Abstract
The isoprenylcysteine carboxyl methyltransferase (Icmt) from Saccharomyces cerevisiae, also designated Ste14p, is a 26-kDa integral membrane protein that contains six transmembrane spanning segments. This protein is localized to the endoplasmic reticulum membrane where it performs the methylation step of the CAAX post-translational processing pathway. Sequence analysis reveals a putative GXXXG dimerization motif located in transmembrane 1 of Ste14p, but it is not known whether Ste14p forms or functions as a dimer or higher order oligomer. We determined that Ste14p predominantly formed a homodimer in the presence of the cross-linking agent, bis-sulfosuccinimidyl suberate. Wild-type untagged Ste14p also co-immunoprecipitated and co-purified with N-terminal-tagged His10-myc3-Ste14p (His-Ste14p). Furthermore, enzymatically inactive His-Ste14p variants L81F and E213Q both exerted a dominant-negative effect on methyltransferase activity when co-expressed and co-purified with untagged wild-type Ste14p. Together, these data, although indirect, suggest that Ste14p forms and functions as a homodimer or perhaps a higher oligomeric species.
Keywords: Membrane/Enzymes, Protein/Isoprenylation, Protein/Methylation, Protein/Post-translational Modification, Yeast, Icmt, Isoprenylcysteine Carboxyl Methyltransferase, Ste14p, Oligomerization
Introduction
Post-translational modifications of eukaryotic proteins have profound effects on their localization and function. Many eukaryotic proteins are modified at their C termini by key modifications including the addition of an isoprenyl lipid moiety, proteolysis, and methylation (1–5). The consensus sequence for isoprenylation, proteolysis, and methylation is a C-terminal CAAX sequence where C is cysteine, A is an aliphatic residue, and X can be one of several amino acids. This three-step post-translational processing has been recognized as crucial for the proper membrane localization and subsequent biological function of Ras proteins and many other key signal transduction proteins, including the Rho small GTP-binding proteins (2, 4, 6, 7).
The final modification, α-carboxyl methyl-esterification of the newly exposed cysteine carboxyl group is performed by an isoprenylcysteine carboxyl methyltransferase (Icmt)3 (4, 8–18). Ste14p from Saccharomyces cerevisiae is the founding member of the Icmt family. A 239-amino acid enzyme with a molecular mass of 26 kDa, Ste14p has six transmembrane-spanning segments and is localized to the endoplasmic reticulum membrane (13, 19). In yeast, carboxyl methylation has been shown to be important for both the proper cellular localization of RAS proteins and the formation of active a-factor mating pheromone (9, 13, 14).
Ste14p possesses a tandem 31GXXXGXXXG39 motif located in transmembrane segment one. The GXXXG motif has been shown to be important in the dimerization of several integral single and multispanning membrane proteins (20, 21) including glycophorin A (22, 23), the ABC transporter ABCG2 (24), yeast ATP synthase (25), human carbonic anhydrase (26), Helicobacter pylori vacuolating toxin (27), and yeast α-factor receptor (28). The spacing of the glycine residues allows them to be positioned on the same face of the helix, and it is believed that the small size of glycine provides a flat surface that permits close packing of the interacting helix. This close packing also allows for van der Waals interactions between surrounding residues in the two helices (22, 29).
To date, little is known about the oligomerization state of Icmt enzymes or the functional consequences of such an interaction. Given the presence of the tandem GXXXGXXXG motif and our prior anecdotal observations of dimers on SDS-PAGE gels, we sought to determine whether Ste14p forms a homodimer or higher oligomeric state utilizing various biochemical techniques including chemical cross-linking, co-purification, and co-immunoprecipitation. In this study we have demonstrated for the first time that Ste14p interacts to form predominantly a homodimer and that this dimerization maybe important for enzyme activity.
EXPERIMENTAL PROCEDURES
Materials
Restriction enzymes were purchased from New England Biolabs (Beverly, MA). The Gibbs Laboratory (Purdue University) synthesized and provided the N-acetyl-S-farnesyl-l-cysteine (AFC). N-Dodecyl-β-d-maltopyranoside (DDM) was purchased from Anatrace, Inc. (Maumee, OH). S-Adenosyl-l-[methyl-14C]methionine and Protein A-Sepharose CL-4B beads were purchased from GE Healthcare. Escherichia coli DH5α subcloning efficiency cells, the anti-Myc monoclonal antibody, the goat anti-mouse IgG, and the goat anti-rabbit IgG were purchased from Invitrogen. The SM1188, SM1058, and SM3495 yeast strains, pSM802 plasmid, and the anti-Ste14 polyclonal antibody were gifts from Dr. S. Michaelis (The Johns Hopkins University School of Medicine). The bis-sulfosuccinimidyl suberate homobifunctional cross-linking agent (BS3) (11.4 Å spacer arm) was purchased from Pierce. Micrococcal nuclease was purchased from Worthington Biochemical Corp. (Lakewood, NJ), and aprotinin was purchased from MP Biomedical (Irvine, CA). All other materials and reagents were purchased from Fisher.
Cloning
Untagged STE14 was expressed under the 3′-phosphoglycerate kinase (PGK) promoter in a plasmid containing the LEU2 selection marker (pRS425-PGK-STE14). pRS425-PGK-STE14 was constructed by ligating the PGK promoter and STE14 gene excised from the plasmid pSM703 with enzymes XhoI and SacII and inserted into plasmid pSM803. Site-directed mutagenesis was performed to create the His-Ste14p-L81F and His-Ste14p-E213Q mutants. Each PCR product containing the mutation was digested sequentially with EagI and SacII and ligated into the His-Ste14p expression plasmid pCHH10m3N (30). All constructs were sequenced bidirectionally.
