Abstract
Erk-5, a member of the MAPK superfamily, has a catalytic domain similar to Erk1/2 and a unique C-terminal domain enabling binding with transcription factors. Aberrant vascularization in the Erk5-null mice suggested a link to angiogenesis. Ectopic expression of constitutively active Erk5 blocks endothelial cell morphogenesis and causes HIF1-α destabilization/degradation. However the mechanisms by which endogenous Erk5 regulates angiogenesis remain unknown. We show that Erk5 and its activating kinase MEK5 are the upstream mediators of the anti-angiogenic signal by the natural angiogenesis inhibitor, pigment epithelial-derived factor (PEDF). We demonstrate that Erk5 phosphorylation allows activation of PPARγ transcription factor by displacement of SMRT co-repressor. PPARγ, in turn is critical for NFκB activation, PEDF-dependent apoptosis, and anti-angiogenesis. The dominant negative MEK5 mutant and Erk5 shRNA diminished PEDF-dependent apoptosis, inhibition of the endothelial cell chemotaxis, and angiogenesis. This is the first evidence of Erk5-dependent transduction of signals by endogenous angiogenesis inhibitors.
Keywords: ERK, NF-κB Transcription Factor, PPAR, Signal Transduction, Transcription Regulation, Angiogenesis, Natural Inhibitors, SMRT (NCoR2), Transcriptional Complexes
Introduction
Extracellular signal-regulated kinase 5 (Erk5)2 is a member of the MAP kinase superfamily, which includes extracellular signal-regulated kinases (Erk1/2), c-Jun N-terminal kinases (JNK1–4), p38 kinases, and Erk5/BMK1 (big mitogen-activated protein kinase). Some of the MAP kinases play important roles in the process of angiogenesis, the growth of the new capillaries. For example, mice deficient for the p38-α that die in utero are attributed to abnormalities in placental angiogenesis (1, 2). Mice null for p38 also display anemia, caused by the deficient production of erythropoietin (Epo), which can act as an inducer of angiogenesis (3, 4). Because Epo expression is increased in response to low oxygen levels, it is not unlikely that hypoxic stress may activate p38 to enhance Epo mRNA synthesis; a similar effect has been observed in hepatoma cells (4).
Erk5 constitutes a separate class of MAP kinases. Whereas its catalytic domain is homologous to that of Erk1/2, the Erk5 C-terminal domain is unique and enables its physical association with transcription factors from the myocyte enhancer factor-2 (MEF2) family (5, 6). On the other hand, Erk5 interacts with p38, which is also capable of activation of MEF2C (7). Mice deficient for Erk5 display striking angiogenic defects in the placenta, yolk sack, and in the brain. Erk-5-null mice also have heart abnormalities, including defective myocardial walls and disorganized trabeculae (8). Not surprisingly, the mice with a knock-out of the Erk5 upstream activating kinase, MEKK3 or of Erk5 target, transcription factor, MEF2C, have similar defects in angiogenesis (2, 9). Whereas angiogenesis defects in p38-null mice are largely similar, the lack of cardiac abnormalities suggests that Erk5 and p38 regulate cardiac development via distinct pathways (1).
Developmental defects in the Erk5 knock-out embryos occur at the time when the embryonic vasculature becomes exposed to increasing laminar flow and shear stress. Because shear stress can activate Erk5 (10), it is likely that Erk5 functions as a sensor and conveyor of the proper physiological responses to mechanical stress in the course of embryonic development.
Among the transcription factors regulated by Erk5 are hypoxia-inducible factor 1-α (HIF), MEF2C (10), lung Krüppel-like factor (LKLF) (7), and peroxisome proliferator-activated receptor γ (PPARγ) (11). Phosphorylation by Erk5 reduces the stability of HIF proteins and therefore VEGF production. The excessive levels of VEGF-A in the Erk5−/− embryos at embryonic day 9.5, especially under hypoxia, are likely to compromise vascular integrity by reducing pericyte investment and causing fenestration of the capillaries (8, 12, 13). Indeed, endothelial cells in Erk5-null animals appear both rounded and disorganized. Moreover, the investment of new vessels by the pericytes in Erk5 KO mice is severely attenuated, suggesting the failure to mature, similar to the immature state of the tumor microvasculature. Thus the lack of Erk5 activity in the vascular stroma contributes to the general destabilization of embryonic vasculature.
Erk5 binding to MEF2C transcription factor under hypoxic conditions activates the expression of the LKLF gene, whose product, another transcription factor, LKLF contributes to T-cell activation (7). In endothelial cells, Erk5 binds to the PPARγ inactive complexes with its co-repressor silencing mediator for retinoic acid receptor and thyroid hormone receptor (SMRT) or nuclear co-repressor 2 (NCoR2) via the PPARγ ligand binding region. Phosphorylation in response to shear stress results in unfolding of the Erk5 transactivation domain, which causes SMRT release and thus facilitates PPARγ activation (11).
Here we report the discovery that the natural inhibitor of angiogenesis can cause Erk5 activation in vascular endothelium and thereby block angiogenesis. We found, that pigment epithelial-derived factor (PEDF) induced Erk5 phosphorylation in remodeling endothelial cells. PEDF, a potent anti-angiogenic factor, blocks angiogenesis by causing endothelial cell apoptosis specifically in the remodeling vasculature (14). PEDF has been previously shown to up-regulate mRNA encoding CD95L, a ligand for the death receptor, CD95/Fas (15). CD95 surface presentation is limited to the activated, remodeling endothelium, thus enabling the selective susceptibility to the PEDF anti-angiogenic action. Our recent study demonstrates that PEDF drives CD95L expression via NFκB-dependent transcription, which is critical for PEDF-dependent apoptosis and anti-angiogenesis (35).
