Abstract
Activation of the epidermal growth factor receptor (EGFR) is a key signaling event that promotes cells to move and cover wounds in many epithelia. We have previously shown that wounding activates the EGFR through activation of the Src family kinases (SFKs), which induce proteolytic shedding of epidermal growth factor-like ligands from the cell surface. A major goal in wound healing research is to identify early signals that promote motility, and here we examined the hypothesis that members of the focal adhesion kinase family are upstream activators of the SFKs after wounding. We found that focal adhesion kinase is not activated by wounding but that a different family member, Pyk2 (PTK2B/RAFTK/CAKβ), is activated rapidly and potently. Pyk2 interaction with c-Src is increased after wounding, as determined by co-immunoprecipitation experiments. Disruption of Pyk2 signaling either by small interfering RNA or by expression of a dominant negative mutant led to inhibition of wound-induced activation of the SFKs and the EGFR, and conversely, overexpression of wild-type Pyk2 stimulated SFK and EGFR kinase activities in cells. In wound healing studies, Pyk2 small interfering RNA or dominant negative inhibited cell migration. These results show that activation of Pyk2 is an early signal that promotes wound healing by stimulating the SFK/EGFR signaling pathway.
Keywords: Cell/Epithelial, Cell/Migration, Growth Factors, Phosphorylation/Phosphotyrosine Signals/Receptors, Receptors/Tyrosine Kinase, Tissue/Organ Systems/Epithelium, Src
Introduction
Wounding induces profound changes in adjacent epithelial cells, which include rearrangement of the actin cytoskeleton, internalization of cell junction proteins, and redistribution of membrane phospholipids (1–4), and they subsequently acquire a phenotype that allows them to migrate rapidly (5, 6). Activation of the epidermal growth factor receptor (EGFR)2 tyrosine kinase (7, 8) is an obligatory signaling event that stimulates a multitude of downstream effectors to induce these changes in many epithelia, including the gut, airway, epidermis, and cornea (9–14). Understanding the molecular mechanisms of wound-induced EGFR activation is central to understanding how epithelia acquire a motile phenotype.
The proximal event for activation of the EGFR is proteolytic release of transmembrane ligands, such as heparin-binding EGF-like growth factor, by a mechanism that resembles transactivation of the EGFR by G-protein coupled receptors (15–20). Recent data suggest that the Src family kinases (SFKs) control EGFR ligand shedding after wounding (5, 21). The SFKs are ubiquitously expressed non-receptor tyrosine kinases that regulate cytoskeletal dynamics, cell/matrix interactions, and transduction of extracellular signals (22). The SFKs contain Src homology 2 and 3 domains that bind to consensus phosphotyrosine and proline-rich domains, respectively, which can lead to activation by loss of inhibitory intramolecular interactions (23).
We have recently found that SFKs and the EGFR are activated rapidly in cells within 250 μm of the wound edge (5). In addition, extracellular ATP is released from cells and can induce activation of the EGFR further from the wounds. However, extracellular ATP signaling is not necessary for full induction of movement of the epithelial cell sheet after wounding (5, 6).
Understanding the early events in the signaling pathway that leads to EGFR activation remains a significant challenge, and the purpose of this work was to identify the immediate activator of the SFKs. The focal adhesion kinase (FAK) family of non-receptor tyrosine kinases includes FAK and Pyk2 (proline-rich tyrosine kinase 2) (also known as RAFTK (related adhesion focal tyrosine kinase) and CAKβ (cell adhesion kinase β)). They can be activated by a variety of stimuli that are likely to occur at wound edges, such as mechanical stretching, integrin/extracellular matrix interactions, and Ca2+ signaling (24–26). Here we examined the possible role of the FAK family in activating the SFKs and subsequently the EGFR.
EXPERIMENTAL PROCEDURES
Materials
Antibodies against the EGFR, the EGFR phosphorylated on tyrosine 1173, Pyk2 phosphorylated on tyrosine 402, FAK phosphorylated on tyrosine 397, and c-Src (mouse monoclonal antibody used for immunoprecipitation) were from Santa Cruz Biotechnology; antibodies against SFK phosphorylated on tyrosine 416, SFK non-phosphorylated on tyrosine 416, and c-Src (rabbit polyclonal used for immunoblotting) were from Cell Signaling Technology; antibodies against Pyk2 and Fyn were from BD Biosciences; antibodies against myristoylated alanine-rich C-kinase substrate phosphorylated on serines 152 and 156 were from EMD Biosciences; antibodies against paxillin were from Invitrogen; antibodies against β-actin and FLAG were from Sigma. Tyrphostin AG 1478 and Src kinase inhibitor-I were from EMD Biosciences. ATP and grade VII potato apyrase were from Sigma. Cell culture reagents were from MediaTech, and all other supplies and reagents were from ISC BioExpress or ThermoFisher unless otherwise noted.
Cell Culture and Wounding Models
Human corneal limbal epithelial (HCLE) cells have been immortalized by abrogation of p16INK4A/Rb and p53 functions and overexpression of the catalytic subunit of the telomerase holoenzyme (27). HCLE cells were cultured in keratinocyte serum-free medium (Invitrogen) supplemented with 0.3 mm CaCl2, 25 μg/ml bovine pituitary extract, 0.1 ng/ml human recombinant EGF, 50 IU/ml penicillin, and 50 μg/ml streptomycin. At least 16 h prior to experiments, cells were washed and cultured in the same medium without pituitary extract and EGF. For wound signaling experiments, we employed a model for stimulating cell sheets by the acute exposure to free space that has been previously described (15). Briefly, cells are cultured around agarose droplets until they form confluent lanes 1–5 cells wide and are then acutely stimulated or “wounded” by the removal of the agarose droplets. This wounding model minimizes signaling from cell damage and allows biochemical isolation of wound edge cells (5). For wound healing assays, cells were cultured to confluence around a single agarose strip (15) and were induced to differentiate into a stratified epithelium (27) by culturing in Dulbecco's modified Eagle's medium/F-12 (1:1) with 10% newborn calf serum. After two (adenovirus experiments) or three (siRNA experiments) days, the agarose strip was removed, and cultures were allowed to heal for 18 h before fixation and staining with either Gentian violet or immunofluorescence reagents (see below). Micrographs of cultures were obtained before and after wounding, and wound areas were quantified using the region tracing utility in MetaMorph® software (Universal Imaging).
