction where they become stably arrested. To examine this possibility, we incubated replication forks with the Rad51 paralog complexes and removed samples at various time points for analysis by EM.
We monitored the location of complex binding (i.e. duplex or junction) as a function of time. The number of BCDX2-bound DNAs remained constant over the 60 min (Table 1). Analysis of the location of BCDX2 revealed that 73% were bound specifically at the junction at 2 min (Table 1). This number increased to 84% at 60 min.
TABLE 1.
Time course of BCDX2 and CX3 complexes binding to DNAs containing junctions
BCDX2 or CX3 complexes were incubated with replication fork DNAs for 2, 10, or 60 minutes, fixed, and prepared for EM as previously described. The percentage of junction-bound or duplex-bound molecules was expressed as a percentage of total bound molecules. More than 100 DNAs were examined per time point.
| Protein complex | Time (min) | Overall % bound molecules | % Bound to junction DNA | % Bound to duplex DNA | n = |
|---|---|---|---|---|---|
| BCDX2 | 2 | 52 | 73 | 27 | 106 |
| 10 | 30 | 74 | 26 | 162 | |
| 60 | 48 | 84 | 16 | 103 | |
| CX3 | 2 | 46 | 93 | 7 | 101 |
| 10 | 60 | 75 | 25 | 210 | |
| 60 | 22 | 91 | 9 | 102 |
The percentage of molecules bound by CX3 reached a peak at 10 min with 60% of molecules bound (n = 101) (Table 1). Like, BCDX2, 93% of those molecules bound by CX3 were bound at the junction at 2 min (Table 1). The absence of duplex DNA binding at early time points suggests that complexes must slide very quickly along the duplex DNA, or that they do not need to bind duplex DNA to find junctions.
Binding to Replication Fork DNAs with Varying Amounts of ssDNA at the Junction
Rad51 paralogs bind efficiently to ssDNA, but not dsDNA (19, 20, 25). The replication fork templates used in Fig. 2 contain 25 nt of ssDNA at the base of the replication fork junction (28). Therefore, it is possible that ssDNA may be responsible at least in part, for localizing the Rad51 paralogs to the replication fork structures. In an effort to determine the extent that ssDNA contributes to protein binding on these templates, we examined the binding of BCDX2 and CX3 to replication fork templates containing different sized ssDNA gaps at the base of the replication fork junction. To create these DNA templates, plasmids containing a N.BbvCIA nicking site at the 5′-end of a 400-bp G-less cassette were nicked and strand displacement reactions were carried out using the Klenow fragment (exo-) in the absence of dCTP. Strand displacement is therefore stopped at the end of the G-less cassette when it encounters the first guanine. Primers were designed that anneal to the base of the displaced arm leaving a 1-, 5-, 15-, or 25-nt ssDNA gap. The arms were converted to dsDNA by primer extension using Klenow fragment (exo-) thus generating a 400-bp arm on a 3.4-kb duplex circle (see Ref. 28 and “Experimental Procedures”).
Scoring greater than 200 molecules per replication fork template we observed little evidence that ssDNA contributed significantly to the binding of these junctions. The percentage of junction bound molecules was consistent regardless of the length of ssDNA at the base of the fork.
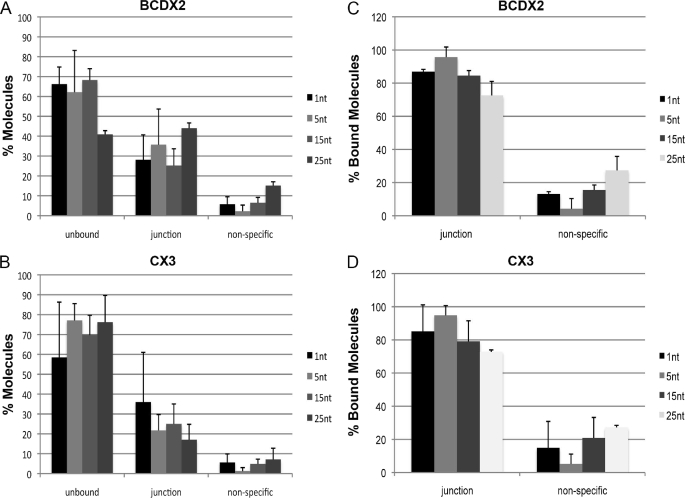
Overall 42 ± 20% of the DNA was bound by BCDX2 for replications forks containing a 1nt gap, compared with templates containing longer ssDNA gaps of 5, 15, or 25 nt that had 23 ± 8%, 30 ± 10%, and 24 ± 13% of the replications forks bound by BCDX2 respectively (Fig. 4A). The same was true for CX3 complexes. Overall 34 ± 9% of the DNA was bound by CX3 for replications forks containing a 1-nt gap. In comparison templates containing longer gaps of 5, 15, or 25 nt had 38 ± 21%, 32 ± 6%, and 59 ± 25% of the replication forks bound by CX3, respectively (Fig. 4B).