Yeast Strains
STE14 gene expression plasmids were transformed individually or together into SM1188, a Δste14 strain (STE14–3::TRP1, MATa trp1 leu2 ura3 his4 can1) by the modified Elble method (31). Transformation efficiency was increased by the addition of a 50 mm final concentration of dithiothreitol. Strain designations are shown in Table 1. Synthetic complete medium lacking uracil (SC−URA), leucine (SC−LEU), or both uracil and leucine (SC−URA−LEU) were used to culture all strains at 30 °C, except SM1058 and SM1188, which were grown on yeast complete medium (1% (w/v) Bacto-yeast extract, 2% (w/v) Bacto-peptone, 2% (w/v) glucose).
TABLE 1.
S. cerevisiae strains used in this study
| Name | Genotype | Reference |
|---|---|---|
| SM1058 | Mata trp1 leu2 ura3 his4 can1 | Ref. 10 |
| SM1188 | Ste14-4::TRP1, isogenic to SM1058 | Ref. 10 |
| CH2704a | 2μ URA3 PPGK-His10-myc3N-STE14 | Ref. 30 |
| CH2714a | 2μ URA3 PPGK-His10-myc3N | Ref. 30 |
| CH2866a | 2μ LEU2 PPGK-STE14 | This study |
| CH2865a | 2μ URA3 PPGK-His10-myc3N-STE14 + 2μ LEU2 PPGK-STE14 | This study |
| CH2870a | 2μ URA3 PPGK-His10-myc3N-STE14-L81F | This study |
| CH2867a | 2μ URA3 PPGK-His10-myc3N-STE14-L81F + 2μ LEU2 PPGK-STE14 | This study |
| CH2781a | 2μ URA3 PPGK-His10-myc3N-STE14-E213Q | This study |
| CH2899a | 2μ URA3 PPGK-His10-myc3N-STE14-E213Q + 2μ LEU2 PPGK-STE14 | This study |
| SM3495a | 2μ URA3 STE14 | Ref. 13 |
a Strains are isogenic to SM1188. PGK-phosphoglycerate kinase promoter, His10-10 histidine residues, myc3-three myc epitopes, N-tags located on the amino terminus.
Crude Membrane Preparations from Yeast Cells
Crude membranes were prepared as described previously (30). Briefly, yeast cells were cultured in SC−URA, SC−LEU, or SC−URA−LEU medium to mid-log phase (2.0 A600/ml). The yeast cells were harvested by centrifugation, and the cell pellet was resuspended in lysis buffer plus protease inhibitors (0.3 m sorbitol, 10 mm Tris-HCl, pH 7.5, 0.1 m NaCl, 5 mm MgCl2, 1% aprotinin, and 2 mm 4-(2-aminoethyl)benzenesulfonyl fluoride) and incubated on ice for 15 min. The cells were rapidly frozen twice by submersion in liquid N2 and thawed at room temperature. The cells were lysed by passing twice through a French press at 18,000 p.s.i. The cellular mixture was then centrifuged at 500 × g, and the supernatant was collected and incubated on ice for 20 min after the addition of micrococcal nuclease (50 units/ml). The supernatant was then centrifuged at 300,000 × g for 30 min at 4 °C. After centrifugation, the supernatant was aspirated, and the membrane pellet was resuspended in lysis buffer containing 10% glycerol. The membrane preparation was separated into aliquots, frozen on dry ice, and stored at −80 °C. Coomassie Plus protein assay reagent (Pierce) was used to determine the protein concentration.
BS3 Cross-linking Analysis
Reactions contained 80 μg of crude membrane protein or 2.5 μg of pure protein plus either 0.8 or 0.4 mm BS3 (11.4 Å spacer arm), respectively, in 10 mm MOPS, pH 7.0. Samples were incubated at room temperature for 20 min and terminated by the addition of 1× nonreducing SDS-PAGE sample buffer (0.5 m Tris-HCl, pH 6.8, 30% sucrose (w/v), 10% sodium dodecyl sulfate (w/v), and 0.1% bromphenol blue). The samples were resolved on a 10 or 7.5% SDS-PAGE gel, and the His-Ste14p proteins were detected by immunoblot analysis.
Purification of His-Ste14p
His-Ste14p, His-Ste14 (L81F), and His-Ste14p (E213Q) were purified as previously described (30). Briefly, 25 mg of crude membrane protein were solubilized in lysis buffer containing 20 mm imidazole and 1% DDM (w/v) for 1 h at 4 °C. The solubilization mixture was then centrifuged at 300,000 × g for 30 min to remove the insoluble fraction. The solubilized protein in the resulting supernatant was then incubated with Talon metal affinity beads (500 μl of a 50:50 suspension) for 1 h and washed 4 times. The first two washes utilized Buffer A containing (0.3 m sorbitol, 10 mm Tris-HCl, pH 7.5, 0.1 m NaCl, 5 mm MgCl2,1% aprotinin, 2 mm 4-(2-aminoethyl)benzenesulfonyl fluoride, 1 mm dithiothreitol, 1% DDM (w/v), and 40 mm imidazole). The third wash was with Buffer B that contained 500 mm KCl added to Buffer A, and the fourth wash was Buffer C, which contained the same components as Buffer B but a reduced detergent concentration of 0.1% DDM (w/v). After the fourth wash, the purified protein was eluted from the column by the addition of 1 m imidazole in a buffer containing (0.3 m sorbitol, 10 mm Tris-HCl, pH 7.5, 0.1 m NaCl, 5 mm MgCl2, 1% aprotinin, 2 mm 4-(2-aminoethyl)benzenesulfonyl fluoride, 1 mm dithiothreitol, and 0.1% DDM (w/v)), and the fractions were concentrated by centrifugation at 5000 × g using an Amicon Ultra-15 concentrator (Millipore Co., Billerica, MA) for 40 min at 4 °C. The protein concentration was determined by an Amido Black protein assay (32).