Western Blotting and Immunoprecipitation
Following stimulation, cells were washed in ice-cold phosphate-buffered saline (171 mm NaCl, 10.1 mm Na2HPO4, 3.35 mm KCl, and 1.84 mm KH2PO4, pH 7.2) and lysed in either SDS (1% in H2O) or immunoprecipitation buffer (50 mm Tris-Cl, 260 mm NaCl, 0.02% NaN3, 5 mm EDTA, 1% Triton X-100, 0.5 mm Na3VO4, 50 mm NaF, 1 tablet per 10 ml of protease inhibitor mixture (Roche Applied Science), and 5 μm pepstatin A, pH 7.5). Protein contents of extracts, determined with the BCA assay (ThermoFisher), were normalized prior to SDS-PAGE. For immunoprecipitation, ∼250 μg of cellular protein was incubated with 30 μl of protein A-Sepharose slurry and 1 μg of antibody on an end-over-end rotator at 4 °C overnight. Bead pellets were washed twice with immunoprecipitation buffer and three times with immunoprecipitation buffer with Triton X-100 reduced to 0.1% before the addition of SDS-PAGE sample buffer. Multiple exposures of Western blots were collected, and densitometry of appropriate images was performed using QuantityOne software (Bio-Rad).
siRNA Transfection and Adenoviral Infection
10 nm siRNA duplexes targeting Pyk2 were transfected using siPORTTM NeoFXTM transfection reagent (Applied Biosystems) according to manufacturer's protocol. Cells were then reseeded after 2 days at experimental densities and used 4 days after transfection. Multiple Pyk2 siRNA oligonucleotides were used: CACAUGAAGUCCGAUGAGAdTdT (Sigma) and a Pyk2 SMARTpool that contains at least four duplexes of undisclosed sequence (Millipore). As a control siRNA, we used MISSION® siRNA Universal Negative Control 1 (Sigma). FLAG-tagged Pyk2 and FLAG-tagged PRNK cloned into adenovirus vectors were from Dr. Joseph C. Loftus (Mayo Clinic, Scottsdale, AZ). Adenovirus encoding tet-OFF (used as adenovirus control) was from Dr. Ora A. Weisz (University of Pittsburgh, Pittsburgh, PA), and adenovirus encoding green fluorescent protein was from Dr. Michael P. Czech (University of Massachusetts, Worcester, MA). Preliminary experiments using green fluorescent protein-encoding virus helped to determine the multiplicity of infection necessary for achieving expression in 90–100% of cells. For signaling studies, cells were infected at a multiplicity of 10 for 1 h and were used the following day. For wound healing studies, cells were infected at a multiplicity of 20 in a small volume (1 × 105 cells/μl) for 1 h with gentle agitation every 15 min. After washing and centrifugation, cells were seeded and used 2 days later.
Immunofluorescence Microscopy
The cells were fixed for 30 min by adding one-tenth volume of 37% formaldehyde to the cells. Cells were then processed for immunofluorescence analysis as described previously (15). F-actin was visualized with Alexa Fluor® 633 phalloidin. Images were captured with a Nikon TE2000E microscope with a ×20 oil objective (numerical aperture 0.75) or an Olympus IX81 confocal microscope equipped with a ×60 oil objective (numerical aperture 1.4). Images were generated and analyzed using Fluoview® software (Olympus).
Statistical Analysis
All experiments reported were performed a minimum of three times with similar results. For some figures, representative Western blots are shown below densitometry from at least four replicates. Quantitative data were plotted (means ± S.D.) and analyzed by t test or one-way analysis of variance, followed by Bonferroni's multiple comparisons test using Prism (GraphPad Software).
RESULTS
Pyk2, but Not FAK, Is Activated after Wounding
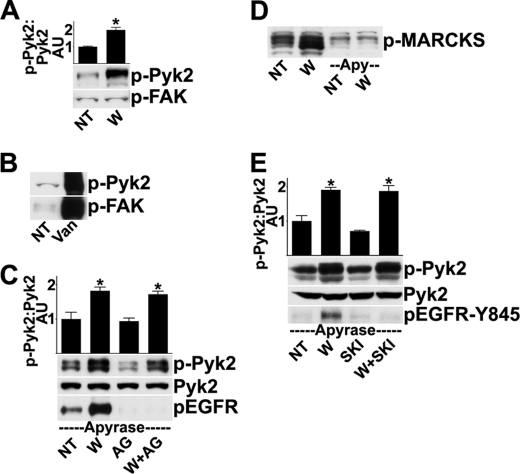
The EGFR is activated within minutes after wounding sheets of HCLE cells in a process that is triggered by SFK activation (5, 21). To examine the role of the FAK family, we first tested whether FAK or Pyk2 is activated after wounding, using a wounding model that detects signaling in cells near wound edges (see “Experimental Procedures” and Ref. 15). Pyk2 was activated 5 min after wounding as determined by Western blotting for its major autophosphorylation site on Tyr-402 (Fig. 1A). In contrast, activation of FAK was not detected. Pyk2 and FAK activation were observed after treatment of cells with a 100 nm concentration of the phosphatase inhibitor sodium orthovanadate (Fig. 1B), indicating that the blotting procedure was capable of detecting both kinases when autophosphorylated. Because FAK was not activated detectably after wounding, we concluded that FAK signaling is not likely to regulate SFK activity in this context, and we therefore focused on Pyk2.
FIGURE 1.
Pyk2, not FAK, is activated rapidly after wounding. A, HCLE cells were given no treatment (NT) or were wounded (W) and incubated for 5 min, and cell extracts were analyzed by Western blotting for Pyk2 phosphorylated on Tyr-402 (p-Pyk2) or for FAK phosphorylated on Tyr-397 (p-FAK). Due to various splicing and post-translational events, the antibody for phosphorylated Pyk2 detects multiple bands, all of which are used in analysis. The ratio of phospho-Pyk2/Pyk2 (not shown) was determined by densitometry. *, significant differences from unwounded groups (p < 0.001). Columns are means, and error bars are S.D. B, as a positive control for the antibodies, 100 nm Na3VO4 was added to the cells for 30 min prior to analysis (Van). C, in the presence of 25 units/ml apyrase, cells received no additional treatment (NT) or were wounded (W) and incubated for 5 min. Where indicated, cells were treated with 100 nm tyrphostin AG 1478 (AG) for 30 min prior to and during wounding. Western blots were probed for phospho-Pyk2 and were then stripped and reprobed for total levels of Pyk2 and EGFR phosphorylated on Tyr-1173 (p-EGFR). The ratio of phospho-Pyk2/Pyk2 was determined by densitometry. *, significant differences from unwounded groups (p < 0.001). D, cells were treated as indicated, and Western blots were probed for phosphorylated myristoylated alanine-rich C-kinase substrate (p-MARCKS), which is a marker of protein kinase C activity (29, 30). E, cells were treated as in C, except 1 μm of Src kinase inhibitor-I (SKI) was added for 30 min prior to and during wounding, where indicated. The blots were analyzed with an antibody that recognizes the EGFR phosphorylated on Tyr-845 (p-EGFR-Y845). Densitometry was performed as in C. Apy, apyrase.