FIGURE 4.
Quantitative analysis of BCDX2 and CX3 bound replication forks containing different amounts of ssDNA at the fork junction. Large numbers of DNA molecules were surveyed to determine protein-free and protein-bound fractions on replication forks containing 1, 5, 15, or 25 nt of ssDNA at the junction of the fork. A, replication forks containing 1-, 5-, 15-, or 25-nt gaps bound by BCDX2 or B, CX3 were examined from two-three independent experiments. Molecules were scored as unbound, bound at base of the three-way forked junction, or bound nonspecifically (defined as along the arm or along the circular portion of the template). Error bars are ± S.D. C and D, the protein-bound fractions were scored for specific junction binding and nonspecific binding (defined as binding elsewhere on the DNA) for BCDX2 and CX3, respectively. Error bars represent ± S.D.
BCDX2 continued to bind exclusively to the junction of the replication forks with 87 ± 1% (n = 267) of junctions bound at this site for replications forks containing a 1-nt gap and 96 ± 6% (n = 233) and 85 ± 3% (n = 266) for replications forks with longer gaps of 5nt and 15 nt gaps, respectively (Fig. 4C). Similar results were obtained for CX3. Binding of CX3 to replication forks with a 1-nt gap resulted in 85 ± 16% (n = 211) of replication forks bound exclusively at the junction. Replication forks containing longer gaps of 5 nt and 15 nt had 95 ± 6%, (n = 217) and 79 ± 12%, (n = 228) of CX3 protein bound specifically at the junction (Fig. 4D). Therefore, we conclude that the small amount of ssDNA present at the base of the replication fork junctions does not contribute significantly to the recognition or specific binding of Rad51 paralog complexes to forked junctions.
Structure of Free and DNA-bound BCDX2 and CX3 Complexes
Closer examination of the tungsten-shadowed BCDX2 and CX3 protein complexes bound to DNA templates suggested that they formed a large multimeric protein complex with reasonably uniform size and shape (Figs. 2 and 5). In an effort to explore the structure of these complexes in more detail we used a combination of techniques including negative staining EM, and a technique we described previously as glycerol spray/low voltage EM (30). This latter EM preparative method is an adaptation of that described independently by Erickson and Branton (31, 32). It has the advantage of controlled drying and not exposing proteins to chemical fixatives, air-drying, or strong metal salts. These techniques allow us to obtain better resolution of structures formed by the protein complexes.
FIGURE 5.
Binding of BCDX2 or CX3 to Holliday Junction DNA. CX3 (A) or BCDX2 (B) was incubated with Holliday junction templates, mounted onto carbon-coated copper grids, and rotary shadowcast with tungsten for visualization by EM. Images are shown in reverse contrast. The bar is equivalent to 50 nm.
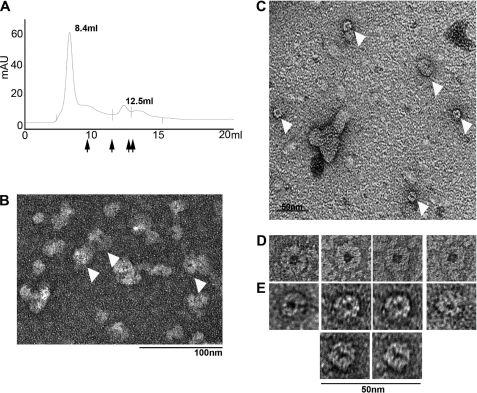
Using these approaches we were able to visualize fields of uniform CX3 complexes (Fig. 6D and supplemental Fig. S1). Measurements of CX3 dimensions by tungsten-shadowing, glycerol spray, and negative staining EM suggest that the rings have a diameter ∼16 nm (Table 2). The observation that almost all of the CX3 molecules observed by glycerol spray and negative staining techniques lay in a single orientation is consistent with a flat protein structure that is too narrow to lay on the EM support on its edge (Fig. 6 and supplemental Fig. S1). As such side profiles of the protein complex were not often visible by the techniques used in this study. Furthermore, we were able to visualize a central pore in the CX3 complex by negative staining EM.