In this study we found that Erk5 activation by PEDF was critical for its anti-angiogenic action; a dominant-negative mutant of the Erk5-activating kinase, MEK5(A) (16) opposed PEDF anti-angiogenic action in vitro and in vivo, whereas the constitutively active MEK5 mutant, MEK5(D) (16) facilitated PEDF anti-angiogenic action. We found that PEDF increased phosphorylation of Erk5 associated with PPARγ, a transcription factor that has been recently identified as one of the molecular mediators of the PEDF angioinhibitory effect (17). Erk5 phosphorylation led to the release of SMRT transcriptional repressor from PPARγ transcriptional complexes. Surprisingly, NFκB activation by PEDF was critically dependent on PPARγ, as it was blocked by the synthetic PPARγ antagonist, T0070979. This is the first study that links Erk5 to the action of the naturally occurring angiogenesis inhibitor and demonstrates the importance of the endogenous Erk5 activation for blocking angiogenesis.
MATERIALS AND METHODS
Cells and Reagents
Human dermal capillary endothelial cells (HMVEC, Lonza, Allendale, NJ) were grown on a gelatinized surface in MCDB131 medium (Sigma), 5% fetal bovine serum, and SingleQuots supplements (Clonetics, North Brunswick, NJ). Recombinant human bFGF and VEGF were from R&D Systems (Minneapolis, MN). PEDF was purchased from Bioproducts (West Palm Beach, FL). PEDF 34-mer peptide (18) was synthesized to order (GenScript, Piscataway, NJ). The PPARγ inhibitor T0070979 was from Calbiochem.
Expression Vectors
Constitutively active (CA) and dominant-negative (DN) MEK5 mutants, MEK(D) and MEK(A), respectively (16, 19), cloned in pCMV5(HA)3 expression vector were provided by Dr. J-D. Lee (Scripps Research Institute, La Jolla, CA). Both inserts were excised by MluI/XhoI digestion and cloned in similarly digested pGIPZ lentiviral vector (Open Biosystems, Huntsville, AL). Precloned shRNAmir for Erk-5 in pGIPZ lentiviral vector was also from Open Biosystems.
Lentiviral Infection
HMVECs were infected with the lentiviruses at MOI 50. Cells were plated in 6-well plates the day prior to infection. Infection was carried out in MDCB131 medium supplemented with 1% fetal bovine serum and 8 μg/ml polybrene. After 4–5 h, the infection medium was replaced with HMVEC growth medium. Infected cells were passaged after 24–48 h. Infection efficiency was ∼95–98% as was estimated by GFP expression.
Western Blotting
EC whole cell extracts were prepared with M-PER reagent (Thermo Fisher, Pittsburgh, PA) per manufacturer's instructions. Proteins were separated by SDS-PAGE, transferred to polyvinylidene difluoride membranes, probed, and developed with an ECL kit (Amersham Biosciences). For pIκBα detection, ECs were pretreated with 10 μm MG132 proteasome inhibitor (Calbiochem). The following antibodies were used: phospho-ERK5 rabbit pAb, phospho-IκBα mouse mAb Ser32/36 were from Cell Signaling Technology, Carlsbad, CA); NFκB p65 rabbit pAb and PPARγ mouse mAb (Santa Cruz Biotechnology); tubulin mAb (Thermo Scientific, Waltham, MA); GAPDH horseradish peroxidase-conjugated mAb (Sigma). SMRT polyclonal antibody was raised against the peptide REELRRTPELPLAPRPLKE derived from the mouse SMRT protein sequence.
Immunoprecipitation
Cell extracts precleared with protein A/G-agarose beads (Santa Cruz Biotechnology) (1 h, 4 °C) were incubated overnight (4 °C) with indicated antibodies; the complexes were washed with lysis buffer, resolved by SDS-PAGE, and analyzed by Western blot.
Electrophoretic Mobility Shift Assay (EMSA)
Nuclear extracts were isolated as described previously (20). The cells were lysed in hypotonic buffer (20 mm HEPES-KOH, pH 7.9, 1.5 mm MgCl2, 25% glycerol, 50 mm NaCl, 0.2 mm EDTA, 0.5 mm dithiothreitol, and 0.2 mm phenylmethylsulfonyl fluoride), the nuclei pelleted, lysed in hypertonic buffer (20 mm HEPES-KOH, pH 7.9, 1.5 mm MgCl2, 25% glycerol, 420 mm NaCl, 0.2 mm EDTA, 0.5 mm dithiothreitol, and 0.2 mm phenylmethylsulfonyl fluoride) and cleared by centrifugation. EMSA was performed using NushiftTM PPARγ kit (Active Motif, Carlsbad, CA) according to the manufacturer's protocol. The biotinylated probe containing the PPRE consensus sequence was added to the nuclear extracts in binding buffer 0.05 mg/ml poly (dI-dC)) with or without unlabeled specific (wild-type PPRE) or nonspecific (mutant PPRE) competitor (15 min at room temperature). The samples were resolved by a 4% non-denaturing PAGE.
Real-time RT-PCR
Total RNAs were extracted and reverse transcribed with a SuperScript kit (Promega, Madison, WI). Q-PCR was performed with iQ SYBRGreen Supermix (Bio-Rad) using MJ Research Chromo4 thermocycler with primers 5′-GAGTTCATGCTTGTGAAGGATGC-3′ (forward) and 5′-CGATATCACTGGAGATCTCCGCC-3′ (reverse).
Immunostaining
Cells on coverslips were fixed in methanol/acetone (1:1) blocked (1 h at room temperature, 4% donkey serum) and incubated overnight (4 °C) with p65 antibodies (C20, Santa Cruz Biotechnology) and Cy3-conjugated secondary antibodies (Jackson Immunoresearch, West Grove, PA). Images obtained by fluorescent microscopy (Nikon Diaphot 2000) were quantified with MetaMorph software package (Molecular Devices, Sunnyvale, CA).