To eliminate effects of extracellular ATP, which can cause activation of the SFKs and the EGFR (5), 25 units/ml apyrase was included in the medium (Fig. 1C) to hydrolyze extracellular ATP to undetectable levels (5). As a further test of the efficacy of the treatment, we noted that inclusion of apyrase blocked activation of protein kinase C (Fig. 1D), which is induced by ATP released from cells after wounding (28–30). In the following experiments, apyrase was included in the incubations to eliminate signaling by ATP.
In some systems, Pyk2 is activated downstream of the EGFR (31–33). However, activation of Pyk2 was not reduced in the presence of a 100 nm concentration of the EGFR tyrosine kinase inhibitor tyrphostin AG 1478, which effectively blocked EGFR autophosphorylation (Fig. 1C). Pyk2 contains tyrosines that can be phosphorylated by the SFKs, and some studies indicate that SFK-mediated phosphorylation regulates Pyk2 activity (34–36). However, wound-induced Pyk2 autophosphorylation was unaffected by treatment with a 1 μm concentration of the SFK inhibitor Src kinase inhibitor-I (SKI) (Fig. 1E). The SFK inhibitor blocked EGFR phosphorylation of Tyr-845, which is catalyzed by the SFKs, demonstrating the efficacy of the inhibitor (Fig. 1E). Together, these results indicate that Pyk2 is activated independently of the EGFR and the SFKs.
Activation and Association of the SFKs with Pyk2 after Wounding
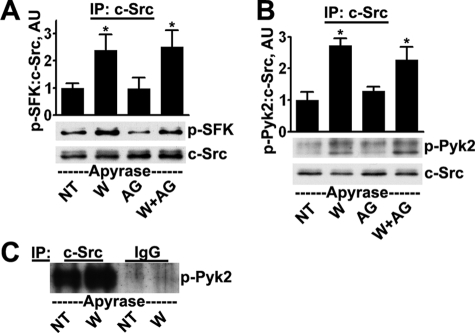
We were able to immunoprecipitate the major SFK isoforms c-Src and Fyn from our cells and analyze their activation states with an antibody that recognizes activated forms of the SFKs. Both c-Src (Fig. 2A) and Fyn (supplemental Fig. 1) were activated within 5 min after wounding. The SFKs may be activated downstream of the EGFR (37); however, the presence of 100 nm tyrphostin AG 1478, which effectively blocked EGFR activity (not shown), had no effect on c-Src or Fyn activation, indicating that c-Src and Fyn are activated upstream of the EGFR.
FIGURE 2.
c-Src is activated and interacts with Pyk2 after wounding. Cells received no treatment (NT) or were wounded (W) and incubated for 5 min, all conditions in the presence of 25 units/ml apyrase. Where indicated, cells were treated with 100 nm tyrphostin AG 1478 (AG) for 30 min prior to and during wounding. The extracts were precipitated with c-Src antibodies, which removed ∼90% of the c-Src present in the extracts (not shown), and the precipitates were subjected to Western blotting for SFK phosphorylated on Tyr-419 (p-SFK) and subsequently for total levels of c-Src (A) and Pyk2 phosphorylated on Tyr-402 (p-Pyk2) and total levels of c-Src (B). Ratios were determined by densitometry. *, significant differences from unwounded groups (p < 0.01). Columns are means, and error bars are S.D. C, lysates were subjected to immunoprecipitation (IP) with anti-c-Src antibodies or with non-immune mouse IgG; immunoprecipitates were subjected to Western blotting for phospho-Pyk2. Overexposed film shows the absence of signal from the non-immune control.
The SFKs can be activated by binding to Pyk2 after it is autophosphorylated (26). Analysis of immunoprecipitates of c-Src showed the presence of increased levels of Pyk2 after wounding, suggesting an increased level of association of the two molecules (Fig. 2B). The specificities of the signals were confirmed by performing mock precipitations with non-immune IgG (Fig. 2C). The interaction did not depend on EGFR activation because co-immunoprecipitation was not blocked by the presence of tyrphostin AG 1478 (Fig. 2B). A similar analysis with c-Fyn was not possible because the signals were too faint.
Pyk2 Mediates SFK and EGFR Activation after Wounding
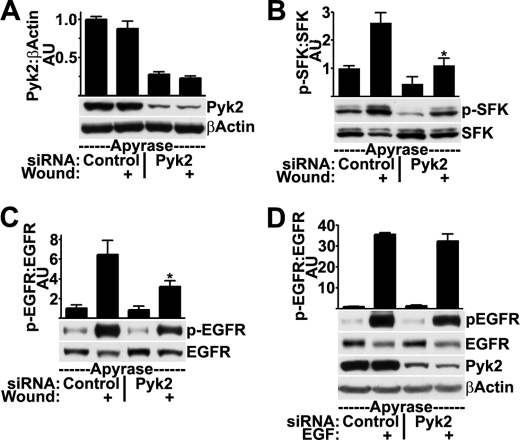
We initially examined the effects of reducing Pyk2 expression with siRNA prior to wounding. Cells were transfected with a 10 nm concentration of a pool of Pyk2-targeted siRNA oligonucleotides before wounding. Pyk2 siRNA transfection caused 70% reduction in Pyk2 expression compared with controls and resulted in inhibition of wound-induced SFK and EGFR activation (supplemental Fig. 2). As a confirmation and to better control for off-target effects (38), we repeated these studies using a 10 nm concentration of a single Pyk2-targeted siRNA oligonucleotide. The Pyk2 siRNA caused ∼75% reduction in Pyk2 expression (Fig. 3A) and significantly inhibited SFK (Fig. 3B) and EGFR (Fig. 3C) activation after wounding. Treatment of Pyk2 siRNA-transfected cells with EGF yielded EGFR activation equivalent to that yielded by treatment of control-transfected cells (Fig. 3D), indicating that the siRNA did not interfere with the normal function of the EGFR.
FIGURE 3.
