Abstract
Wall teichoic acid (WTA) glycopolymers are major constituents of cell envelopes in Staphylococcus aureus and related Gram-positive bacteria with important roles in cell wall maintenance, susceptibility to antimicrobial molecules, biofilm formation, and host interaction. Most S. aureus strains express polyribitol phosphate WTA substituted with d-alanine and N-acetylglucosamine (GlcNAc). WTA sugar modifications are highly variable and have been implicated in bacteriophage susceptibility and immunogenicity, but the pathway and enzymes of staphylococcal WTA glycosylation have remained unknown. Revisiting the structure of S. aureus RN4220 WTA by NMR analysis revealed the presence of canonical polyribitol phosphate WTA bearing only α-linked GlcNAc substituents. A RN4220 transposon mutant resistant to WTA-dependent phages was identified and shown to produce altered WTA, which exhibited faster electrophoretic migration and lacked completely the WTA α-GlcNAc residues. Disruption of a gene of unknown function, renamed tarM, was responsible for this phenotype. Recombinant TarM was capable of glycosylating WTA in vitro in a UDP-GlcNAc-dependent manner, thereby confirming its WTA GlcNAc-transferase activity. Deletion of the last seven amino acids from the C terminus abolished the activity of TarM. tarM-related genes were found in the genomes of several WTA-producing bacteria, suggesting that TarM-mediated WTA glycosylation is a general pathway in Gram-positive bacteria. Our study represents a basis for dissecting the biosynthesis and function of glycosylated WTA in S. aureus and other bacteria.
Keywords: Bacteriophage, Carbohydrate Biosynthesis, Carbohydrate Structure, Cell Wall, Glycosylation, Staphylococcus aureus, GlcNAc-transferase, Wall Teichoic Acid
Introduction
The bacterial cell envelope represents an interface for the interaction of bacterial cells with host molecules, bacteriophages, other bacteria, and inanimate molecules or surfaces. The properties of bacterial cell surfaces are largely governed by proteins and glycopolymers, both of which have highly variable and often strain-specific compositions and roles (1, 2). Although many bacterial surface proteins have been studied in detail, our knowledge on structures, biosynthetic pathways, and functions of cell wall glycopolymers is still incomplete. Nevertheless, it has become clear that many bacterial cell wall glycopolymers are crucial for bacterial cell wall maintenance, fitness, or virulence and represent promising targets for vaccines or antimicrobial compounds (2).
Staphylococcus aureus and most other Gram-positive bacteria express teichoic acid polymers at their surfaces that are either linked to membrane lipids (lipoteichoic acid) or to peptidoglycan (wall teichoic acid (WTA)2) (3). WTA is composed of phosphate-containing repeating units with a very variable and often species-specific composition. Most S. aureus strains express polyribitol phosphate (Rbo-P)-type WTA, which is composed of repetitive d-ribitol units connected by 1,5-phosphodiester bonds (4). The repeating units can be further substituted with d-alanine at C2-OH and/or with 2-acetamido-2-deoxy-d-glucopyranose (GlcNAc) at C4-OH via α- or β-glycosidic linkages whose relative abundance varies between individual strains (5, 6). WTA accounts for a major portion of the total dry weight of the staphylococcal cell wall (7) but does not play an essential role for S. aureus viability under laboratory conditions (8, 9), although important roles in biofilm formation and autolysin control have been documented (10, 11). Of note, WTA is of pivotal importance during host colonization and infection inasmuch as it facilitates attachment of the bacteria to WTA binding epithelial or endothelial cell receptors (8, 12) and protects the bacterial cells from bactericidal agents such as skin antimicrobial fatty acids (13). Moreover, alanyl residues in teichoic acid play important roles in resistance to cationic antimicrobial host defense factors, antibiotics, and bacteriocins (14, 15). In contrast, WTA sugar modifications have been implicated in the ability of WTA to elicit specific antibody responses (16–18) and binding of bacteriophages to S. aureus (19, 20), Bacillus subtilis (21, 22), or Listeria species (23).
Most steps of WTA backbone biosynthesis, which occurs on the universal lipid carrier undecaprenyl phosphate (C55-P), have recently been elucidated in B. subtilis 168 (24–26) and S. aureus (27, 28), and many of the involved enzymes have been characterized in vitro. The same holds true for the pathway of WTA and lipoteichoic acid alanylation, which consists of four proteins that activate d-alanine by ATP hydrolysis, link it to the dedicated carrier protein DltC, translocate it across the cytoplasmic membrane, and connect it with WTA or lipoteichoic acid repeating units (29, 30). In contrast, WTA glycosylation has hardly been investigated. Nevertheless, WTA β-GlcNAc-transferase activity (EC 2.4.1.70) has been detected in crude cell extracts of S. aureus strain Copenhagen in the 1960s (31), and a S. aureus strain H mutant 52B2 deficient in WTA GlcNAc-transferase activity has been described (20), the genetic basis of which has remained unknown.
Here we report on the identification and characterization of a novel S. aureus protein TarM that is responsible for the glycosylation of WTA with α-GlcNAc and exhibits UDP-GlcNAc-dependent WTA GlcNAc-transferase activity in vitro. Moreover, we show that TarM bears a point mutation in the phage-resistant mutant S. aureus strain 52B2 that leads to a truncated and inactive TarM. Our study paves the way for elucidating the function of the sugar residues carried by WTA in S. aureus-bacteriophage and host interaction.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Media
Escherichia coli strains Top10 or DH5α were used in cloning experiments. E. coli BL21(DE3) was used as a host for expression of recombinant protein. Unless otherwise noted, bacteria were grown in BM broth (1% Tryptone, 0.5% yeast extract, 0.5% NaCl, 0.1% K2HPO4, 0.1% glucose) or tryptic soy broth (Oxoid) supplemented with appropriate antibiotics at concentration of 2.5 μg/ml (erythromycin), 10 μg/ml (chloramphenicol), or 100 μg/ml (ampicillin).
Transposon Mutagenesis of S. aureus Strain RN4220
The transposon plasmid pBTn has been described recently (32). The features of this temperature-sensitive E. coli/S. aureus shuttle vector includes a mini-transposon with an erythromycin resistance cassette flanked by inverted repeats from the horn fly transposon and a xylose-inducible transposase himar1, which can mobilize the mini-transposon into the chromosome with no bias for any specific sequence.
To construct a transposon library, S. aureus RN4220 was transformed with pBTn followed by mobilizing the mini-transposon into the genome upon xylose induction of the transposase and by curing the transposon plasmid via temperature shifts to nonpermissive conditions as described recently (32). Briefly, an overnight culture of pBTn-transformed S. aureus RN4220, grown at permissive temperature (30 °C) in tryptic soy broth supplemented with 0.5% xylose, chloramphenicol, and erythromycin, was diluted 1:100 into fresh tryptic soy broth with 0.5% xylose and erythromycin and incubated at 42 °C for 24 h. This procedure was repeated twice with and once without antibiotic. Then the overnight cultures were centrifuged at 5000 × g for 10 min, and the cells were resuspended in 15% glycerol for storage as a mutant library at −80 °C. Because of insertion of the mini-transposon, which carries the erythromycin resistance cassette into the genome, and curing of pBTn, which bears a chloramphenicol resistance cassette as selection marker in S. aureus, the putative transposon mutants should be erythromycin-resistant and susceptible to chloramphenicol. The presence of this resistance pattern in the mutant library was verified.
