Abstract
The classical nuclear factor κB (NF-κB) signaling pathway is under the control of the IκB kinase (IKK) complex, which consists of IKK-1, IKK-2, and NF-κB essential modulator (NEMO). This complex is responsible for the regulation of cell proliferation, survival, and differentiation. Dysregulation of this pathway is associated with several human diseases, and as such, its inhibition offers an exciting opportunity for therapeutic intervention. NEMO binding domain (NBD) peptides inhibit the binding of recombinant NEMO to IKK-2 in vitro. However, direct evidence of disruption of this binding by NBD peptides in biological systems has not been provided. Using a cell system, we expanded on previous observations to show that NBD peptides inhibit inflammation-induced but not basal cytokine production. We report that these peptides cause the release of IKK-2 from an IKK complex and disrupt NEMO-IKK-2 interactions in cells. We demonstrate that by interfering with NEMO-IKK-2 interactions, NBD peptides inhibit IKK-2 phosphorylation, without affecting signaling intermediates upstream of the IKK complex of the NF-κB pathway. Furthermore, in a cell-free system of IKK complex activation by TRAF6 (TNF receptor-associated factor 6), we show that these peptides inhibit the ability of this complex to phosphorylate downstream substrates, such as p65 and inhibitor of κBα (IκBα). Thus, consistent with the notion that NEMO regulates IKK-2 catalytic activity by serving as a scaffold, appropriately positioning IKK-2 for activation by upstream kinase(s), our findings provide novel insights into the molecular mechanisms by which NBD peptides exert their anti-inflammatory effects in cells.
Keywords: Cytokine, Fibroblast, Inflammation, NF-κB, Peptide Interactions, Protein Kinases, Protein Phosphorylation, Signal Transduction
Introduction
The transcription factor nuclear factor κB (NF-κB)3 is responsible for the regulation of a multitude of diverse, context-dependent cellular outcomes, including cell proliferation differentiation and survival (1). A number of stimuli, including proinflammatory cytokines and engagement of innate immune receptors and antigen receptors, as well as various environmental stimuli trigger NF-κB activation (1). In most cell types, NF-κB family members in their resting state are retained in the cytosol as homo- or heterodimers in complexes with inhibitors of κBs (IκBs) (2). Upon stimulation, IκBs are rapidly phosphorylated, ubiquitinated, and subsequently degraded via the proteasome complex (2, 3). Free NF-κB dimers then translocate to the nucleus and up-regulate the expression of various target genes. NF-κB activation is transient and tightly regulated. Consistently dysregulated and prolonged NF-κB-mediated responses are associated with several human diseases, including autoimmune disorders and certain cancers (4). Accordingly, the suppression of NF-κB-mediated cell signaling offers an exciting opportunity for therapeutic intervention.
IκB phosphorylation is carried out by the IκB kinase (IKK) complex. This complex consists of two catalytically active subunits, IKK-1 and IKK-2, that share 50% sequence similarity, and a regulatory subunit referred to as NF-κB essential modulator (NEMO). NEMO is a primarily helical protein containing significant stretches of coiled-coil structure as well as a zinc finger domain (5). NEMO-deficient cells show neither increased IKK activity nor NF-κB DNA binding activity in response to several proinflammatory stimuli, including tumor necrosis factor-α (TNF-α), interleukin-1β (IL-1β), and lipopolysaccharide (LPS) (6). However, the actual role of NEMO in IKK activation has not been fully elucidated. It has been proposed that NEMO can recruit IKK complexes to activated receptors, where IKKs are activated by oligomerization, conformational changes, or trans-autophosphorylation (7–9). NEMO may also orient the catalytic subunits for targeted phosphorylation of the activation loop by an upstream kinase or enable the IKKs to interact with their substrates.
The specific molecular mechanisms of NEMO-IKK interactions involve a C-terminal hexapeptide core sequence present on both IKK-1 and IKK-2 (LDWSWL) termed the NEMO binding domain (NBD) (10), with more recent studies expanding this domain to include additional amino acids spanning this core sequence (5, 11). Peptides corresponding to the IKK-2 NBD have been found to disrupt the association of recombinant NEMO with recombinant IKK-1 or IKK-2 in vitro (12). The cell-permeable versions of these peptides blocked NF-κB activation in various cellular and animal models of inflammation. Consequently, several studies have revealed reduced osteoclastogenesis and joint destruction in collagen-induced arthritis models, bowel injury in colitis models, and alleviation of pathology in murine models of inflammation-induced brain injury and Parkinson disease (13–17). Importantly, continuous administration of NBD peptides in animal models did not reveal any overt signs of toxicity (14). Interestingly, although NBD peptides inhibited NF-κB reporter gene activity in response to proinflammatory stimuli, they did not inhibit basal reporter gene activity (10). Together, these studies emphasize the importance of NBD peptides in IKK complex activation and NF-κB signaling and suggest that NEMO-IKK-2 interactions can be targeted for therapeutic intervention. Blocking inflammation-induced activation of IKK/NF-κB signaling while sparing basal levels of signaling necessary for normal cell maintenance is expected to result in good efficacy without significant adverse effects.
Mutational analysis of the core IKK-2 NBD sequence identified Asp738, Trp739, and Trp741 within the peptide as critical residues for NEMO binding. Mutant NBD peptides containing alanine replacements at any of these positions lost the inhibitory effects exhibited by the wild-type peptides (10, 12). The recently solved structure of NEMO-IKK peptides provided data consistent with the mutational study results (5). Structural features of the interactions may prove to be critical for the rational design of inhibitors targeting the NEMO-IKK-2 interactions.
Despite the abundant literature on NBD peptides, direct evidence for their role in disrupting the native IKK complex in the isolated state or in cells is still lacking. In this study, we expanded on previous reports to show that NBD peptides inhibited inflammation-induced but not basal cytokine production. We also provided evidence for sequential events of IKK-2 activation during inflammatory responses by showing that the anti-inflammatory effects of NBD peptides occurred before, not after, the IKK complex had been activated by cytokines. Furthermore, we demonstrate that disruption of the IKK complex by NBD peptides resulted in release of IKK-2 from the complex. Taken together, this study provides novel and intriguing data regarding the role of NBD peptides in the regulation of the NEMO-IKK-2 signaling.