FIGURE 6.
Visualization of BCDX2 and CX3 ring-shaped complexes by negative staining electron microscopy. A, representative elution profile of BCDX2 complex from Superdex 200 column. The major peak contained all four Rad51 paralogs. Arrows indicate elution of thyroglobulin (669 kDa), ferritin (440 kDa), catalase (232 kDa), and aldolase (158 kDa). B, representative field of BCDX2 complexes visualized by mounting the peak fraction onto a charged carbon grids followed by staining with 2% uranyl acetate as described under “Experimental Procedures.” The bar is 100 nm. C, negatively stained BCDX2 fractions with rings of mixed size diameter. The bar is 50 nm. D and E, panel of rings observed by negative staining of the BCDX2 and CX3 fractions, respectively. The bar is 50 nm.
TABLE 2.
Summary of measurements from BCDX2 and CX3 protein complexes using three EM preparative methods
The diameter of BCDX2 or CX3 complexes was measured from digital images of glycerol-sprayed, negatively stained, and tungsten-shadowed complexes using Gatan digital micrograph software. The diameter is reported ± S.D.
| Protein complex | EM method | Diameter ± S.D. | n = |
|---|---|---|---|
| nm | |||
| BCDX2 | Glycerol spray | 21 ± 4 | 118 |
| BCDX2 | Negative staining | 28 ± 7 | 51 |
| BCDX2 | Tungsten shadowing | 28 ± 9 | 22 |
| CX3 | Glycerol spray | 16 ± 3 | 106 |
| CX3 | Negative staining | 21 ± 7 | 17 |
| CX3 | Tungsten shadowing | 19 ± 4 | 51 |
Fields of uniform BCDX2 complexes were also observed by negative staining and glycerol spray EM (Fig. 6 and data not shown). In this case the rings were larger than that observed for CX3 complexes (Table 2). Again, most molecules were observed in a single orientation indicating that the molecules are flat and unable to lie on the supports in different orientations. Comparisons of the size of over a hundred BCDX2 complexes suggested that they had a diameter of ∼21 nm (Table 2).
We occasionally observed smaller ring-shaped protein complexes in the background of negatively stained BCDX2 samples (Fig. 6, C and E). These contained a central pore and had a diameter more similar to that observed with CX3 complexes. Thus it is possible that the larger BCDX2 complex dissociates into smaller subcomplexes during preparation of the BCDX2 complex for negative staining EM, or alternatively that there is a small percentage of heterodimers or monomers in the preparation.
To examine this possibility we used size exclusion chromatography to isolate the whole complex from possible subcomplexes or monomers prior to preparation for EM. The BCDX2 complex eluted as a single peak near the void volume of the column (Fig. 6A). This fraction was mounted for EM and revealed ring-shaped complexes identical to those observed in previous preparations (Fig. 6B). A second peak with a mass of ∼160 kDa contained only two of the four Rad51 paralogs from the BCDX2 complex. This suggests that the BCDX2 preparation contained a low level of BCDX2 subcomplexes that may account for the occasional smaller ring structures. In addition this confirms that the ring structures observed by EM contain the entire BCDX2 complex.
Interestingly, the elution of the complexes near the void volume is indicative of either a very large protein complex or a complex with an asymmetrical structure (see “Discussion”). The latter is consistent with our EM data in which the complexes are observed in a single orientation consistent with a flat ring-shaped particle rather than a globular protein.
DISCUSSION
In this study we used EM to demonstrate that Rad51B-Rad51C-Rad51D-Xrcc2 (BCDX2) and Rad51C-Xrcc3 (CX3) complexes bind strongly to Holliday junction and replication fork structures. Furthermore, BCDX2 and CX3 were seen to bind to the junction of replication forks and the intersection of the four duplex arms of the Holliday junction with exceptionally high specificity for the junctions relative to the much longer regions of duplex DNA or DNA ends. This work also provides the first visualization of BCDX2 and CX3 structures free in solution and bound to DNA and suggests that both BCDX2 and CX3 form flat ring-shaped complexes.
