Abstract
Surfactant protein D (SP-D) is an innate immune collectin that recognizes microbes via its carbohydrate recognition domains, agglutinates bacteria, and forms immune complexes. During microbial infections, proteases, such as elastases, cleave the carbohydrate recognition domains and can inactivate the innate immune functions of SP-D. Host responses to counterbalance the reduction of SP-D-mediated innate immune response under these conditions are not clearly understood. We have unexpectedly identified that SP-D could interact with protein fractions containing ovomucin and ovomacroglobulin. Here, we show that SP-D interacts with human α2-macroglobulin (A2M), a protease inhibitor present in the lungs and serum. Using enzyme-linked immunosorbent assays, surface plasmon resonance, and carbohydrate competition assays, we show that SP-D interacts with A2M both in solid phase (KD of 7.33 nm) and in solution via lectin-carbohydrate interactions under physiological calcium conditions. Bacterial agglutination assays further show that SP-D·A2M complexes increase the ability of SP-D to agglutinate bacteria. Western blot analyses show that SP-D, but not A2M, avidly binds bacteria. Interestingly, intact and activated A2M also protect SP-D against elastase-mediated degradation, and the cleaved A2M still interacts with SP-D and is able to enhance its agglutination abilities. We also found that SP-D and A2M can interact with each other in the airway-lining fluid. Therefore, we propose that SP-D utilizes a novel mechanism in which the collectin interacts with protease inhibitor A2M to decrease its degradation and to concurrently increase its innate immune function. These interactions particularly enhance bacterial agglutination and immune complex formation.
Keywords: Carbohydrate-binding Protein, Innate Immunity, Lung, Pattern Recognition Receptor, Protease Inhibitor, Surface Plasmon Resonance (SPR), alpha-2-Macroglobulin, Bacterial Agglutination, Collectin, Surfactant Protein D
Introduction
Microorganisms that enter the body will encounter the components of the innate immune system, which mount the first line of defense against pathogens. Both soluble and membrane-associated innate immune pattern-recognition receptors recognize the pathogen-associated molecular patterns present on various microbes (1–4). Most microbial surfaces are coated with an array of carbohydrates, and soluble pattern-recognition receptors such as collectins specifically recognize those carbohydrate patterns. Although each microorganism has a unique array of structural components, collectins can recognize a variety of sugars and have the flexibility to bind different molecular patterns present on most microbes; therefore, they work as broad spectrum, high specificity opsonins (1, 3, 5). Recognition results in opsonization and/or agglutination of the microbes thereby increasing the formation of immune complexes, reducing the growth and multiplication of microbial pathogens, and facilitating clearance (1, 4).
Surfactant protein D (SP-D)2 is a well characterized innate immune collectin (1, 5, 6) that is present in lung surfactant and many other mucosal surfaces (7–9). It is a hydrophilic, soluble pattern-recognition receptor composed of a disulfide bond forming an amino-terminal segment, a fibrillar collagen-like region, trimerizing hydrophobic neck domain, and a globular head that contains a carbohydrate recognition domain (CRD). These 43-kDa SP-D monomers form stable trimers and further assemble as an X or large asterisk-like oligomers (1, 3). The CRDs of SP-D interact with a variety of carbohydrate moieties present on the surfaces of microbes in a calcium-dependent manner (6, 10). The oligomeric forms of the protein allow SP-D to generate many low affinity interactions between its CRDs and the carbohydrate structures on the surface of the pathogens, thereby allowing the collectin to increase its avidity to effectively agglutinate several microbes. For example, SP-D is able to bind to bacterial cell wall components such as peptidoglycan and lipoteichoic acid, which are the primary ligands present on Gram-positive bacteria (2). SP-D also interacts with the lipopolysaccharide present on Gram-negative bacteria such as Escherichia coli and promotes agglutination (10–13). However, during infection, proteolytic enzymes such as elastases are secreted into the lungs both by newly recruited neutrophils (14) and bacterial pathogens (15). These proteases cleave the CRDs and neutralize the functions of collectins (14–16). How the host counteracts the reduction in the innate immune functions under these situations is not clearly understood.
During bacterial infection and lung inflammation, large amounts of serum proteins and immune cells enter the lungs (17–19). α2-Macroglobulin (A2M) is present in low concentrations in the normal lungs (0.09–2.02 μg/ml) (18, 19), and its concentration increases more than 100-fold during infection and inflammation (10.9–220 μg/ml) (18–20). The bronchoalveolar lavage fluid (BALF) concentration of A2M can range between 0.03 and 8.12 μg/ml in cystic fibrosis patients (20). Native A2M is a 720-kDa glycoprotein composed of four identical subunits linked as pairs by disulfide bonds (21–23). When A2M interacts with thiol, serine, aspartic acid, and metalloproteases, it allows cleavage of its “bait region” present within the tetrameric cage (24). The conformational change induced by cleavage of the thiol ester bond or proteolysis traps the enzyme within its cage (25, 26). The conformational change also renders the A2M to be recognized by the A2M receptor (LRP-1, CD91) (27, 28). Each subunit of A2M has eight N-linked glycosylation sites that decorate the surface of the protein, and the serum collectin mannose-binding lectin recognizes the mannose residues present on A2M (22). However, its functional consequences are not clearly understood.
We have serendipitously identified that SP-D could bind to chromatographic fractions that contain ovomucin and ovomacroglobulin. Therefore, we hypothesized that SPD would bind to human A2M and that A2M would protect the collectin from proteolytic degradation by elastase. This study establishes the interaction between these two proteins. Interestingly, we have also identified that A2M enhances the function of SP-D, particularly to agglutinate bacteria. Both intact and cleaved A2M support this novel function. Therefore, our data suggest that the host may have a unique mechanism to boost the SP-D-mediated innate immune response via A2M-SP-D interactions during bacterial infection.
EXPERIMENTAL PROCEDURES
Reagents
All reagents were obtained from Sigma unless otherwise stated. Custom antibodies against SP-D(GXY)8(n/CRD) (henceforth referred to as SP-D(n/CRD) were generated in rabbits by Cocalico Biologicals, Inc. (Reamstown, PA). Polyclonal rabbit IgG was purified by Na2SO4 precipitation and protein G affinity chromatography as described by the manufacturer (GE Healthcare). Briefly, the IgG that bound to the affinity column was eluted with glycine buffer (pH 2.5), and the eluate was collected in vials containing 1 m Tris buffer (pH 7.4). Antibodies were further purified by Superose 6 (10 × 300 mm; GE Healthcare) gel filtration column chromatography in phosphate-buffered saline and then stored frozen in aliquots at −20 °C.
SP-D
Native SP-D was purified from therapeutic BALF from a pulmonary alveolar proteinosis patient as described previously (29, 30). Briefly, BALF supernatant was incubated with maltose-agarose beads in the presence of 10 mm CaCl2 (TSC: 20 mm Tris-HCl (pH 7.4), 150 mm NaCl, 10 mm CaCl2) to capture the SP-D. Nonspecifically bound components were removed by washing the beads with 1 m NaCl in the binding buffer (20 mm Tris-HCl (pH 7.4), 1 m NaCl, 10 mm CaCl2). SP-D was then eluted using a Tris buffer containing manganese chloride (20 mm Tris-HCl (pH 7.4), 100 mm MnCl2). SP-D was further purified using a Superose 6 (10 × 300 mm) gel filtration column on an AKTA fast protein liquid chromatography system (GE Healthcare) in HSE (50 mm HEPES (pH 7.4), 150 mm NaCl, 2 mm EDTA). The recombinant neck and head domain of SP-D, SP-D(n/CRD), was expressed in E. coli and purified as described previously (31).
Ovalbumin and A2M Purification
OVA (grade III, 500 μg) was injected into the Superdex 200 size exclusion column (10 × 300 mm, GE Healthcare) on the AKTA fast protein liquid chromatography system in HSE buffer (50 mm HEPES (pH 7.4), 150 mm NaCl, 2 mm EDTA) and fractions were collected. A fraction eluted at 15.8 ml was further purified by injection in low salt HEPES buffer (50 mm HEPES (pH 7.4), 25 mm NaCl, 2 mm EDTA) into a Mono Q anion exchange column (5 × 50 mm; GE Healthcare) and eluted using a salt gradient in 50 mm HEPES and 2 mm EDTA (25–1000 mm NaCl, over 30 min at a flow rate of 0.5 ml/min). Fractions from both purifications were subjected to SDS-PAGE under reducing and denaturing conditions and stained with Bio-Safe Coomassie Blue (Bio-Rad). Selected bands on the gels were analyzed by mass spectrometry at the Advanced Protein Technology Centre at the Hospital for Sick Children.