EXPERIMENTAL PROCEDURES
Reagents
Dulbecco's modified Eagle's medium (DMEM), l-glutamine, sodium pyruvate, penicillin/streptomycin, HEPES, and trypsin were obtained from Invitrogen. Fetal bovine serum (FBS) originated from SAFC Biosciences (Lenexa, KS), and recombinant human IL-1β was from R&D Systems (Minneapolis, MN). Polyclonal anti-IKK-1/2 antibodies (Abs), anti-IRAK-1 Abs, anti-JNK1/2 Abs, anti-p65 Abs, and anti-IKK-2 Abs were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA); anti-phospho-TAK-1 Abs, anti-phospho-HSP27 Abs, anti-phospho-JNK Abs, and anti-phospho-IKK-1(Ser180)/IKK-2(Ser181) Abs were from Cell Signaling Technologies (Danvers, MA). Anti-IκBα monoclonal antibodies (mAbs) were made in house. Anti-NEMO mAbs were purchased from BD Pharmingen (San Jose, CA), and anti-tubulin Abs were obtained from Abcam Inc. (Cambridge, MA). Complete protease inhibitor mixture was from Roche Applied Science, and phosphatase inhibitor mixtures, dimethyl sulfoxide (DMSO), LPS, dithiothreitol, Tween 20, bovine serum albumin, and Protein A-agarose beads were from Sigma. The protein assay kit was purchased from Bio-Rad. NuPage sample buffer, Novex native Tris/glycine sample buffer, Tris/glycine SDS-polyacrylamide gels, and SimplyBlue SafeStain were from Invitrogen. ImmEdge pen was obtained from Vector Laboratories (Burlingame, CA) and DuoLinkTM 100 Detection Kit 563 from OLINK Bioscience (Uppsala, Sweden). Antennapedia homeodomain-conjugated IKK-2 11-mer NBD peptides (TALDWSWLQTE and TALDASALQTE) were purchased from Calbiochem. IKK-2 45-mers (PAKKSEELVAEAHNLCTLLENAIQDTVREQDQSFTALDWSWLQTE and PAKKSEELVAEAHNLCTLLENAIQDTVREQDQSFTALDASALQTE) were purchased from American Peptide Inc. (Sunnyvale, CA). The IKK-2-selective inhibitor, PHA-408 (18, 19), and IRAK-4 inhibitor, compound 44 (20), were synthesized at Pfizer (St. Louis, MO). MSD blocking buffer A originated from Meso Scale Discovery (Gaithersburg, MD); LANCE Europium-W1024 labeled anti-glutathione S-transferase (GST) Abs and Alphascreen glutathione donor beads and anti-FLAG acceptor beads were obtained from PerkinElmer Life Sciences. Streptavidin-AlexaFluor647 conjugate was purchased from Invitrogen.
Peripheral Blood Mononuclear Cell Cultures
Human whole blood was collected from healthy donors in sodium-heparinized tubes (BD Biosciences), and peripheral blood mononucleated cells (PBMC) were isolated by Ficoll separation. Cells were washed in Dulbecco's phosphate-buffered saline, resuspended in DMEM containing 5% endotoxin-free FBS and 10 units/ml penicillin/streptomycin, and plated at 2.5 × 105 cells/well in 96-well tissue culture plates. Cells were pretreated with increasing concentrations of either NBD peptides or PHA-408 for 2 h prior to stimulation with vehicle or 10 ng/ml LPS for 18 h (1% DMSO, final concentration). Basal or LPS-induced production of TNF-α and IL-6 were measured by MSD technology. IC50 values were determined using a data analysis program (Pfizer, St. Louis, MO). Cytotoxicity was assessed using the Alamar Blue assay from Promega (Madison, WI).
Cultures of Synovial Fibroblasts Derived from Patients with Rheumatoid Arthritis
Synovial fibroblasts derived from patients with rheumatoid arthritis (RASF) were purchased from Cell Applications (San Diego, CA). Adherent RASF were isolated via enzymatic digestions from primary synovial tissues isolated after knee synovectomy and were cultured in DMEM high glucose, containing 15% defined bovine serum (HyClone Laboratories, Logan, UT) and 50 μg/ml gentamicin. For cytokine analysis, 1.5 × 104 cells/well were plated in a 96-well plate and were allowed to attach overnight. The growth media were replaced with fresh DMEM containing 1% serum, and the cells were pretreated with increasing concentrations of either NBD peptides or PHA-408 for 2 h prior to stimulation with 1 ng/ml IL-1β for 18 h. Secreted IL-6 was measured by MSD technology. IC50 were determined using a data analysis program (Pfizer), and cytotoxicity was assessed using the Alamar Blue assay.
Immunoprecipitation and Western Blot Analysis
RASF were preincubated for 30 min with NBD peptides prior to simulation with 1 ng/ml IL-1β for 10 min at 37 °C. Cells were lysed in a buffer containing 20 mm Tris, pH 7.6, 150 mm NaCl, 0.5% Nonidet P-40, 100 μg/ml phenylmethylsulfonyl fluoride, and protease and phosphatase inhibitors, and protein concentrations were determined by the Bradford method. The IKK complex was immunoprecipitated from 50 μg of total cell lysates after a 1-h incubation with anti-NEMO mAbs followed by a 1-h incubation with Protein A-Sepharose beads. The pellets were washed, and the proteins were denatured by the addition of reducing sample buffer and were resolved on Tris/glycine SDS-polyacrylamide gels. The proteins were transferred to polyvinylidene difluoride membranes, which were then incubated with primary Abs for 2 h at 25 °C followed by a 1-h incubation with secondary Abs. The results were visualized using the Li-Cor Odyssey Infrared Imaging System (Li-Cor, Lincoln, NE). For native PAGE, 30 μg of total cell lysates were added directly to Novex native Tris/glycine sample buffer and resolved on Tris/glycine PAGE. Western blot analysis was performed as described above. To determine the composition of the IKK complex resolved by native PAGE, gels were stained using SimplyBlue SafeStain dye, and a region of the gel that corresponded to ∼150–250 kDa was excised. Proteins were extracted in 2× reducing SDS sample loading buffer using a Dounce homogenizer and resolved on Tris/glycine PAGE, and Western blot analysis was carried out as described above.
In Situ Proximity Ligation Assay (PLA)
RASF cells were plated at 2.5 × 104 cells/well (8-well culture slides, BD Falcon (Bedford, MA)) in DMEM containing 10% FBS, 1% l-glutamine, 1% sodium pyruvate, 100 units/ml penicillin, 100 μg/ml streptomycin, and glucose and were incubated overnight at 37 °C in a 5% CO2 atmosphere. Cells were fed with serum-free DMEM and preincubated with either 1% DMSO or NBD peptides dissolved in DMSO (1% DMSO, final concentration) for 30 min. Cells were treated with 1 ng/ml IL-1β for 15 min, washed with Tris-buffered saline containing 0.05% Tween 20 (TBST), fixed with 4% paraformaldehyde, and then permeabilized with 0.1% Triton X-100. The chamber wells were removed, and hydrophobic barriers were made between individual wells using an ImmEdge pen. Nonspecific binding was blocked by incubating cells for 30 min at 37 °C with a blocking solution provided by the manufacturer and incubated overnight at 4 °C with anti-IKK-2 Abs and anti-NEMO Abs raised in different species. Cells incubated with either anti-IKK-2 Abs or anti-NEMO Abs were used as negative controls. The proximity ligation assay (fluorescence signal generated at 40 nm only when two PLA probes are in close proximity) was conducted using DuoLinkTM 100 Detection Kit 563. Briefly, after incubation with primary Abs, samples were incubated for 1 h at 37 °C with a pair of secondary Abs conjugated with oligonucleotides (PLA probes). Subsequently, hybridization, ligation, amplification, and detection were performed according to the manufacturer's protocol. Preparations were mounted in DuoLink mounting medium containing the Hoechst nuclear dye and photographed at ×20 magnification using a Nikon E800M fluorescence microscope. The signals were analyzed using the BlobFinder imaging software developed by the Centre for Image Analysis, Uppsala University (Uppsala, Sweden). An average analysis was made from all signals being counted in one image and then divided by the number of cells in that image, thus giving the average signals/cell.