Endothelial Cell Chemotaxis Assay (21)
EC starved in serum-free medium supplemented with 0.1% bovine serum albumin (BSA, Sigma), were harvested, suspended at 1.5 × 106/ml and plated on the lower side of gelatinized membranes (8-μm pores, Nuclepore, Florham Park, NJ) in inverted modified Boyden chambers (Applied Biosystems, Foster City, CA). The cells were allowed 2 h to adhere at culture conditions (37 °C, 5% CO2). The test substances were diluted in MCDB131, 0.1% BSA and added to the upper wells, the cells incubated for an additional 3–4 h. The membranes were recovered fixed, stained, and mounted. Migrated cells were counted in 10 random high power (×400) fields per well. Each sample was tested in quadruplicate. Basic FGF (20 ng/ml), or VEGF (200 pg/ml) used to induce chemotaxis, served as positive controls, MCDB131 + 0.1% BSA was considered a negative control (random migration).
Apoptosis Assay
Apoptosis was measured using the TUNEL assay kit (Chemicon, Temecula, CA) as recommended by the manufacturer. Confluent HMVECs were grown on coverslips and treated overnight with PEDF or the 34-mer peptide, with or without protective bFGF (20 ng/ml) in MCDB131 supplemented 0.2% fetal bovine serum. The cells were counterstained with propidium iodide (PI); 3–5 ×10 fields were analyzed using an epi-fluorescent microscope: total cell number (PI-positive cells) and TUNEL-positive cells were quantified using MetaMorph software (Molecular Devices, Sunnyvale, CA), and % apoptotic cells calculated.
Scratch Wound Assay
Endothelial cells were grown to confluence in 24-well plates. A scratch wound was inflicted with a sterile pipette tip and identical areas photographed at 0 and 6 h of treatment. The wound area was measured in at least 10 independent ×10 fields per well and statistical significance determined by one-way ANOVA.
Animals
6–8-week-old female athymic nude mice (nu/nu) (Harlan, Indianapolis, IN) were kept in pathogen-free environment; autoclaved water and γ-irradiated commercial diet were given ad libitum. The mice were handled under sterile conditions, according to institutional guidelines and in compliance with the Guide for Care and Use of Laboratory Animals (National Institutes of Health).
Matrigel Assay (22)
Matrigel (BD Biosciences, San Jose, CA) was supplemented with VEGF (200 ng/ml), with or without the 34-mer PEDF peptide (100 μm). EC expressing GFP, CA-MEK5, or DN-MEK5 were mixed into Matrigel (105 cells/ml) and injected subcutaneously in the median abdominal area of nude mice. After 10–13 days, the plugs were excised and snap-frozen in liquid nitrogen.
Immunohistochemistry
Frozen Matrigel sections (5 μm) were fixed in sequential changes of acetone, acetone/chloroform, and acetone at −20 °C; the sections were stained with anti-CD31 rat anti-mouse antibody (4 μg/ml, BD Biosciences, San Jose, CA) followed by the donkey anti-rat Rhodamin Red-X conjugate (1:200 dilution, Jackson ImmunoResearch, West Grove, PA) to detect blood vessels. Morphometric analysis: microvascular density (MVD) was measured on digital images with MetaMorph software as a CD31-positive area and averaged between 6–10 random ×10 fields.
RESULTS
PEDF Induced Erk5 Activation
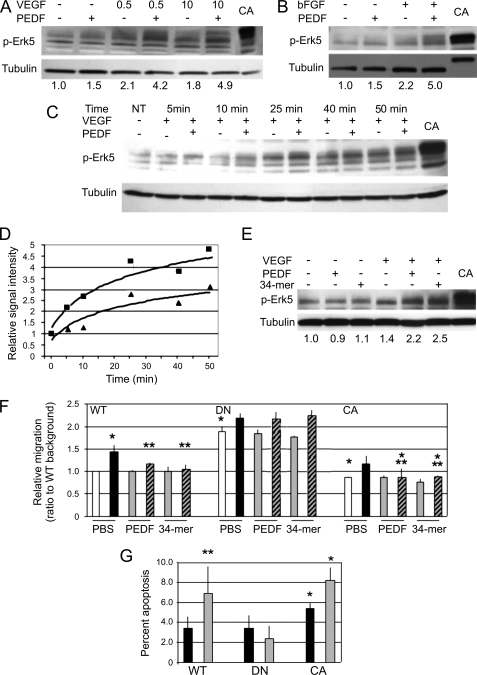
Treatment of the human microvascular endothelial cells (HMVECs) with anti-angiogenic concentration of PEDF (10 nm) caused increased Erk5 activation (phosphorylation) in a time-dependent manner. Erk5 activation by PEDF could be detected as early as after 5 min of treatment and lasted up to 1 h. This increase could be observed only if the endothelial cells were preactivated with bFGF (10 ng/ml) or VEGF (Fig. 1, A–C). Phosphorylation of Erk5 increased in a time-dependent manner, whereas, consistent with earlier studies some activation was noted in the presence of VEGF alone. However, in the presence of PEDF, Erk5 phosphorylation was significantly enhanced (Fig. 1, C and D). We have tested anti-angiogenic and neurotrophic PEDF fragments for the ability to induce Erk5 activation. The anti-angiogenic PEDF fragment, 34-mer (18) had a similar effect (Fig. 1E), suggesting that Erk5 activation may be important for PEDF-dependent anti-angiogenesis.
FIGURE 1.