Similarly, A2M (>98% purity; Sigma) was injected into the Superose 6 size exclusion column (10 × 300 mm, GE Healthcare) on the AKTA fast protein liquid chromatography system in HS buffer (50 mm HEPES (pH 7.4), 150 mm NaCl), and fractions were collected. Fractions from the purification were subjected to SDS-PAGE under reducing and denaturing conditions and stained with Silver Stain Plus (Bio-Rad).
Co-purification of SP-D and A2M
To determine whether SP-D and A2M interact in a physiological setting, SP-D was purified from the BALF as described above. Samples from multiple stages of the purification (i.e. before purification, manganese pool, and Superose 6 elution) were subjected to SDS-PAGE under reducing and denaturing conditions and then transferred to nitrocellulose membrane. The membrane was probed for both SP-D and A2M (anti-A2M; AbD Serotec, Planegg, Germany) followed by chemiluminescent detection.
Methylamine Cleavage of A2M
A2M (minimum 98% purity, Sigma) (2 μg) was incubated with different concentrations (100 to 10 mm) of methylamine in Tris buffer (50 mm Tris (pH 8.0), 85 mm NaCl) for 2 h at room temperature to determine the optimal methylamine concentration (32). Samples were subjected to native PAGE without reducing or denaturing conditions on a 5% (w/v) discontinuous gel in Tris-glycine buffer (25 mm Tris-base, 192 mm glycine) (33). Using the optimal concentration (10 mm) of methylamine, a larger quantity of A2M (500 μg) was cleaved for 2 h at room temperature and then passed through a PD10 desalting column (SephadexTM G-25M, 1.45 × 50 mm, GE Healthcare) in HS buffer (50 mm HEPES (pH 7.4), 150 mm NaCl) to remove excess methylamine and to exchange the buffer.
Deglycosylation of A2M
PNGase F (0.05 units) (from Elizabethkingia meningosepticum, recombinant, expressed in E. coli (Sigma)) was added to A2M (70 μg) and reaction buffer (50 mm Tris-HCl (pH 8.0). The mixture was incubated at 37 °C for 72 h.
Binding of SP-D to OVA or A2M in Solid Phase Assays
Different amounts of OVA (grade III), size column-purified OVA, size column-purified A2M, deglycosylated, or whole A2M in sodium carbonate buffer (15 mm Na2CO3, 35 mm NaHCO3 (pH 9.6)) were coated onto 96-well adsorbent microtiter plates (Maxisorp, Nunc, Rochester, NY) starting with 20 μg/well and following a 2-fold serial dilution. Bovine serum albumin (BSA) was also coated as a negative control, and in some assays, mannan was used as a positive control. Nonspecific binding was blocked using 5% (w/v) BSA in Tris-buffered saline with Tween 20 (Bio-Rad) and CaCl2 (TBST-Ca2+: 20 mm Tris-HCl (pH 7.4), 150 mm NaCl, 0.05% (v/v) Tween 20, 5 mm CaCl2). SP-D (0, 0.5, and 1 μg/ml) was then added to the wells in TBST-Ca2+ and incubated for 1.5 h at 37 °C. Wells were then washed with TBST-Ca2+ buffer, and binding was detected using biotinylated anti-SP-D (1 μg/ml polyclonal IgG) followed by incubation with horseradish peroxidase-conjugated streptavidin (Pierce). The plates were developed with 3,3′,5,5′-tetramethylbenzidine substrate (Bio-Rad); the reaction was stopped with 0.5 m H2SO4, and absorbance was read at 450 nm using Molecular Devices VersaMax microplate reader and SoftMax Pro software (Molecular Devices, Sunnyvale, CA).
Effect of Divalent Cations on SP-D-A2M Interactions
A2M was coated onto ELISA plates at 1 μg/well in carbonate buffer (pH 9.6), and wells were blocked as in the previous binding assays. Wells were then washed with TBST containing 5 mm CaCl2, MgCl2, MnCl2, or EDTA and then were incubated with 1 μg/ml SP-D in TBST with the corresponding buffers for 1.5 h at 37 °C. Binding was detected as above using biotinylated anti-SP-D and horseradish peroxidase-conjugated streptavidin in the corresponding buffers. Similar experiments were conducted to determine the optimal calcium concentration for binding. Buffers containing CaCl2 from 0 to 10 mm were used in each step.
Carbohydrate Inhibition Assays
A fixed amount of A2M (1 μg/well) was coated onto plates as described above, and free surfaces were blocked with BSA. SP-D (1 μg/ml) in the presence of 25 mm d-galactose, d-maltose, d-mannose, or d-fucose was then added, and the plates were incubated at 37 °C for 1.5 h. Detection was as in the first binding assay described above using in TBST-Ca2+ as the buffer.
Surface Plasmon Resonance (SPR)
The ProteOn XPR36 protein interaction array system (Bio-Rad) was used to evaluate the binding between SP-D and A2M. SP-D or SP-D(n/CRD) in acetate buffer (pH 4.5) was immobilized on a GLC chip (Bio-Rad) at concentrations of 0, 5, 10, and 20 μg/ml SP-D or 25 and 50 μg/ml SP-D(n/CRD) via the amine coupling method, as per the manufacturer's instructions. A2M or methylamine-cleaved A2M was passed through a PD10 desalting column to exchange the buffer into HSC-T buffer (50 mm HEPES, 150 mm NaCl, 5 mm CaCl2, 0.005% (v/v) Tween 20) and was then passed over the chip at concentrations of 200–0 μg/ml in a 2-fold dilution at a flow rate of 25 μl/min for 2 min and allowed to disassociate for another 2 min by flowing HSC-T over the chip. The association rate (on-rate; ka), dissociation rate (off-rate; kd), and the dissociation constant (KD = ka/ kd) were calculated by the ProteOn Manager software using the Langmuir binding model for kinetic analysis. The oligomeric molecular masses of A2M, SP-D, and SP-D(n/CRD) were assumed to be 720, 540, and 55.5 kDa, respectively. The monomeric molecular masses for A2M, SP-D and SP-D(n/CRD) were assumed to be 179, 43, and 18.5 kDa, respectively.
Bacterial Agglutination
E. coli Y1088 was grown on LB agar, and then single colonies were grown up in 4 ml of LB broth overnight at 37 °C. Bacteria were then spun down at 1000 × g for 5 min and resuspended in TS (20 mm Tris-HCl (pH 7.4), 150 mm NaCl) and washed in the same buffer three times to remove any soluble bacterial components. Bacteria were suspended in TSC, and the absorbance at 600 nm (A600) was adjusted to 1. Bacteria were then mixed with A2M (0, 5, or 50 μg/ml) for 1 h at 37 °C and then mixed with SP-D (1 μg/ml), and the A600 was read every 15 min for 2 h. Alternatively, SP-D and A2M were preincubated for 1 h at 37 °C before being mixed with the bacteria. Bacteria were also mixed with A2M alone and SP-D alone. This experiment was also repeated using identical amounts of methylamine cleaved A2M, SP-D, and E. coli.
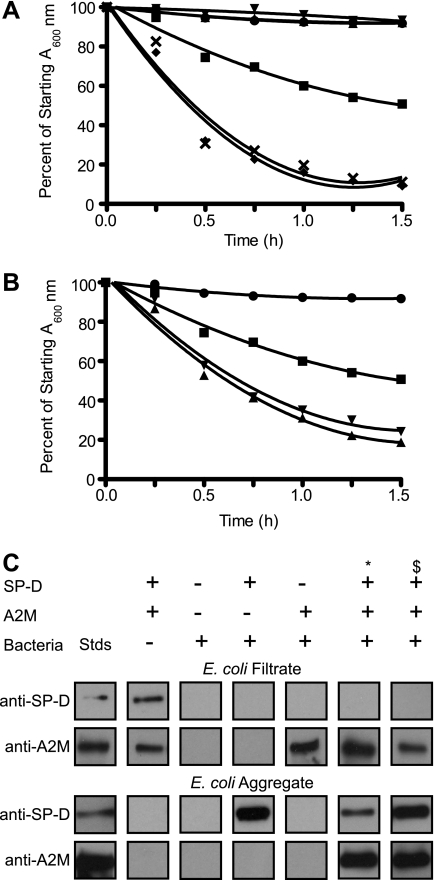
After the agglutination assay, the bacterial samples taken at the 2-h incubation period were passed through a 0.1-μm filter using a Mini-Extruder (Avanti Polar Lipids, Inc., Alabaster, AL) to effectively separate the immune complexes (aggregate) from the free proteins (filtrate). The aggregate was then pushed off the filter in the opposite direction using TSC. Samples were then subjected to SDS-PAGE under reducing and denaturing conditions and transferred to nitrocellulose membrane. After blocking, membranes were incubated with anti-SP-D or anti-A2M, followed by anti-rabbit IgG-horseradish peroxidase and chemiluminescence detection.