In Vitro Methyltransferase Vapor Diffusion Assay
Reactions were performed as described previously (9, 30) with minor modifications. Reactions contained 5 μg of crude membrane preparations, 200 μm AFC, 20 μm S-adenosyl-l-[methyl-14C]methionine ([14C]SAM) (50–60 mCi/mmol), and 100 mm Tris-HCl, pH 7.5, in 60 μl. The reactions were incubated at 30 °C for 30 min and terminated by the addition of 50 μl of 1 m NaOH, 1% SDS (v/v). The reaction (100 μl) was then spotted onto pleated filter papers that were wedged into the neck of vials containing 10 ml of scintillation fluid and allowed to diffuse at room temperature for 2.5 h. The filters were discarded, and the base labile [14C]methyl groups transferred were measured by a Packard Tri-Carb 1600CA liquid scintillation counter. Background counts from the empty vector control, CH2714, were subtracted from the total overall counts of all three samples. Reactions with purified protein contained ∼0.3 μg of purified protein reconstituted with 200 μm AFC and 100 μg of E. coli polar lipids by dilution in 100 mm MES, pH 7.0, vortexed, and incubated on ice for 10 min before use in the methyltransferase assay (30, 33).
SDS-PAGE and Immunoblot Analysis
Unless otherwise specified, protein samples were incubated in a 65 °C heat block for 20–30 min after the addition of 2× SDS sample buffer (0.5 m Tris-HCl, pH 6.8, 30% sucrose (w/v), 10% sodium dodecyl sulfate (w/v), 3.5 m 2-mercaptoethanol, and 0.1% bromphenol blue (w/v). The proteins were separated by 10% SDS-PAGE and transferred to a 0.2-μm PROTRAN nitrocellulose membrane at 100 V for 1 h. The nitrocellulose membrane was blocked at room temperature for 1 h in 20% (w/v) nonfat dry milk in phosphate-buffered saline with Tween 20 (137 mm NaCl, 2.7 mm KCl, 4 mm Na2HPO4, 1.8 mm KH2PO4, and 0.05% (v/v) Tween 20 pH 7.4) (PBST). The membrane was then incubated for 3 h at room temperature with primary antibody (1:1000 α-Ste14p) diluted in 5% (w/v) dry milk in PBST. The membrane was washed 3 times in PBST and then incubated for 1 h at room temperature with a goat α-rabbit IgG secondary antibody (1:10,000) conjugated to HRP diluted in 5% (w/v) dry milk in PBST. The membrane was washed three times with PBST, and the proteins were detected using SuperSignal West Pico enhanced chemiluminescence (Pierce).
Immunoprecipitation of Ste14p
One hundred μg of crude membrane protein expressing His-Ste14p constructs were added to 400 μl of RIPA buffer (10 mm Tris-HCl, pH 7.4, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 1 mm EDTA, and 1% aprotinin). The proteins were immunoprecipitated overnight at 4 °C with gentle rotation in the presence of ∼3 μg of anti-Myc antibody that recognizes the Myc epitope at the N terminus of His-Ste14p. After the incubation, 40 μl of a 50% (v/v) mixture of protein A-Sepharose beads resuspended in RIPA buffer was added to each sample and allowed to incubate for 2.5 h at 4 °C with gentle rotation. Each sample was then centrifuged at 13,000 × g for 1 min and washed 3 times with 1× RIPA buffer. The immune complexes were eluted from the beads by the addition of 2× SDS-PAGE sample buffer and heated at 65 °C for 30 min. The entire sample was separated by 10% SDS-PAGE and analyzed by immunoblotting as described above with an α-Ste14p polyclonal antibody (1:500) and a goat α-rabbit IgG secondary antibody (1:10,000) conjugated to HRP. Protein bands were visualized by SuperSignal Pico enhanced chemiluminescence (Pierce).
RESULTS
Chemical Cross-linking Revealed His-Ste14p, and Untagged Ste14p Can Form Homodimers and Higher Order Homooligomers
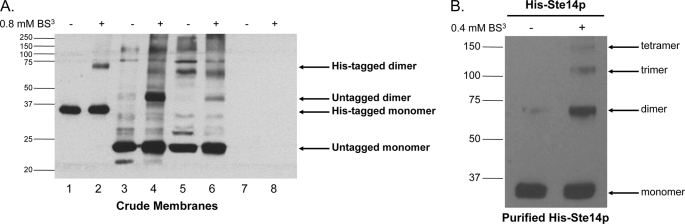
To determine whether Ste14p was capable of forming dimers or higher order complexes, crude membrane preparations expressing His-Ste14p or untagged Ste14p were treated with 0.8 mm BS3, a homobifunctional amine-reactive cross-linking reagent, with an 11.4-Å spacer arm, and subjected to SDS-PAGE and immunoblot analysis with a α-Ste14 polyclonal antibody that recognizes the last 42 C-terminal amino acids of Ste14p (13). In crude membranes, His-Ste14p formed a dimer in the presence of BS3 (Fig. 1A, lanes 1 and 2). Untagged Ste14p expressed at varying levels in yeast on a 2μ plasmid behind either the strong PGK promoter (Fig. 1A, lanes 3 and 4) or the endogenous STE14 promoter (Fig. 1A, lanes 5 and 6) also formed dimers in crude membrane preparations (Fig. 1A, lanes 3–6). Crude membranes from a Δste14 strain transformed with the empty vector (CH2714) were used as a negative control (Fig. 1A, lanes 7 and 8). These data suggest that the N-terminal 10 histidine residues and three Myc epitopes were not responsible for the interaction. Co-migration of a nonspecific protein band with the Ste14p dimer precluded our ability to show dimerization of endogenous Ste14p in a wild-type strain.
FIGURE 1.