Erk5 is activated by PEDF and is critical for PEDF angioinhibitory functions. A–D, Erk5 activation by PEDF. HMVECs were activated with VEGF at indicated doses (A) or bFGF (10 ng/ml, B) and treated with PEDF at an anti-angiogenic concentration of 10 nm. Total cell lysates were analyzed by Western blotting for phosphorylated Erk5 (p-Erk5). HMVECs expressing constitutively active MEK5 (CA) were used as a positive control. The membrane was stripped and probed for tubulin to assess loading. The results of densitometry analysis performed with ImageJ software are shown below. The results were normalized to the ratio between p-Erk5 and tubulin in untreated control samples. C and D, time-dependent induction of Erk5. The cells were treated with VEGF and PEDF (10 nm) for the indicated time periods, p-Erk5 was measured by Western blot (C), quantitative analysis was performed by densitometry and normalized against p-Erk/tubulin ratio in the untreated control (D). E, Erk5 activation by PEDF anti-angiogenic epitope. HMVECs were activated with VEGF (0.5 ng/ml) and treated fro 20 min with 20 nm PEDF or 20 nm 34-mer, as indicated. Densitometry analysis was performed as in A. F, Erk5 is required for PEDF blockade of the HMVEC chemotaxis. HMVECs were transfected with the dominant-negative and constitutively active MEK5 (DN and CA, respectively) or vector control (WT). Chemotaxis was induced with 20 ng/ml bFGF and blocked with 10 nm PEDF or with 10 nm 34-mer. The results were normalized to the WT background migration. *, p < 0.05; **, p < 0.01; ***, p < 0.0001. □, BSA control; ■, bFGF; gray, PEDF or the 34-mer; hatches, PEDF or the 34-mer + bFGF. G, role of Erk5 in PEDF-dependent apoptosis. HMVECs generated as in F were transferred in a serum-free medium, treated with protective bFGF (20 ng/ml) and 20 nm PEDF where indicated. *, p < 0.05; ***, p < 0.0001.
Erk5 Was Essential for the Angiogenesis Blockade by PEDF
To determine, if Erk5 was essential for PEDF anti-angiogenic signaling, we utilized the dominant-negative (DN-MEK5) and constitutively active (CA-MEK5) mutants of the Erk5-activating kinase, MEK5. We infected HMVECs with the lentiviral vectors expressing MEK5 mutants and empty vector to generate HMVEC population expressing CA-MEK5, DN-MEK5, and control population expressing GFP alone (HMVEC-CA, HMVEC-DN, and HMVEC-GFP, respectively). The resultant cells were tested in the endothelial cell chemotaxis assay. In HMVECs transfected with DN-MEK5 the inhibition of HMVEC chemotaxis by PEDF was significantly attenuated (Fig. 1F, p < 0.05). Similarly, DN-MEK5 dramatically relieved the inhibition of bFGF-induced chemotaxis by the 34-mer PEDF peptide (Fig. 1F). Moreover, HMVEC-DN showed significantly higher baseline migration, whereas MEK-CA expression mimicked the inhibitory effect of PEDF (Fig. 1F).
PEDF blocks angiogenesis by selectively inducing apoptosis of the activated endothelial cells (14). We therefore tested the effect of DN-MEK5 on PEDF-induced apoptosis. As was shown previously, bFGF protected HMVECs from stress-induced apoptosis and PEDF-induced cell death in the presence of protective bFGF. Interestingly, CA-MEK5 appeared to sensitize HMVECs to stress-induced apoptosis (p < 0.005), thus mimicking the effect of PEDF. DN-MEK5 had no effect on apoptosis under serum deprivation (this effect did not reach statistical significance, Fig. 1G) but completely abolished PEDF-induced cell death and CA-MEK5 caused a mild increase in PEDF-dependent apoptosis (p < 0.05) (Fig. 1G).
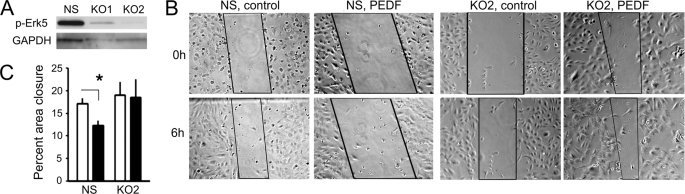
To eliminate the possibility of MEK5 affecting angiogenesis independent of Erk5, we have generated Erk5 knock-down cells using lentivirally expressed shRNAmir (Fig. 2A) and tested their ability to respond to PEDF in a scratch wound assay. In EC infected with non-silencing shRNA, PEDF treatment caused 30–35% reduction of the wound closure in a 6-h assay. In contrast, Erk5 knock-down resulted in similar rates of the wound closure in the absence and in the presence of PEDF (Fig. 2, B and C).
FIGURE 2.
Erk5 knock-down abrogates PEDF anti-angiogenic activity. A, Erk5 knock-down. We used two distinct shRNAmir cloned in pGIPZ lentiviral vectors. Western blot of the total cell lysates showed 70 and 90% knock-down (KO1 and KO2, respectively). KO2 population has been selected for further analysis. B and C, scratch wound assay was performed on confluent endothelial cell monolayer. B, representative images of the scratch wounds at 0 and 6 h are shown and the areas of the wound used for the quantitative measurements are marked. C, rate of the endothelial cell migration into the wound surface was measured as percent wound closure. □, control; ■, 20 nm PEDF. Asterisk indicates statistically significant difference (p < 0.05 by one-way ANOVA).