Protease Inhibition Assays
SP-D(n/CRD) (2 μg) was incubated with A2M (0–62.5 μg/ml in a 2-fold serial dilution) in TSC buffer (20 mm Tris-HCl (pH 7.4), 150 mm NaCl, 3 mm CaCl2) overnight at 37 °C. Pseudomonas aeruginosa elastase (0.5 unit; Calbiochem) was then added, and the mixture was incubated for 2 h at 37 °C. Samples were then subjected to SDS-PAGE under reducing and denaturing conditions, and gels were stained with Bio-Safe Coomassie Blue. Alternatively, SP-D(n/CRD) (5 μg) was incubated with either intact or methylamine-cleaved A2M (100 μg/ml) in TSC buffer for 1 h at 37 °C and then was incubated with elastase (0.05 unit) for another 2 h at 37 °C. Samples were then subjected to SDS-PAGE as above.
Statistical Analysis
The mean differences were determined by Student's t test. When multiple means are involved, the values were compared with control mean using Dunnett's test. For agglutination assays, nonlinear, second order polynomial regression lines were fit by the least squares method and compared by a sum of squares F-test. The data were analyzed using Prism (GraphPad Software, La Jolla, CA).
RESULTS
SP-D Binds to Fractions Containing Ovomucin and Ovomacroglobulin
As part of another study, we investigated whether SP-D bound the allergen ovalbumin. We separated a grade III OVA preparation on a size exclusion column and then used the resulting eluate in an ELISA-style binding assay (supplemental Fig. S1). We found that grade III OVA is not very pure and produced more than one protein peak. SP-D did not bind to the fractions containing the major OVA peak but did bind avidly to early eluting fractions containing high molecular weight proteins (supplemental Fig. S1, a and b). A sample from under the OVA peak of supplemental Fig. S1a (fraction at 15.8 ml) was further purified via Mono Q ion chromatography, and a sample of pure OVA was tested for SP-D binding (supplemental Fig. S1, c and d). SP-D can bind to the unpurified OVA that contained minor contaminants but not to highly purified OVA (p < 0.01). Protein bands from supplemental Fig. S1b were analyzed via mass spectrometry, and the proteins in the SP-D binding fractions were identified as ovomucin and ovomacroglobulin. Because SP-D binds ovomacroglobulin-containing fractions, we then examined whether the human homolog, α2-macroglobulin, could interact with SP-D.
SP-D Binds Human A2M with High Affinity
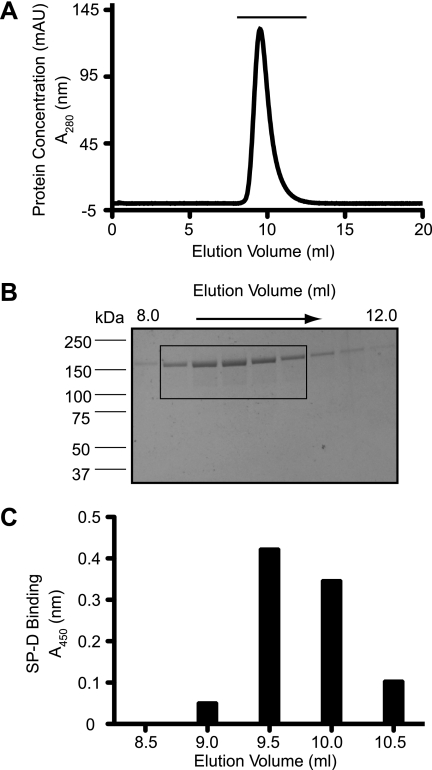
To determine whether SP-D binds A2M, we first performed ELISA-style binding experiments. To be certain about the purity of A2M, and to show that SP-D was binding only to A2M, we size-fractionated a sample of A2M on a size exclusion column (Fig. 1A). Silver staining revealed no other protein bands (Fig. 1B). Fractions from the purification were tested for SP-D binding in an ELISA-style assay. We found that SP-D specifically bound to fractions containing A2M from under the peak (Fig. 1C). ELISA-style binding assays further showed that SP-D (0.5 μg/ml) bound immobilized A2M in a concentration-dependent manner (Fig. 2A) and that it recognizes even small amounts of A2M (0.5 μg/well). Therefore, we consider that SP-D binds well to A2M.
FIGURE 1.
Commercial human A2M has no detectable contaminants. A, human A2M (100 μg; Sigma) was separated on a Superose 6 size exclusion column. There is only one protein peak present, representing the A2M. Fractions of 0.5 ml were collected and then subjected to SDS-PAGE followed by silver staining (fractions from below the horizontal line are represented in B). B, silver-stained SDS-polyacrylamide gel showing proteins from fractions collected at 8.16 ml through 12.16 ml. Each fraction was 0.5 ml. Only A2M from under the peak is visible on the gel. C, SP-D binds only to fractions containing A2M. Fractions from the separation (samples from every 0.5 ml of eluate, boxed lanes in B) were coated on ELISA plates. SP-D was incubated with these samples in the presence of calcium (5 mm), and the amounts of SP-D that bound to these fractions were determined by standard ELISA procedure. SP-D binding profile corresponds to the fractions containing A2M. mAU, milliabsorbance unit.
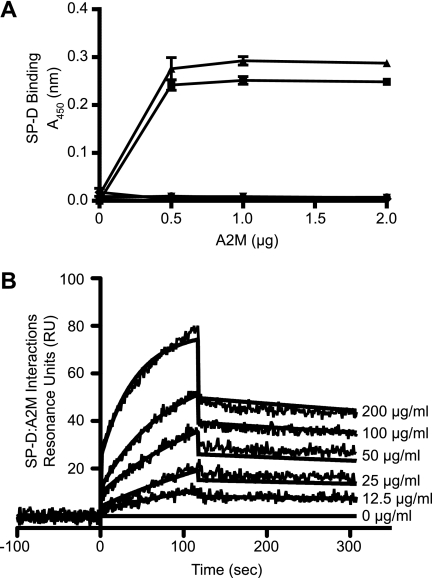
FIGURE 2.
SP-D binds human A2M. A, SP-D binds immobilized A2M. Varying concentrations of A2M and BSA (20 μg/ml, diluted 2-fold) were immobilized onto microtiter plates, and SP-D binding was measured by an ELISA-style assay (A450 nm). ●, A2M; ▾, BSA; ■, A2M + 0.5 μg/ml SP-D; ▴, A2M + 1 μg/ml SP-D; ♦, BSA + 1 μg/ml SP-D. SP-D binds A2M, but not BSA, in a concentration-dependent manner. Control graphs overlap each other and lie close to the x axis. Results are representative of three independent experiments. Error bars indicate S.E. B, SPR analysis shows that immobilized SP-D binds A2M. Surface plasmon resonance sensorgram shows 20 μg/ml SP-D immobilized on a GLC chip binding to soluble A2M (200 μg/ml in a 2-fold serial dilution) in the presence of 5 mm CaCl2. A2M was washed out at 120 s. The KD value for this interaction is 7.33 × 10−9 m indicating interactions can occur even in the nanomolar range. Curves were modeled using the Langmuir binding model. Results are representative of three separate experiments.
To determine the binding kinetics, we used SPR. The SPR analysis was conducted on three different concentrations of immobilized SP-D and six different concentrations of A2M (Fig. 2B). The kinetic parameters for the interaction between A2M and SP-D, assuming the multimeric forms, are as follows: association (on-rate; ka) is 8.33 ± 0.56 × 104 m−1 s−1; the dissociation rate (off-rate; kd) is 6.11 ± 0.26 × 10−4 s−1, and hence, the dissociation constant (KD) is 7.33 × 10−9 m. The data indicate that these two proteins can bind to each other even in nanomolar concentrations. The nanomolar concentrations for SP-D and A2M correspond to 3.95 and 5.28 μg/ml, respectively. The KD value for SP-d-mannan interaction (mannan being a preferential binding partner of SP-D) is also in the nanomolar range (data not shown). These results show that immobilized forms of SP-D and A2M will recognize each other with high affinity.