Isolation of Phage-resistant Mutants
To isolate phage-resistant mutants, an aliquot of the transposon mutant library was infected with phage 80 at a multiplicity of infection higher than 100. After incubation at room temperature for 30 min, the cells were centrifuged at 5000 × g for 10 min and plated out on BM agar containing erythromycin. Single colonies of the surviving mutants were propagated by streaking them repeatedly out on BM agar three to five times, and phage susceptibility of those clones was further verified by soft agar assay with a phage panel including phage K, known as a lytic broad host range phage (33), and phages 11, 52A, and 80, which belong to phage serogroup B (34). Briefly, the phage lysate were spotted on soft agar plates containing bacterial cells and incubated overnight at 37 °C. Macro-plaques formed on the bacterial lawns indicate that the test strain is susceptible to the corresponding phage. The phage-resistant mutants were treated with mitomycin, which is known to induce prophages, and the supernatants were tested for the release of phages to exclude that the mutants were phage-resistant because of lysogenesis. To identify the insertion site of the transposon in the phage resistant mutants, total DNA was isolated, purified with the NucleoSpin® tissue kit (Macherey-Nagel, Düren), and sequenced with primer erm5B (32), which anneals to the erythromycin resistance cassette of the mini-transposon.
Extraction and Purification of WTA
WTA was extracted with diluted NaOH as described previously (28) with minor modifications. Briefly, S. aureus strains were grown overnight at 37 °C in 250 ml of BM, spun down at 5000 × g for 10 min, washed once with 50 ml of buffer 1 (50 mm MES, pH 6.5), and resuspended in 30 ml of buffer 2 (4% w/v SDS, 50 mm MES, pH 6.5). After sonication for 15 min to get rid of loosely cell surface-associated molecules, the samples were cooked in a boiling water bath for 1 h. Then the cells were collected by centrifugation at 12,000 × g for 15 min, and the pellet was washed once with 50 ml of buffer 3 (2% NaCl, 50 mm MES, pH 6.5) followed by 3–5× washing with warm buffer 1 (∼ 50 °C). The samples were then treated with proteinase K at a concentration of 20 μg/ml in a buffer consisting of 20 mm Tris, pH 8.0, and 0.5% w/v SDS. After overnight digestion at 50 °C, the samples were washed 3–5× with 50 ml of warm buffer 1.
To release WTA from peptidoglycan, the pellets were resuspended in 0.1 m NaOH and rotated at room temperature for at least 12 h. The extraction was stopped by centrifugation at 14,000 × g for 20 min. Because the alanyl group of the WTA repeating unit is labile at alkaline conditions, the WTA samples were devoid of the alanyl group. The supernatant was stored as crude WTA sample after neutralization with 0.1 m HCl.
For further purification of WTA, a batch adsorption protocol with the anion exchanger DEAE-Sephadex A25 (GE Healthcare) was developed based on an established WTA purification procedure (4). Briefly, before loading on the DEAE-Sephadex A25 matrix, the crude WTA sample (∼2 μmol of phosphate) was passed through a PD-10 gel filtration column (GE Healthcare) with 20 mm Tris-HCl, pH 7.2, as elution buffer. The fraction eluted in the void volume was added to 3 ml of 50% DEAE-Sephadex A25 matrix slurry pre-equilibrated with the same buffer (20 mm Tris-HCl, pH 7.2). After rotation at room temperature for 1 h, the matrix was spun down at 1000 × g for 3 min and incubated with washing buffer (20 mm Tris-HCl, pH 7.2, 50 mm NaCl) for 30 min. Then, the WTA was eluted by adding 10 ml of elution buffer (20 mm Tris-HCl, pH 7.2, 0.3 m NaCl) and rotating at room temperature for 0.5 h. To increase the yield, the elution step was repeated once or twice. The eluted fractions were then combined and dialyzed against water with a Spectra/Por®3 dialysis membrane with a Mr cutoff of 3.5 kDa (VWR International GmbH, Darmstadt) followed by concentration to 1–2 ml in a SpeedVac concentrator at 45 °C. The resulting pure WTA samples were stored at −20 °C for further analysis.
Lectin Overlay Analysis of WTA with Wheat Germ Agglutinin (WGA)
WTA samples corresponding to 100 nmol of phosphate were spotted on nitrocellulose membranes and air-dried. The dot blot was then developed with biotinylated WGA followed by incubation with horseradish peroxidase (HRP)-conjugated streptavidin (streptavidin-HRP) and detection by enhanced chemiluminescence (ECL) reaction.
Briefly, WTA dot blots were blocked in 1× Roti block solution (Carl Roth GmbH & Co. Karlsruhe, Germany) with TBST (25 mm Tris, 0.15 m NaCl, 0.05% Tween 20, 3 mm KCl, pH 7.4) for 1 h at room temperature. After briefly rinsing the membranes three times in TBST, the blots were further incubated for 2 h with biotinylated WGA (B-1025, Vector Laboratories) diluted at 1:1000 in buffer TBST-Ca2+ (TBST containing 1 mm CaCl2). Before ECL detection, the blots were rinsed with buffer TBST-Ca2+ 3 times for 5 min and then incubated with streptavidin-HRP (R&D Systems, Minneapolis) with a dilution of 1:400 in TBST-Ca2+ for 1 h. The SuperDetect chemiluminescent substrate (Tebu-Bio Laboratories, Offenbach, Germany) was used for ECL according to the manufacturer's instruction.
PAGE Analysis of WTA
For PAGE analysis of WTA samples, a 26% and 0.75-mm-thick resolving gel was cast as described recently (28). The WTA samples (∼100 nmol of phosphate per lane) were loaded on gels and resolved in a Bio-Rad Protean II Xi electrophoresis cell for 24 h using a constant current of 40 mA/gel and a running buffer containing 0.1 m Tris base and 0.1 m Tricine, pH 8.2.
WTA bands were visualized using the Alcian blue silver staining method (35). Briefly, the gels were fixed and stained with 0.005% (w/v) Alcian blue in EAW solution (40% ethanol and 5% acetic acid in water) at room temperature for several hours until the WTA bands became visible. To remove unspecific background staining, the gels were incubated in EAW solution for 30 min. To intensify the staining pattern, the Bio-Rad silver stain kit (Bio-Rad) was used according to the manufacturer's instruction after oxidation of the gels with 0.7% NaIO4 for 20 min.
Recombinant Expression of tarM
The open reading frame of tarM was amplified and subcloned into the pET28a vector (Merck) at the NdeI and SacI sites for expression in E. coli BL21(DE3) with a His6 tag fused in-frame to the N terminus of TarM. After 4 h, induction of an early log-phase culture with isopropyl β-d-1-thiogalactopyranoside at a concentration of 0.5 mm, the biomass was harvested by centrifugation (5000 × g, 10 min). Then the cells were resuspended in a lysis buffer containing 0.04 mm Tris-HCl, pH 8, and 200 μg/ml lysozyme and disrupted by sonication. After centrifugation at 16,000 × g for 20 min at 4 °C, the supernatant was stored as the crude TarM-containing cell extract. The cell lysate prepared from mock-transformed cells (transformed with empty pET28 vector) was used as a control. Expression of recombinant TarM was monitored by SDS-PAGE and immunoblot analysis with a monoclonal anti-His tag-specific antibody (Penta-His Antibody, Qiagen, Hilden, Germany) according to standard protein detection techniques.