IKK-2 Kinase Activity Assay
RASF were plated at 1.5 × 104 cells/well (96-well plates) in DMEM containing 15% FBS and 50 μg/ml gentamicin and incubated overnight. Cells were refreshed with DMEM containing 1% FBS, pretreated for 1 h with 2% DMSO (final concentration) or the inhibitors prior to treatment with 1 ng/ml IL-1β for 10 min. Cells were lysed with MSD lysis buffer (150 mm NaCl, 20 mm Tris, pH 7.5, 1 mm EDTA, 1 mm EGTA, 1% Triton X-100, phosphatase inhibitors I and II, and protease inhibitors). Separate plates were set up for measuring cytotoxicity using the Alamar Blue assay. Kinase activity was done using MSD high bind 96-well plates coated with anti-phospho-IκBα Abs as described previously (19). Briefly, the plates were blocked at room temperature for 1 h with MSD blocking buffer A. Kinase activity was measured by incubating 20 μg of cell lysates with 1 μm ATP and 5 μm biotinylated IκBα peptide in 30 μl of reaction buffer containing 25 mm HEPES, 2 mm MnCl2, 2 mm MgCl2, and 10 mm NaF for 90 min. The plates were washed twice with TBST, incubated with MSD sulfatag streptavidin for 90 min, and washed twice again with TBST. The plates were read using MSD sector imager 6000. To test the effects of the inhibitors on IKK-2 activity after cells had been treated for 10 min with 1 ng/ml IL-1β, bulk lysates were incubated with the inhibitors for 1 h, and kinase activity was measured as described above.
Time-resolved Fluorescence Resonance Energy Transfer Binding Assay
To monitor the binding of NEMO and NBD peptides, 4 nm GST-NEMO-(2–200) and 1.75 nm biotinylated IKK-1/2 chimeric 44-mer peptide (biotin-C6-PAKKSEELVAEAHNLCTLLENAIQDTVREQGNSMMNLDWSWLTE-NH2 in NEMO binding buffer (50 mm HEPES (pH 7.4), 0.01% bovine serum albumin, 0.0005% Tween 20, and 1 mm dithiothreitol) were incubated for 30 min at room temperature in a 10-μl volume. The readout reagents consisting of 4 nm LANCE Europium-W1024-labeled anti-GST Abs and 16 nm Streptavidin-AlexaFluor647 conjugate in NEMO binding buffer were added to the reaction in a 10-μl volume and incubated for 2 h at room temperature before reading the fluorescence resonance energy transfer signal on an Envision 2102 Multilabel plate reader (PerkinElmer Life Sciences). For competition studies, competitor molecules (unlabeled IKK-2 11-mer or IKK-2 45-mer) were titrated into the binding mixture and incubated for 30 min, and the signal was detected as described above. Reported IC50 values are the mean IC50 ± S.E. of four (n = 4) independent experiments.
Alphascreen Binding Assay
To monitor NEMO and IKK-2 interactions, 2 nm GST-NEMO-(2–200) and 2 nm IKK-2-FLAG in NEMO binding buffer were incubated for 30 min at room temperature in a 10-μl volume. The readout reagents consisting of a 40-mg/ml/bead mixture of glutathione donor beads and anti-FLAG acceptor beads diluted in NEMO binding buffer were added to each reaction and incubated overnight at room temperature before reading the Alphascreen signal on an Envision 2102 Multilabel plate reader (PerkinElmer Life Sciences). For competition assays, competitor molecules (unlabeled peptides as described above) were titrated into the binding mixture and incubated for 30 min, and the signal was read as described above.
TRAF6 Cell-free Activation Assay
HeLa cells grown to confluence in T162 flasks were washed, scraped in PBS, and pelleted, and the hypotonic lysis buffer (20 mm HEPES, pH 7.4, 10 mm KCl, 1.5 mm MgCl2, 1 mm EGTA, 1 mm phenylmethylsulfonyl fluoride, 0.5 mm dithiothreitol, 1× Complete) was added as described previously (21). Cells were further lysed in Dounce homogenizer on ice and centrifuged at 16,000 × g for 10 min at 4 °C, and protein concentrations were determined by the Bradford method. The reaction was carried out with or without the inhibitors in a buffer containing 50 mm Tris-HCl, pH 7.5, 5 mm MgCl2, 0.1 μm okadeic acid, ATP, recombinant human TRAF6 (TNF receptor-associated factor 6), and 1 μg of cell lysates. Samples were incubated for 60 min at 30 °C, and the reaction was stopped by adding an equal volume of sample buffer. Western blot analysis was performed as described above.
Statistical Analysis
Samples were run in triplicate. Statistical significance was assessed using an unpaired Student's t test.
RESULTS
NBD Peptides Inhibit Inflammation-induced but Not Basal Cytokine Production
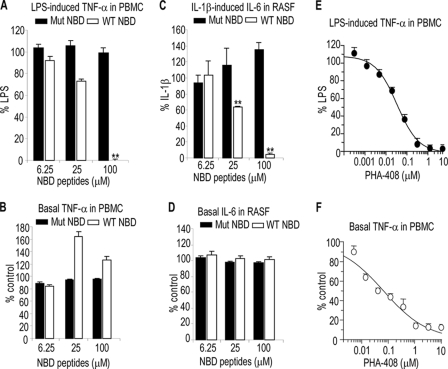
Consistent with previously published studies (10), antennapedia-conjugated wild type (WT) NBD peptides (IKK-2 11-mer) but not mutant NBD peptides completely blocked LPS-induced production of TNF-α and IL-6 in PBMC (Fig. 1A) (data not shown) or IL-1β-induced IL-6 production in RASF (Fig. 1C). However, neither WT nor mutant NBD peptides had an inhibitory effect on the production of these cytokines in basal conditions (Fig. 1, B and D and data not shown). Interestingly, WT NBD peptides caused a slight increase in basal cytokine production, data that are consistent with previous findings on NF-κB reporter gene activity (10). In contrast, the selective IKK-2 kinase inhibitor, PHA-408 (18, 19), inhibited both LPS-induced and basal TNF-α production in PBMC with equal potency (IC50 ∼ 40–70 nm) (Fig. 1, E and F). Basal cytokine levels represented ∼10% of cytokine production induced by LPS or IL-1β in the two cell types tested (data not shown). Furthermore, WT but not mutant NBD peptides inhibited cytokine-induced release of glycosaminoglycans from bovine nasal cartilage explant cultures (data not shown). None of the inhibitory effects described here were associated with cytotoxicity (data not shown). Taken together, these data suggest that the mechanisms of action of NBD peptides are distinct from those of ATP kinase inhibitors of IKK-2.