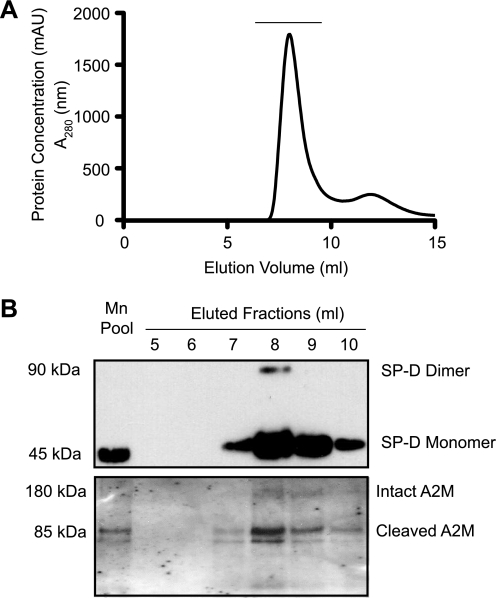
SP-D and A2M Co-purify from BALF
A2M is present in the lungs and can be detected in BALF (18–20). SP-D present in the airways is routinely purified from the BALF (29, 30). Therefore, to determine whether SP-D and A2M interact in vivo, a co-purification of the two proteins was performed. In this procedure, SP-D and SP-D·A2M complexes present in the BALF were captured by a maltose affinity matrix in the presence of calcium. Then, SP-D and SP-D·A2M complexes were selectively eluted by manganese, which preferentially dissociate SP-D-carbohydrate interactions. The eluate was further separated in a gel filtration column (Fig. 3A). Western blot analyses confirm that SP-D and A2M were indeed co-purified from this procedure (Fig. 3B). Notably, most of the A2M detected in these fractions was already activated and cleaved by proteases. Therefore, we consider that SP-D and A2M interact in solution in a physiological setting.
FIGURE 3.
SP-D and A2M co-purify from BALF. A, SP-D present in the BALF was captured by maltose-agarose affinity chromatography in the presence of calcium ions, competitively eluted with manganese ion, and the proteins present in the manganese elution pool (Mn Pool) were further separated by a Superose 6 gel exclusion column. B, Western blot showing that SP-D and A2M co-purify from BALF. Fractions underlined in A (from elution at 5–10 ml, 1-ml fractions) were subjected to Western blots for SP-D and A2M. The major protein present under this peak was SP-D. Intact and cleaved A2Ms were also detected in these fractions. Some surfactant protein A (SP-A) and gp-340 were also seen in the same fractions (data not shown).
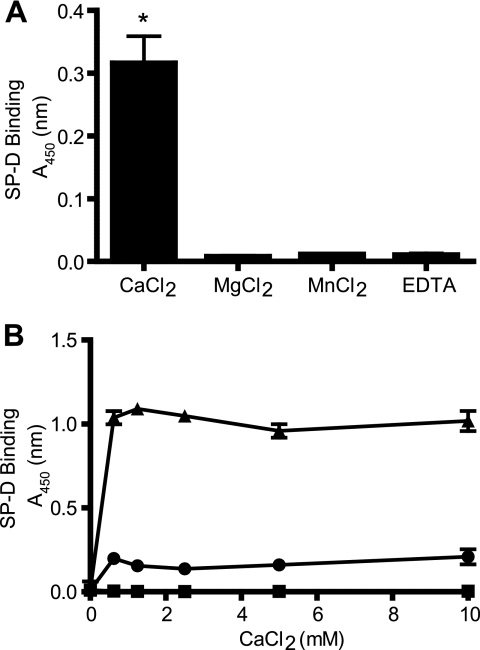
SP-D Binds A2M in a Calcium-dependent Manner
SP-D is known to bind most of its ligands in a calcium-dependent manner (6). Therefore, to determine whether SP-D-A2M interactions also occur in the presence of calcium, ELISA-style binding assays were conducted. SP-D bound well to immobilized A2M in TBST containing 5 mm CaCl2 (Fig. 4A) but not in the presence of other divalent cations (MgCl2 and MnCl2) at the same concentrations. There was also no binding observed in the presence of EDTA, indicating the requirement for the divalent cation calcium (p < 0.01). Subsequently, ELISA-style binding assays were conducted to ascertain the optimal calcium concentration required for SP-D-A2M interactions (Fig. 4B). Different concentrations of calcium (0–10 mm) were used in the buffers, and SP-D-A2M interactions occurred and reached a plateau at 0.625 mm CaCl2. Although the values were high, similar results were observed for SP-D-mannan interactions. Therefore, SP-D-A2M interactions are calcium-dependent and occur at physiological concentrations of this cation.
FIGURE 4.
SP-D binds A2M in a calcium-dependent manner. A, ELISA-style binding assay shows the requirement of calcium ions for SP-D-A2M interaction. A2M was immobilized onto microtiter plates at a concentration of 1 μg/ml. SP-D (0.1 μg/well) was added to blocked wells in a buffer containing 5 mm CaCl2, MgCl2, MnCl2, or EDTA. SP-D bound A2M only in the presence of CaCl2. Results are representative of three independent experiments. SP-D binds well to A2M in the presence of calcium compared with magnesium, manganese, or EDTA containing buffer conditions (*, p < 0.01). B, ELISA-style binding assay shows the optimal CaCl2 concentration required for SP-D-A2M interaction. A2M (2.5 μg/well), BSA (2.5 μg/well), and mannan (0.025 μg/well) were immobilized onto microtiter plates. SP-D (1 μg/ml) was added in TBST containing CaCl2 (0–10 mm in a 2-fold serial dilution). SP-D bound A2M and the positive control mannan, but not to BSA, at calcium concentrations starting at 0.625 mm. ▴, mannan; ●, A2M; ■, BSA. Results are representative of three independent experiments. Error bars indicate S.E.
Carbohydrate Recognition Domain of SP-D Recognizes A2M
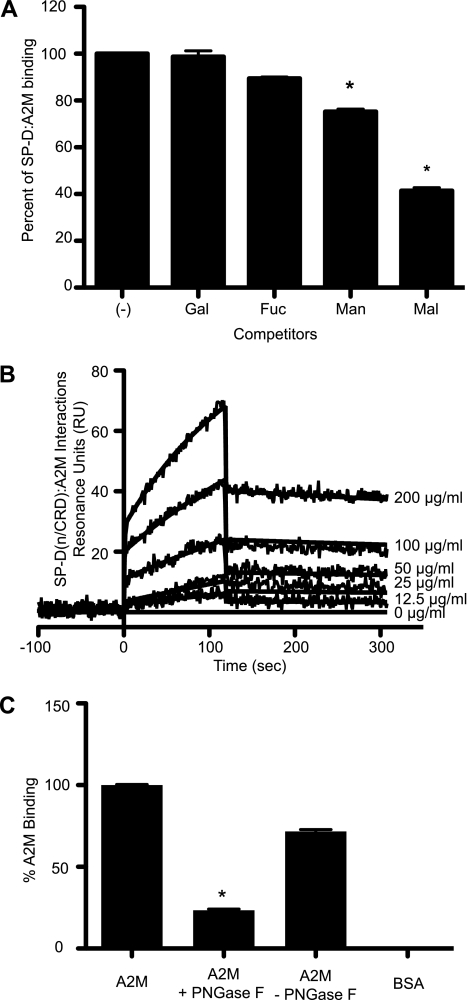
The CRDs of SP-D are known to interact with glycoproteins via protein-protein or protein-carbohydrate interactions (31, 34). To determine the type of interactions occurring, carbohydrate competition assays were performed. In the presence of 25 mm concentrations of d-mannose and d-maltose, to which SP-D binds avidly, the binding of SP-D to A2M decreased as the concentration of the sugars increased (data not shown). At 25 mm concentrations of mannose and maltose, there was significant inhibition of the interactions between SP-D and A2M (Fig. 5A) (p < 0.01). The effect was not seen as strongly with d-galactose and is barely seen with d-fucose. SP-D does not bind well to d-fucose, and thus this sugar was used as a negative control. As expected, d-fucose does not compete SP-D-A2M interactions. Therefore, these results collectively indicate that the CRD of the SP-D is responsible for the interactions with A2M.
FIGURE 5.
