Abstract
THAP1 is a sequence-specific DNA binding factor that regulates cell proliferation through modulation of target genes such as the cell cycle-specific gene RRM1. Mutations in the THAP1 DNA binding domain, an atypical zinc finger (THAP-zf), have recently been found to cause DYT6 dystonia, a neurological disease characterized by twisting movements and abnormal postures. In this study, we report that THAP1 shares sequence characteristics, in vivo expression patterns and protein partners with THAP3, another THAP-zf protein. Proteomic analyses identified HCF-1, a potent transcriptional coactivator and cell cycle regulator, and O-GlcNAc transferase (OGT), the enzyme that catalyzes the addition of O-GlcNAc, as major cellular partners of THAP3. THAP3 interacts with HCF-1 through a consensus HCF-1-binding motif (HBM), a motif that is also present in THAP1. Accordingly, THAP1 was found to bind HCF-1 in vitro and to associate with HCF-1 and OGT in vivo. THAP1 and THAP3 belong to a large family of HCF-1 binding factors since seven other members of the human THAP-zf protein family were identified, which harbor evolutionary conserved HBMs and bind to HCF-1. Chromatin immunoprecipitation (ChIP) assays and RNA interference experiments showed that endogenous THAP1 mediates the recruitment of HCF-1 to the RRM1 promoter during endothelial cell proliferation and that HCF-1 is essential for transcriptional activation of RRM1. Together, our findings suggest HCF-1 is an important cofactor for THAP1. Interestingly, our results also provide an unexpected link between DYT6 and DYT3 (X-linked dystonia-parkinsonism) dystonias because the gene encoding the THAP1/DYT6 protein partner OGT maps within the DYT3 critical region on Xq13.1.
Keywords: Cell/Cycle, Diseases/Neurological, Protein/Protein-Protein Interactions, Transcription/Target genes, Transcription/Zinc finger, THAP1, Dystonia
Introduction
Dystonias are neurological diseases characterized by involuntary muscle contractions which result in twisting, repetitive movements, and abnormal postures (1, 2). At least six primary forms, where dystonia is the only neurologic feature, have been described (1, 2), but the disease gene has been discovered for only two of these, DYT1 and DYT6 (3, 4). Early onset DYT1 generalized dystonia is caused by mutations in the gene encoding TorsinA (TOR1A), a member of the AAA+ family of ATPases, which may function as a chaperone in the nuclear envelope and endoplasmic reticulum (3). Recently, mutations in THAP1 have been identified as a cause of mixed-onset DYT6 primary torsion dystonia (4–6). THAP1 is the prototype of a previously uncharacterized family of cellular factors (> 100 distinct members in the animal kingdom), defined by the presence at their amino terminus of the THAP-zinc finger (THAP-zf),3 an atypical zinc-dependent sequence-specific DNA binding domain (7–9). THAP1 recognizes a consensus DNA target sequence of 11 nucleotides (THABS for THAP1 binding sequence) considerably larger than the 3–4 nucleotide motif typically recognized by classical C2H2 zinc fingers (8). Although THAP1 biological roles are not completely understood, data supporting an important function in cell proliferation and cell cycle pathways have been provided (10). THAP1 was found to be involved in the regulation of endothelial cell proliferation and G1/S cell cycle progression, and RRM1, a pRb/E2F cell cycle target gene involved in S-phase DNA synthesis, was identified as the first direct target gene of THAP1 (10). In addition to cell proliferation, THAP1 may also play roles in cell survival and/or apoptosis because it has been shown to interact with Par-4, a proapoptotic factor linked to prostate cancer and neurodegenerative diseases, including Parkinson disease (11).
Several distinct mutations in THAP1 were found in DYT6 dystonia patients and most of them mapped to the DNA binding THAP-zf, suggesting the mutations may cause disease by disrupting the DNA binding activity of THAP1 (4, 5). Indeed, one of the THAP1 missense mutant proteins (F81L) was functionally analyzed and found to exhibit strongly reduced DNA binding affinity (4). Together, these findings supported the possibility that transcriptional dysregulation, because of mutations in THAP1, might contribute to the phenotype of DYT6 dystonia. Interestingly, transcriptional dysregulation because of reduced expression of TAF-1 (RNA polymerase II TATA-box-binding protein-associated factor 1) has previously been proposed to cause another form of dystonia, DYT3 or X-linked dystonia-parkinsonism (12, 13).
In this study, we report that THAP1 shares sequence characteristics, in vivo expression profiles and cellular partners with THAP3, a previously uncharacterized member of the THAP-zf protein family. Using functional proteomics, we show that HCF-1, a potent transcriptional coactivator and cell cycle regulator (14–19), and OGT, the enzyme that catalyzes the addition of O-GlcNAc (20), are major cellular partners of THAP3. THAP1 also associates with HCF-1 and OGT in vivo and mediates recruitment of HCF-1 to the RRM1 promoter during endothelial cell proliferation. Our results provide an unexpected link between THAP1/DYT6 and OGT, the product of a gene (OGT) mapped within the DYT3 critical interval on Xq13.1 (12, 21), suggesting an unexpected link between DYT6 and DYT3 dystonias.
EXPERIMENTAL PROCEDURES
Cell Culture and RNA Interference
Hela cells, Hela Tet-Off cells (Clontech), and Hela HLR cells (Stratagene) were grown in Dulbecco's modified Eagle's medium. Knockdown of THAP1 and HCF-1 expression in primary human endothelial cells (HUVECs) was performed using ON-TARGET plus SMARTpool and individual siRNA duplexes (Dharmacon, Lafayette, CO) as previously described (10).