Assay for in Vitro WTA GlcNAc-transferase Activity
WTA GlcNAc-transferase assays were carried out at 37 °C for 4 h with non-glycosylated WTA as the sugar acceptor and UDP-GlcNAc as the sugar donor. Each reaction had a total volume of 200 μl containing 0.02 m Tris-HCl buffer, pH 8, 10 mm MgCl2, non-glycosylated WTA (corresponding to 100 nmol of phosphate), 2 mm UDP-GlcNAc, and 200 μg of recombinant crude TarM or control lysates. The reaction was stopped by heating at 99 °C for 10 min. The enzymatic products were extracted two times with water-saturated phenol and once with phenol-chloroform-isoamyl alcohol (25:24:1, v/v/v) to remove proteins before spotting on nitrocellulose membranes and analysis by WGA lectin overlay as described above.
Cloning and Sequencing of the tarM Gene from Strain H and Its Derivative 52B2
Shaw et al. (20) reported in 1970 a phage-resistant mutant 52B2, derived from the parental strain H by treatment with mutagenic chemicals. To find out whether there is a mutation in tarM of 52B2, the genes from strain H and mutant 52B2 were PCR-amplified with Phusion® hot start high fidelity DNA polymerase (F-540L, New England Biolabs), subcloned in the pCR4Blunt-TOPO vector (K2875-20, Invitrogen), and sequenced.
Construction of tarM Complementation Vectors
Wild-type or mutant tarM genes were PCR-amplified from genomic DNA of the corresponding strains. The amplicons were subcloned into the E. coli/S. aureus shuttle vector pRB474 (36) at the EcoRI and BamHI sites, and the resulting plasmids were designated pRB474-tarM, pRB474-H-tarM, and pRB474-52B2-tarM.
General and Analytical Chemistry Methods
The composition of pure WTA samples was determined by methanolysis of the samples with 2 m HCl, MeOH at 85 °C for 45 min followed by acetylation using acetic anhydride and pyridine (1:1, v/v) at 85 °C for 30 min and detection by gas-liquid chromatography (GLC) and GLC-mass spectrometry. The systems used were a Hewlett-Packard 5880 instrument equipped with a SPB-5 capillary column (30 m × 0.25 mm, film thickness 0.25 μm) and applying a temperature gradient of 120 (kept for 3 min) to 260 °C at 3 °C/min for GLC or a Hewlett-Packard 5989 instrument equipped with a HP-5MS capillary column (30 m × 0.25 mm, film thickness 0.25 μm) and applying the same temperature gradient for GLC-mass spectrometry. The absolute configuration was determined by GLC by comparison with authentic standards of the acetylated (S)-2-butanol glycoside derivative after butanolysis (2 m HCl, (S)-2-butanol at 85 °C for 2 h) and acetylation (acetic anhydride and pyridine (1:1, v/v)) (37). Phosphate content and hexosamine quantification were achieved photometrically as previously described elsewhere (38, 39). Alternatively, released inorganic phosphate was measured with a QuantiChromTM phosphate assay kit (DIPI-500) (Biotrend Chemical GmbH, Cologne. Germany) upon drying and then digesting WTA in 40 μl of 70% (w/v) HClO4 at 165 °C for 2 h.
NMR Spectroscopy
NMR experiments were carried out in D2O at 300 K. All one-dimensional (1H and 13C) and two-dimensional homo- (1H,1H correlation spectroscopy (COSY), total correlation spectroscopy (TOCSY), and rotating-frame nuclear Overhauser enhancement spectroscopy (ROESY)) and heteronuclear 13C,1H single quantum correlation distortionless enhancement by polarization transfer (HSQC-DEPT), heteronuclear multiple bond correlation experiment (HMBC), and 1H,31P multiple quantum correlation-total correlation spectroscopy (HMQC-TOCSY)) experiments were recorded with a Bruker DRX Avance 600 MHz spectrometer (operating frequencies of 600.03 MHz for 1H NMR, 150.89 MHz for 13C NMR, and 242.90 for 31P) using standard Bruker software. COSY, TOCSY, and ROESY were recorded using data sets (t1 by t2) of 2048 by 512 points, and 16 scans were acquired for each t1 value in the case of COSY and TOCSY, whereas 32 scans were acquired in the case of ROESY. The TOCSY experiment was carried out in the phase-sensitive mode with mixing times of 100 ms. The 13C,1H correlations were measured in the 1H-detected mode via HSQC-DEPT with proton decoupling in the 13C domain acquired using data sets of 2048 by 512 points and 32 scans for each t1 value. The HMBC spectra were acquired using data sets of 2048 by 512 points and 96 scans for each t1 value. The 1H,31P-HMQC-TOCSY experiment was recorded using a data set of 2048 by 512 points (16 scans for each t1 value) using a mixing time of 100 ms. Chemical shifts are reported relative to internal acetone (δH 2.225; δC 31.50).
In Silico Analysis of TarM
The gene context of tarM was analyzed at the CMR website (Comprehensive microbial resources, J. Craig Venter Institute). To identify proteins homologous to TarM, the protein data base UniProtKB (released on October 25, 2009 at European Bioinformatics Institute) was searched with TarM as query sequence. The TarM homologues were aligned by ClustalW program.
RESULTS
WTA of S. aureus Strain RN4220 Has a Canonical 1,5-Rbo-P Structure and Is Substituted with α-GlcNAc
The glycosylation patterns of WTA have been analyzed in S. aureus strains Copenhagen (5) and MN8m (6), but the WTA structure of an S. aureus strain, whose genome sequence is available and which is amenable to recombinant DNA techniques, has hardly been addressed. Jenni and Berger-Bächi (40) confirmed the production of Rbo-P WTA in S. aureus NCTC8325, but they did not address the question if GlcNAc is linked via α- or β-glycosidic bonds, which is of potential relevance for immunogenicity (16–18, 41) and host interaction of S. aureus WTA (2).
To provide a basis for elucidating the pathway of WTA glycosylation in S. aureus, WTA of strain RN4220, a descendent of NCTC8325, was isolated and analyzed. Purification of WTA was performed by DEAE Sephadex batch adsorption chromatography upon releasing the WTA polymers from cell walls by treatment with NaOH. Compositional analysis by GC revealed the presence of mainly ribitol and hexosamine. The hexosamine was identified as d-GlcN, and the GlcNAc/phosphate ratio was found to be 0.63. It should be noted that GlcNAc is transformed to glucosamine (GlcN) under the conditions used for WTA hydrolysis. Alanine could not be detected, as the WTA alanyl group is labile at alkaline pH and was lost during extraction with diluted NaOH.