FIGURE 1.
Inhibition of inflammation-induced but not basal cytokine production by NBD peptides. PBMC were preincubated with the indicated concentrations of NBD peptides (A and B) or PHA-408 (E and F) for 2 h prior to stimulation with LPS (A and E) or vehicle (B and F) for 18 h. RASF were preincubated with the indicated concentrations of NBD peptides (C and D) for 2 h prior to stimulation with IL-1β (C) or vehicle (D) for 18 h. Filled and open columns represent mutant (Mut) NBD peptides and WT NBD peptides, respectively. Data are expressed as percentage of LPS- or IL-1β-stimulated cells or untreated cells (Basal). The data represent the mean ± S.D. of at least three independent determinations from one representative experiment. The experiments were performed at least twice with similar results. **, p < 0.005, significant differences compared with LPS or IL-1β-stimulated cells. NBD peptides used were IKK-2 11-mer.
NBD Peptides Inhibit IL-1β-induced IKK-2 Phosphorylation and Activation in RASF
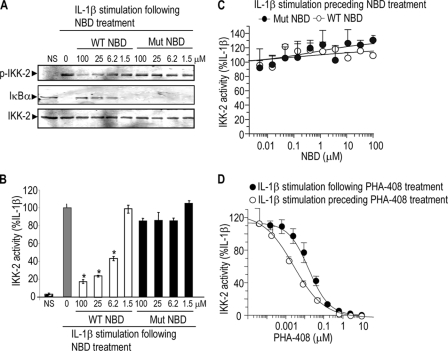
IKK-2 is activated by phosphorylation at Ser181 in the kinase activation loop (22, 23). To gain further insights into the mechanisms of action of NBD peptides, RASF were pretreated with NBD peptides prior to treatment with vehicle or IL-1β. Cell lysates were used both for Western blot analysis, probing for phosphorylation of IKK-2 and loss of IκBα, and to measure directly IKK kinase activation by monitoring phosphorylation of the substrate IκBα peptide. The data indicated that WT but not mutant NBD peptides inhibited IKK-2 phosphorylation at Ser181 and IκBα degradation (Fig. 2A) as well as the ability of IKK to phosphorylate IκBα (Fig. 2B) in a concentration-dependent manner. Interestingly, if RASF were first stimulated with IL-1β and the lysates were then incubated with NBD peptides, the peptides failed to inhibit IKK activity (Fig. 2C), IKK-2 phosphorylation, and IκBα degradation (supplemental Fig. S1). In contrast, PHA-408 inhibited IKK activity when it was preincubated with RASF prior to IL-1β stimulation or added to lysates from IL-1β-stimulated cells (comparable IC50 in the 30–60 nm range) (Fig. 2D). Together, these data indicate that NBD peptides inhibit NF-κB signaling by preventing IKK-2 activation via phosphorylation.
FIGURE 2.
Inhibition of IL-1β-induced IKK-2 phosphorylation and activity in RASF by NBD peptides. RASF were stimulated for 10 min with 1 ng/ml IL-1β following cell treatment for 1 h with NBD peptides (A and B) or PHA-408 (D, closed circles) or were stimulated for 10 min with 1 ng/ml IL-1β preceding the treatment of cell lysates for 1 h with NBD peptides (C) or PHA-408 (D, open circles). Cell lysates from the same experiment (A and B) or separate experiments (C and D) were used for Western blot analysis (A) or kinase activity (B–D). The data (B–D) represent the mean ± S.D. of at least three independent determinations. The experiments were repeated at least twice with similar results. *, p < 0.05, significant differences compared with IL-1β-stimulated cells. NS, nonstimulation. NBD peptides used were IKK-2 11-mer. Mut, mutant.
NBD Peptides Do Not Affect Signaling Intermediates Upstream of the IKK Complex
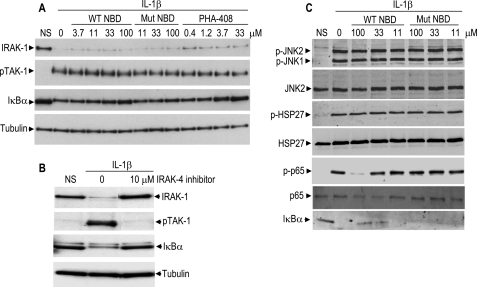
Based on the findings that NBD peptides inhibited IKK-2 phosphorylation, we hypothesized that these peptides affect upstream components of this pathway in a feedback mechanism. Stimulation of RASF with IL-1β resulted in the disappearance of the band corresponding to IRAK-1 (interleukin-1 receptor-associated kinase-1), phosphorylation of TAK-1 (transforming growth factor-β-activated kinase-1), and degradation of IκBα (Fig. 3A). Similar to the IKK-2 inhibitor, PHA-408, WT NBD peptides inhibited IκBα degradation in a concentration-dependent manner but had no effect on IRAK-1 fate and TAK-1 phosphorylation. Mutant NBD peptides failed to protect IκBα from degradation and to affect IRAK-1 fate or TAK-1 fate. In contrast, the IRAK-4 inhibitor, compound 44 (20), inhibited signaling of all components of the pathway that were assayed, IRAK-1, TAK-1, and IκBα (Fig. 3B). We also wanted to establish that the NBD peptide mode of action was specific to the IKK-NF-κB pathway. NBD peptides at concentrations that inhibited p65 phosphorylation at Ser536 and IκBα degradation had no effect on JNK and p38 MAPK pathways (Fig. 3C), results that are consistent with previous reports (10). Collectively, these data suggest that NBD peptides act specifically at the IKK complex to disrupt IKK-2 phosphorylation by upstream kinase(s), including TAK-1.
FIGURE 3.
Lack of inhibitory effects of NBD peptides on NF-κB signaling intermediates upstream of the IKK complex and MAPK pathway. RASF were treated with the indicated concentrations of NBD peptides (A and C) or PHA-408 (A) or IRAK-4 inhibitor (B) for 30 min, followed by stimulation with 1 ng/ml IL-1β for 10 min. Cell lysates were analyzed by Western blot. Tubulin was used as a control for protein loading. NS, nonstimulation. NBD peptides used were IKK-2 11-mer. Mut, mutant.