Previous attempts by others to visualize CX3 protein complexes by EM were problematic revealing only dispersed and irregular structures with occasional ring like structures in the background (25). Thus the structure of the CX3 complex could not be determined. Using the glycerol spray/low voltage and negative staining EM we were successful at visualizing fields of uniform CX3 complexes, which revealed a flattened ring-shape structure. Size analysis revealed that the CX3 complex has a diameter of ∼16 nm. This is consistent with that of a higher oligomer of CX3 protein rather than protein monomers or heterodimers. This work also provides the first EM visualization of the BCDX2 complex as a flat ring-shaped structure. Its size is larger than that of CX3 with a diameter of ∼21 nm.
The occasional observation of smaller rings in the background of BCDX2 complexes by negative staining leads us to consider the model in which the BCDX2 complex may be composed of smaller rings that are sometimes dissociated during EM preparation. Consistent with this idea, ring structures have been reported previously for Xrcc2-Rad51D subcomplex (33). The size and shape of Xrcc2-Rad51D rings were consistent with the size of rings observed in the background of negatively stained BCDX2 complexes (33).
In agreement with previous reports CX3 and BCDX2 complexes eluted from size exclusion chromatography in a 1:1 (CX3) or 1:1:1:1 (BCDX2) ratio (19, 20, 25). This suggests that each ring complex contains an equal number of each paralog. In addition, the observation that most CX3 and BCDX2 complexes are observed in a single orientation on the EM support in both glycerol spray and negative staining EM preparations suggest that the complexes are flat rings with narrow side to small to support themselves. This latter finding may explain why Rad51 paralog complexes elute near the void volume of SEC columns. Mass analysis using gel filtration is based on the assumption that the protein complex is globular. Therefore, it is not unexpected that the complexes eluted in fractions consistent with a larger mass. Indeed, a complex that is not globular may have a significantly altered elution and such structures are often eluted in the void volume.
The visualization of affinity purified complexes before and after the SEC strongly suggests that the predominant structure formed by the CX3 and BCDX2 complexes are flat ring-shaped structures. However, it remains possible that other structural states of the complexes may exist within the population. However, we did not identify any such molecules after analyzing fields of BCDX2 or CX3 complexes by EM.
Previous studies have demonstrated that both BCDX2 and CX3 bind preferentially to ssDNA consistent with their role in the early stages of HRR (20, 25). However, Rad51B and the BCDX2 complex have been shown to bind to branched structures including Y-shaped molecules and Holliday junctions as demonstrated by electrophoretic mobility shift assays (EMSA) (20, 23, 34). Visualization of binding to Holliday junctions and forked DNA structures by EM however, has up until now, been lacking. The EM images in this report provide the first visualization of BCDX2 and CX3 complexes binding to Holliday junctions and replication forks. They reveal that BCDX2 and CX3 bind predominantly to structured components of DNA substrates, rather than being localized at duplex regions or at free ends. The Holliday junction structures contain four intersecting 575-bp DNA duplexes, and the replication fork is created within a 3.4-kb circular duplex DNA. Therefore, there are large numbers of potential binding sites available for binding. But neither complex bound to the duplex DNA regions of these templates. We did, however, observe a high binding specificity for the junction of the replication fork and the intersection of the four duplex DNA arms in the Holliday junctions. It remains possible that the complexes recognize ssDNA at the junction of the replication fork or through transient breathing of the Holliday junction structure. However, decreasing the ssDNA regions at the base of the replication forks to as little as 1nt had little impact in the overall binding or the specificity of the complexes for the junctions of the DNAs. To test this further we incubated the templates with a competitor DNA containing ssDNA at one end and still observed preferential binding of CX3 and BCDX2 to the junction structures (data not shown). Therefore we conclude that ssDNA does not contribute significantly to the recognition or specific binding of these complexes to replication fork structures.
There are several possible mechanisms for recognizing junction structures including scanning the DNA and binding tightly to junctions when they are encountered. Or alternatively, a bind and release mechanism where complexes bind directly to the junction without preloading or scanning duplex DNA. In the first model, scanning could be slow or fast. In the case of slow scanning, a large number of duplex bound DNAs would be anticipated at early time points that eventually transition to junction bound DNAs as the complexes encounter junction structures. We did not observe this by EM. This suggests that CX3 and BCDX2 may scan the duplex DNA quickly and bind to junctions rapidly or may use a bind and release mechanism to load directly onto the junctions without the need to scan duplex DNA.