Immunoblot of His-Ste14p cross-linked chemically with BS3. A, 80 μg of crude membrane protein were incubated with 0.8 mm BS3 for 20 min at room temperature. The reaction was terminated by the addition of SDS-PAGE sample buffer without reducing agents and then heated for 20 min at 65 °C. Various amounts of crude membrane proteins were separated by 10% SDS-PAGE, and His-Ste14p was detected using a Ste14p polyclonal antibody (1:1,000) and a HRP-conjugated secondary goat anti-rabbit antibody (1:10,000) as follows: lane 1, 1 μg of His-Ste14p (CH2704); lane 2, 1 μg of His-Ste14p (CH2704) + BS3; lane 3, 10 μg of untagged Ste14p (CH2866; 2μ PGK promoter); lane 4, 10 μg of untagged Ste14p (CH2866; 2μ PGK promoter) + BS3; lane 5, 50 μg of SM3495 (untagged Ste14p; 2μ STE14 promoter); lane 6, 50 μg of SM3495 (untagged Ste14p; 2μ STE14 promoter) + BS3; lane 7, 50 μg of CH2714 (negative control); lane 8, 50 μg of CH2714 (negative control) + BS3. Protein bands were visualized using enhanced chemiluminescence (ECL). B, purified His-Ste14p (2.5 μg) was incubated with 0.4 mm BS3 for 20 min, and the reaction was terminated by the addition of SDS-PAGE sample buffer without reducing agents. The reactions were heated for 30 min at 65 °C, 0.1 μg of purified protein was separated by 7.5% SDS-PAGE, and the protein bands were visualized by immunoblot analysis as described above.
To further demonstrate the formation of homooligomers, purified His-Ste14p was subjected to similar cross-linking analysis (Fig. 1B). Purified His-Ste14p formed predominantly homodimers in the presence of BS3 in addition to smaller amounts of higher order oligomers including trimers and tetramers. Furthermore, small amounts of His-Ste14p homodimer were observed in the absence of BS3 (Fig. 1B), suggesting a robust interaction between subunits.
His-Ste14p Co-immunoprecipitated with Untagged Ste14
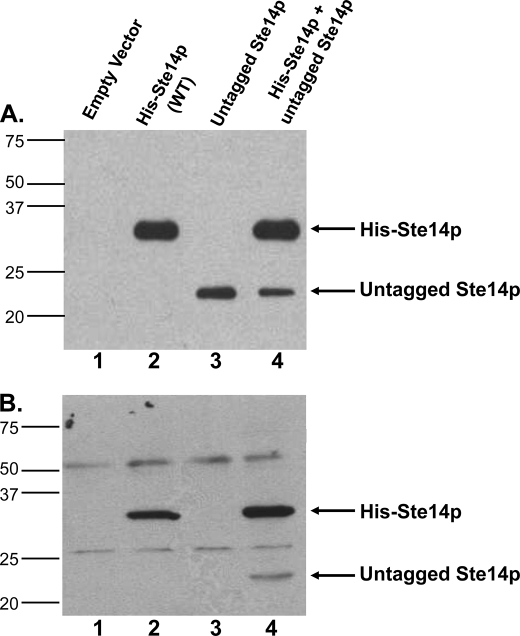
Co-immunoprecipitation experiments were performed to demonstrate that Ste14p exists as a homodimer or oligomer in cells. His-Ste14p was first co-expressed with untagged wild-type Ste14p in a Δste14 strain, SM1188 (30) (Fig. 2A). His-Ste14p migrated at a molecular mass of ∼35 kDa, due to the addition of the 10 histidine residues and 3 Myc epitopes at the N terminus of the proteins (Fig. 2A, second lane). Untagged Ste14p migrated at ∼23 kDa and was expressed at levels ∼30–35% that of His-Ste14p when expressed alone (Fig. 2A, third lane) or together with His-Ste14p (Fig. 2A, fourth lane), as quantified by the Nucleovision imaging system equipped with a high sensitivity CCD camera. Even though the expression of both proteins was driven by the PGK promoter in a high copy number episomal plasmid, overexpression of His-Ste14p utilized the URA3 selection marker, and overexpression of the untagged protein utilized the LEU2 selection marker (Table 1). It has been demonstrated previously that in selective medium, LEU2 plasmids are maintained less well than their URA3 counterparts, resulting in fewer plasmids per cell and, thus, lower protein expression (34). We hypothesize that the possible differences in the numbers of plasmids per cell accounts for the lower expression level of untagged Ste14p. Alternatively, the His-Ste14p may be inherently more stable than the untagged protein.
FIGURE 2.
Expression and co-immunoprecipitation of untagged Ste14p with His-Ste14p. A, 2 μg of crude membrane protein expressing tagged or untagged Ste14p variants were subjected to 10% SDS-PAGE and immunoblot analyses. First lane, empty vector; second lane, His-Ste14p; third lane, untagged-Ste14p; fourth lane, His-Ste14p + untagged-Ste14p. Proteins were detected with a Ste14p polyclonal antibody (1:500) and HRP-conjugated goat anti-rabbit secondary antibody (1:10,000), and the bands were visualized using ECL. B, 100 μg of protein from crude membrane preparations were incubated in 1× RIPA buffer plus ∼3 μg of anti-Myc monoclonal antibody overnight at 4 °C with gentle rotation. 40 μl of 50% (vol:vol) protein A-Sepharose beads in 1× RIPA buffer were added to each tube and incubated another 2.5 h at 4 °C with gentle rotation. The bead-immuno-protein complexes were processed as described under “Experimental Procedures,” and the entire sample was separated by 10% SDS-PAGE. Immunodetection was with a Ste14p polyclonal antibody (1:500) and a goat anti-rabbit secondary antibody (1:10,000) conjugated to HRP. Protein bands were visualized by ECL. WT, wild type.
His-Ste14p was then immunoprecipitated from crude membrane preparations using an α-Myc monoclonal antibody followed by immunoblotting with an α-Ste14 polyclonal antibody (Fig. 2B). Crude membranes from the Δste14 strain transformed with the empty vector were used as the negative control. Untagged Ste14p was not immunoprecipitated when expressed alone (Fig. 2B, third lane) but was detected when co-expressed with His-Ste14p as evidenced by the ∼23-kDa molecular mass band (Fig. 2B, fourth lane). These data support our hypothesis that Ste14p forms an oligomeric species.