Plasmid Constructions
The full-length coding region of human THAP3 was cloned in-frame with an 82-bp linker encoding Flag/HA tags in pTRE-Tight expression vector (Clontech). The resulting vector was co-transfected with pRep-Hygro vector into Hela Tet-Off cells and stable transformants were obtained after 4 weeks of selection in 200 μg/ml hygromycin. Human THAP1 ORF was cloned into expression vector pFlag-CMV5a (Sigma) to generate pFlag-THAP1 expression vector. GST, GST-THAP11–213, GST-THAP3162–239 fusion proteins were produced using pGEX-2T prokaryotic expression vector (Amersham Biosciences), and purified as previously described (11). GST-THAP3H178A, -THAP3Y180A, and -THAP3HBM (DHSY/AAAA) mutants were obtained from wild-type GST-THAP3162–239 using PCR. Gal4-THAP3 and Gal4-THAP3HBM mutant expression vectors were generated by inserting the corresponding full-length THAP3 fragments, generated by PCR, into pCMVGT vector downstream of the Gal4-DB (amino acids 1–147). The THAP3HBM mutant was also cloned into pTRE-Tight expression vector. For two-hybrid assays, the open-reading frames of human THAP-zf proteins were amplified by PCR from full-length clones (Open Biosystems) and inserted into the pGADT7 expression vector (Clontech).
Sequence Analysis
Multiple sequence alignments of the THAP-zf proteins and their HBMs were generated with ClustalW according to the Blosum matrix and colored with Boxshade. Prediction of coiled-coil domains was performed with COILS and PAIRCOIL programs.
Immunoaffinity Purification and Mass Spectrometry Analysis
Nuclear extracts from induced Hela-t-THAP3 cells were prepared as previously described (22) with minor modifications. Nuclei were incubated for 30 min in buffer B: 20 mm Tris pH 7.4; 400 mm NaCl, 5 mm MgCl2, 10 mm β-mercaptoethanol, 0,5% Nonidet, 1 mm phenylmethylsulfonyl fluoride, and Complete protease inhibitor mixture (Roche). The salt concentration was adjusted to 150 mm NaCl, and nuclear extracts were loaded onto a 4-ml 10–40% glycerol gradient in 150 mm NaCl buffer B, centrifuged at 50,000 rpm (200,000 × g) for 4 h. Fractions corresponding to a peak determined after immunoblotting were pooled and purified by immunoprecipitation with anti-FlagM2 antibody-conjugated agarose beads (Sigma), which were washed with 150 mm NaCl-containing IP buffer and eluted with a solution at 500 μg/ml of Flag peptide. As a control, a mock purification was performed from uninduced HeLa cells. Samples from induced and non-induced HeLa cells were separated by SDS-PAGE, and submitted for proteomic analysis (see supplemental information).
Immunoprecipitation and Western Blot Analyses
Nuclear extracts prepared in buffer B (20 mm Tris, pH 7,4; 400 mm NaCl, 5 mm MgCl2, 10 mm β-mercaptoethanol, 0,5% Nonidet, 1 mm phenylmethylsulfonyl fluoride, and Complete protease inhibitor mixture, Roche), were incubated for 16 h at 4 °C with anti-HA mAb (clone HA-7; Sigma; A 2095), control Ig or polyclonal anti-HCF-1 antibodies (15), and precipitated proteins were captured with Protein G-Sepharose beads (eBioscience). Alternatively, Flag-tagged protein complexes were captured with anti-Flag (M2) agarose beads (Sigma; A 2220). After extensive washing, bound proteins were eluted in 2× SDS loading buffer and analyzed by Western blot with mAbs to HA or Flag epitope tags (Sigma), or rabbit antiserum to HCF-1 (15) or OGT (AL28; a generous gift of Dr. Gerald Hart), followed by horseradish peroxidase-conjugated goat anti-mouse or anti-rabbit Ig (1/10000; Promega). Blots were developed with an enhanced chemiluminescence kit (GE Healthcare). Rabbit anti-THAP1 (10) and mouse anti-tubulin-α (Sigma) antibodies (1/1000) were used for some Western blot analyses.
Yeast Two-hybrid and GST Pulldown Assays
HF7c and Y190 strains were transformed with pGAL29 encoding the HCF-1 kelch domain (amino acids 3–455) fused to the GAL4 DNA binding domain, and subsequently retransformed with pGADT7 or the GADT7-THAP clones. HF7c cotransformant strains were patched on SD-Leu, Trp, and SD-Leu/Trp/His media. Y190 cotransformant strains were assayed for β-galactosidase using the Gal-Screen System (Applied Biosystems). GST pulldown assays were performed as previously described (11) using immobilized GST, GST-THAP1, GST-THAP3, or GST-THAP3HBM mutant proteins, and 35S-labeled HCF1 kelch domain (amino acids 3–455) generated in vitro with the TnT-coupled reticulocyte lysate system (Promega, Madison, WI).
Reporter Gene Assays
Hela HLR cells, which contain a Firefly luciferase chromatin-integrated reporter gene under the control of five GAL4 binding sites, were co-transfected with 50 ng of pCMVGT-Gal4-THAP3 or -Gal4-THAP3HBM constructs and 50 ng of Renilla luciferase construct (pRL-CMV, Promega), with or without 1 μg of HCF-1-V5 expression vector (15) using JetPEI reagent (2 μl/well, Polyplus Transfection). After 48h, Firefly and Renilla luciferase activities were assayed with the Dual luciferase assay system (Promega). Firefly luciferase activities were normalized to Renilla luciferase to control for transfection efficiency.