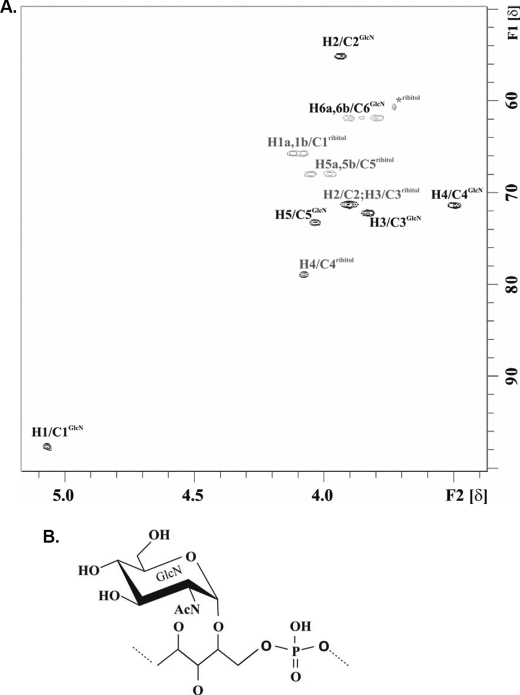
The structural details of WTA repeating units were established by NMR spectroscopy. The 1H NMR spectrum revealed an anomeric signal at δ 5.07 that could be assigned to α-GlcNAc (Fig. 1A). The α-configuration was confirmed by 1JC1,H1 174.5 Hz. The complete analysis of the GlcNAc ring was easily performed using the COSY and TOCSY experiments. The analysis of ribitol was complicated because of the overlap of signals in COSY and TOCSY experiments. The complete assignment could be achieved by a HSQC-DEPT experiment (Fig. 1A). Chemical shifts for both GlcNAc and ribitol are listed in Table 1. The N-acetylation was confirmed by the signals at δ 2.08/23.32 (1H/13C), which correlated in the HMBC experiment with a carbon signal at δ 175.75, corresponding to the carboxyl carbon. This carbon showed also a correlation with H2 of GlcNAc in the HMBC spectrum. In the 1H,31P HMQC-TOCSY experiment, a strong phosphorus signal could be observed at δ 0.98 that correlated strongly to those of H1b and H5a of ribitol, thus confirming the phosphate substitution. The position of the α-GlcNAc linkage in WTA of strain Copenhagen has been described to be at C4 of ribitol (5). This same linkage was observed in our case and confirmed by a cross-peak between H1 of GlcNAc and H4 of ribitol in the ROESY experiment as well as by cross-peaks between H1 GlcNAc and C4 of ribitol and H4 of ribitol and C1 of GlcNAc in the HMBC spectrum. Of note, C4 was assigned in analogy to data of other studies (5, 6). However, because positions 2 and 4 of ribitol are equivalent in this case, further analysis will be necessary to clarify this point. Taken together our data demonstrate that S. aureus RN4220 produces canonical Rbo-P WTA in which GlcNAc is α-linked to position 4 (or 2) of the ribitol residue (Fig. 1B).
FIGURE 1.
Structural analysis of WTA from S. aureus strain RN4220. A, shown is part of heteronuclear 13C,1H single quantum correlation distortionless enhancement by polarization transfer (HSQC-DEPT) spectrum of WTA from S. aureus strain RN4220. The spectrum was measured in D2O at 300 K. B, shown is the proposed schematic structure of WTA repeating units of S. aureus RN4220. d-Alanine has been shown to be linked to C2 (5) but was not detectable in our samples because WTA was released from cell walls at alkaline conditions where the d-alanine esters are lost.
TABLE 1.
1H and 13C chemical shifts of WTA from RN4220
| Unit | 1/1′ | 2 | 3 | 4 | 5/5′ | 6/6′ |
|---|---|---|---|---|---|---|
| Rbo | ||||||
| 1H | 3.97/4.05 | 3.90 | 3.90 | 4.08 | 4.08/4.12 | |
| 13C | 67.92 | 71.31 | 71.31 | 78.93 | 65.72 | |
| α-GlcNAc | ||||||
| 1H | 5.07 | 3.93 | 3.83 | 3.49 | 4.03 | 3.79/3.90 |
| 13C | 97.61 | 55.08 | 72.22 | 71.39 | 72.22 | 61.82 |
Inactivation of tarM in S. aureus Strain RN4220 Leads to Resistance to Serogroup B Phages
Cell wall phosphate content and WTA polymerase activity in the previously described phage-resistant S. aureus 52B2 mutant are similar to those of the parental strain, suggesting that this mutant produces WTA but lacks GlcNAc modification (20). In addition, it has been demonstrated that both α-GlcNAc and β-GlcNAc residues can allow adsorption and infection by phages 80 and 52A (42, 43), which belong to phage serogroup B. These pioneering studies suggested that it should be possible to identify the WTA GlcNAc-transferase of S. aureus by screening transposon mutant libraries for resistance to serogroup B phages.
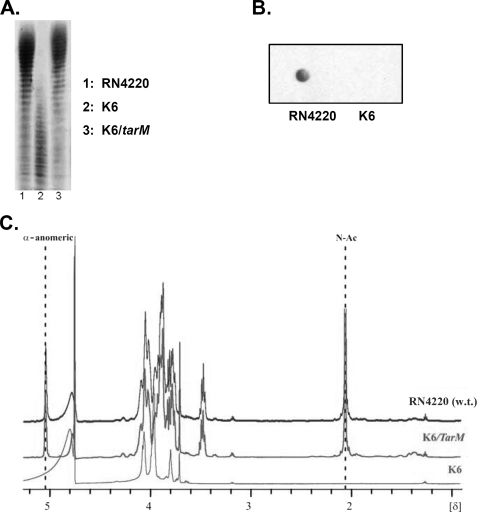
We chose S. aureus strain RN4220 for this approach because it is free of capsule and prophages (44), which might interfere with the screening procedure. Strain RN4220 was mutagenized with a mariner-based transposon, which enables fully random insertion of an erythromycin resistance cassette into the chromosome (45). By screening the transposon library with phage 80, we isolated a mutant K6, which was resistant to all tested temperate serogroup B phages including phage 11, 80, and 52A, whereas susceptibility to the unrelated lytic phage K was retained (Fig. 2B). Analysis of the genomic DNA sequence flanking the transposon revealed that the transposon had integrated into the open reading frame SACOL1043, which encodes a protein of unknown function with similarity to glycosyltransferases and which we renamed tarM. The gene was disrupted at 1119 bp downstream of the start codon (Fig. 2A). The tarM mutant K6 exhibited no major changes in growth kinetics, microscopic appearance, or antibiotic susceptibility patterns compared with the parental strain (data not shown). Of note, tarM is not located within one of the known WTA gene clusters. The adjacent genes all have unknown functions and have not been implicated in cell wall biosynthesis (Fig. 2A).
FIGURE 2.
Genetic context of tarM and complementation of the phage-resistant mutant K6. A, shown is the phage-resistant mutant K6 has a transposon (Tn) insertion mutation in open reading frame SACOL1043 (tarM), which encodes a putative glycosyltransferase. The arrow indicates the putative direction of transcription, and the hairpin loop indicates the predicted transcription terminator. B, shown is the soft agar assay for phage susceptibility of RN4220 wild type, tarM mutant K6, and complemented mutant (K6/tarM). In contrast to the wild type (left), the K6 mutant (middle) was resistant to serogroup B phage 52A, 80, and 11, and the phage susceptibility of the mutant could be restored by complementation with a plasmid vector expressing tarM (right). The lytic phage K, which does not belong to the serogroup B of temperate phages, was used as a control.