Knockdown of Pyk2 with siRNA inhibits SFK and EGFR activation after wounding but not after stimulation with EGF. HCLE cells transfected with 10 nm control or Pyk2 siRNA were treated with apyrase and were wounded as indicated. Western blots were probed for total levels of Pyk2 and β-actin (A), SFK phosphorylated (p-SFK) and non-phosphorylated (SFK) on Tyr-419 (B), and EGFR phosphorylated on Tyr-1173 (p-EGFR) and total levels of EGFR (C). Ratios were determined by densitometry. *, significant decrease from wounded controls (p < 0.001). D, HCLE cells transfected with control or Pyk2 siRNA were treated with 10 ng/ml EGF for 5 min where indicated prior to Western blotting and densitometry. Columns are means, and error bars are S.D.
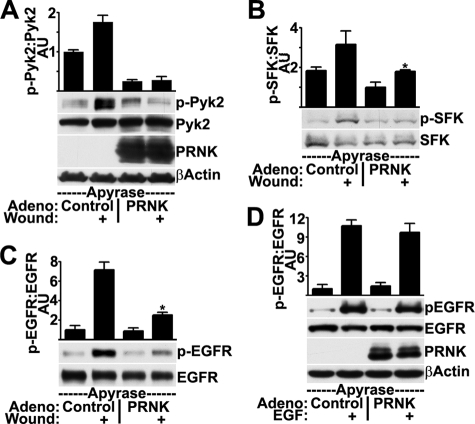
As a complement to these studies, we tested whether Pyk2 is necessary for wound-induced SFK and EGFR activation by wounding sheets of cells that express Pyk2-related non-kinase (PRNK), which is a truncated version of Pyk2 that consists of the C-terminal 228 amino acids and lacks the kinase domain. It can act in a dominant negative fashion to inhibit endogenous Pyk2 activation and signaling (39). PRNK (∼30 kDa) was clearly expressed in HCLE cells infected with an adenovirus coding for PRNK, and its presence blocked Pyk2 activation after wounding as expected (Fig. 4A). Importantly, activation of SFKs (Fig. 4B) and the EGFR (Fig. 4C) were also significantly inhibited by the presence of PRNK. Inhibition of Pyk2 signaling by either siRNA (Fig. 3B) or PRNK (Fig. 4B) reduced SFK activities in the unwounded condition. This indicates the presence of a basal level of stimulation of signaling by Pyk2, similar to that seen previously (40, 41). EGF-induced EGFR activation was not inhibited by PRNK expression (Fig. 4D), indicating that PRNK did not affect the EGFR directly.
FIGURE 4.
Expression of a Pyk2 dominant negative mutant inhibits SFK and EGFR activation after wounding but not after stimulation with EGF. HCLE cells infected with control adenovirus or adenovirus coding for PRNK were incubated with apyrase and were wounded as indicated. Western blots were probed for Pyk2 phosphorylated on Tyr-402 (p-Pyk2) and total levels of Pyk2, PRNK, and β-actin (A), SFK phosphorylated (p-SFK) and non-phosphorylated (SFK) on Tyr-419 (B), and EGFR phosphorylated on Tyr-1173 (p-EGFR) and total levels of EGFR (C). Ratios were determined by densitometry. *, significant decrease from wounded controls (p < 0.001). D, HCLE cells infected with control adenovirus or adenovirus coding for PRNK were treated with 10 ng/ml EGF for 5 min where indicated prior to Western blotting and densitometry. Columns are means, and error bars are S.D.
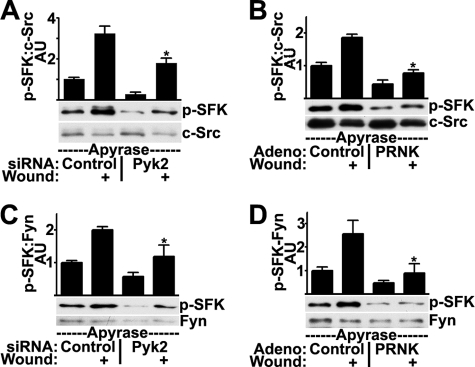
We also examined the effects of blocking Pyk2 signaling on the individual SFK isoforms. C-Src activation was inhibited when Pyk2 signaling was disrupted by either siRNA (Fig. 5A) or PRNK (Fig. 5B). Similarly, activation of the Fyn isoform was inhibited by either Pyk2 siRNA (Fig. 5C) or PRNK (Fig. 5D). Therefore, although we could not determine the degree to which Fyn and Pyk2 interact after wounding, Pyk2 signaling does appear to be necessary for Fyn activation. Together, the results from the studies with siRNA and PRNK indicate that Pyk2 mediates activation of the SFKs and the EGFR after wounding.
FIGURE 5.
Disruption of Pyk2 signaling by siRNA or PRNK inhibits wound-induced activation of c-Src and Fyn. HCLE cells transfected with 10 nm control or Pyk2 siRNA (A and C) or infected with control adenovirus or adenovirus coding for PRNK (B and D) were treated with apyrase and were wounded as indicated. c-Src immunoprecipitates were immunoblotted for SFK phosphorylated on Tyr-419 (p-SFK) and for total levels of c-Src (A and B). Fyn immunoprecipitates were immunoblotted for SFK phosphorylated on Tyr-419 and for total levels of Fyn (C and D). Ratios were determined by densitometry. *, significant decrease from wounded controls (p < 0.001). Columns are means, and error bars are S.D.
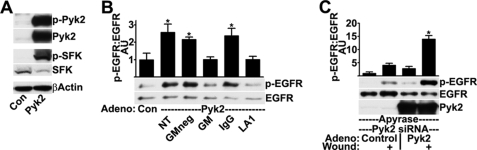
After wounding, EGFR activation is induced by the SFK-mediated proteolytic shedding of transmembrane ligands, such as heparin-binding EGF-like growth factor or amphiregulin (15, 16). To test whether increasing Pyk2 signaling promotes SFK and EGFR activation, we infected cells with adenovirus encoding wild-type Pyk2, which caused highly increased levels of activated, phosphorylated Pyk2 and, as expected, highly increased levels of activated SFKs (Fig. 6A). Importantly, Pyk2 overexpression also induced significant increases in the levels of activated EGFR (Fig. 6B), in agreement with a role for Pyk2 as an upstream activator.
FIGURE 6.