Chromatin Immunoprecipitations (ChIPs), qPCR Assays, and Northern Blot
ChIP-qPCR assays were performed as previously described (10) using 5 μg of antibodies specific for THAP1 (10) or HCF-1 (15). Sequences of the primers are available in the supplemental information. QuantiTect Primer Assays (Qiagen) were used for analysis of HCF-1, RRM1, and GAPDH expression by qPCR. For Northern blot analysis, blots of poly(A)+ RNA from adult mouse and human tissues were hybridized according to the manufacturer's instructions (Clontech), using random-primed human THAP3 and THAP1 cDNA probes.
RESULTS
THAP3 Shares Similarities with THAP1
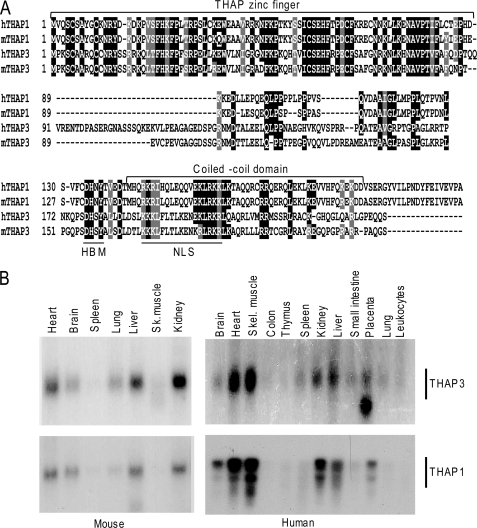
Human THAP-zf proteins are not known to share homologies outside the THAP-zf. However, we observed that, among the 12 human THAP-zf proteins (7), THAP3, shares sequence similarities with THAP1 not only within the THAP-zf (7) but also within the carboxyl-terminal domain, which contains in both proteins, a conserved bipartite nuclear localization sequence (NLS), and a coiled-coil domain predicted by COILS and PAIRCOIL programs (Fig. 1A). Northern blot analyses revealed that THAP3 and THAP1 also share striking similarities in their expression profile both in mouse and human tissues (Fig. 1B). Together, these findings indicated THAP3 and THAP1 exhibit similarities not only in their primary structure and sequence but also in their expression pattern in vivo.
FIGURE 1.
THAP3 shares primary structure and in vivo expression pattern with THAP1/DYT6. A, multiple sequence alignment of human THAP3 and THAP1 and their mouse orthologues. Black boxes indicate identical residues, whereas shaded boxes show similar amino acids. B, Northern blots containing 2 μg/lane poly(A)+ RNA from adult tissues were probed with 32P-labeled THAP3 and THAP1 cDNA probes. Major transcripts of about 1.3 kb (THAP3) and 2.4 kb (THAP1) were detected in both mouse and human tissues. The human THAP1 Northern blot has previously been presented (10) and is shown here only for comparison with THAP3.
THAP1 and THAP3 Associate with HCF-1 and OGT
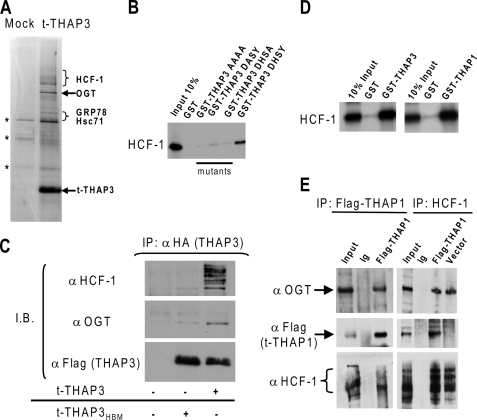
To identify THAP3-interacting partners in vivo, nuclear extracts were prepared from Hela-t-THAP3 cells, conditionally expressing a carboxyl-terminal Flag/HA-tagged THAP3, and fractionated in a glycerol gradient. A predominant THAP3-containing complex of ∼0.6 MDa (supplemental Fig. S1) was identified, immunoprecipitated from the corresponding fractions with anti-Flag antibody, and analyzed by nanoLC-MS/MS mass spectrometry (Fig. 2A). The purification was reproducible, as several THAP3-interacting proteins were repeatedly identified in three independent affinity purification/proteomic analyses (Table 1). Strikingly, many of the THAP3-interacting proteins corresponded to differentially processed forms of HCF-1. Numerous peptides were identified within both N- and C-terminal parts of HCF-1, including many peptides bearing O-glycosylated residues within the HCF-1 basic domain (supplemental Fig. S2). Interestingly, the human OGT enzyme (20) and protein chaperones with demonstrated O-GlcNAc lectin activity, such as the HSP70-like proteins Hsc71 and GRP-78 (23), were also reproducibly identified in the THAP3-associated protein complexes (Fig. 2A and Table 1). The association of HCF-1 and OGT with THAP3 was further confirmed by co-immunoprecipitation assays and immunoblot analyses of nuclear extracts fractionated in glycerol gradients (supplemental Fig. S1). Together, these proteomic analyses indicated that O-GlcNAcylated-HCF-1, O-GlcNAc transferase OGT, and O-GlcNAc lectins constitute the core components of the THAP3-associated protein complexes in human Hela cells.