NBD Peptides Weakly Inhibit the Binding of Recombinant NEMO and IKK-2 in Vitro
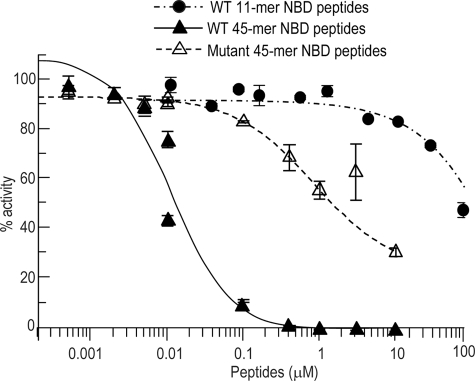
An Alphascreen-based assay was used to investigate the ability of NBD peptides to disrupt the interactions of purified recombinant NEMO and IKK-2. A truncated version of NEMO was used that included all regions necessary for interactions with IKK-2 (5, 24, 25). NEMO-(2–200) was expressed as a GST fusion protein, and a FLAG tag was added to the C terminus of full-length IKK-2. The addition of the tag served as an aide for purification as well as providing convenient epitopes to establish the Alphascreen-based assay. GST-NEMO-(2–200) and IKK-2-FLAG but not mutant NBD peptides (W739A and W741A) produced a significant signal, demonstrating specific interactions between the two proteins (data not shown). Having established that the signal was indeed due to the NEMO-IKK-2 interactions at the NBD region, we next wanted to determine if NBD peptides could disrupt these interactions. The addition of the longer WT NBD peptides (IKK-2 45-mer) blocked the interactions in a concentration-dependent manner (Fig. 4). These peptides had no effect on the signal generated by a control GST-FLAG fusion construct used to monitor nonspecific binding (supplemental Fig. S2). The version mutant of IKK-2 45-mer containing W739A and/or W741A possessed significantly weaker activity with IC50 of >1 μm, confirming the importance of these two residues in generating a high affinity interaction. The minimally sized WT NBD peptides (IKK-2 11-mer) blocked this interaction at much higher concentrations, exhibiting weak inhibitory effects (40%) at 100 μm.
FIGURE 4.
Differential potency of NBD-containing peptides. Purified recombinant GST-NEMO-(2–200) (2 nm) was incubated with purified recombinant C terminus FLAG-tagged full-length IKK-2 (2 nm). The ability of WT or mutant (W739A and W741A) NBD peptides to disrupt NEMO-IKK-2 binding was measured by Alphascreen. The data represent the mean ±S.E. of four independent experiments.
The interactions of the NBD containing peptides with GST-NEMO-(2–200) were also examined by time-resolved fluorescence resonance energy transfer. The IKK-2 45-mer and the shorter NBD peptides competed for NEMO binding, with IC50 in the 1–7 nm range and >100 μm, respectively. These results were consistent with those produced by the Alphascreen assay. We also performed fluorescence polarization and Biacore experiments to generate apparent binding affinities for the IKK-2 NBD-containing peptides with NEMO-(2–200). The results obtained were comparable with those from the Alphascreen- and time-resolved fluorescence resonance energy transfer-based assays (supplemental Table 1). Overall, the data were in agreement with those in the literature (5). Together, these data provided evidence that IKK-2 residues outside of the consensus NBD core sequence contributed to the formation of a high affinity interaction with NEMO. The weak potency of NBD peptides (11-mer) in inhibiting the binding of recombinant NEMO and IKK-2 led us to explore alternative systems that might better reflect the in vivo situation in order to study these interactions.
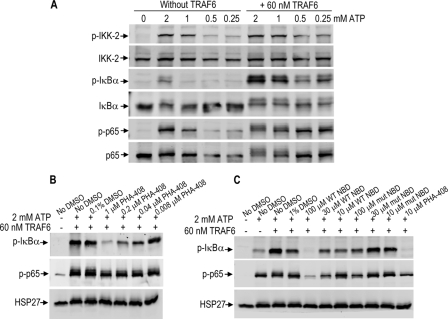
NBD Peptides Inhibit TRAF-6-dependent Activation of the IKK Complex in a Cell-free System
To expand on the findings that TRAF6 activates the IKK complex in a cell-free system (21), experiments were carried out with HeLa cell lysates and various concentrations of TRAF6. Based on dose findings studies (data not shown), 60 nm TRAF6 was chosen for the subsequent studies as the optimal concentration to activate the IKK complex. Next, cell lysates were incubated with increasing concentrations of ATP in the presence of 60 nm TRAF6. The addition of ATP alone dose-dependently increased phosphorylation of IKK-2 and p65. However, IκBα phosphorylation and to a lesser extent p65 phosphorylation were dependent on the addition of exogenous TRAF6 (Fig. 5A). This system was validated pharmacologically using the selective IKK-2 inhibitor, PHA-408. The addition of PHA-408 inhibited IκBα phosphorylation and to a lesser extent p65 phosphorylation (Fig. 5B). Similarly, WT NBD peptides inhibited IκBα and p65 phosphorylation in a concentration-dependent manner, whereas the mutant NBD peptides exhibited only a slight inhibitory effect at 100 μm (Fig. 5C). Although not quantitative, these results suggest that the native IKK complex in cells is more sensitive to the inhibitory effects of NBD peptides as compared with the complex formed in vitro by the purified recombinant NEMO and IKK-2 proteins.
FIGURE 5.
Disruption of TRAF-6-dependent NEMO-IKK-2 interactions in cell-free system by NBD peptides. A, HeLa cell extracts were prepared and treated with 0, 0.25, 0.5, 1, or 2 mm ATP with or without 60 nm exogenous TRAF6. B and C, HeLa cell extracts were treated with 0.1 or 1% DMSO (vehicle), PHA-408, WT, or mutant NBD peptides (0.1 or 1% DMSO, final concentration) in the absence or presence of 2 mm ATP and 60 nm exogenous TRAF6 and were analyzed by Western blot. The experiments were repeated three times with similar results. HSP27 was used as a control for protein loading. NBD peptides used were IKK-2 11-mer.
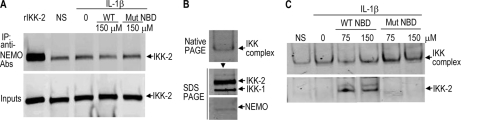
NBD Peptides Release IKK-2 from an IKK Complex in Cells
To determine the effects of NBD peptides on NEMO-IKK-2 interactions in cells, RASF were preincubated with NBD peptides for 30 min prior to treatment with IL-1β for 10 min. The IKK complex was immunoprecipitated from cell lysates using anti-NEMO Abs. Western blot analysis of the immunoprecipitated material using anti-IKK-2 Abs (Fig. 6A) revealed that the peptides at the concentrations that completely inhibited NF-κB signaling (Fig. 2, A and B) and IKK complex activity in the TRAF6 assay (Fig. 5C) had no significant effect on the relative concentration of IKK-2 in the complex. Based on these data and despite a previous demonstration by immunoprecipitation that NBD peptides inhibited NEMO-IKK-2 binding (10), a more sensitive assay was needed to monitor the effects of NBD peptides on the native complex.