Overexpression of wild-type Pyk2 stimulates SFK and EGFR activation. HCLE cells infected with control adenovirus (Con) or adenovirus coding for Pyk2 were subjected to Western blotting for Pyk2 phosphorylated on Tyr-402 (p-Pyk2), total levels of Pyk2, SFK phosphorylated (p-SFK) and non-phosphorylated (SFK) on Tyr-419, and β-actin (A) and EGFR phosphorylated on Tyr-1173 (p-EGFR) and total levels of EGFR (B). 30 min before lysis, cells were incubated with 10 μm GM 6001-negative control (GMneg) or GM 6001 (GM) or 2 h before with 20 μg/ml non-immune IgG (IgG) or the EGFR ligand-binding domain antibody LA1 (LA1). Ratios were determined by densitometry. *, significant increase from controls (p < 0.001). C, HCLE cells transfected with 10 nm Pyk2 siRNA were infected with control adenovirus or adenovirus coding for Pyk2 and then were treated with apyrase and were wounded as indicated. Western blots were probed for EGFR phosphorylated on Tyr-1173 (p-EGFR), for total levels of EGFR and for total levels of Pyk2. Columns are means, and error bars are S.D.
To address the mechanism of EGFR activation induced by Pyk2 overexpression, we first incubated cells with the general matrix metalloprotease inhibitor GM 6001. Treatment with GM 6001 but not a structurally similar control compound inhibited EGFR activation (Fig. 6B). GM 6001 did not inhibit activation by exogenous ligand (15), and the result is consistent with the notion that EGFR activation is the result of a proteolytic event. Also, the anti-EGFR antibody LA1, which blocks activation of the EGFR by ligands, inhibited EGFR activation, whereas control antibodies did not. These data indicate that Pyk2 regulates EGFR activation by triggering the proteolytic release of transmembrane ligands.
Knockdown of Pyk2 by siRNA inhibited wound-induced EGFR activation (Fig. 3C). As a control, we found that EGFR activation after wounding was enhanced in siRNA-transfected cells after overexpression of Pyk2 (Fig. 6C, compare with Fig. 3C). Taken together, these data indicate that Pyk2 signaling promotes SFK and EGFR activation after wounding.
We subsequently examined whether reactive oxygen species (ROS), which are produced rapidly after wounding epithelial cell sheets (42), are upstream of Pyk2 activation. We observed that incubation of cells with the ROS inhibitors N-acetyl cysteine or butylated hydroxyanisole did not abrogate Pyk2 activation after wounding, although they inhibited activation by H2O2, which generates ROS (supplemental Fig. 4). This suggests that ROS do not stimulate wound-induced Pyk2 activation.
Pyk2 Activation Is Necessary for Full Induction of Cell Motility
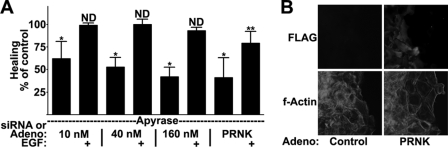
Because Pyk2 regulates SFK and EGFR activation, we expected that modulation of Pyk2 signaling would alter healing rates in our cell culture wound healing assay (see “Experimental Procedures”). Transfection with a single oligonucleotide resulted in a dose-dependent reduction of healing with a maximal 60% reduction of healing (Fig. 7A). The effect of Pyk2 siRNA on healing was not due to differences in cell division or death because similar cell densities were observed in control and Pyk2 siRNA-transfected cultures (supplemental Fig. 3A). Importantly, treatment with 10 ng/ml EGF during healing resulted in similar healing rates in control and Pyk2 siRNA-transfected conditions (Fig. 7A). This shows that the effects of the siRNA are not due to effects on downstream signaling by the EGFR or on the basic motility machinery.
FIGURE 7.
Pyk2 activation is necessary for full induction of cell motility. A, HCLE cells were transfected with control or Pyk2 siRNA or were infected with control or PRNK-expressing adenovirus prior to wound healing assays in the presence of 25 units/ml apyrase and 10 ng/ml EGF (EGF) as indicated. The percentage change in healing from corresponding untreated or EGF-treated control was plotted. *, p < 0.001 (significant difference from untreated controls); **, p < 0.05 (significant difference from EGF-treated controls); ND, no significant difference from EGF-treated controls (p > 0.05). Columns are means, and error bars are S.D. B, immunofluorescence microscopy of cells treated as in A. Images were acquired with a ×20 objective. All cells stained for F-actin, but only PRNK-expressing cells stained for FLAG.
Infection of cells with PRNK-expressing adenovirus reduced wound healing by 60% compared with infection with control virus (Fig. 7A). The effect of PRNK expression on healing was not due to differences in cell division or death because similar cell densities were observed in control and PRNK-infected cultures (supplemental Fig. 3B). The PRNK mutant used was tagged with the FLAG epitope; in areas that contained a few uninfected cells, the FLAG-negative cells had migrated further than the FLAG-positive cells (Fig. 7B). The major part of the reduction of motility caused by PRNK was recovered by the addition of exogenous EGF, but motility was still 20% below controls under these conditions. This is probably due to the numerous protein/protein interactions that are expected to be disrupted by the dominant negative approach. Pyk2 and PRNK interact with paxillin (43), a component of focal adhesions, and we monitored localization of paxillin by immunofluorescence confocal microscopy. In both FLAG-negative and FLAG-positive cells, paxillin was observed in small puncta near the base of the cell that co-localized with termini of actin fibers (supplemental Fig. 3C), indicating that PRNK expression does not cause any major perturbation of focal adhesions. Because inhibition of Pyk2 signaling both by siRNA and the dominant negative PRNK resulted in inhibition of healing, we conclude that full induction of cell motility in epithelial cells depends on Pyk2 and its downstream signaling to the SFKs and the EGFR.
DISCUSSION
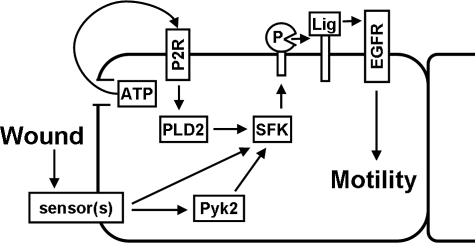
Activation of the SFKs and the EGFR in cells near the wound edge is crucial for induction of cell movement (10, 44), but the signals that trigger activation of the kinases after wounding are still only partially known. In this paper, we have identified Pyk2 as an upstream regulator of wound-induced SFK and EGFR activation. Evidence for this conclusion includes the following: 1) Pyk2 was activated rapidly after wounding, and pharmacological inhibition of signaling by the SFKs or the EGFR did not abrogate Pyk2 activation; 2) association of c-Src and Pyk2 was increased after wounding; 3) overexpression of wild-type Pyk2 stimulated EGFR activation; and 4) inhibition of Pyk2 signaling with siRNA or a dominant negative mutant inhibited SFK and EGFR activation and healing of wounds in cell sheets. These observations, combined with previously published reports from our laboratory and other laboratories, have led to development of the working model for mechanisms of wound-induced EGFR activation near the epithelial wound edge shown in Fig. 8. Wounds are detected by some type of sensors, whose identities at present are unknown, and trigger activation of Pyk2, which subsequently binds and stimulates SFK activity. In a parallel signaling response, ATP is released from cells at the wound edge, through either cell breakage or an unidentified channel in the cell membrane, and stimulates SFK activity by binding to purinergic receptors that signal through phospholipase D. Once activated, the SFKs mediate the proteolytic release of EGFR ligands that bind to and activate the EGFR (5, 10, 15, 21, 45, 46).