Examination of the DNA-bound complexes revealed flat ring-shaped molecules similar to free protein. Indeed, one can visualize the different orientations of the flat disc in its bound state in some EM images (Figs. 2 and 5). It is unclear what the manner of DNA binding is for the BCDX2 or CX3 complexes. It is possible that the DNA threads through the pore of the complexes. This has been observed for other ring-shaped proteins for example the SV40 large T antigen, the T7 gp4, and E. coli DNA B (35). Alternatively the DNA could wrap around the outside of the ring. Clearly further EM analysis of these complexes is required to clarify how these complexes interact with DNA. Such information would clearly facilitate models of how these complexes facilitate DNA repair and replication.
The finding that both BCDX2 and CX3 bind to Holliday junctions and replication forks is consistent with their role in the latter stages of HRR (21). Given that both early and late acting roles have been proposed for Rad51 paralogs, it is possible that different subcomplexes have different functional roles. Alternatively, association with other proteins may regulate the function of the Rad51 paralog complexes. For example in vivo interactions with other proteins could influence the equilibrium of different subcomplexes and/or modulate the behavior of these complexes. Possible candidates for interacting partners include the GEN1 resolvase and BLM helicase, both of which have been associated with Rad51 paralogs (21, 22). Consistent with this idea the Rad51D-Xrcc2 subcomplex has been shown to stimulate the activity of BLM helicase on Holliday junction templates (22).
It will be important to determine the binding preferences for each of the Rad51 paralog complexes and establish if associations with interacting proteins modulate their binding preferences or function. EM provides a novel platform for investigating such interactions and may reveal the mode of action of these protein complexes on Holliday junctions and replication forks.
Supplementary Material
Acknowledgment
We thank Dr. Stephen West for the gift of the human Rad51 paralog-expressing constructs.
This work was supported, in whole or in part, by National Institutes of Health Postdoctoral Fellowship Grant GM077900 (to S. A. C.) and Grants GM31819 and ES013773 (to J. D. G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- HRR
- homologous recombination repair
- DSB
- double strand breaks
- ds
- double-stranded
- ss
- single-stranded
- EM
- electron microscopy
- moi
- multiplicity of infection
- HJ
- Holliday junction.
REFERENCES
- 1.Symington L. S. (2002) Microbiol. Mol. Biol. Rev. 66, 630–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jang Y. K., Jin Y. H., Shim Y. S., Kim M. J., Yoo E. J., Choi I. S., Lee J. S., Seong R. H., Hong S. H., Park S. D. (1996) Mol. Gen. Genet 251, 167–175 [DOI] [PubMed] [Google Scholar]
- 3.Mazin A. V., Bornarth C. J., Solinger J. A., Heyer W. D., Kowalczykowski S. C. (2000) Mol. Cell 6, 583–592 [DOI] [PubMed] [Google Scholar]
- 4.Benson F. E., Baumann P., West S. C. (1998) Nature 391, 401–404 [DOI] [PubMed] [Google Scholar]
- 5.Shinohara M., Gasior S. L., Bishop D. K., Shinohara A. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 10814–10819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuan S. S., Lee S. Y., Chen G., Song M., Tomlinson G. E., Lee E. Y. (1999) Cancer Res. 59, 3547–3551 [PubMed] [Google Scholar]
- 7.Cousineau I., Abaji C., Belmaaza A. (2005) Cancer Res. 65, 11384–11391 [DOI] [PubMed] [Google Scholar]
- 8.Alexeev A., Mazin A., Kowalczykowski S. C. (2003) Nat. Struct. Biol. 10, 182–186 [DOI] [PubMed] [Google Scholar]
- 9.Thacker J., Zdzienicka M. Z. (2004) DNA Repair 3, 1081–1090 [DOI] [PubMed] [Google Scholar]