SP-D binds A2M through its CRD in a carbohydrate-dependent manner. A, binding of SP-D to A2M can be inhibited by competition with mannose and maltose. A2M (5 μg/well) was coated onto microtiter plates. SP-D (5 μg/ml) was then mixed with 25 mm sugars (galactose (Gal), fucose (Fuc), mannose (Man), or maltose (Mal)) and added to the plates in TBST + 5 mm CaCl2. In the presence of maltose and mannose, SP-D bound significantly less to A2M compared with the condition with no inhibitor (−) control (*, p < 0.01). This indicates that CRDs of SP-D are involved in binding to A2M. B, SPR analysis shows that immobilized SP-D(n/CRD) binds A2M. Surface plasmon resonance sensorgram shows 50 μg/ml SP-D(n/CRD) immobilized on a GLC chip binding to A2M (200 μg/ml in a 2-fold serial dilution) in the presence of 5 mm CaCl2. A2M was washed out at 120 s. The KD for this interaction is 1.57 × 10−8 m. Curves were modeled using the Langmuir binding model. C, A2M was deglycosylated using the enzyme PNGase F as per the manufacturer's instructions at 37 °C for 72 h. Samples of A2M, A2M + PNGase F and reaction buffer, A2M and reaction buffer alone, and BSA were coated on ELISA plates. The ability of SP-D to bind to these samples was tested in the presence of 5 mm CaCl2. Deglycosylation of A2M significantly reduces SP-D-A2M interactions (*, p < 0.01), further suggesting that SP-D binds A2M via carbohydrate moieties present on the A2M. Error bars indicate S.E.
To further confirm the involvement of the CRD of SP-D in recognizing A2M, SPR analysis was performed using SP-D(n/CRD). We used two different concentrations of SP-D(n/CRD) (25 and 50 μg/ml) and six concentrations of A2M (2-fold serial dilution from 200 μg/ml). SP-D(n/CRD)-A2M interactions occurred in a concentration-dependent manner (Fig. 5B), as was seen with the full-length SP-D, but higher concentrations of SP-D(n/CRD) were required for the binding to occur. This is because the native SP-D molecule has multiple subunits, whereas SP-D(n/CRD) only contains a trimeric subunit. These results suggest that SP-D recognizes A2M via its globular head domain. The ka is 2.42 ± 0.30 × 104 m−1 s−1; the kd is 3.80 ± 0.19 × 10−4 s−1, and therefore, the KD is 1.57 × 10−8 m (Fig. 5B). As expected, this value is 1 order of magnitude higher than the KD value for native SP-D-A2M interactions (Fig. 2B).
SP-D Does Not Bind Well to Deglycosylated A2M
To directly confirm that SP-D binds to the carbohydrate moieties present on A2M, we conducted a deglycosylation experiment. A2M was deglycosylated using the bacterial enzyme PNGase F, which cleaves N-linked glycans. SP-D bound well to A2M but not to BSA (negative control). Binding of SP-D to deglycosylated A2M was significantly reduced compared with A2M that had only been incubated in the reaction buffer for 72 h (p > 0.01) (Fig. 5C). Therefore, we infer that SP-D recognizes carbohydrate moieties present on A2M.
A2M Enhances the Ability of SP-D to Agglutinate Bacteria
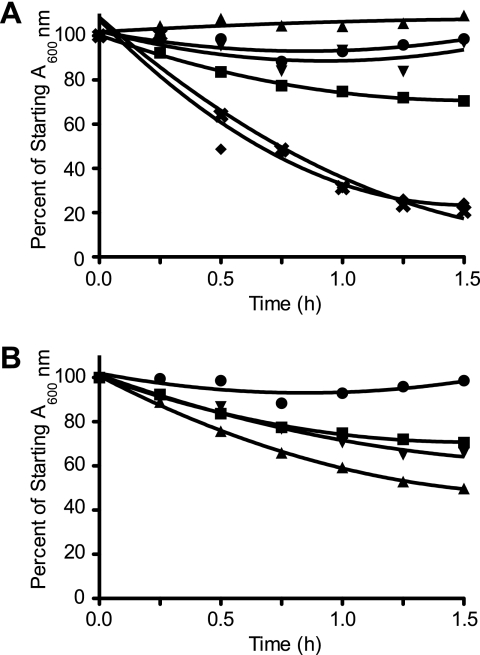
SP-D is known to agglutinate bacteria (3). To show the biological relevance of SP-D-A2M interactions, we used bacterial agglutination assays. As expected, SP-D agglutinated the E. coli in a time-dependent manner (Fig. 6A), but A2M alone was not able to cause agglutination. When SP-D was preincubated with A2M, prior to being added to E. coli, the agglutination occurred more effectively than the SP-D alone control (Fig. 6A). Adding A2M to the bacteria prior to SP-D addition also showed increased agglutination (Fig. 6B), but its effect was lower, suggesting that pre-formation of SP-D·A2M complexes increased the ability of SP-D to agglutinate bacteria. Pre-agglutinating bacteria with SP-D prior to A2M addition did not increase agglutination (data not shown). These results showed that SP-D avidly agglutinated Gram-negative bacteria, such as E. coli, and that A2M increased the biological activity of SP-D.
FIGURE 6.
A2M enhances the ability of SP-D to agglutinate E. coli, and SP-D, but not A2M, binds E. coli. A, SP-D, but not A2M, is able to agglutinate bacteria and cause a reduction in the A600 nm. A2M (50 or 5 μg/ml) or SP-D (1 μg/ml) was added to 200 μl of E. coli suspension in the presence of 5 mm CaCl2. The addition of A2M premixed with SP-D greatly enhances the agglutination of bacteria. A2M (50 or 5 μg/ml) and SP-D (1 μg/ml) were pre-mixed for 1 h at 37 °C in the presence of 5 mm CaCl2 prior to being added to 200 μl of bacterial suspension. ●, bacteria alone; ■, bacteria + 1 μg/ml SP-D; ▴, bacteria + 50 μg/ml A2M; ▾, bacteria + 5 μg/ml A2M; ♦, bacteria + 50 μg/ml A2M and 1 μg/ml SP-D premixed; ×, bacteria and 5 μg/ml A2M + 1 μg/ml SP-D premixed. The bacteria and bacteria + A2M alone conditions are overlapping with each other on the graph. All values are significantly different from the SP-D alone condition (p < 0.01). B, pre-mixing the bacteria with A2M enhances E. coli agglutination by SP-D, but not as much as pre-mixing the SP-D and A2M prior to addition to the bacterial suspension. A2M (50 or 5 μg/ml) was mixed with the bacteria suspension for 1 h at 37 °C prior to SP-D (1 μg/ml) addition in the presence of 5 mm CaCl2. ●, bacteria alone; ■, bacteria + 1 μg/ml SP-D; ▴, bacteria pre-mixed with 50 μg/ml A2M then 1 μg/ml SP-D; ▾, bacteria pre-mixed with 5 μg/ml A2M then 1 μg/ml SP-D. All values are significantly different from the SP-D alone condition (p < 0.01). All experiments were conducted at the same time but plotted separately for clarity (A and B). Results are representative of three independent experiments. Error bars indicate S.E. C, bacterial-SP-D·A2M immune complexes from the agglutination assays were separated from the filtrate. The presence of SP-D and A2M in these complexes was determined by Western blot analyses. A2M is present in the filtrate (top 2 panels) and is only present in the aggregate fraction when SP-D is present (bottom 2 panels), suggesting that A2M does not bind to the bacteria but does bind to the SP-D. Conditions: A2M, 50 μg/ml; SP-D, 1 μg/ml; *, A2M and SP-D premixed before being added to bacteria; $, A2M and bacteria premixed prior to SP-D addition.
SP-D, but Not A2M, Binds to These Bacteria
The ability of A2M to increase the biological function of SP-D could occur via different mechanisms. A2M could bind either to bacteria, SP-D, or both. To determine whether A2M directly interacts with bacteria and enhances the agglutination ability of SP-D, we conducted Western blot analysis after separating immune complexes from soluble proteins. When the A2M was incubated with bacteria, all the A2M was seen in the filtrate (Fig. 6C, top panels), whereas all of the SP-D was present in the immune complexes (aggregate) (Fig. 6C, bottom panels). This indicated that SP-D, but not A2M, significantly bound to these bacteria. When SP-D was preincubated with A2M, or present together with SP-D, the protease inhibitor was detected in the immune complexes (Fig. 6C, bottom panels). These results indicated that A2M would not readily recognize bacteria but recognized these immune complexes via SP-D.