FIGURE 2.
THAP3 shares protein partners with THAP1/DYT6. A, analysis of affinity-purified t-THAP3 complex. Proteins purified from induced (t-THAP3) or uninduced (mock) HeLa cells were separated on SDS-PAGE and stained with Coomassie Blue. Proteins identified by mass spectrometry as THAP3-specific partners are indicated on the right. Nonspecific proteins are denoted with an asterisk (*). B, wild-type THAP3 and THAP3HBM mutants fused with GST were incubated with in vitro translated [35S]methionine labeled HCF-1 kelch domain, and HCF-1 binding was analyzed by autoradiography. C, THAP3 HBM is essential for THAP3-HCF-1 interaction in vivo. Flag/HA-tagged THAP3 and THAP3HBM mutant proteins, were expressed in Hela cells, and nuclear extracts were immunoprecipitated with anti-HA antibody. Immunoblotting was performed with anti-HCF-1, anti-OGT and anti-Flag antibodies. D, THAP1 interacts with HCF1 kelch domain in vitro. GST, GST-THAP11–213, and GST-THAP3162–239 fusion proteins were incubated with in vitro translated [35S]methionine-labeled HCF1 kelch domain and binding was revealed by autoradiography. E, THAP1 associates with HCF-1 and OGT in vivo. Nuclear extracts from Hela cells transfected with pFlag-THAP1 or pFlag-CMV5a empty vector were immunoprecipitated with anti-Flag, anti-HCF1 antibodies, or rabbit immunoglobulins (Ig). Immunoprecipitates were analyzed by Western blotting with anti-OGT, anti-Flag, or anti-HCF-1 antibodies.
TABLE 1.
THAP3-associated proteins identified by mass spectrometry
Proteins were identified with the Mascot software by database search in Uniprot. The table shows the protein name, Uniprot accession number (AC), gene name, molecular weight (MW), best Mascot score (if the protein was identified in several bands of the migration lane), number of identified peptides (No. Sequences), total number of MS/MS sequencing scans performed (No. Total MS/MS, for all the peptide ions of the protein, and all the gel bands in which it was identified, reflecting the abundance of the protein in the migration lane), and mean ratio of peptides intensity signal between the lane of immunopurified complex and control lane (Ratio ip/control). Three independent experiments were performed, and proteins displayed in the table were classified as potential specific partners (identified only in the complex lane, or with a Ratio ip/control>10) in at least 2 of 3 experiments.
| Protein name | AC | Gene name | MW | Best score | No. of Sequences | No. of Total MS/MS | Ratio ip/control | No. of Experiments |
|---|---|---|---|---|---|---|---|---|
| Host cell factor | P51610 | HCFC1 | 210,707 | 1180 | 43 | 734 | 78 | 3 |
| THAP domain-containing protein 3 | Q8WTV1 | THAP3 | 27,384 | 904 | 20 | 229 | 191 | 3 |
| UDP-N-acetylglucosamine–peptide N-acetylglucosaminyltransferase 110-kDa subunit | O15294 | OGT | 118,104 | 618 | 35 | 160 | 17 | 3 |
| Heat shock cognate 71 kDa protein (Hsc71) | P11142 | HSPA8 | 71,082 | 743 | 28 | 93 | 16 | 3 |
| 78-kDa glucose-regulated protein precursor (GRP-78) | P11021 | HSPA5 | 72,402 | 709 | 31 | 53 | 24 | 2 |
| Stress-70 protein, mitochondrial precursor | P38646 | HSPA9 | 73,920 | 606 | 27 | 52 | 14 | 2 |
Numerous viral and cellular factors bind the HCF-1 amino-terminal kelch domain, via a tetrapeptide motif ((D/E)HXY), designated the HCF-1-binding motif (HBM) (19, 24, 25). A consensus HBM (DHSY) is also present in THAP3 (Fig. 1A), and GST pulldown assays revealed that wild-type THAP3 binds to HCF-1 in vitro. Mutations of the THAP3-HBM strongly reduced (single amino acid mutants) or completely abrogated (quadruple mutant) the THAP3-HCF-1 interaction in vitro (Fig. 2B). Moreover, co-immunoprecipitation assays in human cells revealed that THAP3, but not the THAP3HBM mutant, is able to precipitate HCF-1 and OGT (Fig. 2C). The association of THAP3HBM with OGT was also significantly reduced, suggesting THAP3 associates with OGT in vivo mainly through its interaction with HCF-1. We also observed that THAP1 contains a consensus HBM (DHNY) (Fig. 1A) and interacts with HCF-1 in two-hybrid (see below) and GST pulldown assays in vitro (Fig. 2D). In addition, THAP1 associates with HCF-1 and OGT in co-immunoprecipitation assays in vivo (Fig. 2E). These later findings indicated THAP3 shares its two major cellular partners with THAP1.
THAP1 and THAP3 Belong to a Large Family of HCF-1 Binding Factors
Comparison of the THAP1 and THAP3 sequences with other human THAP-zf proteins showed that a consensus HBM is also present in human THAP2 (EHSY), THAP5 (EHSY), THAP6 (EHSY), THAP7 (EHSY), THAP9 (DHNY), and THAP11 (DHSY), whereas THAP4 contains two HBM-like sequences (LHSY) (Fig. 3A). The HBMs are always located upstream predicted coiled-coil domains (Fig. 3B) and are found at similar positions in the orthologues of THAP-zf proteins in other species, including zebrafish and xenopus, indicating strong evolutionary conservation (supplemental Fig. S3).