- 10.Takata M., Sasaki M. S., Tachiiri S., Fukushima T., Sonoda E., Schild D., Thompson L. H., Takeda S. (2001) Mol. Cell. Biol. 21, 2858–2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuller L. F., Painter R. B. (1988) Mutat. Res. 193, 109–121 [DOI] [PubMed] [Google Scholar]
- 12.Jones N. J., Cox R., Thacker J. (1987) Mutat Res. 183, 279–286 [DOI] [PubMed] [Google Scholar]
- 13.Pittman D. L., Schimenti J. C. (2000) Genesis 26, 167–173 [DOI] [PubMed] [Google Scholar]
- 14.Shu Z., Smith S., Wang L., Rice M. C., Kmiec E. B. (1999) Mol. Cell. Biol. 19, 8686–8693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deans B., Griffin C. S., O'Regan P., Jasin M., Thacker J. (2003) Cancer Res. 63, 8181–8187 [PubMed] [Google Scholar]
- 16.Bishop D. K., Ear U., Bhattacharyya A., Calderone C., Beckett M., Weichselbaum R. R., Shinohara A. (1998) J. Biol. Chem. 273, 21482–21488 [DOI] [PubMed] [Google Scholar]
- 17.Takata M., Sasaki M. S., Sonoda E., Fukushima T., Morrison C., Albala J. S., Swagemakers S. M., Kanaar R., Thompson L. H., Takeda S. (2000) Mol. Cell. Biol. 20, 6476–6482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sung P., Krejci L., Van Komen S., Sehorn M. G. (2003) J. Biol. Chem. 278, 42729–42732 [DOI] [PubMed] [Google Scholar]
- 19.Kurumizaka H., Ikawa S., Nakada M., Eda K., Kagawa W., Takata M., Takeda S., Yokoyama S., Shibata T. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 5538–5543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masson J. Y., Tarsounas M. C., Stasiak A. Z., Stasiak A., Shah R., McIlwraith M. J., Benson F. E., West S. C. (2001) Genes Dev. 15, 3296–3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y., Masson J. Y., Shah R., O'Regan P., West S. C. (2004) Science 303, 243–246 [DOI] [PubMed] [Google Scholar]
- 22.Braybrooke J. P., Li J. L., Wu L., Caple F., Benson F. E., Hickson I. D. (2003) J. Biol. Chem. 278, 48357–48366 [DOI] [PubMed] [Google Scholar]
- 23.Yokoyama H., Sarai N., Kagawa W., Enomoto R., Shibata T., Kurumizaka H., Yokoyama S. (2004) Nucleic Acids Res. 32, 2556–2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henry-Mowatt J., Jackson D., Masson J. Y., Johnson P. A., Clements P. M., Benson F. E., Thompson L. H., Takeda S., West S. C., Caldecott K. W. (2003) Mol. Cell 11, 1109–1117 [DOI] [PubMed] [Google Scholar]
- 25.Masson J. Y., Stasiak A. Z., Stasiak A., Benson F. E., West S. C. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 8440–8446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee S., Cavallo L., Griffith J. (1997) J. Biol. Chem. 272, 7532–7539 [DOI] [PubMed] [Google Scholar]
- 27.Alani E., Lee S., Kane M. F., Griffith J., Kolodner R. D. (1997) J. Mol. Biol. 265, 289–301 [DOI] [PubMed] [Google Scholar]
- 28.Subramanian D., Griffith J. D. (2005) J. Biol. Chem. 280, 42568–42572 [DOI] [PubMed] [Google Scholar]
- 29.Stansel R. M., de Lange T., Griffith J. D. (2001) EMBO J. 20, 5532–5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Griffith J. D., Lindsey-Boltz L. A., Sancar A. (2002) J. Biol. Chem. 277, 15233–15236 [DOI] [PubMed] [Google Scholar]
- 31.Fowler W. E., Erickson H. P. (1979) J. Mol. Biol. 134, 241–249 [DOI] [PubMed] [Google Scholar]
- 32.Shotton D. M., Burke B. E., Branton D. (1979) J. Mol. Biol. 131, 303–329 [DOI] [PubMed] [Google Scholar]
- 33.Kurumizaka H., Ikawa S., Nakada M., Enomoto R., Kagawa W., Kinebuchi T., Yamazoe M., Yokoyama S., Shibata T. (2002) J. Biol. Chem. 277, 14315–14320 [DOI] [PubMed] [Google Scholar]
- 34.Yokoyama H., Kurumizaka H., Ikawa S., Yokoyama S., Shibata T. (2003) J. Biol. Chem. 278, 2767–2772 [DOI] [PubMed] [Google Scholar]
- 35.Stasiak A. Z., Larquet E., Stasiak A., Müller S., Engel A., Van Dyck E., West S. C., Egelman E. H. (2000) Curr. Biol. 10, 337–340 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.