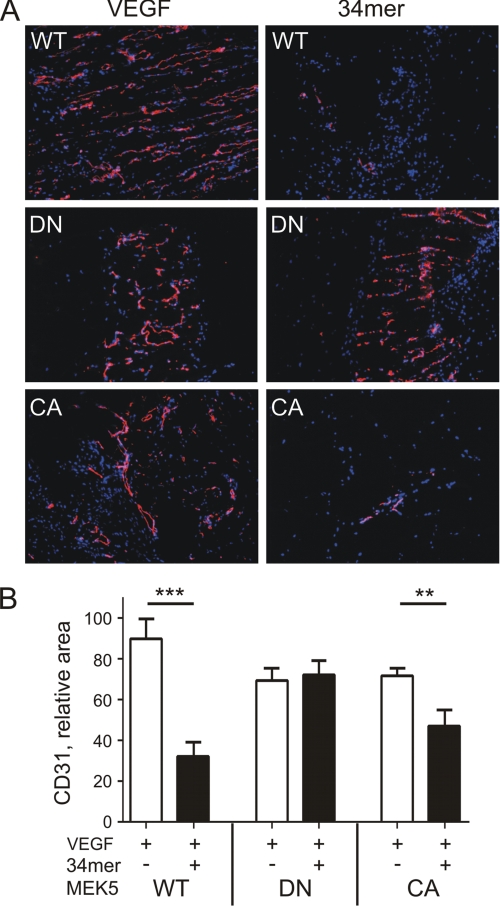
To test whether Erk5 contributes to PEDF anti-angiogenic effect in vivo, we used a Matrigel plug assay. In immune-compromised animals, human endothelial cells can survive and form viable capillaries, which form anastomoses with mouse neovessels (23). When used in Matrigel assay, HMVEC-CA, HMVEC-DN, and HMVEC-GFP formed new capillaries in response to pro-angiogenic VEGF with similar efficiency. However, in the case of HMVEC-DN, PEDF failed to block VEGF-induced angiogenesis (p < 0.05; Fig. 3, A and B). Thus the inhibition of Erk5 phosphorylation appeared critical for PEDF anti-angiogenic action in vitro and in vivo.
FIGURE 3.
Erk5 is critical for PEDF anti-angiogenic action in vivo. A, cryogenic sections of the Matrigel plugs containing control HMVECs (WT), or HMVECs expressing dominant negative or constitutively active MEK5 (DN and CA, respectively). Angiogenesis was induced with 200 ng/ml VEGF and blocked with 100 μm 34-mer, where indicated. The sections were stained for the endothelial cell marker, CD31 (PECAM1). B, morphometric analysis of the digital images of the sections shown in A with MetaMorph software package. Microvascular density was quantified as relative CD31-positive area. **, p < 0.01; **, p < 0.001.
PEDF Requires Erk5 Induction for PPARγ Activation
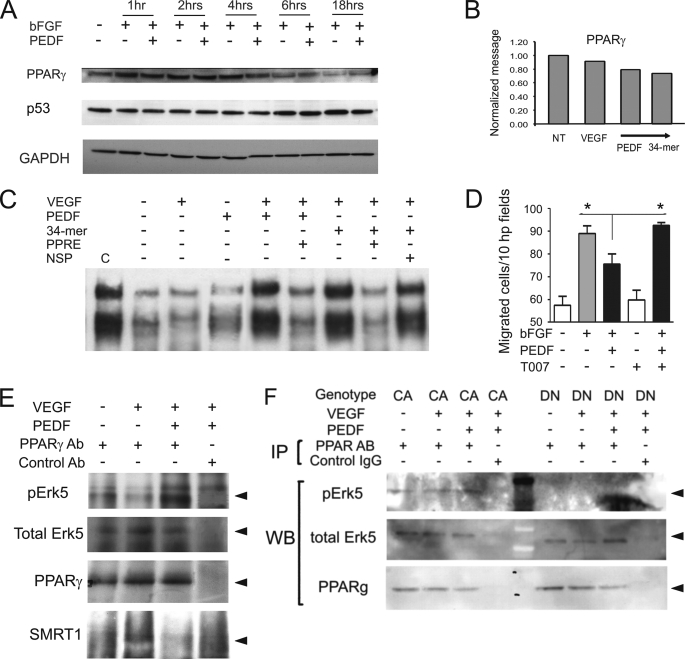
A member of the steroid nuclear receptor superfamily, PPARγ has been recently identified as a critical mediator of PEDF-dependent anti-angiogenic signaling (17, 24). On the other hand, Erk5 has been previously shown to bind and activate PPARγ (11). To investigate, whether PEDF-dependent Erk5 and PPARγ activation are connected, we measured PPARγ levels in HMVEC. In contrast with the published data, we were unable to detect an increase in the levels of PPARγ mRNA or protein (Fig. 4, A and B). However, EMSA with the probe containing PPRE consensus sequence showed increased DNA binding by PPARγ in the activated HMVECs treated with PEDF or the 34-mer (Fig. 4C). In contrast with previous reports, we were unable to detect p53 stabilization upon PEDF treatment (Fig. 4A). However, PPARγ was critical for PEDF anti-angiogenic function because PPARγ antagonist, T0070979 disrupted its ability to block endothelial cell migration (Fig. 4D). Immunoprecipitation analysis showed the increased amount of phosphorylated Erk5 associated with PPARγ in the PEDF-treated HMVECs (Fig. 4E). Interestingly, the amount of total Erk5 associated with PPARγ remained constant suggesting that PPARγ is constitutively associated with Erk5 and activated upon Erk5 phosphorylation. Further analysis of immune complexes between Erk5 and PPARγ showed increased recruitment of SMRT co-repressor in the presence of VEGF alone, which was abolished in the presence of PEDF. Moreover, in HMVEC-CA and in HMVEC-DN the amount of Erk5 precipitated with PPARγ was similar and remained constant regardless of PEDF treatment. Conversely, the baseline amount of p-Erk5 in Erk5/PPARγ complexes was higher in HMVEC-CA, while in HMVEC-DN Erk5 phosphorylation in Erk5/PPARγ complexes was below detection limit (Fig. 4F).