FIGURE 3.
THAP1 and THAP3 belong to a large family of HCF-1-binding THAP-zf proteins. A, multiple alignment of the HBMs found in human THAP-zf proteins. A consensus is shown. B, primary structure of the nine human THAP-zf proteins containing consensus HBMs or HBM-like motifs. The asterisk indicates the position of the HBM, which is always located 4–9 amino acids upstream the predicted coiled-coil domain. C, two-hybrid assay. A yeast strain expressing the GAL4DB-HCF-1 kelch domain was transformed with constructs expressing the indicated human THAP-zf proteins or control protein CREB3. Transformants were patched onto SD-Leu/Trp (LT) and SD-Leu/Trp/His (LTH) plates. D, β-galactosidase assays. The β-galactosidase activity of each strain is expressed as relative light units per second (RLU/s). The result shown for each strain is the average of two independent colonies after subtraction of the background level obtained from the GAL4DB-HCF-1 kelch strain transformed with pGADT7 vector alone.
In agreement with the presence of the HBM, several members of the human THAP-zf protein family were identified in large scale two-hybrid screens with the HCF-1 amino-terminal kelch domain (supplemental Fig. S4). Additional two-hybrid assays (Fig. 3, C and D) revealed that the HCF-1 kelch domain binds to human THAP-zf proteins that contain an HBM (THAP1 to THAP7 and THAP11) or HBM-like motif (THAP4) but not to those which do not possess HBMs (THAP8 and THAP10). Based on our bioinformatics analyses and two-hybrid results, we concluded that THAP-zf proteins represent a large family of HCF-1 binding factors.
THAP3 Recruits HCF-1 to a Chromatin-integrated Promoter for Transcriptional Activation
We asked if interaction of THAP3 with HCF-1 might play a role in transcriptional regulation. Because THAP3 target genes have not yet been identified, we used a chromatin-integrated Gal4-dependent luciferase reporter construct (Hela-HLR cell line). Wild-type THAP3 or THAP3HBM mutant was expressed as Gal4 DNA binding domain-fusion proteins in Hela HLR cells, with or without V5-epitope-tagged HCF-1 (Fig. 4A). ChIP assays revealed that both Gal4-THAP3 and Gal4-THAP3HBM were significantly enriched at the Gal4 binding sites in the Gal4 reporter gene (Fig. 4B). In contrast, ChIP assays with anti-V5 antibodies showed that the V5-epitope-tagged HCF-1 was recruited to the Gal4 binding sites when co-expressed with Gal4-THAP3 but not when co-expressed with Gal4-THAP3HBM (Fig. 4C). Moreover, recruitment of HCF-1 by Gal4-THAP3 resulted in enhanced transcriptional activity of the Gal4-luciferase reporter gene (Fig. 4D), whereas no effect was found with the Gal4-THAP3HBM mutant. We conclude that THAP3, through its HBM, is able to recruit HCF-1 to a chromatin-integrated promoter for transcriptional activation.
FIGURE 4.
THAP3 recruits HCF1 to a chromatin-integrated promoter for transcriptional activation. A, Hela HLR cells harboring a Gal-4-dependent chromatin-integrated luciferase reporter gene were transfected with Gal4-THAP3 or Gal4-THAP3HBM mutant expression vectors, with or without V5-HCF-1 expression vector. Expression of HCF-1 and Gal4-THAP3 fusion proteins was analyzed by Western blotting. B and C, Gal4-THAP3, but not Gal4-THAP3HBM mutant, mediates recruitment of V5-HCF-1 to the chromatin-integrated Gal4-luciferase reporter gene. ChIP assays were performed with anti-Gal4 (B), anti-V5 (C), or irrelevant control antibodies, and the amount of DNA precipitated was quantified by qPCR. Fold enrichment was calculated by dividing the amount of Gal4 promoter precipitated by the different antibodies to the amount of DNA precipitated from the control U2 snRNA gene. Results are shown as means and SDs of three separate data points and are representative of at least two independent experiments. D, recruitment of HCF-1 by Gal4-THAP3 results in enhanced transcriptional activity of the Gal4-luciferase reporter, whereas no effect was found with the Gal4-THAP3HBM mutant. Normalized luciferase activities are shown as means and SDs of three independent transfection experiments.