SP-D Interacts Effectively with Cleaved A2M
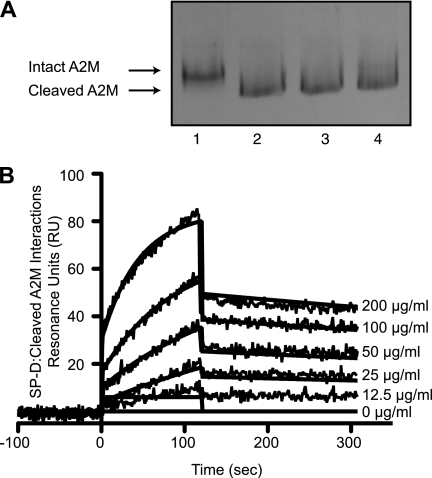
To determine whether the cleaved A2M can still interact with SP-D, we first cleaved A2M with methylamine as described previously (32). Native polyacrylamide gels showed that the methylamine treatment converted the A2M from its slow form (native) to its fast form by cleaving the internal thiol ester bond (Fig. 7A). Binding of SP-D to this A2M was tested by SPR. Identical amounts of the purified A2M and methylamine cleaved A2M were passed over the immobilized SP-D. The changes in SPR response were similar; therefore, both forms of A2M bound to SP-D in a similar manner (Fig. 7B) (KD = 8.54 × 10−9 m for elastase-cleaved A2M versus 7.33 × 10−9 m for intact A2M (Fig. 2B)). These data indicate that SP-D interacts with both forms of A2M with similar kinetics.
FIGURE 7.
Cleaved A2M binds well to SP-D. A, methylamine causes thiol ester cleavage-induced conformational changes in A2M. Incubation with methylamine converted A2M (2 μg) from its slow form (lane 1) to its fast form (lanes 2–4) as can be seen via Coomassie Blue-stained native polyacrylamide gel. Lane 1, A2M; lane 2, A2M + 100 mm methylamine; lane 3, A2M + 40 mm methylamine; lane 4, A2M + 10 mm methylamine. B, surface plasmon resonance sensorgram shows immobilized SP-D (20 μg/ml) on a GLC chip binding to methylamine-cleaved A2M (200 μg/ml in a 2-fold serial dilution) in the presence of 5 mm CaCl2. A2M was washed out at 120 s. The KD for this interaction is 8.54 × 10−9 indicating interactions will occur in the nanomolar range. The KD for intact A2M was 7.33 × 10−9 m (Fig. 2B). Curves were modeled using the Langmuir binding model. These results confirm that SP-D would bind cleaved A2M similar to that of intact A2M.
Intact and Activated A2M Protects SP-D from Elastase-mediated Degradation
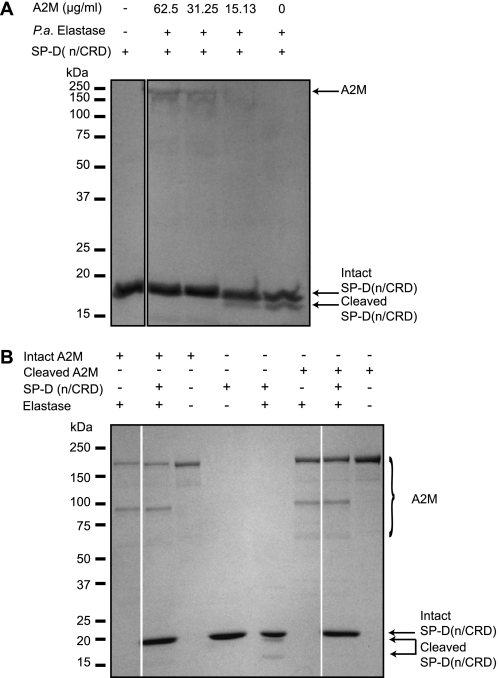
To determine whether A2M protects the CRDs of SP-D against elastase-mediated cleavage, we incubated SP-D(n/CRD) with different concentrations of A2M and a fixed amount of elastase (0.5 units). A2M protected the CRDs of the SP-D from elastase digestion in a concentration-dependent manner (Fig. 8A) with 100% protection being seen down to concentrations of 31.25 μg/ml. We also compared the effectiveness of cleaved A2M in protecting SP-D. Both forms of A2Ms protected SP-D (Fig. 8B). Therefore, we consider that the association between A2M and SP-D can help to protect SP-D from protease degradation.
FIGURE 8.
A2M protects SP-D(n/CRD) from cleavage by elastase. A, intact A2M fully protects 5 μg of SP-D(n/CRD) from elastase (0.5 units) cleavage up to 32.25 μg/ml within 2 h. B, methylamine cleaved A2M also protects SP-D(n/CRD) from cleavage by elastase. SP-D(n/CRD) (5 μg) was incubated with elastase (0.05 units) in the presence of either methylamine cleaved or intact A2M (100 μg/ml). Both the intact and the cleaved A2M protect SP-D(n/CRD) from degradation by elastase.
Cleaved A2M Can Also Enhance SP-D-mediated Bacterial Agglutination
To determine whether the A2M can also enhance the innate immune potential of SP-D after undergoing the conformational change produced by protease cleavage, we converted A2M to its cleaved form. Interestingly, the cleaved A2M was able to support similar E. coli agglutination activity as of the intact A2M (Fig. 9). These results showed that even the cleaved A2M had the ability to increase the innate immune potential of SP-D by enhancing its ability to agglutinate bacteria.
FIGURE 9.
Cleaved A2M retains its ability to enhance SP-D-mediated E. coli agglutination. A, SP-D, but not cleaved A2M, is able to agglutinate bacteria and cause a reduction in the A600 nm. Cleaved A2M (50 or 5 μg/ml) or SP-D (1 μg/ml) was added to 200 μl of E. coli suspension in the presence of 5 mm CaCl2. The addition of cleaved A2M premixed with SP-D greatly enhances the agglutination of bacteria. Cleaved A2M (50 or 5 μg/ml) and SP-D (1 μg/ml) were pre-mixed for 1 h at 37 °C in the presence of 5 mm CaCl2 prior to being added to 200 μl of bacterial suspension. ●, bacteria alone; ■, bacteria + 1 μg/ml SP-D; ▴, bacteria + 50 μg/ml cleaved A2M; ▾, bacteria + 5 μg/ml cleaved A2M; ♦, bacteria + 50 μg/ml cleaved A2M and 1 μg/ml SP-D premixed; ×, bacteria + 5 μg/ml cleaved A2M and 1 μg/ml SP-D premixed. All values are significantly different from the SP-D alone condition (p < 0.01). B, pre-mixing the bacteria with cleaved A2M enhanced E. coli agglutination by SP-D, but not as much as pre-mixing the SP-D and cleaved A2M prior to addition to the bacterial suspension. Cleaved A2M (50 or 5 μg/ml) was mixed with the bacteria suspension for 1 h at 37 °C prior to SP-D (1 μg/ml) addition in the presence of 5 mm CaCl2. ●, bacteria alone; ■, bacteria + 1 μg/ml SP-D; ▴, bacteria pre-mixed with 50 μg/ml cleaved A2M then 1 μg/ml SP-D; ▾, bacteria pre-mixed with 5 μg/ml cleaved A2M then 1 μg/ml SP-D. E. coli experiments were conducted at the same time but plotted separately for clarity (A and B). All values are significantly different from the SP-D alone condition (p < 0.01).
DISCUSSION
Regulation of SP-D-mediated innate immune function in the presence of proteases is not clearly understood. In this study, we have investigated the interactions between the innate immune collectin SP-D and a protease inhibitor A2M. We discovered this interaction after identifying that SP-D does not bind to ovalbumin but rather to the fractions containing contaminating proteins (ovomucin and ovomacroglobulin) found in OVA preparations (supplemental Fig. S1). We found that SP-D binds human A2M in a concentration-, calcium-, and carbohydrate-dependent manner (Figs. 1–5). A2M protects SP-D from proteolytic degradation (Fig. 8), and SP-D-A2M interactions enhance the ability of SP-D to agglutinate bacteria (Figs. 6 and 9). Notably, SP-D recognizes both cleaved and intact A2M (Fig. 7), and both forms of this protease inhibitor increase the bacterial agglutination ability of SP-D (Figs. 6 and 9). Therefore, we have discovered a heretofore unknown interaction and an intriguing biological function for A2M to enhance SP-D-mediated innate immunity during bacterial infection.