Wild-type His-Ste14p and Inactive Variants Are Co-expressed with Untagged Ste14p
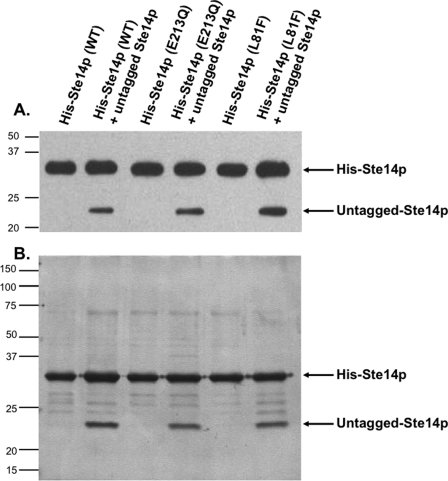
To explore the functional significance of His-Ste14p dimerization, two variants of His-Ste14p (L81F and E213Q) that were previously determined to retain negligible enzymatic activity (19) were also expressed in a Δste14 strain either alone or together with untagged Ste14p (Table 1). The resulting yeast strains were tested for expression of His-Ste14p and untagged Ste14p by immunoblot analysis using a Ste14p polyclonal antibody (Fig. 3A). The His-Ste14p variants were all expressed at similar levels. Similar to what was observed with wild-type His-Ste14p, untagged Ste14p was also expressed at a lower level when co-expressed with either of the His-tagged variants.
FIGURE 3.
Co-expression and co-purification of untagged Ste14p with His-Ste14p and inactive His-Ste14p variants L81F and E213Q. A, expression analysis is shown. 2 μg of crude membrane protein expressing tagged or untagged Ste14p variants were subjected to 10% SDS-PAGE and immunoblot analyses. Proteins were detected with a Ste14p polyclonal antibody (1:500) and HRP-conjugated goat anti-rabbit secondary antibody (1:10,000), and the bands were visualized using ECL. B, co-purification is shown. 25 μg of crude membrane protein was solubilized in 1% (w/v) DDM for 1 h and centrifuged at 300,000 × g for 30 min at 4 °C to remove un-solubilized protein. The supernatant was incubated with Talon metal affinity resin, and the resin was washed as described under “Experimental Procedures.” The purified proteins were eluted with 1 m imidazole, and the fractions were combined and concentrated. 1 μg of purified protein was subjected to 10% SDS-PAGE analysis, and the proteins were visualized by silver nitrate staining. WT, wild type.
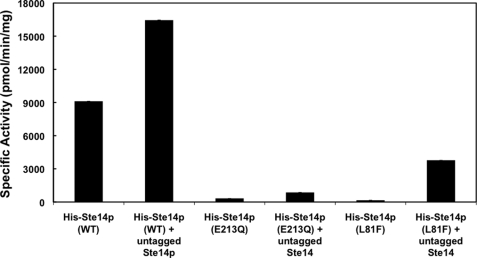
Crude membranes (5 μg) from each of these strains were subsequently tested for methyltransferase activity in vitro using 200 μm AFC as the substrate and 20 μm [14C]SAM as the methyl donor (Fig. 4) (9). Upon normalizing for expression levels using a Nucleovision imaging system equipped with a high sensitivity CCD camera, the activity of untagged Ste14p was higher than that of wild-type His-Ste14p (Fig. 4). These data suggest that the N-terminal tags negatively affect the function of the enzyme. The two His-Ste14p variants, L81F and E213Q, showed negligible activity. Although these two variants were inactive in vitro, His-Ste14p (L81F) retained some function in vivo as determined by a highly sensitive a-factor halo assay, whereas His-Ste14p (E213Q) did not (Ref. 19 and data not shown) (Fig. 4). It was not relevant to normalize the activity levels for the co-expressed proteins because we do not know the relative contributions of each enzyme in a mixture to the total activity. We did determine, however, the specific activities of the co-expressed proteins without normalization. The membranes co-expressing wild-type His-Ste14p and untagged wild-type Ste14p demonstrated the highest activity (755 ± 19 pmol/min/mg). The membranes co-expressing E213Q or L81F His-Ste14p and untagged wild-type Ste14p demonstrated specific activities of 422 ± 21 and 354 ± 3 pmol/min/mg, respectively. The fact that the E213Q and L81F His-Ste14p strains co-expressing untagged Ste14p retained activity is likely the result of the expression of active homodimers of the wild-type protein. Statistically, both homo- and heterodimers of the untagged wild-type enzyme would form, resulting in active protein.
FIGURE 4.
In vitro methyltransferase activities of Ste14p variants in crude membranes. 5 μg of crude membrane protein were incubated with 200 μm AFC and 20 μm [14C]SAM for 30 min at 30 °C in a total volume of 60 μl. The reactions were terminated by the addition of 50 μl of 1 m NaOH/1% SDS (v/v), and the activity was quantified by the vapor diffusion assay as described under “Experimental Procedures.” Each reaction was performed three times in duplicate, and error bars represent ±S.D.). Activities were normalized to the expression level of each construct using immunoblots containing varying amounts of each protein. The protein bands were visualized with ECL as described in Fig. 3 and quantified using a Nucleovision imaging system equipped with a high sensitivity cooled CCD camera. WT, wild type.
Untagged-Ste14p Co-purified with Wild-type and Inactive Variants of His-Ste14p
As demonstrated above, a mixture of dimers will necessarily be expressed in our co-transformed strains. To isolate only the His-Ste14p containing complexes, the His-Ste14p proteins expressed alone or together with untagged Ste14p were purified and assayed for purity by silver nitrate staining (Fig. 3B). The ∼23-kDa untagged-Ste14p protein co-purified with His-Ste14p, His-Ste14p (L81F), and His-Step14p (E213Q). The ability of untagged Ste14p to co-immunoprecipitate and co-purify with His-Ste14p and its variants strongly suggests that Ste14p forms a homodimer or perhaps a higher order oligomer.