FIGURE 4.
Erk5 in the PPARγ transcription complexes is phosphorylated in response to PEDF. A, HMVECs were treated for the indicated time periods with the combinations of bFGF (10 ng/ml) and PEDF (10 nm). PPARγ and P53 levels were measured by Western blot. B, total RNA was extracted and PPARγ mRNA measured by real-time RT-PCR after 6 h of treatment. C, PPARγ mobility shift assay. HMVECs were treated with VEGF ± PEDF or the 34-mer, nuclear extracts were collected and subjected to EMSA with biotinylated PPRE probe. Non-biotinylated wild-type and non-biotinylated mutant PPRE probes were used as specific and nonspecific competitors (PPRE and NSP, respectively). C indicates positive control. D, PPARγ is necessary for PEDF inhibitory activity. HMVECs chemotaxis up the bFGF gradient (20 ng/ml across the membrane) was blocked by PEDF (20 nm) alone or in the presence of T0070907 (100 nm) Asterisks indicate statistically significant differences (p < 0.05, calculated by one-tailed Student's t test). E, wild-type HMVECs (MEK5-WT) were treated with PEDF, VEGF, or VEGF+PEDF. Untreated cells served as a negative control. Cell lysates were collected and subjected to immunoprecipitation with PPARγ antibody followed by Western blot with antibodies for p-Erk5, total Erk5, and SMRT. The input was controlled by Western blot with PPARγ antibody. F, HMVECs expressing MEK5-DN or MEK5-CA, were treated with VEGF (1 ng/ml) and PEDF (10 nm). Protein extracts were precipitated with PPARγ antibody and analyzed by Western blot for p-Erk5 and total Erk5. The input was controlled by Western blot with PPARγ antibody.
PPARγ Was Critical for the PEDF-driven NFκB Activation
We have recently ascribed NFκB the role in PEDF anti-angiogenic function whereas p65/p50 heterodimers drive the transcription of CD95L while p50/p50 homodimers simultaneously repress the transcription of the pro-survival molecule cFLIP. In concert, these events cause endothelial cell apoptosis (35). Similar to the other steroid nuclear receptors, PPARγ can regulate the activity of other transcription factors (25). To determine, whether PPARγ and NFκB activation by PEDF are connected, or exemplify the bifurcation of PEDF signaling pathway, we used synthetic PPARγ antagonist T0070979. T0070979 attenuated the increase of phosphorylated IκB caused by PEDF in bFGF-activated HMVECs (Fig. 5A). Pretreatment with T0070979 also abolished the NFκB nuclear localization caused by PEDF (Fig. 5B) suggesting PPARγ is necessary for NFκB activation. Unexpectedly, total NFκB levels were considerably higher in the activated HMVECs treated with PEDF or the 34-mer (Fig. 5C). Pretreatment with MG132, an inhibitor of proteasomal degradation resulted in uniformly high levels of NFκB p65 and p50 regardless of PEDF treatment (data not shown) suggesting that NFκB stability is increased in the presence of PEDF.
FIGURE 5.
PPARγ is critical for PEDF-dependent NFκB activation. PPARγ antagonist interferes with IκB-α phosphorylation in the presence of PEDF. HMVECs were pretreated 30 min with proteasome inhibitor MG132 (10 μm) and then treated 25 min with the indicated combinations of bFGF (10 ng/ml), PEDF (10 nm), and T0070907 (1 μm). A, whole cell extracts were subjected to Western blot analysis for IκB-α. B, HMVECs plated on coverslips were treated as in A and stained for p65/RelA. NFκB nuclear localization is indicated with white arrows. C, cells were activated with VEGF (0.5 ng/ml) and treated with PEDF or the 34-mer (10 nm). NFκB p65/RelA was measured in the whole cell extracts by Western blot.
DISCUSSION
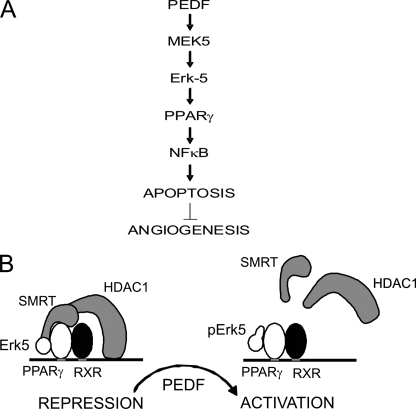
Angiogenesis inhibitors derived from the extracellular matrix (ECM) proteins fall into two main categories. The proteolytic fragments of ECM proteins, mainly collagens, such as endostatin and tumstatin signal predominantly through integrins and focal adhesions. Other secreted matricellular proteins such as thrombospondins and maspin transduce signals via specific cell surface receptors (reviewed in Refs. 26, 27). PEDF belongs to the latter category. Two PEDF receptors have been discovered recently, a membrane-associated phospholipase 2A (28) and a non-integrin laminin receptor (29). Unexpectedly, both have been ascribed functional roles in PEDF anti-angiogenic function (29, 30). Studies by us and others define two main pathways, through which PEDF regulates angiogenesis. In one, PEDF causes the activation of PPARγ, which results in a p53-dependent apoptosis (17). In another, the activation of NFκB and simultaneous JNK-dependent deactivation of NFAT cause endothelial cell killing through the CD95 death cascade (14, 31, 35). Whereas PPARγ activation has been shown to depend on p38 MAP kinase (17), NFAT deactivation is caused by JNK1 and -2 (31). The early upstream kinases in these two pathways remained unknown. It was also unclear, whether the two pathways are independent or interconnected. Our study demonstrates that the two cascades are engaged in a cross-talk. In the VEGF/bFGF-activated endothelial cells, PEDF activates Erk5/BMK1, which is positioned upstream of PPARγ. PPARγ, in turn, appears essential for the activation of NFκB, which up-regulates pro-apoptotic CD95L and dampens survival signaling by displacing NFAT (31, 35). Therefore PEDF causes endothelial cell apoptosis and blocks angiogenesis through sequential activation of MEK5, Erk5, PPARγ, and NFκB (Fig. 6A).