To verify that the phage resistance phenotype is due to disruption of tarM, we transformed mutant K6 with a complementation plasmid pRB474-tarM containing tarM under the control of a constitutive promoter. The complemented mutant was susceptible to serogroup B phage 11, 80, and 52A (Fig. 2B, right), confirming that an intact tarM is required for rendering S. aureus RN4220 susceptible to serogroup B phages, possibly by transferring α-GlcNAc residues to the WTA backbone.
tarM Disruption Leads to Altered WTA That Lacks the α-GlcNAc Residues
To determine the presence or absence of GlcNAc on the WTA backbone of the tarM mutant K6, we extracted and analyzed WTAs from mutant and parental strain. Although the PAGE migration behaviors of WTA from wild-type (Fig. 3A, lane 1) and tarM-complemented mutant strain (lane 3) were almost the same, the tarM mutant WTA migrated much faster (lane 2) and exhibited weaker staining with Alcian blue-silver solution. This finding is in agreement with the anticipated reduction of the WTA mass by almost 50% in the absence of GlcNAc modification, thereby supporting the assumption that disruption of tarM led to the loss of WTA GlcNAc residue.
FIGURE 3.
tarM disruption leads to altered WTA that lacks the α-GlcNAc residues. A, PAGE analysis of WTA is shown. The indicated WTA samples (100 nmol of phosphate/lane) were resolved in polyacrylamide gels and visualized by Alcian blue silver staining. The PAGE migration behaviors of WTA from wild type (lane 1) and tarM-complemented mutant strain (lane 3) were almost the same; the tarM mutant WTA migrated much faster (lane 2) and exhibited weaker staining with Alcian blue-silver solution. B, purified WTA spotted on a nitrocellulose membrane was incubated with GlcNAc binding biotinylated WGA and visualized with streptavidin-HRP. In contrast to wild-type WTA, mutant WTA could not be stained with WGA, indicating the loss of GlcNAc from the K6 WTA backbone. C, 1H NMR spectrum of WTA from RN4220 derivatives is shown. The two characteristic signals for GlcNAc at δ 5.07 and 2.08 (anomeric proton of α-GlcN and methyl group of NAc, respectively) were found and are indicated in the spectra of wild type and the tarM complement (K6/tarM) WTA but were missing in the spectrum of mutant K6 WTA, indicating that disruption of tarM in the mutant K6 leads to the loss of the α-GlcNAc from WTA and that α-GlcNAc modification could be restored by complementation with a plasmid encoding tarM.
Wild-type WTA (Fig. 3B, left) could be stained with WGA that specifically binds to GlcNAc in a dot blot lectin overlay analysis, whereas the K6 mutant WTA did not react with WGA, further supporting the absence of GlcNAc substitution in the WTA of mutant K6.
WTA samples were further purified by ion-exchange chromatography and analyzed by 1H NMR spectroscopy. As clearly demonstrated in Fig. 3C, the two fingerprint signals of α-GlcNAc, i.e. the anomeric proton at δ 5.07 and NAc at δ 2.08 (marked with vertical dotted lines) were present in the spectra of wild-type and tarM-complemented K6 WTA but were completely absent from the NMR spectrum of mutant K6 WTA. In agreement with this finding, the GlcN/phosphate ratio decreased from 0.63 in the case of wild type to 0.04 for K6, whereas this ratio increased again to 0.68 in the complement mutant. These data demonstrate that the disruption of tarM in mutant K6 leads to the loss of α-GlcNAc from WTA and that complementation with tarM restores the WTA α-GlcNAc modification.
Recombinant TarM Has WTA GlcNAc-transferase Activity
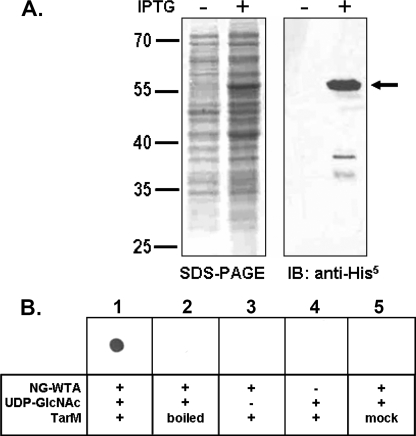
To verify in vitro that TarM represents a WTA GlcNAc-transferase, we produced recombinant TarM with an N-terminal His6 tag by cloning the gene in plasmid pET28a and expressing in E. coli BL21(DE3). Upon isopropyl β-d-1-thiogalactopyranoside induction, a prominent protein band with the expected mass of recombinant TarM (55 kDa) could be observed in SDS-PAGE after Coomassie Blue staining (Fig. 4A, left panel) or Western blot analysis with a monoclonal His tag-specific antibody (right panel), indicating successful recombinant expression of TarM.
FIGURE 4.
In vitro characterization of the enzymatic activity of recombinant TarM. A, expression of recombinant TarM with a calculated molecular mass of 55 kDa was induced by isopropyl β-d-1-thiogalactopyranoside (IPTG), and TarM could be detected (as indicated by an arrow) in SDS-PAGE by Coomassie Blue staining (left) and immunoblot (IB) analysis with an anti-His tag antibody (right). B, WGA lectin overlay analysis of enzymatic products of TarM is shown. The enzymatic reaction system contained the crude TarM-containing E. coli lysates (TarM), non-glycosylated WTA (NG-WTA) as sugar acceptor, and UDP-GlcNAc as sugar donor. The enzymatic product was spotted on the nitrocellulose membrane and analyzed by lectin overlay with biotinylated WGA plus streptavidin-HRP. Cell lysate from E. coli BL21(DE3) transformed with the empty vector were included as mock control (column 5).
The crude TarM-containing cell extract was incubated with non-glycosylated WTA as the sugar acceptor and UDP-GlcNAc as the activated sugar donor. Then the enzymatic product was spotted on nitrocellulose membranes and analyzed by lectin overlay with WGA. As shown in Fig. 4B the enzymatic product could be stained with WGA, indicating the presence of GlcNAc on the WTA polymer (1). However, in samples where the enzyme had been boiled (2) or one of the substrate was missing (3 and 4) or when lysate from E. coli with the empty vector was used (5), the enzymatic product did not react with WGA. These data indicate that recombinant TarM can transfer GlcNAc from the sugar donor UDP-GlcNAc to the acceptor non-glycosylated WTA. Based on our data on in vivo and in vitro activity of TarM, we conclude that TarM is a WTA α- GlcNAc-transferase that does not require further S. aureus proteins to function.