Inactive His-Ste14p Variants L81F and E213Q Exerted a Dominant-negative Effect on Wild-type Ste14p Methyltransferase Activity
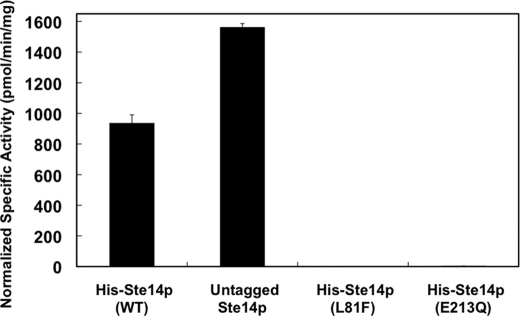
To assess the activity of purified protein preparations of the His-Ste14p variants + untagged-Ste14p, we performed in vitro vapor diffusion methyltransferase assays. Each purified protein mixture was reconstituted in E. coli polar lipid and 100 μm AFC as the methyl acceptor and incubated with 20 μm [14C]SAM as the methyl donor. Purified and reconstituted His-Ste14p + untagged-Ste14p showed higher activity than purified reconstituted His-Ste14p alone, whereas the His-Ste14p variants E213Q and L81F showed little to no activity. Furthermore, the His-Ste14p (E213Q) + untagged Ste14p and His-Ste14p (L81F) + untagged Ste14p complexes retained only ∼5 and ∼20% of wild-type His-Ste14p + untagged Ste14p activity, respectively (Fig. 5). This observed dominant-negative effect on wild-type methyltransferase activity provides evidence, albeit indirect, suggesting that the interaction between Ste14p monomers is important for full function.
FIGURE 5.
In vitro methyltransferase activities of purified His-Ste14p and variants. Purified protein (0.3 μg) was reconstituted in a 3000-fold excess of E. coli polar lipid by 20-fold rapid dilution in 100 mm MES, pH 7.0, and then incubated on ice for 10 min. Reactions contained the reconstituted purified proteins, 200 μm AFC, and 20 μm [14C]SAM and were incubated for 30 min at 30 °C. The reactions were terminated by the addition of 1 m NaOH, 1% SDS, and activity was quantified as described under “Experimental Procedures.” Each reaction was performed three times in duplicate, and error bars represent ±S.D. WT, wild type.
The finding that the L81F mutation in a single Ste14p severely impairs the methyltransferase function of the wild-type His-Ste14p subunit by 80% that of the wild-type complex, but not completely, is consistent with another study that found the introduction of a non-functional H192R mutation in one of the dimeric MalK subunits of the E. coli maltose ABC transporter reduced ATPase activity of a hybrid complex that contained the single substitution and the wild-type subunit to 12% of wild type (35). Although the L81F variant showed little to no in vitro methyltransferase activity, in vivo activity was observed in a highly sensitive a-factor halo assay (19). Given these data, it is likely that the L81F mutation does not completely abolish function and supports low rates of methyltransferase activity that are below the limits of detection of our assay. This hypothesis that the residual activity of L81F was enhanced by co-purification with wild-type Ste14p was corroborated by the fact that the E213Q mutant, which showed no residual halo activity (19), demonstrated only 5% of wild-type activity when co-purified with wild-type Ste14p.
DISCUSSION
To date little is known about the three-dimensional structure of Ste14p or its arrangement in the endoplasmic reticulum membrane. Hydropathy plot analysis of the Ste14p sequence predicted that the protein will have six transmembrane segments and that the majority of the residues will be cytosolically disposed, including the N and C termini (13). This topology was confirmed experimentally in yeast (13, 19). Similar hydropathy plot analysis of the primary sequence of human Icmt (hIcmt) suggested that the protein has a similar core of six transmembrane segments to Ste14p, with two additional membrane spans at the N terminus. This topology was recently demonstrated using live mammalian cells, and the data showed convincingly that hIcmt spans the membrane eight times, and that both N and C termini are exposed to the cytosol (18). The topologies of these two enzymes are remarkably similar, including the existence of a hairpin turn between the last two transmembrane segments (18, 19), suggesting a conserved organization in the membrane. As we have shown here that Ste14p forms a dimer or higher order oligomer, further investigation into the oligomerization states of hIcmt and other Icmts will be important to determine whether this structural organization is common to all members of this enzyme family.
The motif GXXXG, where G is glycine and X is one of the eight most common amino acids found in transmembrane helices including alanine, valine, and isoleucine, is a highly conserved pattern of amino acids found in 80% of proteins that form dimers (36–38). The GXXXG motif was first identified in glycophorin A, a protein that has one transmembrane segment, and it was concluded that the amino acids in this motif create a helix that contains a groove and two ridges. When dimerized, these helices pack tightly with each other, effectively pairing the complementary grooves and ridges (22, 37, 38). Four major sequence patterns emerged from an in-depth study of libraries of GXXXG sequences, namely G(Sm)XXG(Lg)XX(Sm), GXXXGXXXT, (Lg)GXX(Lg)GXX(VI), and G(Lg)XXG(Lg)XX(Sm), where G is glycine, X is any of the amino acids commonly found in transmembrane regions, Sm is a small amino acid (Gly, Ala, Ser), and Lg is a large amino acid (Val, Leu, Ile) (38). Importantly, large numbers of these sequences were found in a SwissProt data base of transmembrane domains (36) suggesting that these sequences are commonly used for naturally occurring helical associations in membrane proteins (38). Numerous other single and multispanning membrane proteins have also been found to contain this motif, including APH-1 and the F0F1-ATPase (20, 21).