FIGURE 8.
A model for mechanisms of EGFR activation at the epithelial wound edge. See “Discussion” for details. P, protease of the MMP or ADAM families; Lig, transmembrane EGFR ligand; PLD2, phospholipase D2; P2R, type-2 purinergic receptor.
In the present study, we have observed partial but not complete inhibition of EGFR activation and wound healing by Pyk2 siRNA or by expression of PRNK. This may be due to incomplete inhibition of Pyk2 signaling. Alternatively, it may suggest the presence of additional mechanisms of SFK and EGFR activation after wounding (Fig. 8). Wounding a sheet of epithelial cells is a complex event that challenges cells with numerous stimuli that have been hypothesized to promote EGFR activation, including exposure to extracellular matrix, physical stretch, loss of polarity, and loss of physical constraints (47–49), and full activation of the EGFR may therefore depend on several signals. The speed of wound re-epithelialization determines clinical outcomes (50–52), so a complete understanding of the mechanisms that induce EGFR activation is essential for understanding how certain pathologies interfere with healing and for developing therapies to improve healing.
The mechanism of wound-induced Pyk2 activation is presently unknown. Our data do not indicate that reactive oxygen species are responsible for activation of Pyk2, but new integrin/matrix adhesions, increased calcium concentrations, and actin cytoskeletal dynamics have all been observed within minutes after wounding and are known to activate Pyk2 (24, 25, 53–55). The extent to which these signals contribute to Pyk2, SFK, or EGFR activation in the present system is unclear.
Some evidence indicates that Pyk2 activation is stimulated by EGFR signaling (31–33), and we have observed Pyk2 activation in HCLE cells treated with EGF,3 suggesting the presence of positive feedback loops in the wounded epithelium. Indeed, inhibition of EGFR signaling with tyrphostin AG 1478 resulted in slight, albeit statistically not significant, reduction of Pyk2 activation after wounding (Fig. 1C).
In this report, we have tested the hypothesis that Pyk2 signaling is necessary for wound-induced EGFR activation by using Pyk2-targeted siRNA oligonucleotides (Fig. 3) and by expression of the Pyk2 dominant negative, PRNK (Fig. 4). Several studies investigating the mechanisms of EGFR transactivation by GPCR agonists have led to suggestions that Pyk2 mediates EGFR transactivation (56–59), although some studies have indicated that Pyk2 and EGFR activation are unrelated events (60, 61). Two studies tested directly whether Pyk2 signaling is necessary for EGFR activation; EGFR transactivation by GPCR agonists was inhibited in Pyk2-deficient fibroblasts (62), and EGFR transactivation by thiazolidinediones was inhibited by Pyk2-targeted siRNA in a liver epithelial cell line (36). Therefore, data presented in this paper add to the mounting evidence for an important connection between Pyk2 and the EGFR. This generates interest in Pyk2 as a therapeutic target for not only wound healing but also EGFR-related pathologies, such as certain cancers and cardiac hypertrophy (41, 63–67).
Most studies are in accord with a positive role for Pyk2 in promoting motility, as we have shown here in HCLE cells. Decreasing Pyk2 expression inhibited motility of vascular smooth muscle cells (68), glioma cells (69), and macrophages (70). Furthermore, Pyk2 overexpression increased motility of glioma (69) and hepatocellular carcinoma (41) cells, and expression of a catalytically active Pyk2 mutant enhanced motility of endothelial (71) and breast cancer (72) cells. However, a few studies are at variance with this conclusion; Pyk2 siRNA stimulated motility of prostate epithelial cells (73), and a kinase-dead Pyk2 mutant had no effect on motility of breast cancer cells (72). Our conclusion that Pyk2 is an upstream activator of the EGFR provides a rationale for a promigratory role in cell motility because the EGFR is widely recognized to stimulate cell migration.
In summary, the results presented here identify Pyk2 activation as a critical early signal that leads to SFK and EGFR activation in cells near the wound edge. By stimulating SFKs and the EGFR, Pyk2 activation is necessary for healing of epithelial wounds.
Supplementary Material
Acknowledgments
We thank Jennifer S. Lozano for technical assistance. We are grateful to Joseph C. Loftus (Mayo Clinic, Scottsdale, AZ) for supplying Pyk2- and PRNK-expressing adenovirus.
This work was supported, in whole or in part, by National Institutes of Health Grants EY013463, EY08098, and T32 EY017271. This work was also supported by grants from Research to Prevent Blindness and the Eye and Ear Foundation (Pittsburgh, PA).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
E. R. Block and J. K. Klarlund, unpublished observations.
- EGFR
- epidermal growth factor receptor
- EGF
- epidermal growth factor
- SFK
- Src family kinase
- FAK
- focal adhesion kinase
- HCLE
- human corneal limbal epithelial
- siRNA
- small interfering RNA
- PRNK
- Pyk2-related non-kinase
- ROS
- reactive oxygen species.