A Point Mutation in tarM Is Responsible for the Phage-resistant Phenotype of S. aureus H Mutant 52B2
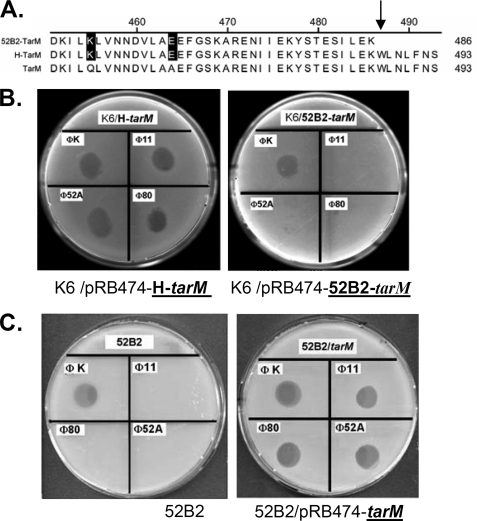
To determine whether the phage resistance phenotype of the previously described mutant 52B2 (20) is also due to mutation of tarM, we sequenced the gene from parental and mutant strains. The deduced amino acid sequence of strain H TarM (H-TarM) was identical to that of RN4220 except for exchanges of Gln-455 and Ala-464 with Lys and Glu residues, respectively (Fig. 5A). Of note, tarM from mutant 52B2 contained a G to A point mutation at position 1461 of the open reading frame, which turns the codon for Trp-487 into a premature stop codon and leads to a truncated TarM that lacks the last seven amino acids (Fig. 5A).
FIGURE 5.
Characterization of TarM from mutant 52B2 derived by chemical mutagenesis of S aureus strain H. A, sequence alignment of TarM from strain H mutant 52B2 (52B2-TarM), strain H wild type (H-TarM), and strain RN4220 wild type (TarM) is shown. A G to A point mutation leads to a seven amino acids deletion at the C terminus of 52B2-TarM as indicated by an arrow. B, shown is a phage soft-agar assay with RN4220 mutant K6 complemented with plasmids encoding H-TarM or 52B2-TarM. Complementation with H-tarM renders K6 susceptible to temperate phages 11, 52A, and 80 but not complementation with 52B2-tarM, indicating the C-terminal seven amino acids of TarM are essential for the in vivo activity of TarM. The lytic phage K, which does not belong to the serogroup B of temperate phages was used as a control. C, complementation of mutant 52B2 with a plasmid encoding the RN4220 TarM restores susceptibility to phages 11, 52A, and 80.
To prove the relevance of the C-terminal deletion for functionality of TarM, the tarM genes from S. aureus H and 52B2 were amplified and cloned in expression vector pRB474, and the resulting plasmids were used to transform the RN4220 tarM mutant K6. Complementation with tarM from strain H rendered K6 sensitive to serogroup B phages (Fig. 5B, left), indicating the capacity of H-TarM to modify WTA with GlcNAc. However, tarM from mutant 52B2 did not restore the phage susceptibility pattern (Fig. 5B, right), indicating that loss of the seven C-terminal amino acids of TarM abolishes its WTA GlcNAc-transferase activity. In agreement with this notion, mutant 52B2 was rendered phage-susceptible again when it was transformed with the plasmid expressing tarM from RN4220 (Fig. 5C, right). Thus, the point mutation in tarM explains the phage resistance phenotype of mutant 52B2, which provides further evidence for the crucial role of TarM in WTA modification with GlcNAc and for the essential function of this modification in the ability of serogroup B phages to infect S. aureus.
DISCUSSION
Although substantial progress has been made in understanding why most Gram-positive bacteria produce WTA or related cell wall glycopolymers, the reasons for the enormous species- and strain-specific differences in WTA structures have remained largely obscure. The enzyme systems involved in biosynthesis of WTA backbones with Rbo-P or glycerol phosphate repeating units and subsequent modification with d-alanine have been elucidated (7, 46, 47) and represent a basis for generating defined panels of mutants or identifying the most promising target enzymes for new antibacterial compounds.
Our study describes a missing module that further completes the picture of the WTA biosynthetic pathway. TarM is the first described enzyme catalyzing the glycosylation of the Rbo-P-type WTA and the first WTA glycosyltransferase whose function has been confirmed in vitro via a recombinant approach. Moreover, we present for the first time a detailed NMR spectrum of Rbo-P-type WTA with α-GlcNAc substitution from S. aureus. Together with the previously published NMR spectrum of Rbo-P WTA with β-GlcNAc substitution from strain MN8m (6) our study forms a solid basis for dissecting the structure and function of glycosylated WTA in S. aureus.
Although WTA modification with d-alanine occurs extracellularly upon translocation of the polymer across the cytoplasmic membrane, it has remained unclear when and how WTA is modified with sugar residues. The fact that TarM lacks a signal peptide and is a soluble protein indicates that it remains in the cytoplasm. Moreover, its dependence on the activated sugar donor UDP-GlcNAc, which is not available in the cell envelope, strongly supports the notion that WTA glycosylation by TarM takes place before the lipid-linked WTA polymer is translocated to the outer leaflet of the cytoplasmic membrane.
TarM was active in vitro in the absence of any other WTA-biosynthetic enzyme, and it acted on polymeric WTA not connected to C55-P (undecaprenyl monophosphate). Thus, interaction with Rbo-P appears to be sufficient for TarM to glycosylate the acceptor substrate. It remains to be analyzed whether TarM acts independently of the rest of the WTA biosynthetic machinery whose components have been shown to form a multienzyme complex in B. subtilis (48) or whether TarM may interact with the WTA polymerase TarL or TarK (27, 28) in vivo.
Of note, the fact that the tarM mutants K6 and 52B2 had no apparent growth defects and produced normal amounts of WTA (data not shown) indicates that the WTA translocase (probably formed by TagG and TagH) and the unknown ligase that links the polymer to peptidoglycan are not affected in their efficiency by the presence or absence of GlcNAc on the repeating units. This represents a difference to the teichoic acid translocase of Streptococcus pneumoniae, which has recently been shown to be unable to translocate polymers lacking the typical phosphocholine substituents (49, 50).
TarM is related to the group 1 family of glycosyltransferases, known activities of which include glucosyltransferases, rhamnosyltransferases, galatosyltransferases, and glucuronosyltransferases (Carbohydrate Active Enzymes data base) (51). Surprisingly, TarM does not share similarity with any mammalian, plant, or bacterial GlcNAc-transferase with experimentally proven activity and seems to represent a new type of GlcNAc-transferase. Moreover, we could identify proteins with clear homology to TarM in several other Gram-positive bacteria (Fig. 6) indicating that TarM is representative of a generally used WTA-biosynthetic enzymes. Of note, related proteins were absent from some bacteria known to produce glycosylated Rbo-P WTA such as Staphylococcus saprophyticus and Listeria monocytogenes, indicating that some bacteria must have alternative ways to glycosylate Rbo-P WTA.
FIGURE 6.
Sequence alignment of TarM and related proteins from other bacterial species. TarM (protein data base UniProtKB ID Q5HH49) was aligned with Bw23 (Protein ID B7ZDM0) from B. subtilis W23, Lgr (Protein ID C2C429) from L. grayi, SepA (Protein ID C5QPJ4) and SepB (Protein ID Q5HLD3) from S. epidermidis, Scap (Protein ID B9CSR7) from S. capitis, Swa (Protein ID C4W875) from S. warneri, Scan (Protein ID B9DJZ6) from Staphylococcus carnosus, and BclA (Protein ID Q5WHX5) and BclB (Protein ID Q5WG79) from B. clausii. Identical and similar amino acids at a given position are shown as black or gray boxes, respectively.