Sequence alignments of the Icmts from 15 species, including Ste14p and hIcmt, reveal a highly conserved GXXXG motif. In Ste14p and hIcmt, this motif is found in the first and third transmembrane segments, respectively. The pattern described above, G(Lg)XXG(Lg)XX(Sm), exactly corresponds to the primary sequence of Ste14p from amino acids 31–39 (31GILLGIFVG39). Given the presence of the third glycine at position 39, this sequence may in fact constitute a tandem GXXXG motif that would create an even larger dimerization surface. In hIcmt, this region corresponds to amino acids 74–78 (74GFVFG78) in transmembrane segment 3, which upon inspection can be further extended to a tandem binding motif from amino acids 70 to 78 (70ACFLGFVFG78). The importance of these residues in oligomerization of either protein remains to be investigated.
As described above, the interaction between proteins containing only one GXXXG motif has been studied extensively (22, 37, 38). Far less is known about proteins containing multiple GXXXG-like dimerization motifs. It has been concluded that multiple dimerization motifs found either in consecutive transmembrane helices or in variable binding motifs that do not exactly match the GXXXG motif, such as G(A/S/T)XXXG(A/S/T), are involved in dimerization and/or oligomeric assembly, exemplified by the α-factor receptor, a G protein-coupled receptor from S. cerevisiae (28). In this and other G protein-coupled receptors that are now thought to oligomerize, such as the γ-opioid receptors (39), dopamine D2 receptors (40), and rhodopsin (41), one transmembrane helix provides a contact site for dimerization, whereas subsequent sites are important for oligomeric assembly. Whereas Ste14p contains only one tandem GXXXG motif in transmembrane segment 1, sequence inspection of hIcmt revealed that each of the two additional N-terminal transmembranes contains a variable GXXXG sequence corresponding to AXXXA (25ASVLA29) and AXXXAXXXA (45ALYVAGLNA53), respectively, which may be present to facilitate higher order oligomerization in the human enzyme.
Dimerization has been shown to be important for catalysis in multiple proteins across multiple species. Our experiments presented here have shown indirectly that Ste14p forms a homodimer or higher order oligomer, and further experimentation will be needed to understand how this arrangement dictates the mechanism of Ste14p and perhaps all Icmts. We are currently exploring the oligomerization state of purified and His-Ste14p by analytical ultracentrifugation, atomic force microscopy, and two-dimensional crystallography. In most cases of protein homodimerization, the dimer is arranged in a head to head or head to tail fashion, and dimer assembly is necessary for allosteric protein interaction and/or active site catalysis. Given the constraints of the membrane and the topology of the enzyme, it is most likely that a Ste14p dimer would be organized in a head to head configuration with the two subunits contributing to one active site or two allosterically controlled active sites. Although we have yet to definitively demonstrate allosteric kinetics with Ste14p in crude or purified enzyme preparations due to the hydrophobic nature of the enzyme and its substrate AFC, this line of inquiry warrants further investigation.
The catalytic mechanism and the residues responsible for the methyltransferase activity of Ste14p or any Icmt are unknown. From data using crude membrane preparations of mammalian Icmts, the proposed kinetic model of methyltransferase activity is an ordered sequential bi-bi reaction (42, 43). The significance of dimer formation will be important for studying this mechanism in depth. Furthermore, a complete understanding of the implications of dimerization on function is especially important for the development of Icmt inhibitors, as hIcmt has been identified recently as an excellent drug target for chemotherapeutic intervention in cancer (5, 44–48).
Footnotes
- Icmt
- isoprenylcysteine carboxyl methyltransferase
- hIcmt
- human Icmt
- AFC
- N-acetyl-S-farnesyl-l-cysteine; DDM-N-dodecyl-β-d-maltopyranoside; SAM-S-adenosyl-l-methionine
- BS3
- bis-sulfosuccinimidyl suberate homobifunctional cross-linking agent
- PGK
- 3′-phosphoglycerate kinase
- MOPS
- 4-morpholinepropanesulfonic acid
- MES
- 4-morpholineethanesulfonic acid
- HRP
- horseradish peroxidase
- RIPA
- radioimmune precipitation assay.
REFERENCES
- 1.Clarke S. (1992) Annu. Rev. Biochem. 61, 355–386 [DOI] [PubMed] [Google Scholar]
- 2.Zhang F. L., Casey P. J. (1996) Annu. Rev. Biochem. 65, 241–269 [DOI] [PubMed] [Google Scholar]
- 3.Young S. G., Ambroziak P., Kim E., Clarke S. (2000) The Enzymes: Protein Lipidation (Tamanoi F., Sigman D. S. eds) Vol. 21, pp. 155–213, Academic Press, San Diego, CA [Google Scholar]
- 4.Hrycyna C. A., Clarke S. (1993) Pharmacol. Ther. 59, 281–300 [DOI] [PubMed] [Google Scholar]
- 5.Gelb M. H., Brunsveld L., Hrycyna C. A., Michaelis S., Tamanoi F., Van Voorhis W. C., Waldmann H. (2006) Nat. Chem. Biol. 2, 518–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casey P. J., Seabra M. C. (1996) J. Biol. Chem. 271, 5289–5292 [DOI] [PubMed] [Google Scholar]
- 7.Deschenes R. J., Stimmel J. B., Clarke S., Stock J., Broach J. R. (1989) J. Biol. Chem. 264, 11865–11873 [PubMed] [Google Scholar]