We have discovered through mass spectrometry analysis of commercial ovalbumin preparations that SP-D binds fractions of protein containing ovomucin and ovomacroglobulin (supplemental Fig. S1). In this study, we show that SP-D interacts with human A2M in a concentration-dependent manner to human A2M (Fig. 2), and the interactions occur in the presence of nanomolar concentrations of these proteins (Fig. 2B). A2M is present in the serum (1–2 mg/ml; 1.4–2.8 μm) (22, 23) and the lungs (0.09–2.02 μg/ml; 0.13–2.8 nm) (18, 19). Notably, the concentration of A2M drastically increases during lung infection, edema fluid accumulation, and inflammation (220 μg/ml; 305.5 nm) (19). SP-D concentration in the lungs is considered to be the 1–5 μg/ml (1.9–9.3 nm) range (1). During infection, SP-D has been reported in the range of 0.012–1.13 μg/ml (0.022–2.09 nm) (35). In the presence of severe infections, degraded fragments of SP-D can be detected suggesting that a constant battle between SP-D and proteases occurs in the lungs (14–16, 36, 37). During certain inflammatory lung conditions, SP-D concentrations increase by a factor of about 10 (38). Supplementing SP-D suppresses lung inflammation (39). We have identified that SP-D and A2M interact in the airway-lining fluid (Fig. 3). Therefore, we consider interaction between SP-D and A2M is biologically relevant to the lungs under normal conditions, as well as during infection and inflammation.
The biological function of SP-D primarily resides in the CRDs of SP-D (1, 3). It is well established that SP-D is a C-type lectin (6, 40) and recognizes carbohydrate ligands via its CRDs in a calcium-dependent manner (10, 41, 42). However, SP-D also interacts with other ligands or glycoproteins such as immunoglobulins (43), decorin (30), gp-340 (34), and microfibril-associated glycoprotein 4 (44) via protein-protein interactions. Our results suggest that SP-D interacts with A2M via its globular head domain in a calcium-dependent lectin-carbohydrate type interaction (Figs. 4 and 5). This mode of interaction is possible because A2M has eight N-linked glycosylation sites on each of its four identical subunits (45). Some of the N-linked glycans present on A2M have been identified as oligomannose structures (Man5–Man7), which make up 8% of the total glycan pool, and the remainder of the glycans terminates with galactose or sialic acid (22). SP-D binds avidly to mannose but not well to galactose or sialic acid (2, 3). We have shown that binding of SP-D to A2M is significantly reduced when the A2M has been deglycosylated (Fig. 5C). A recent report suggests that mannose-binding lectin can recognize Man5–7 glycan moieties present on A2M (22). Considering all the data, we suggest that SP-D recognizes the oligomannose moieties of A2M via lectin-carbohydrate interactions.
The CaCl2 concentration required for this interaction plateaus at about 0.625–1.25 mm (Fig. 4B), and this value falls well within the physiological concentration of calcium (46), suggesting that the interaction between A2M and SP-D is possible in the lungs, and we have demonstrated that SP-D-A2M interactions do occur in BALF (Fig. 3). Under these physiological conditions, SP-D binds avidly to terminal mannosyl residues found in several microbes including bacteria (47). Therefore, A2M could enhance the binding of SP-D to carbohydrates by forming SP-D·A2M complexes. These complexes should increase the valancy and avidity of the SP-D for the carbohydrates so that the collectin could effectively generate bacterial immune complexes.
SP-D has been shown to agglutinate E. coli (48), and our bacterial agglutination studies agree with this notion (Figs. 6 and 9). Interestingly, pre-mixing SP-D and A2M greatly increases the ability of SP-D to agglutinate bacteria (Fig. 6A). SP-D associates with the bacterial aggregate (Fig. 6C, bottom panels) but A2M does not appear in the aggregate fraction unless SP-D is present. SP-D is increased in rats infected with E. coli, and there is a correlation between alveolar-capillary membrane injury and the amount of protein found in the bronchoalveolar lavage fluid (49). However, in some conditions SP-D could decrease as well (35). Because both SP-D and A2M are present in the lungs under normal conditions and during bacterial infection, these two proteins could interact with each other to aid in the agglutination of bacteria by SP-D.
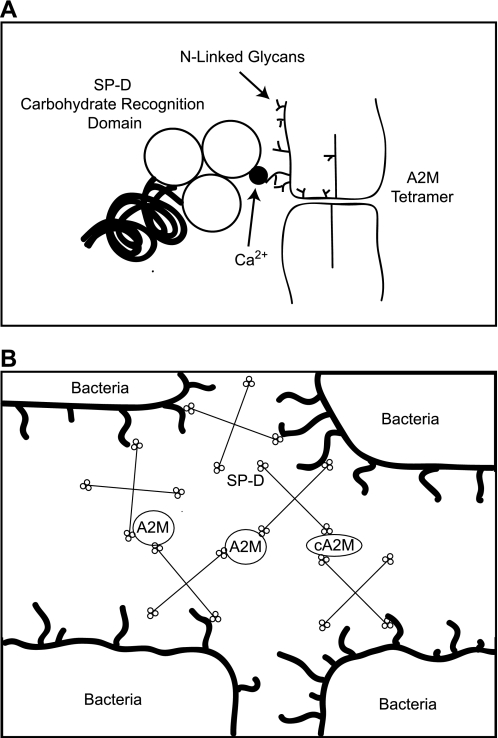
SP-D is also able to bind A2M that has been cleaved by either methylamine (Fig. 7) or elastase (data not shown) and is protected from elastase-mediated degradation by A2M (Fig. 8). The glycosylation status of A2M should be identical to that of uncleaved A2M. During bacterial infection, A2M could protect SP-D from proteases and still increase the ability of SP-D to agglutinate bacteria; therefore, we propose that it is a novel mechanism by which the host maintains effective defense against bacteria when the bioavailability of SP-D is low during high degrees of infection (Fig. 10).
FIGURE 10.
Model summarizing SP-D-A2M interactions and ability of A2M to enhance bacterial agglutinating capacity of SP-D. A, SP-D interacts with A2M through its CRDs in a calcium- and carbohydrate-dependent mechanism. A2M protects SP-D from proteolysis, and the ability of SP-D to interact with the protease inhibitor is not affected by the activation state. B, A2M does not bind bacteria, but both intact (A2M) and activated (cA2M) forms of A2M enhance the agglutinating ability of SP-D. Therefore, A2M-SP-D interactions enhance a biological function of SP-D.
In summary, the data presented here characterize A2M as a new binding partner for SP-D and that the binding characteristics are consistent with the ability of SP-D to bind the protease inhibitor via its lectin-carbohydrate interactions. The SP-D-A2M interactions lead to efficient protection of SP-D from proteases, and interestingly, such interactions increase the biological activity of SP-D. Therefore, we have identified a heretofore unknown function for A2M and a potential mechanism by with the innate immunity is boosted during infection.
Supplementary Material
Acknowledgments
ProteON analyses were performed in the Analytical Laboratory for Bioactive Materials of the Canada Foundation for Innovation-funded center for the study of the complex childhood diseases at the Hospital for Sick Children.
This work was supported by Canadian Institutes of Health Research Grant MOP-84312 (to N. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- SP-D
- surfactant protein D
- A2M
- α2-macroglobulin
- ELISA
- enzyme-linked immunosorbent assay
- SPR
- surface plasmon resonance
- SP-D(n/CRD)
- a recombinant neck and carbohydrate recognition domain of SP-D and eight Gly-X-Y repeats
- BSA
- bovine serum albumin
- PNGase F
- peptide:N-glycosidase F
- BALF
- bronchoalveolar lavage fluid
- OVA
- ovalbumin.