Probing the non-redundant protein data base UniProtKB with TarM, we found the most similar protein (excluding proteins from S. aureus) to be a putative glycosyltransferase with ID number C2C429 from Listeria grayi, whose WTA structure has never been elucidated. This protein shares 32% identity and 60% similarity with TarM over its entire length (Fig. 6). Surprisingly, we could not find TarM homologues in any other Listeria species, indicating that WTA glycosylation is also a species-specific process in the genus Listeria. Nevertheless, several genes involved in WTA glycosylation have previously been identified in different L. monocytogenes serotype 4b strains, whose WTA is composed of repeating units of →4)-β-d-GlcNAc-(1,4-Rbo-P)-(1→ disaccharides, where the GlcNAc can be further substituted with galactose or glucose at the 6-OH or 3-OH groups of GlcNAc, respectively (52). These genes encode the putative glycosyltransferases GltA, GltB (53), and GtcV (54) and a protein of unknown function (GtcA (55)), none of which shares similarity with TarM.
The WTA from B. subtilis strain W23 is a glucose-substituted Rbo-P polymer (22) with a similar structure as the one described in this study for S. aureus strain RN4220 except for the sugar substitution. We identified a TarM homologue with protein data base UniProtKB accession number B7ZDM0 in B. subtilis W23 that shares 29% identity and 60% similarity with RN4220 TarM. Because its gene is located in the vicinity of the WTA and teichuronic acid gene cluster of B. subtilis W23, it is likely to be a glucosyltransferase involved in the transfer of glucose residues to the WTA backbone. The TagE protein (also called GtaA and RodD) of B. subtilis 168 has been implicated in the transfer of glucose residues to glycerol phosphate WTA by indirect genetic evidence (56, 57). The size of TagE is bigger and shares also weak similarity with TarM.
In addition, less well conserved homologues of TarM were found in Staphylococcus epidermidis, Staphylococcus capitis, Staphylococcus warneri, and Bacillus clausii, whose WTA molecules are composed of either glycerol phosphate, of other none Rbo-P repeating units (4), or have not yet been analyzed. Sequence comparison of these TarM homologous proteins yielded similarities over the entire lengths of the proteins (Fig. 6), suggesting that the specificity for the Rbo-P acceptor substrate is governed by only subtle differences in primary or tertiary structure of TarM-related enzymes. Future elucidation of the three-dimensional structure of TarM will help to understand the molecular basis of substrate specificity and catalysis of TarM.
Previous studies have demonstrated that certain S. aureus strains produce WTA with either only α-GlcNAc (strain 3528) or β-GlcNAc (strain Duncan) or with both types of linkages (strain Copenhagen) (17, 41). Of note, these differences have critical consequences for the capacity of WTA to elicit specific immune responses. For example, Juergens et al. (16) found that WTA of S. aureus Copenhagen elicited antibodies in rabbits that were only directed to α-GlcNAc-modified Rbo-P but not to the β-linked epitopes. It is tempting to speculate that the modification of WTA with GlcNAc in different conformations may contribute to the immune evasion strategies of S. aureus that limit the ability of host organisms to raise a protective immune response. In addition, it seems that the WTA GlcNAc residues contribute to staphylococcal adhesion to epithelial and endothelial cells via interaction with host lectin-like receptor in a protein-carbohydrate interactions-involving process (8, 12, 58), which is reminiscent of the mechanism of phage binding to S. aureus cells. Future studies will help to identify the binding partners and roles of the WTA GlcNAc substituents in Staphylococcus-host interaction.
In conclusion, our study closes a critical gap in the knowledge on WTA biosynthesis and represents a good basis for future studies on the role of WTA glycosylation in microbe-host interaction. With the increasing consideration of phages as antimicrobial agents against severe pathogens such as S. aureus, it becomes more and more important to understand the molecular basis of phage adsorption and infection. In this respect our results may be of critical importance for the identification of receptor-binding proteins and lytic enzymes from staphylococcal phages that might be used as antimicrobial agents.
Acknowledgments
We thank Cordula Gekeler, Nadja Gommer, Katharina Jakob, and Herman Moll for technical assistance. We are also grateful to Prof. James T. Park from Tufts University for providing the S. aureus strain H and mutant 52B2.
This work was supported by German Research Foundation Grants TR-SFB34, SFB766, and GKR685 and by the German Ministry of Education and Technology (SkinStaph) (to A. P.) and also in part by the National Institutes of Health Intramural Research Program of NIAIDZIA AI000904-08 (to M. L. and M. O.).
- WTA
- wall teichoic acid
- Tag
- teichoic acid glycerol phosphate
- Tar
- teichoic acid ribitol phosphate type
- Rbo-P
- polyribitol phosphate
- WGA
- wheat germ agglutinin
- MES
- 2-(4-morpholino)-ethane-sulfonic acid
- COSY
- 1H,1H correlation spectroscopy
- TOCSY
- total correlation spectroscopy
- ROESY
- rotating-frame nuclear Overhauser enhancement spectroscopy
- HSQC-DEPT
- 13C,1H single quantum correlation distortionless enhancement by polarization transfer
- HMBC
- heteronuclear multiple bond correlation experiment
- 1H,31P HMQC-TOCSY
- 1H,31P multiple quantum correlation-total correlation spectroscopy
- HRP
- horseradish peroxidase
- GLC
- gas-liquid chromatography
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1.Clarke S. R., Foster S. J. (2006) Adv. Microb. Physiol. 51, 187–224 [DOI] [PubMed] [Google Scholar]
- 2.Weidenmaier C., Peschel A. (2008) Nat. Rev. Microbiol. 6, 276–287 [DOI] [PubMed] [Google Scholar]
- 3.Xia G., Kohler T., Peschel A. (2010) Int. J. Med. Microbiol. 300, 148–154 [DOI] [PubMed] [Google Scholar]
- 4.Endl J., Seidl H. P., Fiedler F., Schleifer K. H. (1983) Arch. Microbiol. 135, 215–223 [DOI] [PubMed] [Google Scholar]
- 5.Sanderson A. R., Strominger J. L., Nathenson S. G. (1962) J. Biol. Chem. 237, 3603–3613 [PubMed] [Google Scholar]
- 6.Vinogradov E., Sadovskaya I., Li J., Jabbouri S. (2006) Carbohydr. Res. 341, 738–743 [DOI] [PubMed] [Google Scholar]
- 7.Neuhaus F. C., Baddiley J. (2003) Microbiol. Mol. Biol. Rev. 67, 686–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weidenmaier C., Kokai-Kun J. F., Kristian S. A., Chanturiya T., Kalbacher H., Gross M., Nicholson G., Neumeister B., Mond J. J., Peschel A. (2004) Nat. Med. 10, 243–245 [DOI] [PubMed] [Google Scholar]