REFERENCES
- 1.Thiery J. P., Sleeman J. P. (2006) Nat. Rev. Mol. Cell Biol. 7, 131–142 [DOI] [PubMed] [Google Scholar]
- 2.Jacinto A., Martinez-Arias A., Martin P. (2001) Nat. Cell Biol. 3, E117–E123 [DOI] [PubMed] [Google Scholar]
- 3.Martin P., Parkhurst S. M. (2004) Development 131, 3021–3034 [DOI] [PubMed] [Google Scholar]
- 4.Friedl P. (2004) Curr. Opin. Cell Biol. 16, 14–23 [DOI] [PubMed] [Google Scholar]
- 5.Block E. R., Klarlund J. K. (2008) Mol. Biol. Cell 19, 4909–4917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farooqui R., Fenteany G. (2005) J. Cell Sci. 118, 51–63 [DOI] [PubMed] [Google Scholar]
- 7.Wells A. (1999) Int. J. Biochem. Cell Biol. 31, 637–643 [DOI] [PubMed] [Google Scholar]
- 8.Jorissen R. N., Walker F., Pouliot N., Garrett T. P., Ward C. W., Burgess A. W. (2003) Exp. Cell Res. 284, 31–53 [DOI] [PubMed] [Google Scholar]
- 9.Hansen L. A., Alexander N., Hogan M. E., Sundberg J. P., Dlugosz A., Threadgill D. W., Magnuson T., Yuspa S. H. (1997) Am. J. Pathol. 150, 1959–1975 [PMC free article] [PubMed] [Google Scholar]
- 10.Zieske J. D., Takahashi H., Hutcheon A. E., Dalbone A. C. (2000) Invest. Ophthalmol. Vis. Sci. 41, 1346–1355 [PubMed] [Google Scholar]
- 11.Repertinger S. K., Campagnaro E., Fuhrman J., El-Abaseri T., Yuspa S. H., Hansen L. A. (2004) J. Invest. Dermatol. 123, 982–989 [DOI] [PubMed] [Google Scholar]
- 12.Goodlad R. A., Wright N. A. (1995) Eur. J. Gastroenterol. Hepatol. 7, 928–932 [DOI] [PubMed] [Google Scholar]
- 13.Puddicombe S. M., Polosa R., Richter A., Krishna M. T., Howarth P. H., Holgate S. T., Davies D. E. (2000) FASEB J. 14, 1362–1374 [DOI] [PubMed] [Google Scholar]
- 14.Myhre G. M., Toruner M., Abraham S., Egan L. J. (2004) Am. J. Physiol. Gastrointest. Liver Physiol. 287, G1213–G1219 [DOI] [PubMed] [Google Scholar]
- 15.Block E. R., Matela A. R., SundarRaj N., Iszkula E. R., Klarlund J. K. (2004) J. Biol. Chem. 279, 24307–24312 [DOI] [PubMed] [Google Scholar]
- 16.Xu K. P., Riggs A., Ding Y., Yu F. S. (2004) Invest. Ophthalmol. Vis. Sci. 45, 4277–4283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blobel C. P. (2005) Nat. Rev. Mol. Cell Biol. 6, 32–43 [DOI] [PubMed] [Google Scholar]
- 18.Daub H., Weiss F. U., Wallasch C., Ullrich A. (1996) Nature 379, 557–560 [DOI] [PubMed] [Google Scholar]
- 19.Ohtsu H., Dempsey P. J., Eguchi S. (2006) Am. J. Physiol. Cell Physiol. 291, C1–C10 [DOI] [PubMed] [Google Scholar]
- 20.Sanderson M. P., Dempsey P. J., Dunbar A. J. (2006) Growth Factors 24, 121–136 [DOI] [PubMed] [Google Scholar]
- 21.Xu K. P., Yin J., Yu F. S. (2006) Invest. Ophthalmol. Vis. Sci. 47, 2832–2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas S. M., Brugge J. S. (1997) Annu. Rev. Cell Dev. Biol. 13, 513–609 [DOI] [PubMed] [Google Scholar]
- 23.Roskoski R., Jr. (2005) Biochem. Biophys. Res. Commun. 331, 1–14 [DOI] [PubMed] [Google Scholar]
- 24.Schlaepfer D. D., Hauck C. R., Sieg D. J. (1999) Prog. Biophys. Mol. Biol. 71, 435–478 [DOI] [PubMed] [Google Scholar]
- 25.Avraham H., Park S. Y., Schinkmann K., Avraham S. (2000) Cell. Signal. 12, 123–133 [DOI] [PubMed] [Google Scholar]
- 26.Dikic I., Tokiwa G., Lev S., Courtneidge S. A., Schlessinger J. (1996) Nature 383, 547–550 [DOI] [PubMed] [Google Scholar]
- 27.Gipson I. K., Spurr-Michaud S., Argüeso P., Tisdale A., Ng T. F., Russo C. L. (2003) Invest. Ophthalmol. Vis. Sci. 44, 2496–2506 [DOI] [PubMed] [Google Scholar]
- 28.Schwiebert E. M., Zsembery A. (2003) Biochim. Biophys. Acta 1615, 7–32 [DOI] [PubMed] [Google Scholar]
- 29.Stumpo D. J., Graff J. M., Albert K. A., Greengard P., Blackshear P. J. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 4012–4016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aderem A. (1992) Cell 71, 713–716 [DOI] [PubMed] [Google Scholar]
- 31.Schauwienold D., Sastre A. P., Genzel N., Schaefer M., Reusch H. P. (2008) J. Biol. Chem. 283, 27748–27756 [DOI] [PubMed] [Google Scholar]
- 32.Park S. Y., Li H., Avraham S. (2007) Cell. Signal. 19, 289–300 [DOI] [PubMed] [Google Scholar]
- 33.Shi C. S., Kehrl J. H. (2004) J. Biol. Chem. 279, 17224–17231 [DOI] [PubMed] [Google Scholar]
- 34.Duong L. T., Lakkakorpi P. T., Nakamura I., Machwate M., Nagy R. M., Rodan G. A. (1998) J. Clin. Invest. 102, 881–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sieg D. J., Ilić D., Jones K. C., Damsky C. H., Hunter T., Schlaepfer D. D. (1998) EMBO J. 17, 5933–5947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dewar B. J., Gardner O. S., Chen C. S., Earp H. S., Samet J. M., Graves L. M. (2007) Mol. Pharmacol. 72, 1146–1156 [DOI] [PubMed] [Google Scholar]
- 37.Abram C. L., Courtneidge S. A. (2000) Exp. Cell Res. 254, 1–13 [DOI] [PubMed] [Google Scholar]
- 38.(2003) Nat. Cell Biol. 5, 489–490 [DOI] [PubMed] [Google Scholar]
- 39.Xiong W. C., Macklem M., Parsons J. T. (1998) J. Cell Sci. 111, 1981–1991 [DOI] [PubMed] [Google Scholar]
- 40.Xie J., Allen K. H., Marguet A., Berghorn K. A., Bliss S. P., Navratil A. M., Guan J. L., Roberson M. S. (2008) Mol. Endocrinol. 22, 2322–2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun C. K., Man K., Ng K. T., Ho J. W., Lim Z. X., Cheng Q., Lo C. M., Poon R. T., Fan S. T. (2008) Carcinogenesis 29, 2096–2105 [DOI] [PubMed] [Google Scholar]