Endogenous THAP1 Recruits HCF-1 to the RRM1 Promoter
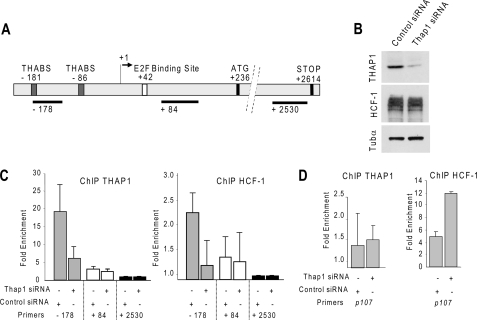
We next asked if HCF-1 might play a role in transcriptional regulation by endogenous THAP1. Using primary human endothelial cells (HUVECs), we have recently shown that THAP1 associates in vivo with consensus THAP1 binding sites found in the RRM1 promoter (10). Because HCF-1 has previously been found to be required for multiple stages of cell cycle progression (18), we determined whether binding of HCF-1 to THAP1 may play a role in transcriptional regulation of RRM1. The RRM1 promoter (Fig. 5A) contains binding sites for both THAP1 and E2F transcription factors. ChIP assays were performed in HUVECs after knockdown of endogenous THAP1 expression with a pool of specific siRNAs (Fig. 5B). ChIP experiments performed in the presence of control siRNAs revealed that THAP1 and HCF-1 are recruited to the promoter region containing the THAP1 binding sites (−178) but not to the 5′-untranslated region containing the E2F site (+84) nor to a site close to the STOP codon (+2630) (Fig. 5C). In cells treated with THAP1 siRNAs, there was a significant reduction of THAP1 association. Interestingly, the reduction in THAP1 binding correlated with a parallel reduction in HCF-1 association with the RRM1 promoter (Fig. 5C and supplemental Fig. S5). In contrast, the association of HCF-1 with the p107 promoter, a promoter bound by E2Fs (19) but not by THAP1, was increased after knockdown of THAP1 (Fig. 5D). These experiments performed in primary human cells indicated that THAP1, rather than E2Fs, is important for HCF-1 recruitment to the RRM1 promoter during endothelial cell proliferation.
FIGURE 5.
Endogenous THAP1 recruits endogenous HCF-1 to the promoter of cell cycle-specific gene RRM1 during cell proliferation in vivo. A, schematic representation of the human RRM1 promoter. The binding sites for THAP1 (THABS) and E2Fs are indicated. The position of the DNA fragments analyzed in ChIP-qPCR assays are shown. B, knockdown of endogenous THAP1 in primary HUVECs. THAP1, HCF-1, and Tubα (loading control) expression levels were analyzed by Western blot. Down-regulation of THAP1 does not modify HCF-1 levels. C, knockdown of THAP1 inhibits recruitment of THAP1 and HCF1 to the RRM1 promoter in vivo. ChIP-qPCR assays with anti-THAP1 or anti-HCF1 antibodies were performed using proliferating primary HUVECs treated with control or THAP1 siRNAs. Immunoprecipitated DNA was quantified in triplicate by qPCR. Fold enrichment was calculated by dividing the amount of the RRM1 promoter −178 and +84 DNA fragments precipitated to the amount of RRM1 + 2530 DNA fragment precipitated (negative control). Results are representative of at least two independent experiments. D, knockdown of THAP1 results in increased recruitment of HCF1 to the p107 promoter in vivo. ChIP-qPCR assays with anti-THAP1 or anti-HCF1 antibodies were performed as described in C.
Endogenous HCF-1 Regulates RRM1 mRNA Levels
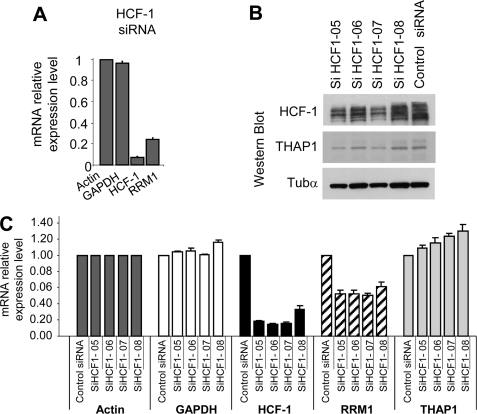
We have previously shown that silencing of endogenous THAP1 in primary HUVECs led to a significant decrease in RRM1 mRNA levels (10). Therefore, we asked whether HCF-1, a potent transcriptional coactivator (24), may also play a role in the regulation of RRM1 expression. Knockdown of endogenous HCF-1 expression with a pool of siRNAs optimized to reduce off-target effects (ON-TARGET-plus SMARTpool siRNAs) resulted in down-regulation of RRM1 mRNA levels (Fig. 6A), indicating HCF-1 is essential for the activation of RRM1 during cell cycle progression. This result was confirmed using 4 individual siRNAs, which similarly reduced HCF-1 levels in HUVECs, as shown by Western blot (Fig. 6B) and qPCR (Fig. 6C) analyses. Importantly, the 4 HCF-1 siRNAs reduced RRM1 mRNA levels (∼50% inhibition), but had no significant effects on mRNA levels of actin, GAPDH, and THAP1. Together, these data provide strong evidence that endogenous HCF-1 modulates RRM1 mRNA levels in primary human endothelial cells.
FIGURE 6.
Endogenous HCF-1 regulates RRM1 expression in primary human endothelial cells. A, knockdown of endogenous HCF-1 with a pool of specific siRNAs reduces RRM1 mRNA levels. RNA was isolated from cells transfected with ON-TARGET-plus SMARTpool HCF-1 siRNAs, 48 h after siRNA transfection, and used for qPCR analysis with the indicated human gene primers (RRM1, HCF-1, and control gene GAPDH). Actin was used as a control gene for normalization. Results are shown as means with S.D. from three separate data points. B and C, knockdown of endogenous HCF-1 with 4 individual ON-TARGET-plus HCF-1 siRNAs. B, HCF-1, THAP1 and Tubα (loading control) expression levels were analyzed by Western blot. C, knockdown of HCF-1 with individual HCF-1 siRNAs results in down-regulation of RRM1 mRNA levels. qPCR analyses were performed as described in A.
Finally, we performed knockdown experiments to address the potential role of OGT in RRM1 regulation. As shown in supplemental Fig. S6, reduction of OGT levels did not affect RRM1 expression in HUVECs. However, this does not preclude a role for OGT in RRM1 regulation in other cell types and for the THAP1/HCF-1/OGT complex in the regulation of other promoters.