FIGURE 6.
Release of IKK-2 from an IKK complex in RASF by NBD peptides. A, RASF were pretreated with 150 μm of WT or mutant (Mut) NBD peptides for 30 min prior to a 10-min treatment with 1 ng/ml IL-1β. The IKK complex was immunoprecipitated (IP) with anti-NEMO mAbs, and the immunoprecipitated material (top) as well as the original cell lysates (Inputs, bottom) were subjected to Western blot analysis. RASF lysates were also subjected to native PAGE and Western blot analysis using anti-NEMO mAbs (B, top). The band corresponding to the IKK complex was excised, and the proteins were extracted in 2× reducing SDS sample loading buffer, resolved using SDS-PAGE, and subjected to Western blot analysis using anti-IKK-1/2 Abs (B, middle) or anti-NEMO mAbs (B, bottom). C, RASF cells were preincubated with the indicated concentrations of WT or mutant NBD peptides for 30 min before stimulation with IL-1β for 10 min, and samples were analyzed by Western blot. Immunoreactive bands were visualized using the Li-Cor Odyssey Infrared Imaging System. The experiments were repeated 2–4 times with similar results. NS, nonstimulation. NBD peptides used were IKK-2 11-mer.
Toward this end, lysates from RASF were subjected directly to native PAGE. Western blot analysis using anti-NEMO Abs revealed an immunoreactive band of ∼150–250 kDa (Fig. 5B, top). When this region of the gel was excised and proteins were extracted and resolved by denaturing SDS-PAGE, Western blot analysis identified IKK-1, IKK-2, and NEMO as the components of the ∼150–250-kDa complex (Fig. 5B, middle and bottom). This experiment was repeated, but this time cells were preincubated with NBD peptides followed by treatment with IL1-β. The data from the native gel show that treatment with WT but not mutant NBD peptides led to the appearance of an IKK-2 band distinct from the complex (Fig. 6C), which was probably released from an IKK complex. The identity of the band migrating above IKK-2 is unknown. Note that the release of IKK-2 did not change the motility of the ∼150–250-kDa IKK complex. Similar data were obtained when unstimulated RASF (no treatment with IL-1β) were treated with WT NBD peptides (data not shown). These novel data suggest that NBD peptides induced IKK-2 release by disrupting NEMO-IKK-2 interactions within a preformed complex.
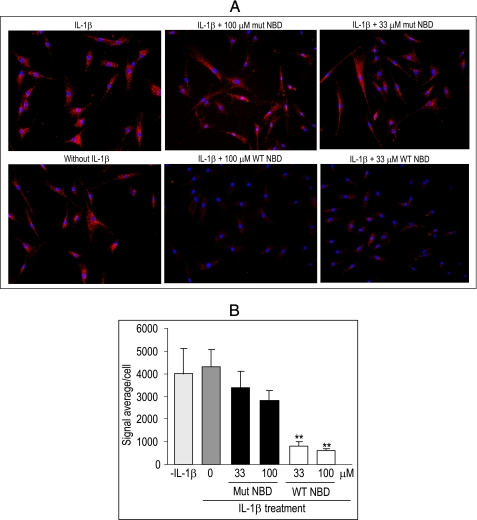
NBD Peptides Disrupt NEMO-IKK-2 Interactions in Cells
The PLA where a fluorescence signal is generated when two PLA probes bound to the targeted molecules are in close proximity has been used successfully to study protein-protein interactions in cells (26). To study NEMO-IKK-2 interactions, fixed RASF were incubated with anti-NEMO Abs and anti-IKK-2 Abs either together or separately. No fluorescence signal was detected when cells were incubated with individual Abs (data not shown). In contrast, strong cell staining was observed when cells were incubated with anti-NEMO Abs and anti-IKK-2 Abs (Fig. 7A). Red dots in PLA represent NEMO-IKK-2 interactions. The intensity of the staining was not different in the absence or presence of IL-1β, data that are consistent with the existence of the IKK complex in unstimulated cells (Fig. 6). Importantly, this staining was inhibited by WT but not mutant peptides, as confirmed by quantitative assessment (Fig. 7B). Consistent with data described above, WT NBD peptides also inhibited NEMO-IKK-2 interactions in unstimulated cells (data not shown). PHA-408 had no effect on NEMO-IKK-2 interactions, as expected (supplemental Fig. S3). These data provide strong evidence that NBD peptides disrupt NEMO-IKK-2 interactions in cells.
FIGURE 7.
Disruption of NEMO-IKK-2 interactions in RASF by NBD peptides. RASF cells were incubated with the indicated concentrations of WT or mutant (mut) NBD peptides for 30 min before stimulation with 1 ng/ml IL-1β for 15 min. NEMO-IKK2 interactions were visualized at ×20 magnification (A) using PLA as detailed under “Experimental Procedures.” The signal was also quantified from an average of nine fields, which was then divided by the numbers of cells in each field, thus giving a signal average/cell (B). The experiments were repeated three times with similar results. The data represent the mean ± S.E. **, p < 0.005, significant differences compared with IL-1β-stimulated cells. NBD peptides used were IKK-2 11-mer.
DISCUSSION
Central to the regulation of the NF-κB pathway is the IKK complex, which consists of two catalytic subunits, IKK-1 and IKK-2, and a regulatory subunit, NEMO. Different models for the regulation of the catalytic subunits by NEMO have been proposed. NEMO may facilitate the recruitment of the IKK complex to activated receptors, where the complex may in turn be activated by oligomerization, conformational changes, or trans-autophosphorylation (7–9, 27). Alternatively, NEMO may position the catalytic subunits for targeted phosphorylation of the activation loop by an upstream kinase (28) or enable the IKKs to interact with IκBs. We found that NBD peptides inhibited IL-1β-induced IKK-2 activation and activity in RASF, data that are consistent with a recent report (16) that was published while these studies were still under completion. We also observed that once the IKK complex in cells had been activated by exposure to cytokines, NBD peptides could no longer block IKK-2-mediated phosphorylation of IκBα. These data suggest that IKK-2 may phosphorylate itself independently of NEMO to maintain signaling and rule out NEMO as the facilitator of IKK interaction with downstream substrates, such as IκBα. Several mitogen-activated protein kinase kinase kinases, including TAK-1, mitogen-activated protein kinase/extracellular signal-regulated kinase kinase (MEK) kinase (MEKK)-1, and MEKK-3 are potential IKK-phosphorylating enzymes (28). We found that in response to IL-1β treatment, TAK-1 phosphorylation in cells correlated well with the activation or fate of the other components of the pathway, including IRAK-1, IκBα and p65. Interestingly, NBD peptides at concentrations that blocked p65 activation had no effect on TAK-1 phosphorylation or the fate of IRAK-1. These findings also rule out a positive feedback mechanism originating from the IKK complex toward upstream intermediates of the pathway, such as TAK-1 and IRAK-1. Thus, our data favor a model where NEMO is required for IKK phosphorylation by upstream kinase(s), such as TAK-1, in response to IL-1β signaling in RASF.