- 9.D'Elia M. A., Pereira M. P., Chung Y. S., Zhao W., Chau A., Kenney T. J., Sulavik M. C., Black T. A., Brown E. D. (2006) J. Bacteriol. 188, 4183–4189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vergara-Irigaray M., Maira-Litrán T., Merino N., Pier G. B., Penadés J. R., Lasa I. (2008) Microbiology 154, 865–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schlag M., Biswas R., Krismer B., Kohler T., Zoll S., Schwarz H., Yu W., Peschel A., Gotz F. (2010) Mol. Microbiol. 75, 864–873 [DOI] [PubMed] [Google Scholar]
- 12.Weidenmaier C., Peschel A., Xiong Y. Q., Kristian S. A., Dietz K., Yeaman M. R., Bayer A. S. (2005) J. Infect. Dis. 191, 1771–1777 [DOI] [PubMed] [Google Scholar]
- 13.Kohler T., Weidenmaier C., Peschel A. (2009) J. Bacteriol. 191, 4482–4484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peschel A., Otto M., Jack R. W., Kalbacher H., Jung G., Götz F. (1999) J. Biol. Chem. 274, 8405–8410 [DOI] [PubMed] [Google Scholar]
- 15.Peschel A., Vuong C., Otto M., Götz F. (2000) Antimicrob. Agents Chemother. 44, 2845–2847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Juergens W. G., Sanderson A. R., Strominger J. L. (1963) J. Exp. Med. 117, 925–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nathenson S. G., Ishimoto N., Anderson J. S., Strominger J. L. (1966) J. Biol. Chem. 241, 651–658 [PubMed] [Google Scholar]
- 18.Torii M., Kabat E. A., Bezer A. E. (1964) J. Exp. Med. 120, 13–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chatterjee A. N., Mirelman D., Singer H. J., Park J. T. (1969) J. Bacteriol. 100, 846–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shaw D. R., Mirelman D., Chatterjee A. N., Park J. T. (1970) J. Biol. Chem. 245, 5101–5106 [PubMed] [Google Scholar]
- 21.Young F. E. (1967) Proc. Natl. Acad. Sci. U.S.A. 58, 2377–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glaser L., Ionesco H., Schaeffer P. (1966) Biochim. Biophys. Acta 124, 415–417 [DOI] [PubMed] [Google Scholar]
- 23.Wendlinger G., Loessner M. J., Scherer S. (1996) Microbiology 142, 985–992 [DOI] [PubMed] [Google Scholar]
- 24.Schertzer J. W., Brown E. D. (2003) J. Biol. Chem. 278, 18002–18007 [DOI] [PubMed] [Google Scholar]
- 25.Bhavsar A. P., Truant R., Brown E. D. (2005) J. Biol. Chem. 280, 36691–36700 [DOI] [PubMed] [Google Scholar]
- 26.Ginsberg C., Zhang Y. H., Yuan Y., Walker S. (2006) ACS Chem. Biol. 1, 25–28 [DOI] [PubMed] [Google Scholar]
- 27.Brown S., Zhang Y. H., Walker S. (2008) Chem. Biol. 15, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meredith T. C., Swoboda J. G., Walker S. (2008) J. Bacteriol. 190, 3046–3056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perego M., Glaser P., Minutello A., Strauch M. A., Leopold K., Fischer W. (1995) J. Biol. Chem. 270, 15598–15606 [DOI] [PubMed] [Google Scholar]
- 30.Debabov D. V., Heaton M. P., Zhang Q., Stewart K. D., Lambalot R. H., Neuhaus F. C. (1996) J. Bacteriol. 178, 3869–3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nathenson S. G., Strominger J. L. (1963) J. Biol. Chem. 238, 3161–3169 [PubMed] [Google Scholar]
- 32.Li M., Rigby K., Lai Y., Nair V., Peschel A., Schittek B., Otto M. (2009) Antimicrob. Agents Chemother. 53, 4200–4210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Flaherty S., Ross R. P., Meaney W., Fitzgerald G. F., Elbreki M. F., Coffey A. (2005) Appl. Environ. Microbiol. 71, 1836–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pantůcek R., Doskar J., Růzicková V., Kaspárek P., Orácová E., Kvardová V., Rosypal S. (2004) Arch. Virol. 149, 1689–1703 [DOI] [PubMed] [Google Scholar]
- 35.Pollack J. H., Neuhaus F. C. (1994) J. Bacteriol. 176, 7252–7259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brückner R. (1992) Gene 122, 187–192 [DOI] [PubMed] [Google Scholar]
- 37.Gerwig G. J., Kamerling J. P., Vliegenthart J. F. (1979) Carbohydr. Res. 77, 10–17 [DOI] [PubMed] [Google Scholar]
- 38.Strominger J. L., Park J. T., Thompson R. E. (1959) J. Biol. Chem. 234, 3263–3268 [PubMed] [Google Scholar]
- 39.Lowry O. H., Roberts N. R., Leiner K. Y., Wu M. L., Farr A. L., Albers R. W. (1954) J. Biol. Chem. 207, 39–49 [PubMed] [Google Scholar]
- 40.Jenni R., Berger-Bächi B. (1998) Arch. Microbiol. 170, 171–178 [DOI] [PubMed] [Google Scholar]
- 41.Nathenson S. G., Strominger J. L. (1962) J. Biol. Chem. 237, 3839–3841 [PubMed] [Google Scholar]
- 42.Chatterjee A. N. (1969) J. Bacteriol. 98, 519–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park J. T., Shaw D. R., Chatterjee A. N., Mirelman D., Wu T. (1974) Ann. N. Y. Acad. Sci. 236, 54–62 [DOI] [PubMed] [Google Scholar]
- 44.Wann E. R., Dassy B., Fournier J. M., Foster T. J. (1999) FEMS Microbiol. Lett 170, 97–103 [DOI] [PubMed] [Google Scholar]
- 45.Bae T., Glass E. M., Schneewind O., Missiakas D. (2008) Methods Mol. Biol. 416, 103–116 [DOI] [PubMed] [Google Scholar]
- 46.Bhavsar A. P., Brown E. D. (2006) Mol. Microbiol. 60, 1077–1090 [DOI] [PubMed] [Google Scholar]
- 47.Swoboda J. G., Campbell J., Meredith T. C., Walker S. (2010) ChemBioChem 11, 35–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Formstone A., Carballido-López R., Noirot P., Errington J., Scheffers D. J. (2008) J. Bacteriol. 190, 1812–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Damjanovic M., Kharat A. S., Eberhardt A., Tomasz A., Vollmer W. (2007) J. Bacteriol. 189, 7105–7111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vollmer W., Tomasz A. (2001) Mol. Microbiol. 39, 1610–1622 [DOI] [PubMed] [Google Scholar]
- 51.Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) Nucleic Acids Res. 37, D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Uchikawa K., Sekikawa I., Azuma I. (1986) J. Biochem. 99, 315–327 [DOI] [PubMed] [Google Scholar]
- 53.Lei X. H., Fiedler F., Lan Z., Kathariou S. (2001) J. Bacteriol. 183, 1133–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spears P. A., Suyemoto M. M., Palermo A. M., Horton J. R., Hamrick T. S., Havell E. A., Orndorff P. E. (2008) Infect. Immun. 76, 4046–4054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Promadej N., Fiedler F., Cossart P., Dramsi S., Kathariou S. (1999) J. Bacteriol. 181, 418–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mauël C., Young M., Margot P., Karamata D. (1989) Mol. Gen. Genet. 215, 388–394 [DOI] [PubMed] [Google Scholar]
- 57.Honeyman A. L., Stewart G. C. (1989) Mol. Microbiol. 3, 1257–1268 [DOI] [PubMed] [Google Scholar]
- 58.Weidenmaier C., Kokai-Kun J. F., Kulauzovic E., Kohler T., Thumm G., Stoll H., Götz F., Peschel A. (2008) Int. J. Med. Microbiol. 298, 505–513 [DOI] [PubMed] [Google Scholar]