REFERENCES
- 1.Palaniyar N., Sorensen G., Holmskov U. (2008) in Animal Lectins: A Functional View (Vasta G. ed) pp. 331–348, Taylor & Francis Ltd, London [Google Scholar]
- 2.Palaniyar N., Nadesalingam J., Reid K. B. (2002) Immunobiology 205, 575–594 [DOI] [PubMed] [Google Scholar]
- 3.Wright J. R. (2005) Nat. Rev. Immunol. 5, 58–68 [DOI] [PubMed] [Google Scholar]
- 4.Areschoug T., Gordon S. (2008) Contrib. Microbiol. 15, 45–60 [DOI] [PubMed] [Google Scholar]
- 5.McCormack F. X., Whitsett J. A. (2002) J. Clin. Invest. 109, 707–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Persson A., Chang D., Crouch E. (1990) J. Biol. Chem. 265, 5755–5760 [PubMed] [Google Scholar]
- 7.Akiyama J., Hoffman A., Brown C., Allen L., Edmondson J., Poulain F., Hawgood S. (2002) J. Histochem. Cytochem. 50, 993–996 [DOI] [PubMed] [Google Scholar]
- 8.Stahlman M. T., Gray M. E., Hull W. M., Whitsett J. A. (2002) J. Histochem. Cytochem. 50, 651–660 [DOI] [PubMed] [Google Scholar]
- 9.Madsen J., Kliem A., Tornoe I., Skjodt K., Koch C., Holmskov U. (2000) J. Immunol. 164, 5866–5870 [DOI] [PubMed] [Google Scholar]
- 10.Wang L., Brauner J. W., Mao G., Crouch E., Seaton B., Head J., Smith K., Flach C. R., Mendelsohn R. (2008) Biochemistry 47, 8103–8113 [DOI] [PubMed] [Google Scholar]
- 11.Wang H., Head J., Kosma P., Brade H., Müller-Loennies S., Sheikh S., McDonald B., Smith K., Cafarella T., Seaton B., Crouch E. (2008) Biochemistry 47, 710–720 [DOI] [PubMed] [Google Scholar]
- 12.Kuan S. F., Rust K., Crouch E. (1992) J. Clin. Invest. 90, 97–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lim B. L., Willis A. C., Reid K. B., Lu J., Laursen S. B., Jensenius J. C., Holmskov U. (1994) J. Biol. Chem. 269, 11820–11824 [PubMed] [Google Scholar]
- 14.Cooley J., McDonald B., Accurso F. J., Crouch E. C., Remold-O'Donnell E. (2008) J. Leukocyte Biol. 83, 946–955 [DOI] [PubMed] [Google Scholar]
- 15.Mariencheck W. I., Alcorn J. F., Palmer S. M., Wright J. R. (2003) Am. J. Respir. Cell Mol. Biol. 28, 528–537 [DOI] [PubMed] [Google Scholar]
- 16.Alcorn J. F., Wright J. R. (2004) J. Biol. Chem. 279, 30871–30879 [DOI] [PubMed] [Google Scholar]
- 17.Craig A., Mai J., Cai S., Jeyaseelan S. (2009) Infect. Immun. 77, 568–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Vyve T., Chanez P., Bernard A., Bousquet J., Godard P., Lauwerijs R., Sibille Y. (1995) J. Allergy Clin. Immunol. 95, 60–68 [DOI] [PubMed] [Google Scholar]
- 19.Baker C. S., Evans T. W., Haslam P. L. (2000) Respiration 67, 533–538 [DOI] [PubMed] [Google Scholar]
- 20.Ratjen F., Hartog C. M., Paul K., Wermelt J., Braun J. (2002) Thorax 57, 930–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Armstrong P. B. (2006) Immunobiology 211, 263–281 [DOI] [PubMed] [Google Scholar]
- 22.Arnold J. N., Wallis R., Willis A. C., Harvey D. J., Royle L., Dwek R. A., Rudd P. M., Sim R. B. (2006) J. Biol. Chem. 281, 6955–6963 [DOI] [PubMed] [Google Scholar]
- 23.Borth W. (1992) FASEB J. 6, 3345–3353 [DOI] [PubMed] [Google Scholar]
- 24.Barrett A. J., Starkey P. M. (1973) Biochem. J. 133, 709–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feldman S. R., Gonias S. L., Pizzo S. V. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 5700–5704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howard J. B., Swenson R., Eccleston E. (1983) Ann. N.Y. Acad. Sci. 421, 160–166 [DOI] [PubMed] [Google Scholar]
- 27.Sottrup-Jensen L., Gliemann J., Van Leuven F. (1986) FEBS Lett. 205, 20–24 [DOI] [PubMed] [Google Scholar]
- 28.Huang W., Dolmer K., Liao X., Gettins P. G. (1998) Protein Sci. 7, 2602–2612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palaniyar N., Nadesalingam J., Clark H., Shih M. J., Dodds A. W., Reid K. B. (2004) J. Biol. Chem. 279, 32728–32736 [DOI] [PubMed] [Google Scholar]
- 30.Nadesalingam J., Bernal A. L., Dodds A. W., Willis A. C., Mahoney D. J., Day A. J., Reid K. B., Palaniyar N. (2003) J. Biol. Chem. 278, 25678–25687 [DOI] [PubMed] [Google Scholar]
- 31.Nadesalingam J., Dodds A. W., Reid K. B., Palaniyar N. (2005) J. Immunol. 175, 1785–1794 [DOI] [PubMed] [Google Scholar]
- 32.Eccleston E. D., Howard J. B. (1985) J. Biol. Chem. 260, 10169–10176 [PubMed] [Google Scholar]
- 33.Barrett A. J., Brown M. A., Sayers C. A. (1979) Biochem. J. 181, 401–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holmskov U., Lawson P., Teisner B., Tornoe I., Willis A. C., Morgan C., Koch C., Reid K. B. (1997) J. Biol. Chem. 272, 13743–13749 [DOI] [PubMed] [Google Scholar]
- 35.Postle A. D., Mander A., Reid K. B., Wang J. Y., Wright S. M., Moustaki M., Warner J. O. (1999) Am. J. Respir. Cell Mol. Biol. 20, 90–98 [DOI] [PubMed] [Google Scholar]
- 36.Hirche T. O., Crouch E. C., Espinola M., Brokelman T. J., Mecham R. P., DeSilva N., Cooley J., Remold-O'Donnell E., Belaaouaj A. (2004) J. Biol. Chem. 279, 27688–27698 [DOI] [PubMed] [Google Scholar]
- 37.von Bredow C., Wiesener A., Griese M. (2003) Lung 181, 79–88 [DOI] [PubMed] [Google Scholar]
- 38.Haley K. J., Ciota A., Contreras J. P., Boothby M. R., Perkins D. L., Finn P. W. (2002) Am. J. Physiol. Lung Cell Mol. Physiol. 282, L573–L584 [DOI] [PubMed] [Google Scholar]
- 39.Ikegami M., Scoville E. A., Grant S., Korfhagen T., Brondyk W., Scheule R. K., Whitsett J. A. (2007) Chest 132, 1447–1454 [DOI] [PubMed] [Google Scholar]
- 40.Gardai S. J., Xiao Y. Q., Dickinson M., Nick J. A., Voelker D. R., Greene K. E., Henson P. M. (2003) Cell 115, 13–23 [DOI] [PubMed] [Google Scholar]
- 41.Håkansson K., Lim N. K., Hoppe H. J., Reid K. B. (1999) Structure 7, 255–264 [DOI] [PubMed] [Google Scholar]
- 42.Crouch E., McDonald B., Smith K., Roberts M., Mealy T., Seaton B., Head J. (2007) Biochemistry 46, 5160–5169 [DOI] [PubMed] [Google Scholar]
- 43.Nadesalingam J., Reid K. B., Palaniyar N. (2005) FEBS Lett. 579, 4449–4453 [DOI] [PubMed] [Google Scholar]
- 44.Lausen M., Lynch N., Schlosser A., Tornoe I., Saekmose S. G., Teisner B., Willis A. C., Crouch E., Schwaeble W., Holmskov U. (1999) J. Biol. Chem. 274, 32234–32240 [DOI] [PubMed] [Google Scholar]
- 45.Sottrup-Jensen L., Sand O., Kristensen L., Fey G. H. (1989) J. Biol. Chem. 264, 15781–15789 [PubMed] [Google Scholar]
- 46.Nielson D. W., Lewis M. B. (1988) Pediatr. Res. 24, 322–325 [DOI] [PubMed] [Google Scholar]
- 47.Ferguson J. S., Voelker D. R., McCormack F. X., Schlesinger L. S. (1999) J. Immunol. 163, 312–321 [PubMed] [Google Scholar]
- 48.Brinker K. G., Martin E., Borron P., Mostaghel E., Doyle C., Harding C. V., Wright J. R. (2001) Am. J. Physiol. Lung Cell Mol. Physiol. 281, L1453–L1463 [DOI] [PubMed] [Google Scholar]
- 49.Russo T. A., Bartholomew L. A., Davidson B. A., Helinski J. D., Carlino U. B., Knight P. R., 3rd, Beers M. F., Atochina E. N., Notter R. H., Holm B. A. (2002) Am. J. Physiol. Lung Cell Mol. Physiol. 283, L655–L663 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.