FIGURE 6.
The results of PEDF-driven Erk5 activation. A, PEDF exposure causes sequential activation of MEK5 and Erk5, which leads to PPARγ activation followed by NFκB activation, endothelial cell apoptosis, and anti-angiogenesis. B, proposed mechanism of PPARγ-dependent induction of NFκB expression. PPARγ/RXR heterodimers recruit SMRT to the p65/RelA promoter: SMRT, in turn, recruits HDAC1, which causes transcriptional repression. When phosphorylated by PEDF, Erk5 displaces SMRT, and thus releases HDAC1 and alleviates trans-repression by PPARγ.
Earlier studies noted increased PPARγ expression in PEDF-treated human umbilical vein endothelial cells (HUVEC) (17). We were unable to observe similar increase. However, DNA binding activity of PPARγ in PEDF-treated HMVECs was increased suggesting an alternative pathway, namely the displacement of SMRT co-repressor, which enables PPARγ transcriptional activity. The discrepancy may be attributed to the different origin of the endothelial cells used in the two studies; large vessel and microvascular endothelium may utilize distinct mechanisms of PPARγ activation. Our findings are, however, consistent with the mechanism of PPARγ activation in HMVECs cells subjected to shear stress (10). We were also unable to detect p53 protein stabilization in the endothelial cells exposed to PEDF, a finding that is consistent with the PEDF ability to block angiogenesis in p53-null mice.3
PPARγ has been previously shown to regulate the activity of other transcription factors, including NFκB (32) Typically, PPARγ activation results in a decreased NFκB activity. The mechanism behind this decrease is not completely elucidated; possible underlying causes include direct PPARγ binding to p65, competition for the co-activators, transfer of the co-repressor molecule NCoR1 to the NFκB upon release from the PPARγ complexes, or PPARγ-dependent blockade of the IκB turnover (33). Neither of the described events seems to take place in our system. In contrast, the blockade of PEDF-dependent NFκB activation by PPARγ antagonists clearly suggests PPARγ is required for NFκB activation by PEDF.
Similar events have been described for the vitamin D-dependent release of NFκB activity in T-helper cells (34). In this system, vitamin D receptor (VDR) mediates the repression of RelB promoter in association with SMRT and HDAC1. The release of SMRT interferes with HDAC1 recruitment and RelB trans-repression. Our study suggests that p65 may be regulated by PEDF in a similar way (Fig. 6B). In the activated endothelial cells PPARγ is associated with unphosphorylated Erk5 and SMRT, which likely recruits HDAC1 to the p65/RelA promoter causing trans-repression. In the presence of PEDF, phosphorylated Erk5 displaces SMRT from PPARγ transcriptional complexes, decreases promoter occupation by HDAC1 and relieves trans-repression. This hypothesis is in agreement with increased total p65/RelA levels in PEDF-treated HMVECs. The blockade of PEDF-dependent NFκB activation by the PPARγ antagonists clearly suggests an upstream position of PPARγ relative to NFκB in the PEDF signaling cascade. In conclusion, we, for the first time, identified Erk5 as a critical mediator of anti-angiogenic signaling, where it activates PPARγ, thus causing increased NFκB levels and activity, which ultimately cause endothelial cell apoptosis and counter angiogenesis.
Acknowledgments
We thank Dr. Tony Hollenberg (Beth Israel Deaconess Medical Center, Boston, MA) for help with the antibody design and production and Philippe Chlenski for technical assistance in preparing the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants NIH R01 HL68033, HL077471 (to O. V. V.), and NIDDK R01DK078125 (to R. N. C.).
N. P. Bouck and O. V. Volpert, unpublished observations.
- Erk
- extracellular signal-regulated kinase
- MAPK
- mitogen-activated protein kinase
- PPAR
- peroxisome proliferator-activated receptor
- CA
- constitutively active
- DN
- dominant-negative
- GFP
- green fluorescent protein
- BSA
- bovine serum albumin
- VEGF
- vascular endothelial growth factor
- TUNEL
- terminal deoxynucleotidyltransferase-mediated dUTP nick end-labeling
- PEDF
- pigment epithelial-derived factor
- SMRT
- silencing mediator for retinoic acid receptor and thyroid hormone receptor
- Epo
- erythropoietin
- FGF
- fibroblast growth factor
- HMVEC
- human dermal capillary endothelial cells.