- 8.Anderson J. A., Hrycyna C. A. (2006) in The Enzymes: Protein Methyltransferases (Clarke S. G., Tamanoi F. eds) Vol. 24, pp. 245–272, Academic Press, San Diego, CA [Google Scholar]
- 9.Hrycyna C. A., Clarke S. (1990) Mol. Cell. Biol. 10, 5071–5076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hrycyna C. A., Sapperstein S. K., Clarke S., Michaelis S. (1991) EMBO J. 10, 1699–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hrycyna C. A., Wait S. J., Backlund P. S., Jr., Michaelis S. (1995) Methods Enzymol. 250, 251–266 [DOI] [PubMed] [Google Scholar]
- 12.Marr R. S., Blair L. C., Thorner J. (1990) J. Biol. Chem. 265, 20057–20060 [PubMed] [Google Scholar]
- 13.Romano J. D., Schmidt W. K., Michaelis S. (1998) Mol. Biol. Cell 9, 2231–2247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sapperstein S., Berkower C., Michaelis S. (1994) Mol. Cell. Biol. 14, 1438–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stephenson R. C., Clarke S. (1990) J. Biol. Chem. 265, 16248–16254 [PubMed] [Google Scholar]
- 16.Stephenson R. C., Clarke S. (1992) J. Biol. Chem. 267, 13314–13319 [PubMed] [Google Scholar]
- 17.Dai Q., Choy E., Chiu V., Romano J., Slivka S. R., Steitz S. A., Michaelis S., Philips M. R. (1998) J. Biol. Chem. 273, 15030–15034 [DOI] [PubMed] [Google Scholar]
- 18.Wright L. P., Court H., Mor A., Ahearn I. M., Casey P. J., Philips M. R. (2009) Mol. Cell. Biol. 29, 1826–1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Romano J. D., Michaelis S. (2001) Mol. Biol. Cell 12, 1957–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Curran A. R., Engelman D. M. (2003) Curr. Opin. Struct. Biol. 13, 412–417 [DOI] [PubMed] [Google Scholar]
- 21.Senes A., Engel D. E., DeGrado W. F. (2004) Curr. Opin. Struct. Biol. 14, 465–479 [DOI] [PubMed] [Google Scholar]
- 22.MacKenzie K. R., Prestegard J. H., Engelman D. M. (1997) Science 276, 131–133 [DOI] [PubMed] [Google Scholar]
- 23.Gerber D., Shai Y. (2001) J. Biol. Chem. 276, 31229–31232 [DOI] [PubMed] [Google Scholar]
- 24.Polgar O., Robey R. W., Morisaki K., Dean M., Michejda C., Sauna Z. E., Ambudkar S. V., Tarasova N., Bates S. E. (2004) Biochemistry 43, 9448–9456 [DOI] [PubMed] [Google Scholar]
- 25.Arselin G., Giraud M. F., Dautant A., Vaillier J., Brèthes D., Coulary-Salin B., Schaeffer J., Velours J. (2003) Eur. J. Biochem. 270, 1875–1884 [DOI] [PubMed] [Google Scholar]
- 26.Whittington D. A., Waheed A., Ulmasov B., Shah G. N., Grubb J. H., Sly W. S., Christianson D. W. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 9545–9550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McClain M. S., Iwamoto H., Cao P., Vinion-Dubiel A. D., Li Y., Szabo G., Shao Z., Cover T. L. (2003) J. Biol. Chem. 278, 12101–12108 [DOI] [PubMed] [Google Scholar]
- 28.Overton M. C., Chinault S. L., Blumer K. J. (2003) J. Biol. Chem. 278, 49369–49377 [DOI] [PubMed] [Google Scholar]
- 29.Schneider D., Engelman D. M. (2004) J. Mol. Biol. 343, 799–804 [DOI] [PubMed] [Google Scholar]
- 30.Anderson J. L., Frase H., Michaelis S., Hrycyna C. A. (2005) J. Biol. Chem. 280, 7336–7345 [DOI] [PubMed] [Google Scholar]
- 31.Elble R. (1992) BioTechniques 13, 18–20 [PubMed] [Google Scholar]
- 32.Schaffner W., Weissmann C. (1973) Anal. Biochem. 56, 502–514 [DOI] [PubMed] [Google Scholar]
- 33.Ambudkar S. V., Lelong I. H., Zhang J., Cardarelli C. (1998) Methods Enzymol. 292, 492–504 [DOI] [PubMed] [Google Scholar]
- 34.Ugolini S., Tosato V., Bruschi C. V. (2002) Plasmid 47, 94–107 [DOI] [PubMed] [Google Scholar]
- 35.Davidson A. L., Sharma S. (1997) J. Bacteriol. 179, 5458–5464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Senes A., Gerstein M., Engelman D. M. (2000) J. Mol. Biol. 296, 921–936 [DOI] [PubMed] [Google Scholar]
- 37.Russ W. P., Engelman D. M. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 863–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Russ W. P., Engelman D. M. (2000) J. Mol. Biol. 296, 911–919 [DOI] [PubMed] [Google Scholar]
- 39.Filizola M., Weinstein H. (2002) Biopolymers 66, 317–325 [DOI] [PubMed] [Google Scholar]
- 40.Guo W., Shi L., Javitch J. A. (2003) J. Biol. Chem. 278, 4385–4388 [DOI] [PubMed] [Google Scholar]
- 41.Liang Y., Fotiadis D., Filipek S., Saperstein D. A., Palczewski K., Engel A. (2003) J. Biol. Chem. 278, 21655–21662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baron R. A., Casey P. J. (2004) BMC Biochem. 5, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi Y. Q., Rando R. R. (1992) J. Biol. Chem. 267, 9547–9551 [PubMed] [Google Scholar]
- 44.Bergo M. O., Gavino B. J., Hong C., Beigneux A. P., McMahon M., Casey P. J., Young S. G. (2004) J. Clin. Invest. 113, 539–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wahlstrom A. M., Cutts B. A., Liu M., Lindskog A., Karlsson C., Sjogren A. K., Andersson K. M., Young S. G., Bergo M. O. (2008) Blood 112, 1357–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Winter-Vann A. M., Baron R. A., Wong W., dela Cruz J., York J. D., Gooden D. M., Bergo M. O., Young S. G., Toone E. J., Casey P. J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 4336–4341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anderson J. L., Henriksen B. S., Gibbs R. A., Hrycyna C. A. (2005) J. Biol. Chem. 280, 29454–29461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blum R., Cox A. D., Kloog Y. (2008) Recent Pat. Anticancer Drug Discov. 3, 31–47 [DOI] [PubMed] [Google Scholar]