DISCUSSION
In this study, we report that dystonia protein THAP1/DYT6 exhibits striking similarities with THAP3, another member of the THAP-zf protein family, in terms of primary structure, in vivo expression pattern and cellular partners. Similarly to THAP3, THAP1 associates with HCF-1, a cell cycle factor and potent transcriptional coactivator, and OGT, the enzyme that mediates O-GlcNAcylation of proteins. THAP3 binding to HCF-1 is mediated by a consensus HBM, a motif that is also found in THAP1. In addition to THAP1 and THAP3, seven other members of the human THAP-zf protein family, contain evolutionary conserved HBMs and possess HCF-1 binding properties, indicating THAP-zf proteins represent a large family of HCF-1-binding factors.
THAP1/HCF-1 Interaction and Transcriptional Regulation during the Cell Cycle
We found that THAP3 and THAP1 are both able to recruit HCF-1 to chromatin-integrated promoters for transcriptional activation. HCF-1 was recruited to the THAP1 binding sites, rather than the E2F site, in the promoter of cell cycle-specific gene RRM1, and endogenous THAP1 was found to be essential for HCF-1 recruitment during endothelial cell proliferation. This is an important result because, although recruitment of HCF-1 on cell cycle-specific promoters has been found to be correlated with the presence of E2F transcription factors (19), the absolute requirement of E2Fs for HCF-1 recruitment has not yet been shown. Knockdown of HCF-1 expression with siRNAs resulted in down-regulation of RRM1 mRNA levels, indicating HCF-1 is essential for the activation of RRM1 during cell cycle progression. Together, these findings suggest THAP1 and E2Fs may cooperate for HCF-1 recruitment on cell cycle specific promoters, with THAP1 playing important roles on some promoters (i.e. RRM1) and E2Fs (19) on other promoters (i.e. p107, cyclin A). They also support the possibility the critical role of HCF-1 in G1/S cell cycle progression (14, 18) may be caused by its interaction with both E2Fs and THAP1.
HCF-1 was initially discovered as a cellular cofactor in the induction of herpes simplex virus transcription and is a potent coactivator for viral VP16 and numerous transcription factors (reviewed in (24, 26)). Accordingly, our results indicate that interaction of THAP1 and THAP3 with HCF-1 plays a role in transcriptional activation. However, the interaction of HCF-1 with Ronin (the mouse orthologue of human THAP11) has recently been proposed to be involved in transcriptional repression in mouse embryonic stem cells (27). Therefore, interaction of THAP-zf proteins with HCF-1 may be important for both transcriptional activation and repression, depending on the THAP-zf protein involved, the cellular context and/or the cell cycle status. This would be similar to what has previously been proposed for the E2F family (19).
An Unexpected Link between DYT6 and DYT3 Dystonias
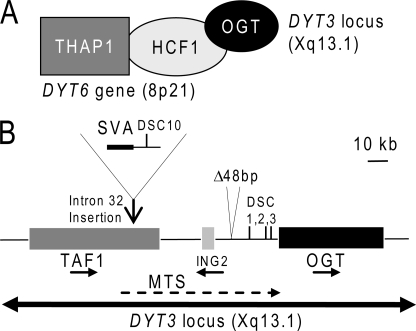
Our data also link THAP1 to OGT, an enzyme that plays an essential role in glucose metabolism by transferring O-GlcNAc to many nucleocytoplasmic proteins (20). The identification of a physical link between THAP1/DYT6 and OGT (Fig. 7A) supports the possibility OGT may play a role in DYT3 dystonia/parkinsonism. Indeed, the OGT gene is mapped within the DYT3 critical interval on Xq13.1 and mutations have been identified in DYT3 patients that may affect the OGT regulatory regions (Fig. 7B). These include 4 disease-specific single nucleotide changes (DSCs), a 48-bp deletion and a SVA retrotransposon insertion in intron 32 of the TAF1 gene (12, 21). The DSCs have been proposed to alter a multiple transcript system (MTS) expressed within the DYT3 critical interval (21). In addition, the retrotransposon insertion has been shown to correlate with reduced TAF1 mRNA levels (12). However, the molecular pathological mechanism in DYT3 dystonia remains unknown and the different mutations within the DYT3 locus may also affect expression of OGT. An intriguing possibility is that OGT and glucose metabolism may play a role in the pathology of both DYT6 and DYT3 dystonias.
FIGURE 7.
A potential link between DYT6 and DYT3 dystonias. A, THAP1/DYT6 associates with HCF-1 and OGT. B, map of the DYT3 critical region on Xq13.1. The different genes (TAF1, ING2, OGT), transcripts (MTS) and mutations (SVA retrotransposon, DSCs, and 48-bp deletion) are indicated.
Supplementary Material
Acknowledgments
We thank Dr. G. Hart (Baltimore, MD) for the gift of anti-OGT antibodies and S. Assbaghi for help with generation of THAP3HBM mutants.
The work was supported by grants from Ligue Nationale contre le Cancer (Equipe labellisée Ligue 2009), INCA, and ANR-Programme Blanc “Regulome.” This work was also supported, in part, by the Intramural Research Program of the National Institutes of Health, Laboratory of Viral Diseases, NIAID.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Methods and Figs. S1–S6.