REFERENCES
- 1.Mudgett J. S., Ding J., Guh-Siesel L., Chartrain N. A., Yang L., Gopal S., Shen M. M. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 10454–10459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang J., Boerm M., McCarty M., Bucana C., Fidler I. J., Zhuang Y., Su B. (2000) Nat. Genet. 24, 309–313 [DOI] [PubMed] [Google Scholar]
- 3.Tamura K., Sudo T., Senftleben U., Dadak A. M., Johnson R., Karin M. (2000) Cell 102, 221–231 [DOI] [PubMed] [Google Scholar]
- 4.Liu C., Shi Y., Han Z., Pan Y., Liu N., Han S., Chen Y., Lan M., Qiao T., Fan D. (2003) Biochem. Biophys. Res. Commun. 312, 780–786 [DOI] [PubMed] [Google Scholar]
- 5.Kasler H. G., Victoria J., Duramad O., Winoto A. (2000) Mol. Cell. Biol. 20, 8382–8389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang C. C., Ornatsky O. I., McDermott J. C., Cruz T. F., Prody C. A. (1998) Nucleic Acids Res. 26, 4771–4777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sohn S. J., Li D., Lee L. K., Winoto A. (2005) Mol. Cell. Biol. 25, 8553–8566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Regan C. P., Li W., Boucher D. M., Spatz S., Su M. S., Kuida K. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 9248–9253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin Q., Schwarz J., Bucana C., Olson E. N. (1997) Science 276, 1404–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan C., Takahashi M., Okuda M., Lee J. D., Berk B. C. (1999) J. Biol. Chem. 274, 143–150 [DOI] [PubMed] [Google Scholar]
- 11.Akaike M., Che W., Marmarosh N. L., Ohta S., Osawa M., Ding B., Berk B. C., Yan C., Abe J. (2004) Mol. Cell. Biol. 24, 8691–8704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sohn S. J., Sarvis B. K., Cado D., Winoto A. (2002) J. Biol. Chem. 277, 43344–43351 [DOI] [PubMed] [Google Scholar]
- 13.Sutton K. M., Hayat S., Chau N. M., Cook S., Pouyssegur J., Ahmed A., Perusinghe N., Le Floch R., Yang J., Ashcroft M. (2007) Oncogene 26, 3920–3929 [DOI] [PubMed] [Google Scholar]
- 14.Volpert O. V., Zaichuk T., Zhou W., Reiher F., Ferguson T. A., Stuart P. M., Amin M., Bouck N. P. (2002) Nat. Med. 8, 349–357 [DOI] [PubMed] [Google Scholar]
- 15.Rossi D., Gaidano G. (2003) Haematologica 88, 212–218 [PubMed] [Google Scholar]
- 16.Kato Y., Kravchenko V. V., Tapping R. I., Han J., Ulevitch R. J., Lee J. D. (1997) EMBO J. 16, 7054–7066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho T. C., Chen S. L., Yang Y. C., Liao C. L., Cheng H. C., Tsao Y. P. (2007) Cardiovasc. Res. 76, 213–223 [DOI] [PubMed] [Google Scholar]
- 18.Filleur S., Volz K., Nelius T., Mirochnik Y., Huang H., Zaichuk T. A., Aymerich M. S., Becerra S. P., Yap R., Veliceasa D., Shroff E. H., Volpert O. V. (2005) Cancer Res. 65, 5144–5152 [DOI] [PubMed] [Google Scholar]
- 19.Kato Y., Tapping R. I., Huang S., Watson M. H., Ulevitch R. J., Lee J. D. (1998) Nature 395, 713–716 [DOI] [PubMed] [Google Scholar]
- 20.Andrews N. C., Faller D. V. (1991) Nucleic Acids Res. 19, 2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Good D. J., Polverini P. J., Rastinejad F., Le Beau M. M., Lemons R. S., Frazier W. A., Bouck N. P. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 6624–6628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Passaniti A., Taylor R. M., Pili R., Guo Y., Long P. V., Haney J. A., Pauly R. R., Grant D. S., Martin G. R. (1992) Lab. Invest. 67, 519–528 [PubMed] [Google Scholar]
- 23.Liss C., Fekete M. J., Hasina R., Lam C. D., Lingen M. W. (2001) Int. J. Cancer 93, 781–785 [DOI] [PubMed] [Google Scholar]
- 24.Ho T. C., Yang Y. C., Chen S. L., Kuo P. C., Sytwu H. K., Cheng H. C., Tsao Y. P. (2008) Mol. Immunol. 45, 898–909 [DOI] [PubMed] [Google Scholar]
- 25.Veliceasa D., Schulze-Hoepfner F. T., Volpert O. V. (2008) PPAR Res. 2008, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nyberg P., Xie L., Kalluri R. (2005) Cancer Res. 65, 3967–3979 [DOI] [PubMed] [Google Scholar]
- 27.Mirochnik Y., Kwiatek A., Volpert O. V. (2008) Curr. Drug Targets 9, 851–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Notari L., Baladron V., Aroca-Aguilar J. D., Balko N., Heredia R., Meyer C., Notario P. M., Saravanamuthu S., Nueda M. L., Sanchez-Sanchez F., Escribano J., Laborda J., Becerra S. P. (2006) J. Biol. Chem. 281, 38022–38037 [DOI] [PubMed] [Google Scholar]
- 29.Bernard A., Gao-Li J., Franco C. A., Bouceba T., Huet A., Li Z. (2009) J. Biol. Chem. 284, 10480–10490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ho T. C., Chen S. L., Yang Y. C., Lo T. H., Hsieh J. W., Cheng H. C., Tsao Y. P. (2009) Am. J. Physiol. Cell Physiol. 296, C273–C284 [DOI] [PubMed] [Google Scholar]
- 31.Zaichuk T. A., Shroff E. H., Emmanuel R., Filleur S., Nelius T., Volpert O. V. (2004) J. Exp. Med. 199, 1513–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duan S. Z., Usher M. G., Mortensen R. M. (2008) Circ. Res. 102, 283–294 [DOI] [PubMed] [Google Scholar]
- 33.Chung J. H., Seo A. Y., Chung S. W., Kim M. K., Leeuwenburgh C., Yu B. P., Chung H. Y. (2008) Ageing Res. Rev. 7, 126–136 [DOI] [PubMed] [Google Scholar]
- 34.Griffin M. D., Dong X., Kumar R. (2007) Arch. Biochem. Biophys. 460, 218–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aurora A. B., Biyashev D., Mirochnik Y., Zaichuk T. A., Sánchez-Martinez C., Renault M. A., Losordo D., Volpert O. V. (2010) Blood, in press [DOI] [PMC free article] [PubMed] [Google Scholar]