Despite the overwhelming data on the efficacy of cell-permeable NBD (IKK-2 11-mer) peptides in suppressing cellular inflammatory processes with IC50 in the 50–100 μm range, the effects of these peptides in blocking NEMO-IKK-2 interactions have not been fully elucidated. In this study, we found that these peptides weakly inhibited recombinant NEMO and IKK-2 binding with IC50 higher than 100 μm in a variety of in vitro biochemical and biophysical assay formats. These data are consistent with previous reports (12, 25). The weak potency of these peptides is expected because IKK-2 residues outside of the consensus NBD core sequence contribute to the formation of a high affinity interaction with NEMO. In agreement with this notion and previous studies (5, 25), we found that IKK-2 45-mer was highly potent in inhibiting NEMO-IKK-2 interactions. It is also possible that the reagents used for the biochemical studies might not accurately mimic the IKK complex, which exists as a four-helix bundle (5), and the short NBD peptides may not bind as well. This hypothesis was tested in the TRAF6-dependent IKK complex activation in a cell-free system, where IKK-1, IKK-2, and NEMO are expected to form a tetrameric holocomplex. Although the data are not quantitative, they show that NBD peptides at 100 μm completely suppressed IκBα and p65 phosphorylation in this system. Thus, as we had hypothesized, the native IKK complex was more sensitive to the inhibitory effects of NBD peptides than the one formed by recombinant NEMO and IKK-2.
Our data demonstrate that treatment of cells with WT but not mutant NBD peptides resulted in the release of IKK-2 from an IKK complex, indicating the disruption of this complex. Unexpectedly, we found no apparent changes in the motility of the ∼150–250 kDa band, which corresponded to the IKK complex consisting of IKK-1, IKK-2, and NEMO. One potential explanation of this finding is that NBD peptides at the concentrations used in this study led to the release of a discrete fraction of IKK-2 from the ∼150–250-kDa complex. Alternatively, IKK-2 could have been released from a different IKK complex, which was not detected by Western blot analysis following the native PAGE. However, since IKK-2 was also released by NBD peptides in unstimulated cells, it is likely that it originated from a basic IKK complex, not from a high order complex that is assembled in response to cell stimulation. Regardless of the source of the IKK-2, these data indicate that NBD peptides disrupted NEMO-IKK-2 interactions in cells. This conclusion was strengthened by imaging data showing that the peptides decreased the signal that is generated when the two proteins are in close proximity. In agreement with the existence of the basic IKK complex in unstimulated cells, NEMO-IKK-2 interactions were also visualized in basal conditions. Although collectively, pharmacology experiments indicate that NBD peptides inhibited NEMO-IKK-2 interactions, it is worth noting that because NBD peptides have been shown to inhibit NEMO-IKK-1 interactions (12), and IL-1β can activate the canonical NF-κB pathway independently of IKK-2 (29, 30), some effects of these peptides on NEMO-IKK-1 interactions cannot be ruled out.
Despite its attractiveness for therapeutic intervention in disease states, the NF-κB pathway is expressed ubiquitously and is involved in normal tissue homeostasis and thus cannot be completely blocked. Concerns for targeting this pathway arose from findings of liver toxicity in NEMO or IKK-2 knock-out mice (6, 31) as well as mice treated with some IKK-2 kinase inhibitors (32). Together, our data demonstrate that one can disrupt the IKK-NEMO complex without affecting basal activity. Our data are consistent with previous reports showing the inhibition of TNF-α-induced but not basal NF-κB reporter gene activity (10). Furthermore, NEMO knock-out mouse embryonic fibroblasts appeared to exhibit normal basal NF-κB DNA binding activity and IL-6 production (6). The mechanisms by which these peptides spare basal NF-κB activity are not completely understood. Because several species of these proteins, including IKK-2 or NEMO homodimers, exist in cells, basal NF-κB functions could be carried out by NEMO-independent mechanisms. Alternatively, because NEMO dimerization and interaction with IKKs are negatively regulated by phosphorylation, as discussed previously (12, 33), it can be speculated that phosphorylation/dephosphorylation events may be differentially regulated, depending on the state of cell activation, and may have implications on the effects of NBD peptides. Whatever the mechanisms may be, they need to be understood because the notion of blocking proinflammatory activation of NF-κB signaling while sparing basal levels of signaling necessary for normal cell maintenance will be key in developing a drug that targets this pathway.
In summary, this study provides new evidence for distinguishing the inhibition of the NF-κB pathway by NBD peptides from the classical ATP competitive IKK-2 inhibitors. Furthermore, we have delineated the steps targeted by NBD peptides and presented direct evidence for the disruption of NEMO-IKK-2 interactions in cells by these peptides.
Supplementary Material
Acknowledgment
We thank Dr. Robert Arch for critical reading of the manuscript.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. S1–S3.
- NF-κB
- nuclear factor κB
- Ab
- antibody
- GST
- glutathione S-transferase
- PBMC
- peripheral blood mononucleated cells
- WT
- wild type
- IκB
- inhibitor of κB
- IKK
- IκB kinase
- NEMO
- NF-κB essential modulator
- NBD
- NEMO binding domain
- IL
- interleukin
- LPS
- lipopolysaccharide
- TNF
- tumor necrosis factor
- PLA
- proximity ligation assay
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- mAb
- monoclonal antibody
- RASF
- rheumatoid arthritis synovial fibroblast(s)
- MAPK
- mitogen-activated protein kinase
- JNK
- c-Jun N-terminal kinase.