- THAP-zf
- THAP-zinc finger
- ChIP
- chromatin immunoprecipitation
- HBM
- HCF-1 binding motif
- HCF-1
- host cell factor-1
- HUVECs
- human umbilical vein endothelial cells
- OGT
- O-GlcNAc transferase
- HA
- hemagglutinin
- GST
- glutathione S-transferase.
REFERENCES
- 1.Breakefield X. O., Blood A. J., Li Y., Hallett M., Hanson P. I., Standaert D. G. (2008) Nat. Rev. Neurosci. 9, 222–234 [DOI] [PubMed] [Google Scholar]
- 2.Müller U. (2009) Brain 132, 2005–2025 [DOI] [PubMed] [Google Scholar]
- 3.Ozelius L. J., Hewett J. W., Page C. E., Bressman S. B., Kramer P. L., Shalish C., de Leon D., Brin M. F., Raymond D., Corey D. P., Fahn S., Risch N. J., Buckler A. J., Gusella J. F., Breakefield X. O. (1997) Nat. Genet. 17, 40–48 [DOI] [PubMed] [Google Scholar]
- 4.Fuchs T., Gavarini S., Saunders-Pullman R., Raymond D., Ehrlich M. E., Bressman S. B., Ozelius L. J. (2009) Nat. Genet. 41, 286–288 [DOI] [PubMed] [Google Scholar]
- 5.Bressman S. B., Raymond D., Fuchs T., Heiman G. A., Ozelius L. J., Saunders-Pullman R. (2009) Lancet Neurol 8, 441–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Djarmati A., Schneider S. A., Lohmann K., Winkler S., Pawlack H., Hagenah J., Brüuggemann N., Zittel S., Fuchs T., Raković A., Schmidt A., Jabusch H. C., Wilcox R., Kostić V. S., Siebner H., Altenmüller E., Münchau A., Ozelius L. J., Klein C. (2009) Lancet Neurol. 8, 447–452 [DOI] [PubMed] [Google Scholar]
- 7.Roussigne M., Kossida S., Lavigne A. C., Clouaire T., Ecochard V., Glories A., Amalric F., Girard J. P. (2003) Trends Biochem. Sci. 28, 66–69 [DOI] [PubMed] [Google Scholar]
- 8.Clouaire T., Roussigne M., Ecochard V., Mathe C., Amalric F., Girard J. P. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 6907–6912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bessière D., Lacroix C., Campagne S., Ecochard V., Guillet V., Mourey L., Lopez F., Czaplicki J., Demange P., Milon A., Girard J. P., Gervais V. (2008) J. Biol. Chem. 283, 4352–4363 [DOI] [PubMed] [Google Scholar]
- 10.Cayrol C., Lacroix C., Mathe C., Ecochard V., Ceribelli M., Loreau E., Lazar V., Dessen P., Mantovani R., Aguilar L., Girard J. P. (2007) Blood 109, 584–594 [DOI] [PubMed] [Google Scholar]
- 11.Roussigne M., Cayrol C., Clouaire T., Amalric F., Girard J. P. (2003) Oncogene 22, 2432–2442 [DOI] [PubMed] [Google Scholar]
- 12.Makino S., Kaji R., Ando S., Tomizawa M., Yasuno K., Goto S., Matsumoto S., Tabuena M. D., Maranon E., Dantes M., Lee L. V., Ogasawara K., Tooyama I., Akatsu H., Nishimura M., Tamiya G. (2007) Am. J. Hum. Genet. 80, 393–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tamiya G. (2009) Lancet Neurol. 8, 416–418 [DOI] [PubMed] [Google Scholar]
- 14.Goto H., Motomura S., Wilson A. C., Freiman R. N., Nakabeppu Y., Fukushima K., Fujishima M., Herr W., Nishimoto T. (1997) Genes Dev. 11, 726–737 [DOI] [PubMed] [Google Scholar]
- 15.Narayanan A., Ruyechan W. T., Kristie T. M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 10835–10840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luciano R. L., Wilson A. C. (2003) J. Biol. Chem. 278, 51116–51124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wysocka J., Myers M. P., Laherty C. D., Eisenman R. N., Herr W. (2003) Genes Dev. 17, 896–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Julien E., Herr W. (2003) EMBO J. 22, 2360–2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tyagi S., Chabes A. L., Wysocka J., Herr W. (2007) Mol. Cell 27, 107–119 [DOI] [PubMed] [Google Scholar]
- 20.Hart G. W., Housley M. P., Slawson C. (2007) Nature 446, 1017–1022 [DOI] [PubMed] [Google Scholar]
- 21.Nolte D., Niemann S., Müller U. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 10347–10352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakatani Y., Ogryzko V. (2003) Methods Enzymol. 370, 430–444 [DOI] [PubMed] [Google Scholar]
- 23.Lefebvre T., Cieniewski C., Lemoine J., Guerardel Y., Leroy Y., Zanetta J. P., Michalski J. C. (2001) Biochem. J. 360, 179–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kristie T. M., Liang Y., Vogel J. L. (2010) Biochim Biophys Acta 1799, 257–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Freiman R. N., Herr W. (1997) Genes Dev. 11, 3122–3127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wysocka J., Herr W. (2003) Trends Biochem. Sci 28, 294–304 [DOI] [PubMed] [Google Scholar]
- 27.Dejosez M., Krumenacker J. S., Zitur L. J., Passeri M., Chu L. F., Songyang Z., Thomson J. A., Zwaka T. P. (2008) Cell 133, 1162–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.