- 42.Nikolić D. L., Boettiger A. N., Bar-Sagi D., Carbeck J. D., Shvartsman S. Y. (2006) Am. J. Physiol. Cell Physiol. 291, C68–C75 [DOI] [PubMed] [Google Scholar]
- 43.Li X., Earp H. S. (1997) J. Biol. Chem. 272, 14341–14348 [DOI] [PubMed] [Google Scholar]
- 44.Yamada T., Aoyama Y., Owada M. K., Kawakatsu H., Kitajima Y. (2000) Cell Struct. Funct. 25, 351–359 [DOI] [PubMed] [Google Scholar]
- 45.Boucher I., Yang L., Mayo C., Klepeis V., Trinkaus-Randall V. (2007) Exp. Eye Res. 85, 130–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yin J., Xu K., Zhang J., Kumar A., Yu F. S. (2007) J. Cell Sci. 120, 815–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moro L., Dolce L., Cabodi S., Bergatto E., Boeri Erba E., Smeriglio M., Turco E., Retta S. F., Giuffrida M. G., Venturino M., Godovac-Zimmermann J., Conti A., Schaefer E., Beguinot L., Tacchetti C., Gaggini P., Silengo L., Tarone G., Defilippi P. (2002) J. Biol. Chem. 277, 9405–9414 [DOI] [PubMed] [Google Scholar]
- 48.Iwasaki H., Eguchi S., Ueno H., Marumo F., Hirata Y. (2000) Am. J. Physiol. Heart Circ. Physiol. 278, H521–H529 [DOI] [PubMed] [Google Scholar]
- 49.Vermeer P. D., Einwalter L. A., Moninger T. O., Rokhlina T., Kern J. A., Zabner J., Welsh M. J. (2003) Nature 422, 322–326 [DOI] [PubMed] [Google Scholar]
- 50.Netto M. V., Mohan R. R., Ambrósio R., Jr., Hutcheon A. E., Zieske J. D., Wilson S. E. (2005) Cornea 24, 509–522 [DOI] [PubMed] [Google Scholar]
- 51.Fini M. E., Stramer B. M. (2005) Cornea 24, S2–S11 [DOI] [PubMed] [Google Scholar]
- 52.Wilson S. A., Last A. (2004) Am. Fam. Physician 70, 123–128 [PubMed] [Google Scholar]
- 53.Sammak P. J., Hinman L. E., Tran P. O., Sjaastad M. D., Machen T. E. (1997) J. Cell Sci. 110, 465–475 [DOI] [PubMed] [Google Scholar]
- 54.Klepeis V. E., Cornell-Bell A., Trinkaus-Randall V. (2001) J. Cell Sci. 114, 4185–4195 [DOI] [PubMed] [Google Scholar]
- 55.Zaidel-Bar R., Ballestrem C., Kam Z., Geiger B. (2003) J. Cell Sci. 116, 4605–4613 [DOI] [PubMed] [Google Scholar]
- 56.Montiel M., Quesada J., Jiménez E. (2007) Cell. Signal. 19, 2138–2146 [DOI] [PubMed] [Google Scholar]
- 57.Burdick A. D., Ivnitski-Steele I. D., Lauer F. T., Burchiel S. W. (2006) Carcinogenesis 27, 2331–2340 [DOI] [PubMed] [Google Scholar]
- 58.Park S. Y., Schinkmann K. A., Avraham S. (2006) Cell. Signal. 18, 1063–1071 [DOI] [PubMed] [Google Scholar]
- 59.Kanno H., Horikawa Y., Hodges R. R., Zoukhri D., Shatos M. A., Rios J. D., Dartt D. A. (2003) Am. J. Physiol. Cell Physiol. 284, C988–C998 [DOI] [PubMed] [Google Scholar]
- 60.Zwick E., Wallasch C., Daub H., Ullrich A. (1999) J. Biol. Chem. 274, 20989–20996 [DOI] [PubMed] [Google Scholar]
- 61.Kodama H., Fukuda K., Takahashi T., Sano M., Kato T., Tahara S., Hakuno D., Sato T., Manabe T., Konishi F., Ogawa S. (2002) J. Mol. Cell Cardiol. 34, 139–150 [DOI] [PubMed] [Google Scholar]
- 62.Andreev J., Galisteo M. L., Kranenburg O., Logan S. K., Chiu E. S., Okigaki M., Cary L. A., Moolenaar W. H., Schlessinger J. (2001) J. Biol. Chem. 276, 20130–20135 [DOI] [PubMed] [Google Scholar]
- 63.Behmoaram E., Bijian K., Jie S., Xu Y., Darnel A., Bismar T. A., Alaoui-Jamali M. A. (2008) Am. J. Pathol. 173, 1540–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Iiizumi M., Bandyopadhyay S., Pai S. K., Watabe M., Hirota S., Hosobe S., Tsukada T., Miura K., Saito K., Furuta E., Liu W., Xing F., Okuda H., Kobayashi A., Watabe K. (2008) Cancer Res. 68, 7613–7620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Roelle S., Grosse R., Buech T., Chubanov V., Gudermann T. (2008) Oncogene 27, 1737–1748 [DOI] [PubMed] [Google Scholar]
- 66.Menashi E. B., Loftus J. C. (2009) Cell Tissue Res. 337, 243–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hirotani S., Higuchi Y., Nishida K., Nakayama H., Yamaguchi O., Hikoso S., Takeda T., Kashiwase K., Watanabe T., Asahi M., Taniike M., Tsujimoto I., Matsumura Y., Sasaki T., Hori M., Otsu K. (2004) J. Mol. Cell Cardiol. 36, 799–807 [DOI] [PubMed] [Google Scholar]
- 68.Soe N. N., Ishida T., Ishida M., Sawano M., Abe K., Miho N., Chayama K., Kihara Y., Yoshizumi M. (2009) J. Atheroscler. Thromb. 16, 230–238 [DOI] [PubMed] [Google Scholar]
- 69.Lipinski C. A., Tran N. L., Menashi E., Rohl C., Kloss J., Bay R. C., Berens M. E., Loftus J. C. (2005) Neoplasia 7, 435–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Owen K. A., Pixley F. J., Thomas K. S., Vicente-Manzanares M., Ray B. J., Horwitz A. F., Parsons J. T., Beggs H. E., Stanley E. R., Bouton A. H. (2007) J. Cell Biol. 179, 1275–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kuwabara K., Nakaoka T., Sato K., Nishishita T., Sasaki T., Yamashita N. (2004) Endocrinology 145, 3324–3330 [DOI] [PubMed] [Google Scholar]
- 72.Zrihan-Licht S., Fu Y., Settleman J., Schinkmann K., Shaw L., Keydar I., Avraham S., Avraham H. (2000) Oncogene 19, 1318–1328 [DOI] [PubMed] [Google Scholar]
- 73.de Amicis F., Lanzino M., Kisslinger A., Calì G., Chieffi P., Andò S., Mancini F. P., Tramontano D. (2006) J. Cell. Physiol. 209, 74–80 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.