REFERENCES
- 1.Hayden M. S., Ghosh S. (2008) Cell 132, 344–362 [DOI] [PubMed] [Google Scholar]
- 2.Chen Z., Hagler J., Palombella V. J., Melandri F., Scherer D., Ballard D., Maniatis T. (1995) Genes Dev. 9, 1586–1597 [DOI] [PubMed] [Google Scholar]
- 3.DiDonato J. A., Mercurio F., Karin M. (1995) Mol. Cell Biol. 15, 1302–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karin M., Greten F. R. (2005) Nat. Rev. Immunol. 5, 749–759 [DOI] [PubMed] [Google Scholar]
- 5.Rushe M., Silvian L., Bixler S., Chen L. L., Cheung A., Bowes S., Cuervo H., Berkowitz S., Zheng T., Guckian K., Pellegrini M., Lugovskoy A. (2008) Structure 16, 798–808 [DOI] [PubMed] [Google Scholar]
- 6.Rudolph D., Yeh W. C., Wakeham A., Rudolph B., Nallainathan D., Potter J., Elia A. J., Mak T. W. (2000) Genes Dev. 14, 854–862 [PMC free article] [PubMed] [Google Scholar]
- 7.Li X. H., Fang X., Gaynor R. B. (2001) J. Biol. Chem. 276, 4494–4500 [DOI] [PubMed] [Google Scholar]
- 8.Poyet J. L., Srinivasula S. M., Lin J. H., Fernandes-Alnemri T., Yamaoka S., Tsichlis P. N., Alnemri E. S. (2000) J. Biol. Chem. 275, 37966–37977 [DOI] [PubMed] [Google Scholar]
- 9.Zhang S. Q., Kovalenko A., Cantarella G., Wallach D. (2000) Immunity 12, 301–311 [DOI] [PubMed] [Google Scholar]
- 10.May M. J., D'Acquisto F., Madge L. A., Glöckner J., Pober J. S., Ghosh S. (2000) Science 289, 1550–1554 [DOI] [PubMed] [Google Scholar]
- 11.Strnad J., McDonnell P. A., Riexinger D. J., Mapelli C., Cheng L., Gray H., Ryseck R. P., Burke J. R. (2006) J. Mol. Recognit. 19, 227–233 [DOI] [PubMed] [Google Scholar]
- 12.May M. J., Marienfeld R. B., Ghosh S. (2002) J. Biol. Chem. 277, 45992–46000 [DOI] [PubMed] [Google Scholar]
- 13.Dai S., Hirayama T., Abbas S., Abu-Amer Y. (2004) J. Biol. Chem. 279, 37219–37222 [DOI] [PubMed] [Google Scholar]
- 14.Jimi E., Aoki K., Saito H., D'Acquisto F., May M. J., Nakamura I., Sudo T., Kojima T., Okamoto F., Fukushima H., Okabe K., Ohya K., Ghosh S. (2004) Nat. Med. 10, 617–624 [DOI] [PubMed] [Google Scholar]
- 15.Davé S. H., Tilstra J. S., Matsuoka K., Li F., Karrasch T., Uno J. K., Sepulveda A. R., Jobin C., Baldwin A. S., Robbins P. D., Plevy S. E. (2007) J. Immunol. 179, 7852–7859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shibata W., Maeda S., Hikiba Y., Yanai A., Ohmae T., Sakamoto K., Nakagawa H., Ogura K., Omata M. (2007) J. Immunol. 179, 2681–2685 [DOI] [PubMed] [Google Scholar]
- 17.Ghosh A., Roy A., Liu X., Kordower J. H., Mufson E. J., Hartley D. M., Ghosh S., Mosley R. L., Gendelman H. E., Pahan K. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 18754–18759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mbalaviele G., Sommers C. D., Bonar S. L., Mathialagan S., Schindler J. F., Guzova J. A., Shaffer A. F., Melton M. A., Christine L. J., Tripp C. S., Chiang P. C., Thompson D. C., Hu Y., Kishore N. (2009) J. Pharmacol. Exp. Ther. 329, 14–25 [DOI] [PubMed] [Google Scholar]
- 19.Sommers C. D., Thompson J. M., Guzova J. A., Bonar S. L., Rader R. K., Mathialagan S., Venkatraman N., Holway V. W., Kahn L. E., Hu G., Garner D. S., Huang H. C., Chiang P. C., Schindler J. F., Hu Y., Meyer D. M., Kishore N. N. (2009) J. Pharmacol. Exp. Ther. 330, 377–388 [DOI] [PubMed] [Google Scholar]
- 20.Buckley G. M., Fosbeary R., Fraser J. L., Gowers L., Higueruelo A. P., James L. A., Jenkins K., Mack S. R., Morgan T., Parry D. M., Pitt W. R., Rausch O., Richard M. D., Sabin V. (2008) Bioorg. Med. Chem. Lett. 18, 3656–3660 [DOI] [PubMed] [Google Scholar]
- 21.Deng L., Wang C., Spencer E., Yang L., Braun A., You J., Slaughter C., Pickart C., Chen Z. J. (2000) Cell 103, 351–361 [DOI] [PubMed] [Google Scholar]
- 22.Delhase M., Hayakawa M., Chen Y., Karin M. (1999) Science 284, 309–313 [DOI] [PubMed] [Google Scholar]
- 23.Mercurio F., Zhu H., Murray B. W., Shevchenko A., Bennett B. L., Li J., Young D. B., Barbosa M., Mann M., Manning A., Rao A. (1997) Science 278, 860–866 [DOI] [PubMed] [Google Scholar]
- 24.Drew D., Shimada E., Huynh K., Bergqvist S., Talwar R., Karin M., Ghosh G. (2007) Biochemistry 46, 12482–12490 [DOI] [PubMed] [Google Scholar]
- 25.Lo Y. C., Maddineni U., Chung J. Y., Rich R. L., Myszka D. G., Wu H. (2008) Biochemistry 47, 3109–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Söderberg O., Gullberg M., Jarvius M., Ridderstråle K., Leuchowius K. J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L. G., Landegren U. (2006) Nat. Methods 3, 995–1000 [DOI] [PubMed] [Google Scholar]
- 27.Agou F., Courtois G., Chiaravalli J., Baleux F., Coïc Y. M., Traincard F., Israël A., Véron M. (2004) J. Biol. Chem. 279, 54248–54257 [DOI] [PubMed] [Google Scholar]
- 28.Takaesu G., Surabhi R. M., Park K. J., Ninomiya-Tsuji J., Matsumoto K., Gaynor R. B. (2003) J. Mol. Biol. 326, 105–115 [DOI] [PubMed] [Google Scholar]
- 29.Solt L. A., Madge L. A., Orange J. S., May M. J. (2007) J. Biol. Chem. 282, 8724–8733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Solt L. A., Madge L. A., May M. J. (2009) J. Biol. Chem. 284, 27596–27608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Q., Van Antwerp D., Mercurio F., Lee K. F., Verma I. M. (1999) Science 284, 321–325 [DOI] [PubMed] [Google Scholar]
- 32.Nagashima K., Sasseville V. G., Wen D., Bielecki A., Yang H., Simpson C., Grant E., Hepperle M., Harriman G., Jaffee B., Ocain T., Xu Y., Fraser C. C. (2006) Blood 107, 4266–4273 [DOI] [PubMed] [Google Scholar]
- 33.Palkowitsch L., Leidner J., Ghosh S., Marienfeld R. B. (2008) J. Biol. Chem. 283, 76–86 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.